Abstract
Epigenetics is a growing field not only in the area of cancer research but recently in stem cells including human embryonic stem cell (hESC) research. The hallmark of profiling epigenetic changes in stem cells lies in maintaining pluripotency or multipotency and in attaining lineage specifications that are relevant for regenerative medicine. Epigenetic modifications including DNA methylation, histone acetylation and methylation, play important roles in regulating gene expressions. Other epigenetic modifications include X chromosome silencing, genomic stability and imprinting and mammalian development. This review attempts to elucidate the mechanism(s) behind epigenetic modifications and review techniques scientists use for identifying each modification. We also discuss some of the trends of epigenetic modifications in the fields of directed differentiation of embryonic stem cells and de-differentiation of somatic cells.
Keywords: epigenetics, embryonic stem cell, somatic cell nuclear transfer, DNA methylation, chromatin modifications
Embryonic stem cells (ESCs)
ESCs derived from the blastocyst are capable of self renewal and can remain in an undifferentiated state for indefinite passages in vitro and also can be coaxed to differentiate to different lineages (Martin 1981). This makes them the favorable candidate for developing cellular therapies against many degenerative diseases such as those outlined in Table 1. Following the first successful derivation of 5 hESC lines by Thomson’s group in 1998 (Thomson et al 1998), more new hESC lines have been created (Guo et al 2007; Zhang et al 2006; Cowan et al 2004; Sidhu et al 2008). To date it is estimated, that more than 414 new hESC lines have been produced worldwide and out of which ∼78 are listed on the National Institute Health (NIH) Registry (Guhr et al 2006). Only ∼179 of these lines are characterized to some extent and available for research. Many of these hESC lines are not clonal and are derived under different culture conditions and propagated on different feeder layers (MEF, STO, fetal muscle, skin and foreskin, adult fallopian tube epithelial cells and also some feeder free/serum free systems), hence comparison of these lines are very difficult (Amit et al 2000; Cowan et al 2004; Amit et al 2003).
Table 1.
A concise list of some degenerative diseases that can be treated with cellular transplantation
| Disease/disorder | Type of cells required for transplantation |
|---|---|
| Alzheimer’s disease | Nerve cells |
| Diabetes | Islet cells |
| Cardiovascular disease | Cardiomyocytes |
| Liver disease | Hepatocytes |
| Multiple sclerosis | Glial cells |
| Osteoarthritis | Chrondrocytes |
| Spinal cord injuries | Nerve cells |
Epigenetic modifications play a significant role in maintaining pluripotency in ESCs and at the same time, very relevant in determining the somatic status of terminally differentiated cells. Accordingly, epigenetic profiles of pluripotent genes such as Nanog and OCT4, in ESCs are maintained.
The study of epigenetics – a cell’s epigenome
Gene and protein expression profiling has long been the benchmark in characterizing cell specialization during development. However, recent emphasis is shifted in favor of epigenetic profiling (also referred to as the ‘epigenome’) than genomic profiling as the former is considered to play a significant role in lineage specifications. The study of epigenetics involves covalent modifications to the architectural structure of both chromatin and DNA, but not to the sequence itself. However, there are other players which regulate gene expression such as binding of DNA proteins. These modifications are heritable (El Kharroubi et al 2001) and often regulate gene expression to a certain extent (Li 2002).
In contrast to gene expression profiles observed in somatic and/or differentiated cells, the ESCs have the potential of activating all gene expression profiles of all cell types from one genome. During mammalian development, almost all cells differentiate without changes to the DNA sequence, however their phenotypes are associated with certain activation (or inactivation) of genes. The differential activation/deactivation of genes depends on the presence and arrangement of functional moieties such as methyl (–CH3) and acetyl (–COCH3) groups, which forms the basis of an ‘epigenetic’ environment around the genomic DNA.
Understanding these complex structures of epigenetic modifications can lead to better understandings of how and when genes are activated or repressed. These patterns are established in early embryonic development and are subject to change throughout development (Li 2002; Reik et al 2001).
The mechanism of DNA methylation
Protein expressions determine cell phenotypes that are translated from mRNA transcripts of genomic DNA. Despite gene expression being influenced by changes in DNA sequence (single nucleotide polymorphisms – SNPs, within protein encoding sequences), epigenetic modifications such as DNA methylation can also affect gene expression (Hattori et al 2004).
DNA methylation forms an important means of epigenetic modifications and was first detected nearly 60 years ago, using chromatography techniques (Hotchkiss 1948). Over the past decade, transcriptional silencing of tumor suppressor genes through abnormal DNA methylation patterns has been established (Jones and Laird 1999). DNA methylation is also involved in other cellular processes such as, genomic stability (Peters et al 2001), X chromosome inactivation (Mohandas et al 1981), genomic imprinting (El Kharroubi et al 2001), chromatin structure (Jones et al 1998) and mammalian development (Reik et al 2001).
In mammalian genomes, DNA methylation occurs exclusively at the 5′ position on a cytosine nucleotide in the context of CG sequences (Bird, 2002). CpG islands are regions of DNA where CG nucleotides are present at significantly higher levels than the rest of the genome, these islands often reside at 5′ ends of all housekeeping and many tissue-specific genes – the promoter region (Gardiner-Garden and Frommer 1987). This methylated 5′ cytosine can act as a 5th DNA base, as different cytosine methylation status affects gene transcription.
Methylated DNA is often correlated to gene repression (Fuks et al 2000), its precise mechanism remains to be elucidated. One such explanation is that transcriptional binding sites become occupied by a group of methyl-CpG binding proteins (MeCP1, MeCP2, MBD1, MBD2) that specifically bind to methylated DNA. This explanation is supported via the findings that the binding of MeCP2, recruits histone deacetylases to repress transcription (Jones et al 1998; Nan et al 1998; Wade 2001; Fuks et al 2003). This is also evident that these 2 epigenetic modifications; DNA methylation and histone deacetylations are inter-related. MBD2 is also known to bind with NuRD, forming a complex; MeCP1 which has gene repressive capabilities (Feng and Zhang 2001).
DNA methyl transferases (DNMT) are enzymes that add methyl groups to DNA. In mammals there are 3 common transferases, DNMT1, DNMT3a, DNMT3b. DNMT1 is a methylation maintenance enzyme and targets newly synthesized DNA via its N-terminal regulatory domain which aims for the replicating foci (Leonhardt et al 1992). Somatic cells are believed to preserve their methylation patterns via this manner. DNMT3a/3b are de novo methylation enzymes and are most active during 2 stages of embryonic development following the 2 rounds of global genomic demethylation (Morgan et al 2005).
Methylation patterns are dynamic, as commonly demonstrated in reprogramming studies and they also change as cells develop physiologically (Freberg et al 2007; Takahashi et al 2007). Demethylation occurs spontaneously as previously demonstrated (Cervoni and Szyf 2001). Ectopically methylated genes transfected into somatic cells showed demethylating activity. Another study also showed demethylase activity via a mammalian protein which has DNA encoding for a methyl-CpG binding domain is also shown (Bhattacharya et al 1999). However demethylation alone is not sufficient to remove the repressive trait of methylation. This encourages scientists to look at epigenetic modifications as a whole, instead of one entity, specifically at histone modifications, as will be discussed later. Mechanisms that prevent de novo CpG methylation are unclear. Several proposed hypotheses include; DNMT3a/3b requires additional CpG binding proteins (CGBP) or other protein interaction. There may also be a constant demethylase activity, as demonstrated in the above mentioned study (Cervoni and Szyf 2001).
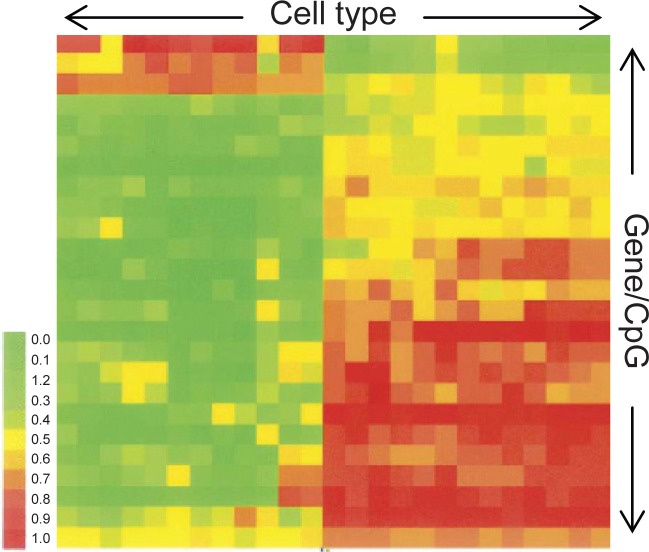
All types of cells would have their own epigenomic signature. This is best depicted using DNA microarrays (for example Illumina’s bead arrays), where an epigenome is expressed in a cluster analysis methylation chart, displaying cell type and CpG methylation levels. A recent study, mapped the methylation status of 23 genes (25 CpG sites) over a range of cell types (Bibikova et al 2006) (Figure 1). From this figure, it is very clear that different cell types are epigenetically (and transcriptionally) divergent from each other, otherwise referred to as being ‘epigenetically unique.’
Figure 1.
Methylation analysis chart. Each column represents a different cell type, while each row is a different genes/CpG site. Each cytosine is graded from being heavily methylated (1) to largely unmethylated (0).
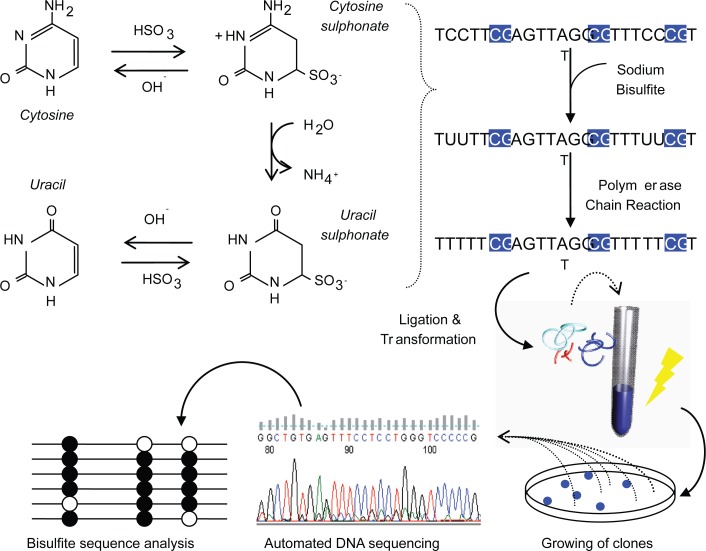
There are many techniques used to study DNA methylation patterns, but the most specific and commonly used, being bisulfite DNA sequencing (Frommer et al 1992). Bisulfite deals structural changes to unmethylated cytosines, while methylated cytosines remain intact. With this change, one can measure the amount of DNA methylation using conventional DNA sequencing procedures (Figure 2). This is a quantitative measure and is generally correlated to gene expression profiles. A most recent study, mapped the entire genome of Arabidopsis thaliana using a shotgun bisulfite approach using a Next-generation sequencer, which provided a map at single base pair resolution of methylated cytosines (Cokus et al 2008).
Figure 2.
Process of bisulfite DNA sequencing. Genomic DNA is treated with sodium bisulfite which deals structural and irreversible changes to a cytosine through denaturation, deamination and desulphonation processes. Taking advantage of these changes, the DNA is PCR-amplified and ligated to plasmid vectors for transformation into Escherichia coli. White colonies are hand picked and plasmid DNA extracted for automated DNA sequencing. Closed circles represent methylated CpGs and open represent unmethylated CpGs.
5′aza-2-deoxycytidine (5azadC) is a global DNA demethylation reagent that clips off methyl groups bound on 5′methyl-cytosines. 5azadC is a deoxy- form of 5-azacytidine and is more readily incorporated into DNA. This causes a more efficient inhibition of methylation than the latter (Momparler et al 1984). Its activity is characterized by the covalent trapping of DNMT which depletes the cell of its enzymatic activity (Juttermann et al 1994). 5azadC is commonly used as a DNA methylation inhibitor and its activity has been correlated with gene expression (Jones and Taylor 1980; Taylor 1993; Juttermann et al 1994; Grassi et al 2003), cellular differentiation (Pinto et al 1989; Choi et al 2004a) and specifically in enhancing hESC differentiation (Xu et al 2002).
Maintaining methylation but not de novo methylation is required for in vitro differentiation, as demonstrated previously (Jackson et al 2004). Briefly, hypomethylated cell lines with Dnmt3a/3b gene knockouts restored their methylation after stable integration of DNMT1 cDNA transgene.
Although DNA methylation has been intensively studied, many questions remain to be answered, including what mechanisms prevent the de novo methylation of normal somatic cells? And the proteomic network of DNMT remains to be elucidated.
Chromatin remodeling and histone modifications
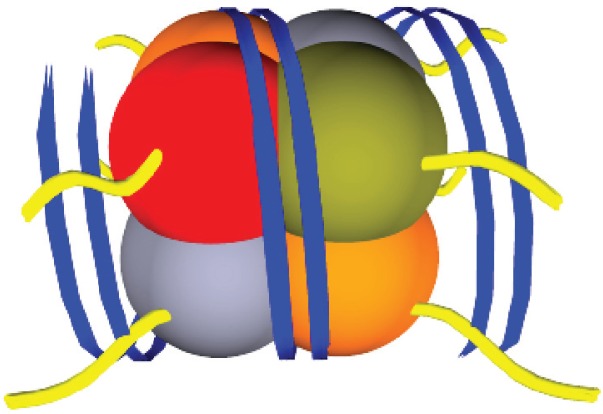
The fundamental unit of chromatin is a nucleosome, which consists of a core of 8 histones; H2A, H2B, H3 and H4 (2 of each). Each core is surrounded by ∼147bp of DNA and is tightly wound around 1.75 turns (Figure 3). There is increasing evidence that transcriptional factors recognize signals given off by histone tail modifications. As there is an association between DNA and histones, it is not surprising that histone tail modifications (acetylation methylation, ubiquitylation and phosphorylation) also affect gene transcription.
Figure 3.
A diagrammatic representation of one chromatin unit. A nucleosome, consisting of 4 histones types; H2A, H2B, H3 and H4 with DNA (blue) tightly wound around the core unit. Histone tails (yellow) protrude from the centre of the histones through the DNA strands (blue).
At the molecular level, the revealing (or hiding) of binding sites that influence gene transcription are outcomes of histone tail modifications. This hiding and revealing of binding sites is determined by overall chromatin structure whether it is relaxed or compact. Acetylation of histone tails removes the positive charge, thus decreasing the affinity between the DNA and histones. This results in a structure called euchromatin and allows easier access of transcriptional factors. In contrast, the result of deacetylation, caused by histone deacetylases (HDACs) is heterochromatin, which results in tightly compacted chromatin and conceals transcriptional DNA binding sites.
Histone tails of H3 holds several amino acids that are notably studied for their correlation with gene expression; these are lysine, arginine, serine and threonine residues. Transcriptionally active genes generally harbors histone H3 lysine 9 acetylation (H3K9ac), H3K4 di-methylation (H3K4me2), tri-methylation (H3K4me3), H3K36me3 and H3K79me3. Transcriptionally repressed genes tend to harbor H3K9me2, H3K9me3, H3K27me3 and histones H4 lysine 20 tri-methylation (H4K20me3) (Dahl and Collas 2007; Freberg et al 2007; Maherali et al 2007). Cell populations expressing high levels of gene(s) are generally enriched with euchromatic markers in their promoter regions as demonstrated in pluripotent genes OCT4, NANOG and heterochromatic markers of somatic gene PAX6 of pluripotent undifferentiated carcinoma cells (Dahl and Collas 2007). A recent study mapped the histone methylation marks in mouse- ESCs, neural progenitor cells and embryonic fibroblasts and highlighted the impact of H3K4me3 and H3K27me3, on transcriptionally active and inactive genes respectively (Mikkelsen et al 2007). Gene promoters which contained both the euchromatic and heterochromatic markers above determine switching cell developmental fates (Bernstein et al 2006).
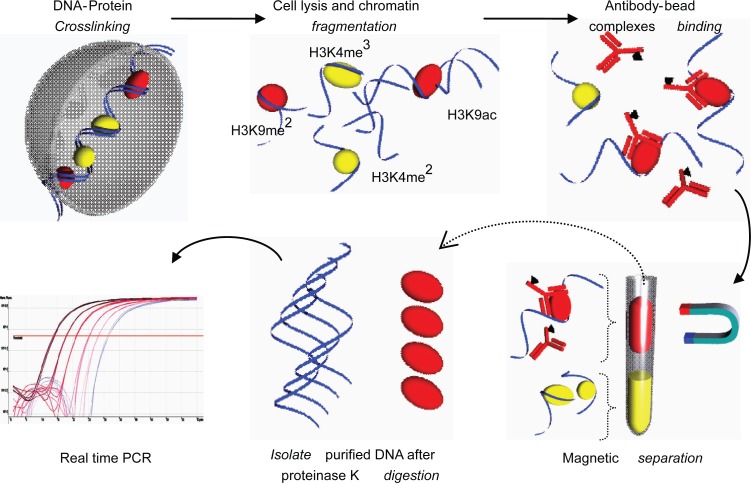
Chromatin immunoprecipitation (ChIP) is a technique used to study chromatin remodeling including histone de/acetylation and de/methylation. Protein-DNA interaction is the basis of this technique and has been used for the past 20 years. Conventional ChIP analysis requires large numbers of starting material; cells and hence, a simplified recipe, Q2ChIP Assay was invented (Dahl and Collas 2007). Briefly, cells are cross linked using sodium butyrate prior to lysis and sonication. Cell lysate is immunoprecipitated and reversed cross linked; unbinding of DNA-histone complexes, DNA is then isolated and used for polymerase chain reaction (PCR) assays (Figure 4).
Figure 4.
Quick and Quantitative Chromatin Immuno-precipitation (Q2ChIP). Cells were harvested and treated with sodium butyrate to allow DNA-protein crosslinking. Cells were lysed and sonicated to produce fragments (∼500 bp). Chromatin fragments were allowed to conjugate to ‘antibody-paramagnetic bead complexes’ (specific for H3K9ac). The solution is magnetically separated and purified fragments are reversed crosslinked and subjected to proteinase K digestion. Isolated DNA is now ready for downstream PCR processes.
Recruitment of histone acetyl transferases (HATs) or presence of histone deactylases (HDAC) inhibitor(s) results in histone acetylation (Cervoni and Szyf 2001). Hyper-acetylated promoter regions correspond to gene activity (Hattori et al 2004). Trichostatin A (TSA) is a commonly used deacetylase inhibitor, and allows re-expression of genes regulated by histone acetylations (Cameron et al 1999), methylation (Ou et al 2007) and also inducing cellular differentiation (Hosseinkhani et al 2007). Other studies have determined common enzymes that demethylate specific lysine residues which generally lead to gene activation/repression (depending on the traits of specific lysine residues): Lysine specific demethylase (LSD1) specifically demethylates H3K4 (Shi et al 2004), Ubiquitously transcribed tetratricopeptide repeat, X chromosome (UTX) and Jumonji domain containing 3 (JMJD3) specifically demethylates H3K27 (Hong et al 2007).
The increase of histone acetylation is associated with a decrease in global methylation (Jackson et al 2004; Ou et al 2007) and a gene’s methylated state is determined by the balance between demethylase activity and state of histone acetylation (Cervoni and Szyf 2001). Treatment of cells with TSA showed re-expression of some methylation-silenced tumor suppressor genes indicating that methylation is the dominant epigenetic suppressor of densely methylated genes (Cameron et al 1999). Another study displays similar results while providing evidence that TSA inhibits DNMT1 (Januchowski et al 2007), which plays the role of 5azadC (as mentioned earlier); demethylation of tumor suppressor genes (Merlo et al 1995).
Lineage specifications
Recently, the study of epigenetic changes during differentiation of ESCs and de-differentiation of somatic cells has revealed some interesting results. More differentiated cells generally harbor more epigenetic modifications as demonstrated in mouse fibroblast cells (fully differentiated cells), where a combination of Trichostatin A (TSA) and 5′aza-2-deoxycytidine (5azadC), was required for OCT4 re-expression, whereas in trophoblast stem cells (less differentiated cells), either 5′azadC or TSA alone can re-activate OCT4 expression (Hattori et al 2004). However, certain cells do retain their ability to switch cell developmental fates through balancing of euchromatic and heterochromatic markers (Mikkelsen et al 2007). One exception to the above rule, applies to germ line cells (non-somatic cells), namely – primordial germ cells (PGCs). PGCs undergoes dynamic methylation changes (Trent 2005) which forms the basis of regulating germ cell-specific gene expression (Maatouk et al 2006). Derivatives of PGCs, such as; arrested metaphase II eggs and mature sperm exhibit hypomethylated major and minor satellite sequences whereas in comparison to somatic cells, are hypermethylated (Yamagata et al 2007). This is one of the best examples using epigenetic modifications in lineage specification. Somatic cells maintain their gene expressions after mitosis through epigenetic mechanisms, a process often referred to as ‘cellular memory.’ However, the cellular memory present in PGCs remains in a ‘reprogrammable’ state with the potential event of totipotency.
Another example of epigenetic lineage specification include specific CpG methylation of promoter regions as previously demonstrated (Takizawa et al 2001). Briefly, a certain CG found in the STAT3 binding element within the GFAP promoter region of neuroepitheilial cells/post-mitotic neurons is highly methylated and is a sign of GFAP suppression. As these cells differentiate into astrocytes, this level of methylation is decreased resulting in the expression of GFAP (Takizawa et al 2001). Therefore methylation of the STAT3 recognition sequence results in GFAP suppression.
Epigenetic modifications of stem cells
Cellular differentiation
There are 3 major stages of epigenetic modifications during gametogenesis and embryonic development. The first occurs in primordial germ cells, where the imprints of genes are erased, shown by the lack of methylation (Seki et al 2007; Maatouk et al 2006). The second stage involves epigenetic acquisition in maturing gametes, namely oocytes (Lucifero et al 2002) and spermatozoan (Oakes et al 2007). The third stage occurs during fertilization and usually involves maintenance of methylation in imprinted genes, while other genes gradually lose their methylation (Mayer et al 2000; Okamoto et al 2004).
As described above, epigenetic modifications are dynamic throughout cellular differentiation. Many recent studies on directed differentiation of ESCs used epigenetic modifying agents 5azadC and TSA to enhance the efficiency of differentiation. Differentiation of ESCs to cardiomyocytes was enhanced with 5azadC and TSA (Xu et al 2002; Hosseinkhani et al 2007). A recent report attempts to describe the dynamics of DNA methylation of differentially methylated regions (DMRs) in 5azadC-induced adipocyte differentiation (Sakamoto et al 2008). Although a majority of associated genes exhibited no change in methylation profiles, a series of 8 out of 65 tissue-dependent DMRs underwent either methylation, demethylation or were transient, from stem cell to mature adipocytes.
In a recent study it is demonstrated that although TSA does not improve astrocyte differentiation, it assisted in the derivation of neurons with normal electrophysiological membrane properties and elongation of dendrites (Balasubramaniyana et al 2006).
Cellular reprogramming and dedifferentiation
Somatic cell nuclear transfer (SCNT) is a process whereby the derivation of cloned animals (Wilmut et al 1997) and donor-matched cell lines (Wakayama et al 2005) is made possible. SCNT is an emerging area in stem cell research and involves transferring nuclei of somatic cells into enucleated oocytes with the aim to derive embryonic stem cells with the same genetic makeup as the ‘donor’ somatic cell. From a clinical perspective, such cells, when transplanted, should not be rejected due to incompatible immune response.
SCNT is also useful for studying drug toxicologies in embryonic cell lines derived from a somatic cell’s genome carrying a certain disease(s) such as those listed in Table 1. This technique has also been used to generate live cloned offspring in various animal species but not in humans (Cibelli et al 1998b; Meng et al 1997; Rideout III et al 2000). However, it remains as an inefficient technique. Many factors influencing SCNT efficiency include; donor cell phenotype (Table 2), developmental stage of recipient oocytes during nuclear transfer (Hall et al 2007), cell cycle stage of donor somatic cell (Kasinathan et al 2001), type of zygote/embryo activation method (Choi et al 2004b) and recently, epigenetic modifications including DNA methylation and histone acetylation (Kishigami et al 2006).
Table 2.
Efficiencies of obtaining live offspring or establishing nt-ESC lines between species across a range of donor cells
Percentage of transferred blastocysts to a surrogate mother.
Percentage of blastocysts giving rise to nt-ESC lines.
Percentage of cleaved embryos developing to morulae/blastocyst stages.
Abbreviations: ESC, embryonic stem cell; ISCI, intracytoplasmic sperm injection.
Epigenetic studies play a significant role in determining reprogramming during SCNT (Byrne et al 2007) and also in a recently developed technology; induced pluripotent stem (iPS) cells, that is, reprogramming somatic cells to pluripotent state by transducing specific pluripotency-associated genes into the somatic cells (Takahashi and Yamanaka 2006; Takahashi et al 2007; Wernig et al 2007; Yu et al 2007). These studies have analysed DNA methylation and histone modifications to confirm their reprogrammed somatic cells have reverted back into a pluripotent state (and even to totipotent status in mice). These epigenetic analyses along with gene expression patterns are essential to specify and confirm the identity of the pluripotent lineage.
As indicated in Table 2, SCNT remains as an inefficient process and many studies are being carried out to improve SCNT efficiency. In a recent study led by Jaenisch (Blelloch et al 2006), it has been demonstrated that less differentiated somatic cells increases SCNT efficiency. It has been shown, by using cells which carry a hypomorphic allele of Dnmt1 (which results in ‘global demethylation’ of the donor genome), higher efficiency could be achieved (65%). This paper may well provide a spark in the future for efficient establishment of nuclear transfer-ESC lines. However higher efficiency of cloning was also achieved by using differentiated cells (namely, granulocytes) compared to that with less differentiated hematopoietic stem cells (Sung et al 2006). Conclusions from both these studies, although correct, encourage us to look at the epigenetic nature of donor cells and how certain somatic epigenomes influences SCNT efficiency.
Bisulfite sequencing and Q2ChIP analyses are useful for studying a limited number of genes at one time. However, it is more convenient to use immunocytochemistry to visually observe global epigenetic patterns and/or dynamics within cell populations. Antibodies specific to epigenetic markers (for eg, 5′methyl-cytosines and H3K9 methylation) is an invaluable tool to study the epigenetic dynamics by observing intensities of fluorescence (Beaujean et al 2004; Yang et al 2006; Seki et al 2007).
SCNT embryos in animals tend to exhibit aberrant epigenetic makers as compared to those produced with traditional IVF technology These embryos tend to exhibit abnormally higher methylation levels during pre-implantation embryogenesis (Dean et al 2001; Beaujean et al 2004). H3K9 acetylation levels in SCNT embryos, on the other hand, were at a lower level than IVF embryos and suggest active histone deacetylase activity. Inhibition of histone deacetylases (HDACs) via trichostatin A, may bring up the levels of acetylation consistent with IVF embryos. These strategies have previously been demonstrated in mice (Enright et al 2003; Kishigami et al 2006) and should be adapted in future studies, for efficient reprogramming of donor nuclei, which is believed to be the major cause for SCNT low efficiency.
Conclusions
Epigenetics is an exponentially growing field in ESC research, especially in cellular reprogramming studies. The aim to establish patient-matched ESC lines is currently hindered by the fact that there are aberrant epigenetic modifications during the reprogramming process and needs to be addressed. Epigenetic modifications are dynamic and directed differentiation studies should aim to address these issues, since growth factors and supplements is not sufficient for the directed differentiation of ESCs. Both DNA methylation and histone modifications are inseparable entities when it comes to cellular differentiation and de-differentiation. Epigenetic changes induced by using specific reagents have the prospects of studying both the cellular differentiation and de-differentiation processes as demonstrated in some previous studies.
Footnotes
Disclosures
The authors have no conflicts of interest to declare.
References
- Amit M, Carpenter MK, Inokuma MS, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–8. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- Amit M, Margulets V, Segev H, et al. Human feeder layers for human embryonic stem cells. Biol Reprod. 2003;68:2150–6. doi: 10.1095/biolreprod.102.012583. [DOI] [PubMed] [Google Scholar]
- Balasubramaniyana V, Boddekea E, Bakelsa R, et al. Effects of histone deacetylation inhibition on neuronal differentiation of embryonic mouse neural stem cells. Neuroscience. 2006;143:939–51. doi: 10.1016/j.neuroscience.2006.08.082. [DOI] [PubMed] [Google Scholar]
- Beaujean N, Taylor J, Gardner J, et al. Effect of limited DNA methylation reprogramming in the normal sheep embryo on somatic cell nuclear transfer. Biol Reprod. 2004;71:185–93. doi: 10.1095/biolreprod.103.026559. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Betthauser J, Forsberg E, Augenstein M, et al. Production of cloned pigs from in vitro systems. Nat Biotech. 2000;18:1055–59. doi: 10.1038/80242. [DOI] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, et al. A mammalian protein with specific demethylase activity form CpG DNA. Nature. 1999;397:579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Chudin E, Wu B, et al. Human embryonic stem cells have a unique epigenetic signature. Genome Res. 2006;16:1075–83. doi: 10.1101/gr.5319906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Wang ZD, Meissner A, et al. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–13. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch RH, Hochedlinger K, Yamada Y, et al. Nuclear cloning of embryonal carcinoma cells. Proc Natl Acad Sci U S A. 2004;101:13985–90. doi: 10.1073/pnas.0405015101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne JA, Pedersen DA, Clepper LL, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myöhänen S, et al. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–87. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- Chen HF, Kuo HC, Chien CL, et al. Derivation, characterization and differentiation of human embryonic stem cells: Comparing serum-containing versus serum-free media and evidence of germ cell differentiation. Hum Reprod. 2007;22:567–77. doi: 10.1093/humrep/del412. [DOI] [PubMed] [Google Scholar]
- Cho S-K, Kim J-H, Park J-Y, et al. Serial cloning of pigs by somatic cell nuclear transfer:Restoration of phenotypic normality during serial cloning. Dev Dyn. 2007;236:3369–82. doi: 10.1002/dvdy.21308. [DOI] [PubMed] [Google Scholar]
- Choi SC, Yoon JY, Shim WJ, et al. 5-azacytidine induces cardiac differentiation of P19 embryonic stem cells. Exp Mol Med. 2004a;36:515–23. doi: 10.1038/emm.2004.66. [DOI] [PubMed] [Google Scholar]
- Choi YH, Love LB, Westhusin ME, et al. Activation of equine nuclear transfer oocytes:methods and timing of treatment in relation to nuclear remodeling. Biol of Reprod. 2004b;70:46–53. doi: 10.1095/biolreprod.103.018200. [DOI] [PubMed] [Google Scholar]
- Cibelli JB, Stice SL, Golueke PJ, et al. Transgenic bovine chimeric offspring produced from somatic cell-derived stem-like cells. Nat Biotech. 1998a;16:642–6. doi: 10.1038/nbt0798-642. [DOI] [PubMed] [Google Scholar]
- Cibelli JB, Stice SL, Golueke PJ, et al. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998b;280:1256–8. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–19. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan CA, Klimanskaya I, McMahon J, et al. Derivation of embryonic stem-cell lines from human blastocysts. New Engl J Med. 2004;350:1353–6. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Dahl JA, Collas P. Q2ChIP, a quick and quantitative chromatin immunoprecipitation assay, unravels epigenetic dynamics of developmentally regulated genes in human carcinoma cells. Stem Cells. 2007;25:1037–46. doi: 10.1634/stemcells.2006-0430. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, et al. Conservation of methylation reprogramming in mammalian development:Aberrant reprogramming in cloned embryos. Proc Natl Acad Sci U S A. 2001;98:13734–8. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kharroubi A, Piras G, Stewart CL. DNA demethylation reactivates a subset of imprinted genes in uniparental mouse embryonic fibroblasts. J Biol Chem. 2001;276:8674–80. doi: 10.1074/jbc.M009392200. [DOI] [PubMed] [Google Scholar]
- Enright BP, Kubota C, Yang X, et al. Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2’-deoxycytidine. Biol Reprod. 2003;69:896–901. doi: 10.1095/biolreprod.103.017954. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–32. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freberg CT, Dahl JA, Timoskainen S, et al. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–53. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AJ, Adams CA, Anderson LS, et al. Development of human cloned blastocysts following somatic cell nuclear transfer with adult fibroblasts. Stem Cells. 2008;26:485–93. doi: 10.1634/stemcells.2007-0252. [DOI] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–31. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A, et al. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–40. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG Islands in vertebrate genomes. J Mol Biol. 1987;196:261–82. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gibbonsa J, Arat S, Rzucidlo J, et al. Enhanced survivability of cloned calves derived from roscovitine-treated adult somatic cells. Biol Reprod. 2002;66:895–900. doi: 10.1095/biolreprod66.4.895. [DOI] [PubMed] [Google Scholar]
- Grassi G, Maccaroni P, Meyer R, et al. Inhibitors of DNA methylation and histone deacetylation activate cytomegalovirus promoter-controlled reporter gene expression in human glioblastoma cell line U87. Carcinogenesis. 2003;24:1625–35. doi: 10.1093/carcin/bgg118. [DOI] [PubMed] [Google Scholar]
- Guhr A, Kurtz A, Friedgen K, et al. Current state of human embryonic stem cell research: an overview of cell lines and their usage in experimental work. Stem Cells. 2006:1–17. doi: 10.1634/stemcells.2006-0053. [DOI] [PubMed] [Google Scholar]
- Guo H, Wei-qiang L, Rui C, et al. Establishment and characterization of two new human embryonic stem cell lines, SYSU-1 and SYSU-2. Chinese Med J. 2007;120:589–94. [PubMed] [Google Scholar]
- Hall VJ, Compton D, Stojkovic P, et al. Developmental competence of human in vitro aged oocytes as host cells for nuclear transfer. Hum Reprod. 2007;22:52–62. doi: 10.1093/humrep/del345. [DOI] [PubMed] [Google Scholar]
- Hamano S, Watanabe Y, Azuma S, et al. Establishment of embryonic stem ES. cell-like cell lines derived from bovine blastocysts obtained by in vitro culture of oocytes matured and fertilized in vitro. J Reprod Dev. 1998;44:297–303. [Google Scholar]
- Hattori N, Nishino K, Y-g Ko, et al. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J Biol Chem. 2004;279:17063–9. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Heindryckx B, Sutter PD, Gerris J, et al. Embryo development after successful somatic cell nuclear transfer to in vitro matured human germinal vesicle oocytes. Hum Reprod. 2007;22:1982–90. doi: 10.1093/humrep/dem106. [DOI] [PubMed] [Google Scholar]
- Hiiragui T, Solter D. Reprogramming is essential in nuclear transfer. Mol Reprod Dev. 2005;70:417–21. doi: 10.1002/mrd.20126. [DOI] [PubMed] [Google Scholar]
- Hong S, Cho Y-W, Yu L-R, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci U S A. 2007;104:18439–444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinkhani M, Hasegawa K, Ono K, et al. Trichostatin A induces myocardial differentiation of monkey ES cells. Biochem Biophys Res Commun. 2007;365:386–91. doi: 10.1016/j.bbrc.2007.02.151. [DOI] [PubMed] [Google Scholar]
- Hotchkiss RD. The quantitative separation of purines, pyrimidines and nucleosides by paper chromatography. J Biol Chem. 1948;175:315–32. [PubMed] [Google Scholar]
- Jackson M, Krassowska A, Gilbert N, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–71. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Januchowski R, Dąbrowski M, Ofori H, et al. Trichostatin A down-regulates DNA methyltransferase 1 in Jurkat T cells. Cancer Lett. 2007;246:313–7. doi: 10.1016/j.canlet.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer-epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs, and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJC, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–91. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of dna methyltransferase rather than DNA demethylation. Proc Natl Acad Sci U S A. 1994;91:11797–801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinathan P, Knott JG, Wang ZD, et al. Production of calves from G1 fibroblasts. Nat Biotech. 2001;19:1176–8. doi: 10.1038/nbt1201-1176. [DOI] [PubMed] [Google Scholar]
- Kishigami S, Mizutani E, Ohta H, et al. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–9. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- Lee J-W, Wu S-C, Tian XC, et al. Production of cloned pigs by whole-cell intracytoplasmic microinjection. Biol Reprod. 2003;69:995–1001. doi: 10.1095/biolreprod.103.015917. [DOI] [PubMed] [Google Scholar]
- Leonhardt H, Page AW, Weier H-U, et al. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–73. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in 1 development. Nat Rev Genet. 2002;3:662–73. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li M, Li Y-H, Hou Y, et al. Isolation and culture of pluripotent cells from in vitro produced porcine embryos. Zygote. 2004;12:43–8. doi: 10.1017/s0967199404002679. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mertineit C, Clarke HJ, et al. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79:530–8. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MRW, et al. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–18. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1 doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mai Q, Yu Y, Li T, et al. Derivation of human embryonic stem cell lines from parthenogenetic blastocysts. Cell Res. 2008;17:1008–19. doi: 10.1038/cr.2007.102. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–8. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- Meng L, Ely JJ, Stouffer RL, et al. Rhesus monkeys produced by nuclear transfer. Biol Reprod. 1997;57:454–9. doi: 10.1095/biolreprod57.2.454. [DOI] [PubMed] [Google Scholar]
- Merlo A, Herman JG, Mao L, et al. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nature Med. 1995;1:686–92. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitalipov S, Yeoman RR, Nusser KD, et al. Rhesus monkey embryos produced by nuclear transfer from embryonic blastomeres or somatic cells. Biol Reprod. 2002;66:1367–73. doi: 10.1095/biolreprod66.5.1367. [DOI] [PubMed] [Google Scholar]
- Mitalipov SM, Nusser KD, Wolf DP. Parthenogenetic activation of rhesus monkey oocytes and reconstructed embryos. Biol Reprod. 2001;65:253–9. doi: 10.1095/biolreprod65.1.253. [DOI] [PubMed] [Google Scholar]
- Mitalipova SM, Zhou Q, Byrne JA, et al. Reprogramming following somatic cell nuclear transfer in primates is dependent upon nuclear remodeling. Human Reproduction. 2007;22:2232–42. doi: 10.1093/humrep/dem136. [DOI] [PubMed] [Google Scholar]
- Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome:evidence for X inactivation by DNA methylation. Science. 1981;211:393–6. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- Momparler RL, Momparler LF, Samson J. Comparison of the antileukemic activity of 5-aza-2′-deoxycytidine, 1-beta-d-arabinofuranosylcytosine and 5-azacytidine against l1210 leukemia. Leukemia Res. 1984;8:1043–9. doi: 10.1016/0145-2126(84)90059-6. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, et al. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:47–58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–9. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Oakes CC, Salle SL, Smiraglia DJ, et al. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev Biol. 2007;307:368–79. doi: 10.1016/j.ydbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, et al. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–9. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Onishi A, Iwamoto M, Akita T, et al. Pig cloning by microinjection of fetal fibroblast nuclei. Science. 2000;289:1188–90. doi: 10.1126/science.289.5482.1188. [DOI] [PubMed] [Google Scholar]
- Ou J-N, Torrisani J, Unterberger A, et al. Histone deacetylase inhibitor trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem Pharm. 2007;73:1297–307. doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Peters AHFM, O’Carroll Dn, Scherthan H, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–37. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Pinto A, Zagonel V, Attadia V, et al. 5-Aza-2-deoxycytidine induces terminal differentiation of leukemic blasts from patients with acute myeloid leukemias. Bone Marrow Transplant. 1989;3:28–32. [Google Scholar]
- Polejaeva IA, Chen S-H, Vaught TD, et al. Cloned pigs produced by nuclear transfer from adult somatic cells. Nature. 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Revazova ES, Turovets NA, Kochetkova OD, et al. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:1–18. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- Rideout WM, III, Wakayama T, Wutz A, et al. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat Genet. 2000;24:109–10. doi: 10.1038/72753. [DOI] [PubMed] [Google Scholar]
- Prather RS, Sims M, First N. Nuclear transplantation in early pig embryos. Biol Reprod. 1989;41:414–8. doi: 10.1095/biolreprod41.3.414. [DOI] [PubMed] [Google Scholar]
- Sakamoto H, Kogoa Y, Ohgane J, et al. Sequential changes in genome-wide DNA methylation status during adipocyte differentiation. Biochem Biophys Res Comm. 2008;366:360–6. doi: 10.1016/j.bbrc.2007.11.137. [DOI] [PubMed] [Google Scholar]
- Seki Y, Yamaji M, Yabuta Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–38. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- Shao H, Wei Z, Wang L, et al. Generation and characterization of mouse parthenogenetic embryonic stem cells containing genomes from non-growing and fully grown oocytes. Cell Biol Int. 2007;31:1336–44. doi: 10.1016/j.cellbi.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Sidhu KS, Ryan JP, Tuch BE. Derivation of a new hESC line, Endeavour-1 and its clonal propagation. Stem Cells Dev. 2008;17:41–52. doi: 10.1089/scd.2007.0055. [DOI] [PubMed] [Google Scholar]
- Suemori H, Tada T, Torii R, et al. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn. 2001;222:273–9. doi: 10.1002/dvdy.1191. [DOI] [PubMed] [Google Scholar]
- Sung LY, Gao S, Shen H, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–8. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:652–5. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Nakashima K, Namihira M, et al. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell. 2001;1:749–58. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Taylor SM. 5-Aza-2′-deoxycytidine: cell differentiation and DNA methylation. Leukemia. 1993;7:3–8. [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–47. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Trent RJ. Molecular Medicine. USA: Elsevier Academic Press; 2005. [Google Scholar]
- Wade PA. Methyl CpG-binding proteins and transcriptional repression. Bioessays. 2001;23:1131–7. doi: 10.1002/bies.10008. [DOI] [PubMed] [Google Scholar]
- Wakayama S, Ohta H, Kishigami S, et al. Establishment of male and female nuclear transfer embryonic stem cell lines from different mouse strains and tissues. Biol Reprod. 2005;72:932–6. doi: 10.1095/biolreprod.104.035105. [DOI] [PubMed] [Google Scholar]
- Walker SC, Shin T, Zaunbrecher GM, et al. A highly efficient method for porcine cloning by nuclear transfer using in vitro-matured oocytes. Cloning Stem Cells. 2002;4:105–12. doi: 10.1089/153623002320253283. [DOI] [PubMed] [Google Scholar]
- Wang L, Duan E, Sung LY, et al. Generation and characterization of pluripotent stem cells from cloned bovine embryos. Biol Reprod. 2005;73:149–55. doi: 10.1095/biolreprod.104.037150. [DOI] [PubMed] [Google Scholar]
- Wells DN, Laible G, Tucker FC, et al. Coordination between donor cell type and cell cycle stage improves nuclear cloning efficiency in cattle. Theriogenology. 2002;59:45–59. doi: 10.1016/s0093-691x(02)01273-6. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–24. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, et al. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–3. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Wolf DP, Thormahlen S, Ramsey C, et al. Use of assisted reproductive technologies in the propagation of rhesus macaque offspring. Biol Reprod. 2004;71:486–93. doi: 10.1095/biolreprod.103.025932. [DOI] [PubMed] [Google Scholar]
- Xu CH, Police S, Rao N, et al. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circulation Res. 2002;91:501–8. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- Yamagata K, Yamazaki T, Miki H, et al. Centromeric DNA hypomethylation as an epigenetic signature discriminates between germ and somatic cell lineages. Dev Biol. 2007;312:419–26. doi: 10.1016/j.ydbio.2007.09.041. [DOI] [PubMed] [Google Scholar]
- Yamazaki W, Ferreira CR, Méo SC, et al. Use of strontium in the activation of bovine oocytes reconstructed by somatic cell nuclear transfer. Zygote. 2005;13:295–302. doi: 10.1017/S0967199405003333. [DOI] [PubMed] [Google Scholar]
- Yang J, Yang S, Beaujean N, et al. Epigenetic marks in cloned rhesus monkey embryos:comparison with counterparts produced in vitro. Biol Reprod. 2007a;76:36–42. doi: 10.1095/biolreprod.106.051383. [DOI] [PubMed] [Google Scholar]
- Yang X-y, Li H, Ma Q-w, et al. Improved efficiency of bovine cloning by autologous somatic cell nuclear transfer. Reproduction. 2006;132:733–9. doi: 10.1530/rep.1.01118. [DOI] [PubMed] [Google Scholar]
- Yang XZ, Smith SL, Tian XC, et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007b;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Yong HY, Hao Y, Lai L, et al. Production of a transgenic piglet by a sperm injection technique in which no chemical or physical treatments were used for oocytes or sperm. Mol Reprod Dev. 2006;73:595–9. doi: 10.1002/mrd.20477. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stojkovic P, Przyborski S, et al. Derivation of human embryonic stem cells from developing and arrested embryos. Stem Cells. 2006;24:2669–76. doi: 10.1634/stemcells.2006-0377. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Yang SH, Ding CH, et al. A comparative approach to somatic cell nuclear transfer in the rhesus monkey. Hum Reprod. 2006;21:2564–71. doi: 10.1093/humrep/del216. [DOI] [PubMed] [Google Scholar]