Abstract
Materiomics is an emerging field of science that provides a basis for multiscale material system characterization, inspired in part by natural, for example, protein-based materials. Here we outline the scope and explain the motivation of the field of materiomics, as well as demonstrate the benefits of a materiomic approach in the understanding of biological and natural materials as well as in the design of de novo materials. We discuss recent studies that exemplify the impact of materiomics – discovering Nature’s complexity through a materials science approach that merges concepts of material and structure throughout all scales and incorporates feedback loops that facilitate sensing and resulting structural changes at multiple scales. The development and application of materiomics is illustrated for the specific case of protein-based materials, which constitute the building blocks of a variety of biological systems such as tendon, bone, skin, spider silk, cells, and tissue, as well as natural composite material systems (a combination of protein-based and inorganic constituents) such as nacre and mollusk shells, and other natural multiscale systems such as cellulose-based plant and wood materials. An important trait of these materials is that they display distinctive hierarchical structures across multiple scales, where molecular details are exhibited in macroscale mechanical responses. Protein materials are intriguing examples of materials that balance multiple tasks, representing some of the most sustainable material solutions that integrate structure and function despite severe limitations in the quality and quantity of material building blocks. However, up until now, our attempts to analyze and replicate Nature’s materials have been hindered by our lack of fundamental understanding of these materials’ intricate hierarchical structures, scale-bridging mechanisms, and complex material components that bestow protein-based materials their unique properties. Recent advances in analytical tools and experimental methods allow a holistic view of such a hierarchical biological material system. The integration of these approaches and amalgamation of material properties at all scale levels to develop a complete description of a material system falls within the emerging field of materiomics. Materiomics is the result of the convergence of engineering and materials science with experimental and computational biology in the context of natural and synthetic materials. Through materiomics, fundamental advances in our understanding of structure–property–process relations of biological systems contribute to the mechanistic understanding of certain diseases and facilitate the development of novel biological, biologically inspired, and completely synthetic materials for applications in medicine (biomaterials), nanotechnology, and engineering.
Keywords: biological materials, hierarchies, multiscale, materiomics, deformation, failure, functional material properties, protein, peptide, universality, diversity
Introduction: motivated by Nature
Advances in imaging methods over the past decades have revealed that biology creates intricate hierarchical structures, which when initiated at nanoscales, result in macro or physiological multifunctional materials to provide structural support, force generation, catalytic properties, or energy conversion.1–5 This is exemplified in a rather wide range of biological materials such as hair, skin, bone, spider silk, or cells, which play important roles in providing key functions to biological systems.6 Ye t conventional engineering methods, focused often on analyses at single or few scales (encompassing such fields as structural analysis and continuum theory, eg, continuum mechanics), typically lack the framework required for the complexities introduced by multiscale interactions, the materials’ discrete hierarchical composition, and structure–property dependencies at all scales as found in many natural materials. Mechanical notions of stress and strain, fracture and plasticity, and toughness and robustness are inherently linked to material behavior at the nanoscale. Similarly, chemical and biological techniques (including such fields as microbiology, proteomics, and condensed matter physics) shed vast insights on nanoscale phenomena, such as the chemical composition of materials or the interactions of residues, but lack the association with mechanical properties. Significant advances have been made in many disciplines and research areas, ranging throughout a variety of scales, from atomistic and molecular to continuum. In tandem, experimental studies have attained nanoscale precision, lending insights into molecular defects and mechanisms. As a result of these advances in disparate fields of science, a fully integrated and holistic paradigm now emerges as a powerful approach that can be broadly applied to elucidate Nature’s design principles, to facilitate the design of novel materials with exceptional material properties, and to understand a variety of diseases from a fundamental point of view.
The study of material properties of biological protein materials has witnessed an exciting development over the past several years, partly due to the emergence of physical science-based approaches in the biological sciences. Specifically, there has been significant effort directed toward the explanation and control of observed macroscopic mechanical and optical behavior of complex polymer composites,7,8 while concurrently the structure of many protein-based (polymeric) materials is being discovered, motivating the design of novel ‘synthetic biological’ materials.9 The rapid expansion in the scope of materials science and engineering has led to incorporation of such fields as experimental and computational biology, biomedical engineering, and genetics in the context of natural and synthetic materials. Recent progress provides insight into biological mechanisms and enables us a peek into how biology works at the ultimate, molecular scale and how this relates to macroscopic phenomena such as cell mechanics, tissue behavior, or functions provided by entire organisms. This has resulted in the cross-disciplinary investigation of protein materials and structures, diseases, as well as the development of novel treatment and diagnostics methods.10–15 There is an accelerated progression and convergence of biology, chemistry, materials science, and engineering, each contributing different aspects of the complexity of Nature’s design. The merger of such perspectives is mutually beneficial: materials scientists have extensive experience in treating structures, processes, and properties of materials systematically and with rigorous mathematical methods, whereas biologists have gained a detailed understanding of biological systems and structures and related functions by utilizing both physiological models and powerful statistical correlations between, for example, genetics and physiology and pathology.
There is accelerating interest in the discovery and understanding of Nature’s structural design rules, in particular for nanoscopic hierarchical molecular structures, and to make them available to engineers to pave the way for tomorrow’s supermaterials (eg, mechanomutable materials, advanced composites, low-density low-energy structural materials, etc.), seamlessly blending synthetic materials with biological systems (eg, tissue and biomedical engineering), and using basic biological systems as templates for design (eg, biomimetic and bio-inspired materials). There is also a surprizing relationship between these material design issues and the understanding (or rather lack thereof) of genetic diseases and disorders, where structural changes are due to mutations on the molecular level which lead to changed chemical and mechanical properties, which in turn lead to a malfunction of the protein network under mechanical load. This type of effort, the linking of mechanisms across multiple scales by using a materials science approach to provide structure–property–process (SPP) links, characterizes the emerging field of materiomics.16 The term materiomics has been proposed with various definitions in the past (see Akita et al,17 Buehler et al,16 Buehler and Keten,18 Fernandes et al,19 and Papenburg et al20), where all of the definitions deal broadly with the complexities of synthesis and function of materials and structures. For example, Akita et al proposes materiomics as the systematic study of material composition and structure to determine material properties of metal/metal oxide catalysts,17 whereas Fernandes et al19 and Papenburg et al20 propose materiomics as dealing with the complexities of tissue engineering. It is evident that both definitions encompass the intricacies of complex materials, yet limit scope to specific material systems. We believe that, although slightly different, the definition of materiomics proposed in earlier works fall under a much broader perspective, encompassing even quantum scales,21,22 as well as natural porous granular materials,23 and thus incorporates a vast array of potential applications in science and engineering.
Materiomics contributes to develop a de novo understanding of material processes and to the potential of exploiting novel concepts in technological innovation. More immediate, materiomics finds applications in elucidating the biological role of materials in biology, for instance, in the progression and diagnosis or the treatment of diseases. Other proponents apply materiomics to help identify new material platforms for tissue engineering applications,19,20 for instance, for the de novo development of biomaterials. Inevitably, materiomics holds the promise for nanoscience and nanotechnology, where material concepts from biology might enable the bottom-up development of new structures and materials or devices.
With this impetus, the field of materiomics attempts to reconcile all aspects of a biological material system – from universal motifs of nanoscale building blocks to macroscale functional properties – with a focus on studying the mechanisms of deformation and failure by utilizing a multiscale computational materials science approach. Figure 1 depicts examples of biological protein material systems that innately require a materiomics framework. The importance of multi-scale interactions, hierarchical structuring, and multifunctionality can also be illustrated by using an analogy of music.24,25 Music, akin to protein materials, is founded by a common basis that can be explained by simple physics: sound. Like the combination of elemental building blocks of carbon, hydrogen, and oxygen that constitute protein materials, the phenomenon of music is fundamentally the combination of traveling sound waves with different frequencies. Such sound waves, however, fail to encapsulate Beethoven’s Symphony No. 9, just as a listing of atoms fails to convey the function of a protein. The ‘function’ of music, be it the aural aesthetic or emotional expression, is a result of multiscale phenomenon of resonance and dissonance, the creation of chords and harmonies, the choice of classical piano or electric guitar; combined in one way the result is Mozart, while in another, the result is the Rolling Stones. The structure of music and protein material (including how it is changed) and the particular observer (eg, the audience or the sensing in a particular physiological environment) are inextricably linked. This is a critical issue in both music and protein materials that is due to the way a particular observer processes and interprets functional properties and how this information is used in feedback loops that can alter the structural makeup at various scales. In biology, this may happen through changes in gene expression (at relatively short timescales) and in evolution (at relatively long timescales). In music, this may happen through alteration of music during composing (which could involve continuous revisions to a piece) or through changes in the way a particular musical piece is played in jam sessions based on the audience’s feedback.
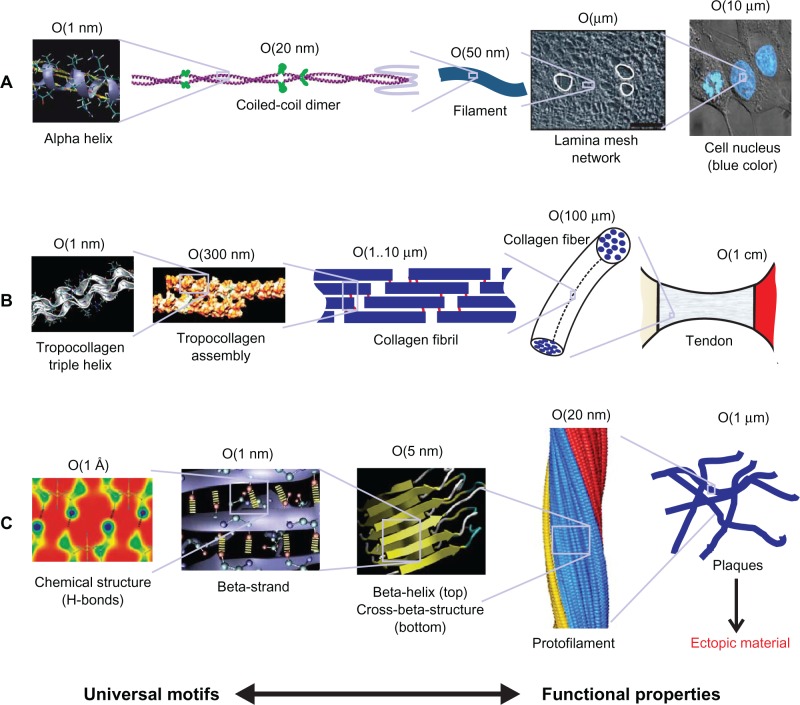
Figure 1.
Simple schematics of three example biological protein materials. A) Intermediate filaments, B) collagenous tissues, and C) amyloid proteins revealing their hierarchical makeup, illustrating potential material candidates benefiting from a materiomic perspective. Materiomics focuses on the development of integrated multiscale material models, focused on mechanical behavior at deformation and failure, fundamentally linked to cross-scale interactions from nano to macro. Copyright © 2010. Buehler and Yung. Adapted with permission from Buehler MJ, Yung YC. How protein materials balance strength, robustness and adaptability. HFSP J. 2010;4(1):26–40.5
Deconstructing the music analogy a little further, we can propose the process of synthesizing orchestra-level music based on four fundamental electronic oscillators (Figure 2). At a fundamental level, our chosen four basic oscillators create sine, square, and other waves, which are considered as raw, unalterable input.26 At the next level, these basic sounds are modulated using envelope generators or filters, employing on-the-fly processing (through Fourier analysis or band-pass filters, eg), which shape the sound of an instrument. The assembly of tones with different duration and pitch over time creates melodies (sometimes referred to as theme or riff), where all tones come from a universal, limited set of harmonics (assembled in octaves). Through the combination of multiple instruments, each of which plays characteristic melodies or riffs, a complex orchestral sound is produced at the largest level. Indeed, while four sound waves differ only minimally in terms of physical properties (frequency, shape, and amplitude), the potential for a great diversity of arrangements is vast. By analogy, sound waves can represent elements, tones can reflect amino acids, protein sequences embody the melody, and their combination can provide the ‘music’ of protein-based structures. Nature has indeed proven to be an adept composer.
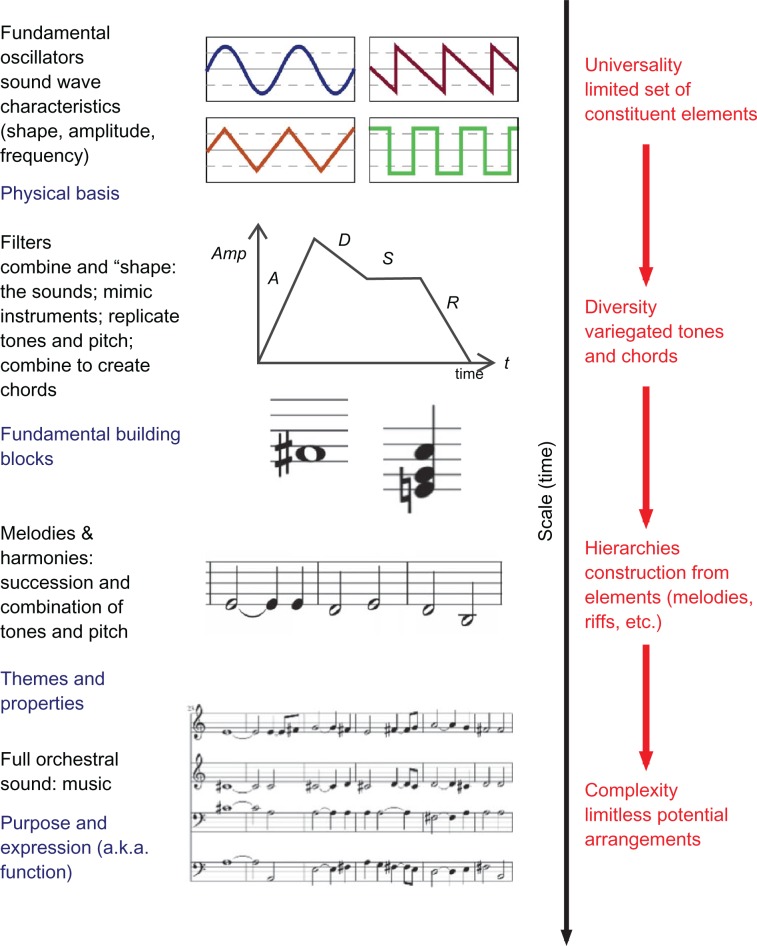
Figure 2.
Illustration of multiscale (or cross-scale) interactions in the case of music, here exemplified for the process of synthesizing orchestra-level music based on four fundamental oscillators (a simplistic model). At a fundamental level, four basic oscillators create unique sound waves characterized by physical properties (ie, shape, amplitude, and frequency). At the next level, these basic sounds are modulated using envelope generators or filters, which shape and mimic the sound of an instrument and construct various tones and pitches. At this level, the fundamental building blocks of music are developed beyond the simple sound waves from which they are composed. The assembly of tones with different duration and pitch over time creates melodies (sometimes referred to as theme or riffs); where all tones come from a universal, limited set of harmonics (assembled in octaves). The theme or type of music is typically dependent on the selection and construction of themes and riffs (ie, the difference between contemporary jazz and classical baroque, for example), which can be thought of as the musical ‘properties’. Through the combination of multiple instruments, each of which plays characteristic melodies or riffs, a complex orchestral sound is produced at the largest level, fulfilling the intended purpose, or musical ‘function’. The example also illustrates how the interplay of diversity and universality provides the fundamental paradigm behind music, resulting in near limitless arrangements from the hierarchical construction of musical elements.
This review article provides an overview of the field of materiomics, including earlier work and future opportunities and intellectual challenges for research. A preliminary discussion to outline the scope and thematic paradigms is provided in Section ‘A synergistic approach: materiomics’, including a definition of the field (Section ‘Definition and scope’), the concept of the materiome (Section ‘Material versus materiome’), underlying material universality and diversity through hierarchies (Section ‘UDP: designing strength from weakness’), and the importance of SPP relations (Section ‘SPP relations: functionality through architecture’). Concepts presented in each section include examples of works in this field. Section ‘Investigative methods: theoretical, computational, and experimental challenges’ provides an overview of investigative methods, including computational and experimental techniques. Section ‘Applied materiomics’ is dedicated to applied materiomics and includes examples where materiomics is currently being utilized with two distinct aims: (i) as a diagnostic tool in the investigation of certain diseases with mechanistic traits which we term pathological materiomics (Section ‘Pathological materiomics: mechanics of disease’) and (ii) as a framework for advanced material design and assembly or materiomic engineering (Section ‘Materiomic engineering: mimicking Nature and materials inspired by biology’). The article concludes in Section ‘Concluding remarks’ with a discussion and an outlook.
The discussions presented in this article are intended to be both a review of current materiomics research as well as a pedagogical discourse. Table 1 presents some definitions of terms introduced and discussed herein. As materiomics is a relatively new field, it behooves us to include discussion to help define and explicate both the intent and scope with analogous examples, illustrating the integrative nature, universality, and benefits and impact of a materiomics approach. The perspectives and overviews presented here are intended to provide a broad overview. Further details can be found in the articles cited.
Table 1.
Summary of a selection of terms and concepts used and/or introduced in this article. The right column of the table provides a brief definition of each term
| Term | Definition |
|---|---|
| Materiomics | The systematic study of the complete material system and the effect on the macroscopic function and failure in a mechanical context, linking processes, structure, and properties at multiple scales, through a materials science perspective, integrating experimental, theoretical, and computational methods. A portmanteau of ‘material’ and the suffix ‘omics’ which refers to ‘all constituents considered collectively’ |
| Materiome | A holistic characterization of a material system, consisting of the material constituents (elemental building blocks and/or structural units), the cross-scale SPP relations (see definition below), and the resulting functionalities/requirements across all levels of hierarchy, from nano to macro |
| Hierarchical system | A system composed of stable, observable subelements that are unified by a superordinate relation. Thereby, lower level details affect higher levels and thus the overall system behavior. A common characteristic of biological materials |
| Complexity | The existence of many interacting components and leads to emerging nonlinear behavior of a system. Complexity in a material system (ie, a complex materiome) necessitates the quantification of cross-scale interactions and mechanisms, which cannot be deduced from general scaling relations |
| UDP | The analysis of materials systems based on the recognition of the universality of structural elements (building blocks) and potential diversity of fundamental functional mechanisms and material behavior |
| SPP relation | The interplay and underlying correlation between a material system’s structure (geometry and material components), resulting properties (stiffness, strength, stability, etc.), and mechanistic processes (including stress transfer, deformation, and eventual failure). The ultimate functionality of the materiome is differentiated from that of the constituent material by the SPP relations |
| Multiscale techniques | Investigative methods, encompassing theoretical, experimental, and computational approaches, which probe material properties across a multitude of length scales. Multiscale techniques aim to establish cross-scale interactions and mechanisms that elucidate SPP relations that supplement material characterization and properties at a single scale level |
| Fine-trains-coarse approach | A bottom-up approach to multiscale model development where parameterization of material behavior at one level (coarse) is fitted from a more sophisticated and robust analysis at a smaller scale (fine), such as fitting molecular force field parameters from quantum mechanical results. Allows efficient computation of subsequent scales with a logical basis in first principles theories |
| Applied materiomics | Practical applications of materiomic techniques and approaches beyond the investigation of material system phenomenon and system characterization. Includes the development of de novo materials or the synthesis and manipulation of biological materials (materiomic engineering), as well as a diagnostic tool for disease and afflictions with mechanistic symptoms (pathological materiomics) |
| Pathological materiomics | The characterization of material properties as manifested for example by genetic disease (eg, point mutations and cellular defects), viral infections (eg, malaria), or injuries/trauma that have a pathological basis in materials behavior, resulting in failure of the material system’s intended function, linking fundamental molecular effects to macroscopic physiological response |
| Materiomic engineering | Materiomic approaches to material system synthesis by utilization of hierarchical structures, self-assembly and/or self-organization processes, and knowledge of the entire materiome of the designed system to explicitly tune mechanistic parameters and behavior, controlling nanoscale components and attain desired macroscopic responses |
A synergistic approach: materiomics
Definition and scope
Materiomics is defined as the systematic study of the complete material system and the effect on the macroscopic function and failure in their mechanical context, linking processes, structure, and properties at multiple scales, from nano to macro, through a materials science perspective, integrating experimental, theoretical, and computational methods. The term has been coined in analogy to genomics – the study of an organism’s entire genome – where, indeed, the suffix ‘omics’ refers to ‘all constituents considered collectively’. The use of a materials science approach to studying biological materials may have broader impact beyond the areas of biological protein materials and biomimetic systems.
Although inspired by genomics, the forerunner of all contemporary ‘omics disciplines,’ the recent explosion and adoption of many omics by researchers (see Greenbaum et al,27 for example) give rise to clarification of the intention of defining materiomics. Traditionally, ‘omics’ is a general term for a broad discipline of science and engineering for analyzing the interactions of biological systems in particular. Such fields are typically characterized by general systems (such as genomics for genes or proteomics for proteins) or processes (eg, interactomics for cellular interactions or mechanomics for stress transfer). Indeed, even the term ‘Omics’ (we use capitalization to denote the field rather the suffix) itself can refer the encompassment of all such bioinformatics research fields to understand all the biological information processing phenomena. Table 2 presents some common ‘omics’ with their associated focus and scope. However, without prudence, the value of a new ‘omic’ could be viewed as self-serving and counterproductive. While the intent of omics in general is the collection of knowledge and information via holistic understanding and integration, the introduction of too many subfields and specialties can promote separation and reductionism of systems and processes under investigations. This is not to devalue the subfields of bioinformatics, where system complexity warrants specialization (eg, characterizing DNA through genomics compared to RNA through ribonomics). Many such fields can be viewed as a hierarchical approach to genomic research. (See Figure 3 for an illustrative example of both genomic and materiomic ‘hierarchies’). To be meaningful beyond a label, new omics should be unifying rather than segregating. With this standpoint, materiomics is neither a subdiscipline of biomaterial engineering, materials science, or mechanics, nor intended to be applied solely to biological systems. Unpresumptuously, materiomics is not introducing a new field of science, but rather encapsulating many fields under a common banner. Just as genomics has motivated research to elucidate biological processes ranging from molecular interactions to complete organisms, it is our hope that the field of materiomics will stimulate extensive research, establishing a hierarchical apex shared between many disciplines promoting integration and collaboration. Indeed, within the biological sciences, the field of genomics has advanced our knowledge base through the successful sequencing of entire genomes. Here, materiomics refers to the general study of a material system’s materiome – the integrated view of the material’s cross-scale interactions that collectively define the material’s properties, function, and purpose.
Table 2.
Some common ‘omics’ with corresponding focus and scope. The brief definitions are meant to provide illustrative descriptions only; citations provided as examples and not intended to be canonical works
| Omic | Focus | Scope |
|---|---|---|
| Omics99 | Analyzing the interactions of biological information in various ‘omes’ | Applied research paradigm to produce knowledge en masse from networks of information via holistic principles and methods |
| Genomics100 | An organism’s entire hereditary information; genome | Determination of entire DNA sequences of organisms, fine-scale genetic mapping including genes, regulatory and noncoding sequences |
| Proteomics101,102 | Protein characterization; protein-coding regions of the genome; proteome | The entire complement of proteins produced by an organism or system, including protein structure, function, and expression |
| Transcriptomics103 | RNA transcripts produced by the genome at any one time; transcriptome | Examines the expression level of RNA in a given cell population, which vary with external environmental conditions, including mRNA, rRNA, tRNA, and noncoding RNA |
| Interactomics104 | Interactions between all macromolecules in a cell; interactome | Analysis and characterization of gene–gene, protein–protein, or protein–ligand interactions; development of molecular interaction maps/networks |
| Mechanomics105,106 | Mechanical systems and processes within an organism; mechanome | General role of force, stress transfer, mechanics, and molecular machinery in biology, encompassing biological motors, mechanical structures, and processes |
| Materiomics [this article/issue] | Material characterization through components, structure, and function; materiome | Analysis of material systems through constitutive components, hierarchical SPP relations, cross-scale interactions, and effects on functionality |
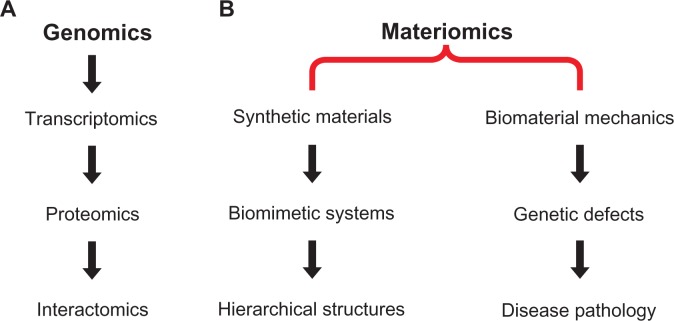
Figure 3.
Example flow of information under genomics and materiomics frameworks. A) Genomics encompasses the entire genetic sequence, which includes specific DNA sequences transcribed to RNA molecules (transcriptomics), in turn, mRNA from a DNA template carry the coding information required for protein synthesis and expression (proteomics), finally, the mapping of protein–protein interactions networks can be characterized by interactomics. It is noted that this is merely one possible flow of information under genomics, with many interactions possible between subdisciplines. B) Two potential paths are given for materiomics. First, of all classes of synthetic materials being developed, a subset may find inspiration from biological materials. From these bio-inspired or biomimetic materials, the motivation may arise from multiscale hierarchical structures, such as those found in spider silk, wood, or bone. Materiomics provides a potential framework for the development of such de novo materials. Second, there is an advancing knowledge base on the mechanical behavior and properties of biomaterials, both at the molecular and system levels (eg, cellular mechanics or soft tissue behavior). At the molecular level, genetic point defects (ie, mutations) can lead to mechanical changes expressed at the macroscale. Such pathology can be quantified and analyzed, leading to new diagnostic and treatment methods for certain diseases. The diverse aims of biomimetic material design and disease pathology can be unified under a materiomic paradigm through the understanding of material systems and functionalities.
In general, materiomics can refer to the study of a broad range of materials, which includes metals, ceramics, and polymers in addition to biological materials such as bone, skin, cells, silk, or ectopic materials (such as amyloids), and the study of interfaces between living and nonliving systems (eg, synthetic tissue scaffolds). Materiomics is thus inherently multidisciplinary (Figure 4), borrowing from fundamental physics and chemistry at the atomistic scale, integrating biological mechanisms at the molecular and cellular level, traversing hierarchical scales, and linking a material’s structure and mechanical properties with its natural requirements and functionalities. Materiomics involves the rigorous understanding of the properties (eg, mechanical, physical, and chemical) and mechanisms (eg, chemomechanical conformation changes, enzymatic processes, and mechanotransduction) of biological matter, which may enable us eventually to integrate concepts from living systems into materials and machine design, seamlessly. Solving these challenging problems may transcend the gap that currently exists between engineering and physical sciences and the life sciences.
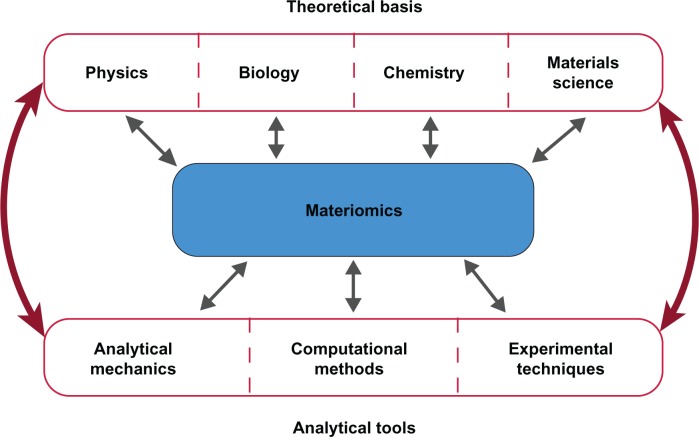
Figure 4.
The study of materiomics has a multidisciplinary theoretical, computational, and experimental foundation resulting from the historical progression of physics, biology, chemistry, and materials science. Each has contributed to the development of nanotechnology and increased knowledge base on the fundamental behavior and functional (eg, mechanical) properties of biological materials, but only through a cross-discipline ‘collective’ approach can the complexity of Nature’s design be understood, and eventually utilized. We thus label this integrated holistic study of Nature’s materials as materiomics. Nature effortlessly incorporates such disparate fields at the nano and molecular scales, requiring a prudent usage of different theory and investigative techniques. Opportunely, analytical techniques such as continuum mechanics, computational methods (including statistical mechanics, molecular dynamics, finite element approaches, etc.), and experimental techniques (such as AFM, nanoindentation or magnetic/optical tweezers) have recently developed the precision and sophistication required to investigate materials throughout a wide range of length and timescales.
Material versus materiome
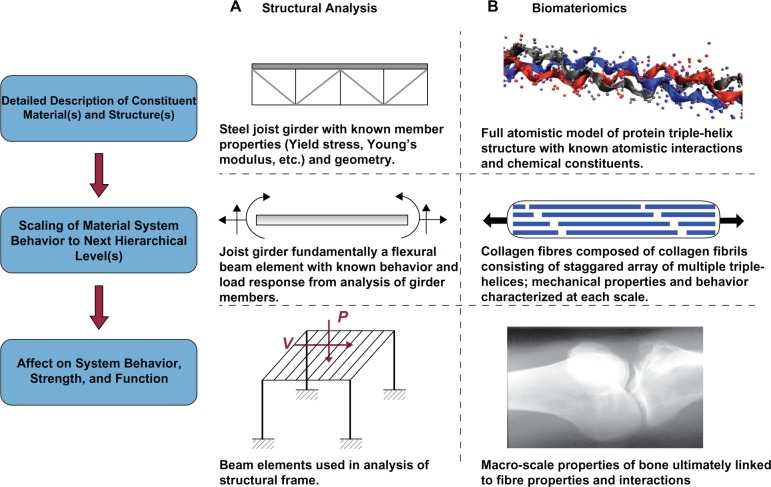
Materiomics, as currently presented, is the study of a system’s materiome or the ‘complete’ material system – its constituents and structure, properties and processes, function, failure, and behavior – in its entirety. The goal is to link the disparate nature of the physical description of a material (ie, components and structure) with the related phenomenological functionalities (ie, strength and robustness). The approach is partially motivated by macroscale engineering techniques such as structural analysis. For example, it is a rather trivial analysis procedure to determine the flexural behavior of a steel joist girder if the geometry and material properties of the truss members are known (see Figure 5). The behavior of an individual joist, in turn, affects the behavior of the system in which it is contained (such as a simple roofing system). If we consider the truss arrangement as the first hierarchy, it is apparent that the mechanical properties of the material used to build the truss, as well as the structure of the truss itself, ultimately affect the mechanical properties and failure of the system. However, at the macroscale, by convention, there is a distinct differentiation between the ‘material’ and the ‘structural system’. Typically, one would not associate conventional material properties such as Young’s modulus or Poisson’s ratio to a roofing frame. At the nanoscale, however, the distinction is not as clear.
Figure 5.
Comparison between the hierarchical component analysis of traditional structural analysis and materiomics, illustrating material constituent descriptions to system level behavior. A) Structural analysis: The flexural behavior of a steel joist girder can be easily determined if the geometry and material properties of the truss members are known. The behavior of an individual joist, in turn, affects the behavior of the system in which it is contained (such as a simple roofing system). If we consider the truss arrangement as the first hierarchy, it is apparent that the mechanical properties of the material used to build the truss, as well as the structure of the truss itself, ultimately affect the mechanical properties and failure of the overall system. B) Biomateriomics: Multiscale hierarchical depiction of bone, a material for which there has been ample research at multiple scale levels, including the triple-helical polypeptide structure, the subsequent formation of collagen fibrils, and the ultimate macroscale system. Unlike a steel frame, however, the system-level (eg, bone) properties are not reduced to the mechanical properties of the first hierarchical level (protein triple-helices). It is apparent that, as the scale of the material is decreased, the distinction between what exactly is labeled as the ‘material’ and the ‘structure’ is simply a matter of perspective, and without a fundamental distinction. Materiomics provides a unifying framework for such hierarchical systems that connote a seamless merger of structure and material throughout all scales.
Consider the hierarchical nature of bone (see Figure 1B) as an example, for which there has been ample research at multiple hierarchical levels, including the triple-helical polypeptide structure, the subsequent formation of collagen fibrils, and the ultimate macroscale system of bone through both computational28 and experimental methods.29–31 Unlike a steel frame, however, the system-level (bone) properties are not reduced to the mechanical properties of the firsta hierarchical level (protein triple-helices). In fact, a literature review results in different Young’s moduli defined for tropocollagen triple-helices,28 collagen fibrils,32 and bone.33 It is apparent that, as the scale of the material is decreased, the distinction between what exactly is labeled the ‘material’ and the ‘structure’ is simply a matter of perspective. Within the context of the materiome, the material and structural information are considered at all hierarchical levels. While a material description of bone can be defined by the macroscale properties such as Young’s modulus or fracture strength, the complete materiome of bone necessarily includes the material information at all levels of hierarchy, the structure– property relations between hierarchies, and the associated functionalities of the system. Figure 6 summarizes the information contained with each hierarchical level of the materiome.
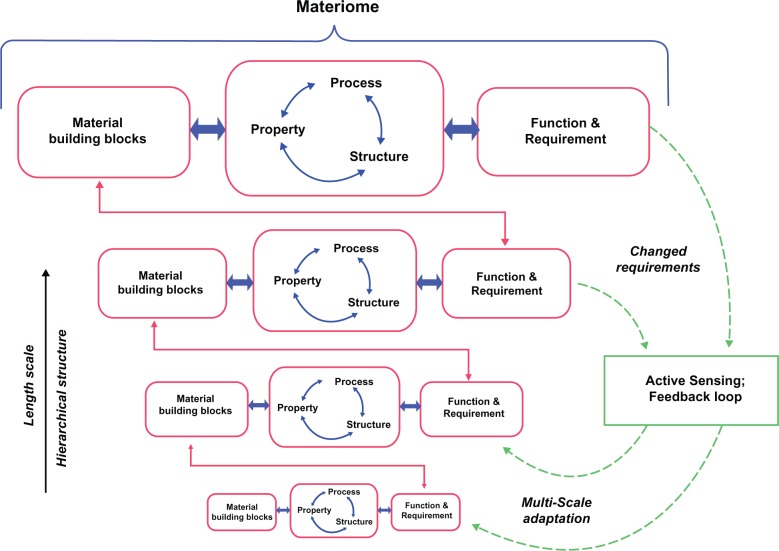
Figure 6.
Schematic representation of materiomic information, consisting of the material constituents (elemental building blocks and/or structural units), the cross-scale SPP relations (structural geometry, stress and strain transfer, and failure mechanisms), and the resulting functionalities/requirements (strength, robustness, toughness, etc.) across all levels of hierarchy.2,38,39 In biological materials hierarchical structures, decentralized processes, material properties, and environmental requirements are brought together in mutual completion. In contrast to a traditional materials science paradigm, relations between ‘external’ functions/requirements and ‘internal’ properties exist on several scales resulting in multifunctionality. Furthermore, as requirements are consistently changing over time (eg, changing loads due to growth or physical activity, changing environment, pathological mechanisms, etc.), continuous adaptation is necessary and is enabled through the existence of active sensing and feedback loops. For example, a macroscale signal such as mechanical strain can induce a change at the gene level (gene expression), which can then induce a cascade of biological mechanisms, including for example, tissue growth or remodeling, or the formation of new structures as in angiogenesis. These mechanisms allow decentralized self-organization and multiscale adaptation of a biological system.
From a cursory perspective, the materiome may be considered merely a ‘multiscale snapshot’ of a material system, that is, simply a catalog of material properties and functions throughout different scales. What such a simple snapshot would lack, however, is the communication and cross-scale interactions that define the functionality of complex materiomic systems. In other words, the materiome provides not only the answer to what the material system is in terms of components, structure, and properties, but also to why the system is the way it is and how it is and/or how it can be manipulated. For example, knowledge of the spatial relationships and interactions of genes and regulatory elements in the cell nucleus are revealing an extensive network of communication within and between chromosomes.34 Such interactions are, not surprisingly, inherently multiscale with nanoscale details exhibited throughout hierarchical levels.14 A simple material description of the chromosome nucleotides and structure is unable to construe such information, as the local environment and material requirement effect gene expression. Such gene expressions continuously change the material but maintain a constant materiome, underlining a clear differentiation of the two concepts. A crucial unresolved issue is the extent to which this organization affects gene function, rather than just reflecting it. By unlocking the complete materiomic information, efforts have been made to utilize gene regulation in the self-assembly and organization structural DNA materials.35–37 Such applications are only possible through the integration of multiscale feedback, chemical interactions, and structural–property relations, which are central to the field of materiomics.
Inspired by biological materials’ hierarchical structures, decentralized processes, material properties, and environmental requirements, materiomics amalgamates the combined effects to mutual completion. In contrast to the traditional paradigm in materials science, relations between ‘external’ functions/requirements and ‘internal’ properties exist on several scales resulting in multifunctionality. Although, requirements are consistently changing (eg, changing loads and changing environment) on several time and length scales, in addition to multifunctionality, robust feedback loops are required and enable decentralized self-organization and self-optimization.
The consideration of the complete materiome of a material system allows a fundamental bottom-up design of purpose-specific materials from the atomistic to the continuum levels. Granted, the understanding of the materiome is still at its infancy, where the role of the relationship between processes, structures, and properties of materials in biological organisms is thus far only partially explored and understood. Approaches in studying the materiome include multiscale simulation methods (eg, molecular dynamics and finite element analysis), multiscale experiments (eg, AFM, optical tweezers, etc.), as well as high-throughput methods based on combination of these techniques. The objective is to ultimately bridge hierarchical levels and piece together not only material properties and structures at the nano-and microscales, but also the ultimate effects on both the mechanical properties and function of the entire material system. A complete understanding of the materiome elucidates not only the cross-scale relations between hierarchies and mechanical properties, but also offers clues how to assemble new materials with disparate and mechanical properties from few constituent building blocks and to identify novel approaches in designing materials that evolve autonomously to adapt to changes in environmental conditions.
With a materiomics perspective, two common themes are frequently encountered that warrant a thorough discussion: (i) the universality–diversity paradigm (UDP) and (ii) SPP relationships.
UDP: designing strength from weakness
The importance of hierarchies is elusive for many material systems. For the sake of argument, one may ask why such complex and redundant hierarchies are even necessary. The simple answer is that hierarchical structures are not required, but they serve to extend the physical design space, while limited to a restricted set of constituent building blocks. In other terms, it provides material scientists and engineers more design parameters to manipulate within the same set of building blocks (ie, structural elements). Nature implements this trick frequently, where a major trait of biological materials is the occurrence of not only hierarchies, but also hierarchies abundant with weak interactions (such as hydrogen bonds), resulting in robust and tough materials that are currently designed. The paradox of a ‘strong’ material being composed of ‘weak’ components has remained difficult to theoretically reconcile. The entire materiome across multiple scales must be introduced, where universal and diverse patterns are unified hierarchically, and the integrated view of it results in a quantitative understanding of how highly functional materials are created based on inferior building blocks.
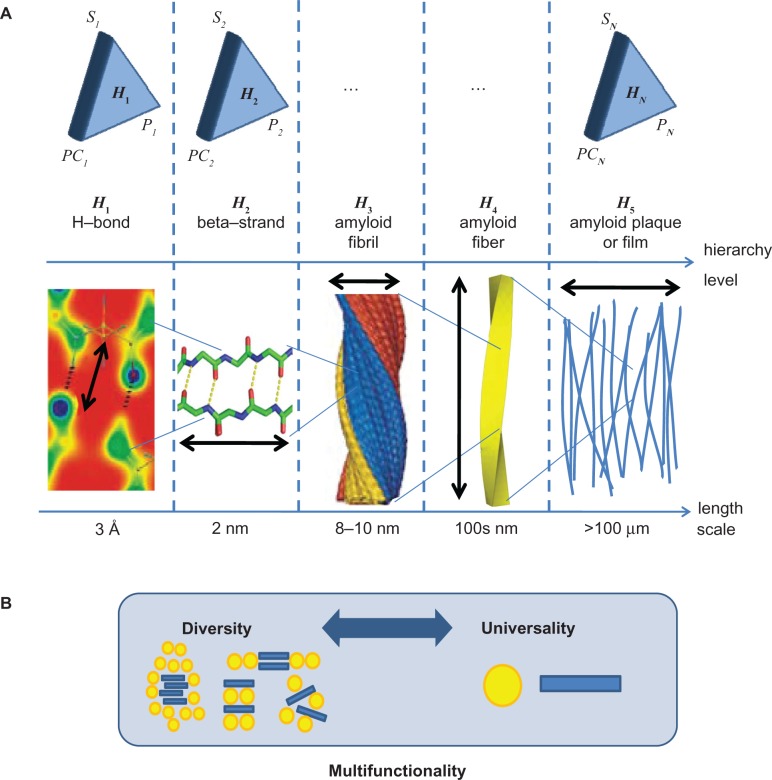
The UPD incorporates the recognition and analysis of biological materials based on the universality and diversity of its fundamental structural elements and functional mechanisms.38,39 For example, proteins constitute the elementary building blocks of a vast variety of biological materials such as cells, spider silk, or bone, where they create extremely robust, multifunctional materials by self-organization of structures over many length and timescales, from nano to macro. Examples of such universal building blocks include α-helices, β-sheets, or tropocollagen molecules. In contrast, other features are highly specific to tissue types, such as particular filament assemblies, β-sheet nanocrystals in spider silk or tendon fascicles. Similarly, cellulosic materials,40 such as wood, grasses, and other green plants, exhibit a wide array of macroscale mechanical properties dependent on the fiber morphology and structure,41,42 yet are composed of similar molecular building blocks (various polysaccharides in both crystalline and amorphous phases43). It is apparent that using only a limited number of components, Nature has produced a broad range of materials with diverse properties and biological functions and created multifunctionality (diversity) by changing structural arrangements of few (universal) constituents rather than inventing new building blocks. The key to achieving diverse properties from a limited set of available building blocks is by expansion of the design space – diversity and optimization through hierarchies (Figure 7).
Figure 7.
UDP. A) General view of hierarchical structural formation from levels H1 to HN. At each hierarchical level there exists a compartmentalized interplay of structure (Si), processes (PCi), and properties (Pi), resulting in a particular hierarchical function/requirement. Each hierarchical level contributes to the system’s entire materiome, as depicted in Figure 6. The lower part shows the hierarchical structure exemplified through amyloid protein material, from weak hydrogen bonding to β-strand structures, fibrils to fibers, and ultimately plaques (see Knowles et al,85 for example). B) Schematic illustration of the interplay of diversity and universality. The integration of diverse and universal features provides the structural basis to achieve multifunctionality without the need to introduce new building blocks. Copyright © 2010. Nature Publishing Group, a division of Macmillan Publishers Limited. Adapted with permission from Buehler MJ. Nanomaterials: strength in numbers. Nat Nanotechnol. 2010;5:172–174.39
A recent materiomics study involved a large number of α-helical elements in all possible hierarchical combinations, while keeping the total material use constant.44 By rearranging the same number of nanoscale elements into hierarchies, the performance of the material in the strength-robustness space was fundamentally changed. Such an investigation demonstrates that the continuous invention of new basic building blocks to obtain changes in performance is unnecessary. The application of universal building blocks in highly diverse architectures might be a strategy that enables adaptation to changes in material requirements by simply adopting new structural arrangements of the same basic constituent components.
This coexistence of universality and diversity as described in the UDP is an overarching feature in biological materials and a crucial component of materiomics. Nature has a unique capacity of creating toughness from weak components, capable of balancing multiple, seemingly incompatible properties such as strength, robustness, and adaptability. Materials like bone, being a nanocomposite of strong but brittle and soft but ductile materials, illustrate this unification of components with disparate properties within a hierarchical structure. Primarily, the utilization of structural hierarchies enables the coexistence of universality and diversity. Indeed, material characteristics such as strength and robustness are contradicting properties that cannot be easily combined within a single scale of traditional materials science, requiring a materiomics approach to reconcile such disparate concepts. Further, such a perspective clearly indicates that structures and processes are amalgamated and cannot be considered alone.
SPP relations: functionality through architecture
As discussed, the UDP illustrates the vast design repertoire found in Nature. However, biological materials have the capacity to integrate multiple, and at times, disparate properties, unaccounted for by simple hierarchical arrangement. The addition of hierarchies is fruitless without consideration of hierarchical function – the structure at each hierarchical level is associated with a specific property and essentially compartmentalized at that scale.
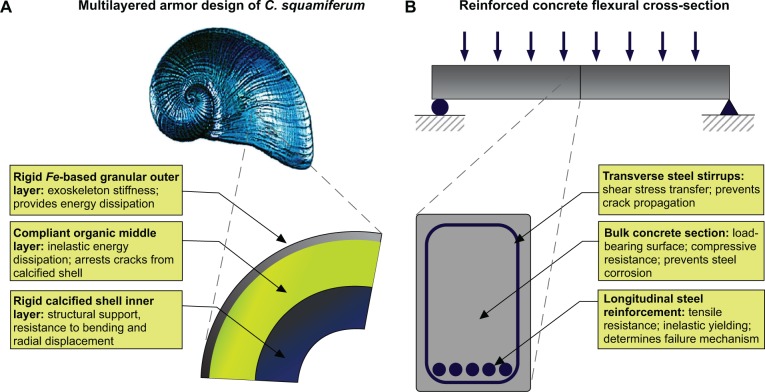
To exemplify, there has been much study on the structure– property relationships of numerous mollusk shell nacreous microstructures,45–47 in which it is generally concluded that the amplification of mechanical properties (increase in strength and toughness) exhibited by biological composites compared to their individual constituent materials is beyond simple rule of mixture formulations. Synthetic structural materials that take advantage of the hierarchical structure–property relationships of such composite systems are increasingly being realized.48,49 A recent study of the shell of a deep-sea hydrothermal vent gastropod50 the material properties of the multilayered shell are dependent on the specific combination of different materials (building blocks), the microstructures, interfacial geometries, gradation, and layering (SPP), which are advantageous for penetration resistance, energy dissipation, mitigation of fracture and crack arrest, reduction of defections, and resistance to bending, and tensile loads (function and requirement). Such investigations help elucidate the gastropod shell materiome, integrating the effect of contrasting material components (compliant organic layers and stiff mineralized platelets) with the mechanical properties of the entire material system. Indeed, each layer of the shell is responsible for distinct and multifunctional roles in mechanical protection (see Figure 8A).
Figure 8.
Natural and engineered examples of SPP relations. A) Schematic of the multilayered functionalities of the shell of Crysomallon squamiferum (adapted from Yao et al).50 each material layer serves distinct functional roles, contributing to the overall mechanical behavior of the shell. B) Cross-section of reinforced concrete flexural member as used in civil engineering. Each structural component is utilized for distinct purpose, the combination of which determines the properties and ultimate function of the flexural member. A materiomics approach facilitates the characterization of such hierarchical structured materials by probing structure–property relations and resulting functionalities.
We can compare the ‘compartmentalized function’ of a gastropod’s shell with a more common macroscale system, a reinforced concrete flexural member (Figure 8B). Here, we consider the materiome of the composite concrete and steel system. Through engineering of the cross-section, the structure and specific placement of the material components serve a distinct mechanical role. The concrete serves as the primary load-bearing medium while protecting the steel elements from corrosion. The longitudinal steel reinforcement is designed to carry tensile stress, while the concrete carries compressive stress. Indeed, the amount of steel ultimately dictates the failure mechanism of the beam (brittle or ductile failure). Finally, the transverse steel stirrups increase the shear capacity of the member while limiting crack propagation. An integrated view of the flexural member’s materiome provides complete view of the materials (concrete and steel), the SPP relations (cross-sectional geometry, stress distributions, and failure mechanisms), and the ultimate function (flexure). Of course, a concrete cross-section is a rather trivial example, not requiring a materiomics perspective. However, it illustrates the subtle interplay between SPP relations and function. For example, based on the geometry of the cross-section alone, one could deduce the function of the member – as a beam subject to a positive bending moment. Alternate functions (such as a cantilever member or as a column) would require variations in the structure but could (potentially) implement the same material components. The structure – not the material – determines the function.
Functionality is ultimately obtained through material architecture. In the case of the concrete beam, an engineer, through analysis of structural requirements, determined macroscale structure. However, the architecture of the gastropod shell, adequately summarized at the microscale as a composite of organic layers and mineralized platelets, explicates only the highest level of hierarchy. Hierarchical sublevels including the protein-based composure of the organic layer(s)51,52 and the properties of the organic-mineral interfaces53 are still being investigated in nacreous materials and are ultimately required for a complete description of the materiome and potential design of de novo synthetic materials.54 We next focus on the techniques currently being implemented to undertake multiscale materiomic investigations.
Investigative methods: theoretical, computational, and experimental challenges
To realize the promising opportunities that arise from an improved understanding of hierarchical protein materials, several critical challenges must be overcome. Up until now, theories describing hierarchical biological materials are still lacking. Virtually no understanding exists about how specific features at distinct scales interact and, for example, participate in mechanical deformation. As materiomics is founded by a combination of multidisciplinary theories and multiscale techniques, approaches that integrate experiment and predictive simulation are essential to this new paradigm of materials research. The behavior of biological materials, in particular their mechanical properties, are intimately linked to the atomic microstructure of the material. Whereas crystalline materials show mechanisms such as dislocation spreading or crack extension, biological materials feature molecular unfolding or sliding, with a particular significance of rupture of chemical bonds such as hydrogen bonds, covalent cross-links, or intermolecular entanglement. Much different mechanisms operate at larger length scales, where the interaction of extracellular materials with cells and of cells with one another, different tissue types and the influence of tissue remodeling become more evident. The dominance of specific mechanisms is controlled by geometrical parameters, the chemical nature of the molecular interactions, as well as the structural arrangement of the protein elementary building blocks, across many hierarchical scales, from nano to macro. Thus, materiomic investigative approaches must also consider multiscale schemes, both experimentally and computationally, to link hierarchical effects and mechanisms.
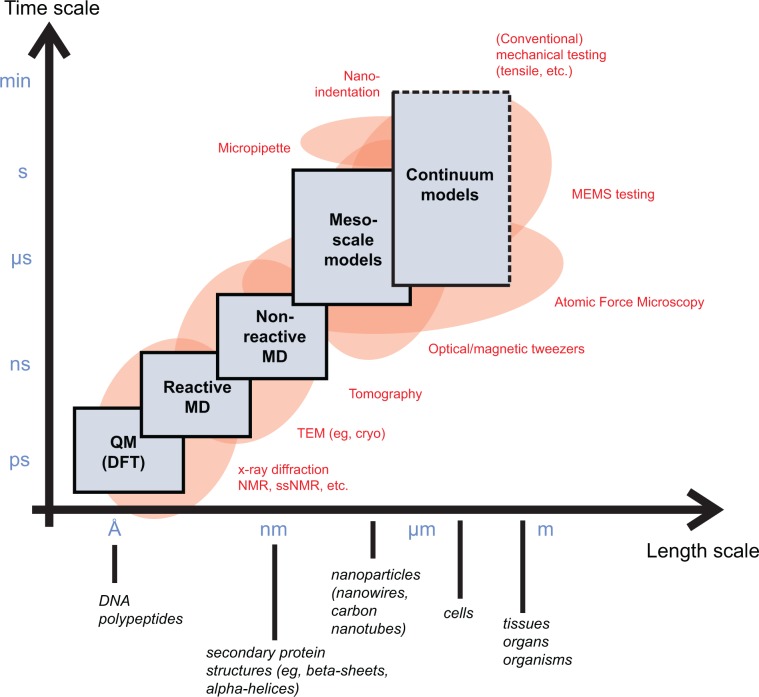
Experimental techniques have gained unparalleled accuracy in both length and timescales, as reflected in development and utilization of atomic force microscope (AFM),46,55,56 magnetic and optical tweezers15,57 or nanoindentation58,59 to analyze biological protein materials and biological molecules.60,61 Single-molecule and single-cell biomechanics assays are necessary to provide critical information and experimental support to develop theoretical materiomic models. At the same time, modeling and simulation have evolved into predictive tools that complement experimental analyzes at comparable length and timescales. Multiscale simulation models for biological materials have become increasingly popular in recent years and have enabled the direct link between experiment and theoretical bottom-up descriptions of materials (Figure 9).
Figure 9.
Overview of computational and experimental methods for materiomic investigations, in particular enabling a multiscale perspective of structures, mechanisms, and properties. Hierarchical coupling of different computational tools can be used to traverse throughout a wide range of length and timescales. Such methods enable one to provide fundamental insight into deformation and fracture phenomena, across various time and length scales. Handshaking between different methods enables one to transport information from one scale to another. Eventually, results of atomistic, molecular, or mesoscale simulation may feed into constitutive equations or continuum models. Experimental techniques, such as the AFM, molecular force spectroscopy, nanoindentation, or magnetic/optical tweezers, now overlap into atomistic and molecular approaches, enabling direct comparison of experiment and simulation. Techniques such as X-ray diffraction, infrared spectroscopy, or NMR provide atomic-scale resolution information about the 3-D structure of protein molecules and protein assemblies. Copyright © 2010. Nature Publishing Group, a division of Macmillan Publishers Limited. Adapted with permission from Buehler MJ, Yung YC. Deformation and failure of protein materials in physiologically extreme conditions and disease. Nat Mater. 2009;8(3):175–188.2
In the field of atomistic-based multiscale stimulation, it is now possible to begin from the smallest scales (considering electrons and atoms) to reach all the way up to macroscopic scales of filaments, fibrils, fibers, and entire tissues, by explicitly considering the characteristic structural features at multiple materiomic hierarchies. Such approaches are possible with the advent of first principles based multiscale simulation techniques (see, for instance, a review article for a broad introduction into this field62). The basic principle underlying these multiscale simulation methods is finer scales train coarser scales.
Even though there are still major challenges ahead of us, this progress now provides one with many opportunities, transforming biomechanics as a discipline through increased integration of computational approaches in scientific research.
Applied materiomics
Irrespective of the challenges still present in a thorough investigation and complete characterization of the materiome as discussed above, current experimental and practical approaches exist that allow the immediate application of materiomics to real problems. This branch of materiomics, termed applied materiomics, is still in its infancy, yet has already demonstrated potential as a valuable basis for material design. Applied materiomics aims beyond the investigation of material system phenomenon and system characterization and includes the development of de novo materials, the synthesis and manipulation of biological materials, as well as a diagnostic tool for disease and afflictions, for example, with mechanistic symptoms. We hence focus discussion on two broad areas of application that are becoming increasingly widespread (throughout different disciplines) and can be encompassed by the common field of applied materiomics: i) pathological materiomics and ii) materiomic engineering.
Pathological materiomics: mechanics of disease
The characterization of material properties for biological materials may also play a crucial role in developing a better understanding of diseases, an application we term pathological materiomics. Injuries and genetic diseases are often caused by structural changes in protein materials (eg, defects, flaws, changes to the molecular structure), resulting in failure of the material’s intended function. These observations may eventually provide explanations of the molecular origin of certain diseases, which exhibit changes in material properties. Additionally, these findings provide evidence that material properties play an essential role in biological systems and that the current paradigm of focusing on biochemistry alone as the cause of diseases is insufficient. It is envisioned that the long-term potential impact of materiomics can be used to predict diseases in the context of diagnostic tools by measuring material properties, rather than focusing on symptomatic chemical readings alone. Such approaches have been explored for cancer and malaria, for instance.12,63 The scale-bridging paradigm of materiomics can potentially emerge as a critical niche in the development of links between failure of biological materials in the context of genetic and infectious disease. Such diseases can be characterized by single point mutations, genetic deficiencies or alterations, or chromosomal aberrations, inherently molecular triggers that lead to dramatic, catastrophic effects at significantly larger scales. A materiomics investigation can be advantageous when such molecular changes are manifested in mechanical behavior and materials phenomenon.
As an illustrative example linking mechanistic response and disease, we can consider the material behavior of human red blood cells in the context of an infectious disease – specifically Plasmodium falciparum malaria. The cause of malaria, a disease that affects nearly 8% of the work population and causing nearly 2–3 million deaths annually,64 is the protoctistan parasite Plasmodium. The parasites penetrate red blood cells and continually change the structure of the cell’s spectrin network.65 Pioneering experimental investigations by Suresh et al have indicated that during the course of a 48-h period after invasion of the red blood cell by the parasite, the effective stiffness of the cell increases by more than a factor of 10.63 The combination of the reduction of deformability and a marked increase in the adhesion of the red blood cells results in obstructed flow of cells through the microvasculature.66 These mechanical factors associated with cell deformability and cytoadherence are considered to be key mechanistic pathways in the pathogenic basis of the disease.67
Current investigative techniques to probe malaria-infected red blood cells have been a combination of experimental and computational methods across a multitude of scales, exemplifying an integrated materiomics framework. The mechanical deformation characteristics of red blood cells have been experimentally determined through such techniques as micropipette aspiration,68 optical tweezers,15,69 and flow studies through microfluidic channels.70 Such biomechanical assays are required to provide critical information regarding disease progression and treatment by monitoring the systematic alterations of cell structure and response. Further, the obtained empirical data may provide the necessary information to parameterize computational models (either independently or in correlation with bottom-up atomistic methods) and facilitate the development of deterministic models.71
In the case of malaria, infection leads to marked changes in the molecular structure of the red blood cell. The experimental investigations cited were not focused on the molecular interactions and triggers initiated by the parasite at the nanoscale, but rather the manifested mechanistic effects, including increased rigidity and cytoadherence, compromised motility and sequestion in microvasculature. With a focus on the entire cell structure, the multiscale bridging previously stressed as critical to a materiomics approach appears to be abandoned. However, red blood cells afflicted with malaria parasites demonstrate the need for the apt consideration of biochemical processes, SPP relationships, and ultimate mechanical response of biological materials within a comprehensive materiomic framework. Indeed, genetic diseases, such as hereditary hemolytic disorders, where the inherited mutations are manifested in the mechanical properties of red blood cells, are being investigated using similar materiomic methods.72 In cases where the disease rises from a genetic mutation, the aim of materiomics is to link the mutations at the nanoscale with the behavior at the macroscale with a bottom-up approach. Such a case involves the investigation of osteogenesis imperfecta (brittle bone disease, often abbreviated as OI).
Osteogenesis imperfecta is a genetic disorder in collagen characterized by mechanically weakened tendon, fragile bones, skeletal deformities, and in severe cases, prenatal death.73 Previous studies have provided a general correspondence between the specific mutation types with phenotypic severity; however, the molecular and mesoscale mechanics by which a single point mutation influences the mechanical behavior of collagenous tissues at multiple length scales remain unknown. Using a materiomics approach, a series of systematic multiscale computational experiments were recently reported, focused on pure collagenous tissue and collagen fibrils to investigate the effect of osteogenesis imperfecta mutations on single molecule properties, changes to intermolecular interactions, and changes to the mechanical properties of collagen fibrils.74,75 The fundamental question addressed is how it is possible that a single point mutation at the level of a single tropocollagen molecule can lead to the failure of macroscopic tissue? It was demonstrated that the mutations cause a fundamental change in stress distribution within the collagen fibrils due to the formation of nanocracks that cause local stress concentrations at the mutations. Ultimately, the molecular scale models predict a softening that the emergence of these stress concentrations in osteogenesis imperfecta could play a role in the physiological effects of the disease as they lead to macroscopically weaker connective tissue, including tendon and in particular bone. The study is a culmination of both multiscale hierarchical constitutive material models for collagenous tissues28,32,76 as well as known clinical and experimental data for the fundamental point mutations of osteogenesis imperfecta.77–79 The integration and reciprocation of computational methods with empirical data are a keystone to materiomics, albeit further work is required to validate the model through quantitative comparison with experiment to link the predictions at larger tissue levels. The benefits of a deterministic mechanistic model from nanoscale point mutations to macroscale effects are evident, and similar approaches could be used for many other diseases in which materials’ failure due to a drastic change of the materiome behavior play a crucial role in disease initiation and progression. The key insight put forth here is that for a comprehensive understanding of disease states such as brittle bone disease, an integrated view of material and structure at multiple scales is critical to link physiological mechanisms and clinical evidence and to develop potential treatment options.
As illustrated, a materiomics approach can be advantageous when pathological conditions ultimately lead to a change in mechanical behavior, providing potential to greatly enhance our understanding of the role of materials’ phenomenon in biological systems through both experimental and computational investigations. Consideration of how mechanical behavior and material properties change in diseases could lead to new pathological insights that expand beyond biochemical signals and interactions. Additionally, conventional models of failure and disease that only consider one level of the material’s structure do not capture the full range of relevant hierarchies and mechanisms and as such remain limited in their ability to describe material breakdown processes associated with disease. Materiomics provides a suitable framework to reconcile the multiscale mechanisms of disease and tissue failure that could prove beneficial for diagnostics and treatment, complementary to current physiological approaches.
Materiomic engineering: mimicking Nature and materials inspired by biology
The ability to design synthetic materials at the same level of complexity of Nature has been a fundamental challenge for science. Only recently has technology evolved to unlock the secrecy of (some) biological self-assembly processes while allowing direct (limited) manipulation of material components at the molecular scale. Synthesis techniques have advances to the point where the complexity of nanostructures that can be fabricated rivals that of naturally occurring structures.80–84 However, the scaling of hierarchical materials from nanoscale building blocks to macroscale functionality is a nontrivial progression. Nature, of course, takes advantage of natural self-assembly over a relatively large timeframe to ‘guide the production’ of material systems. A possible route to the development of bio-inspired and biomimetic systems is the understanding and exploitation of this self-assembly. One such example is the production of nanostructured biofilms from amyloidogenic proteins.85
Amyloids are a class of protein materials that have an innate capacity to form hierarchical structures (Figure 1C). Amyloids can form from diverse protein sequences86 through a process called amyloidogenesis, where proteins lose their native functional configuration (eg, as enzyme or hormone) and form fibers with a characteristic hydrogen bonded cross-β-sheet structure. These characteristic features are universal to the broader family of all amyloids, despite different protein sequences, which has been linked to their high stability, stiffness, and capacity to provide structural templates across length scales.87–89 Diverse features, such as variations in the protein sequence, give rise to biochemical properties specific to particular amyloids.
Knowles et al elegantly exploit these unique properties of amyloids to form multifunctional materials by controlling the interplay of universality and diversity at different levels of hierarchy.85 The assembly procedure results in an intentional hierarchy of length scales: nanometer ordering within the fibrils and micrometer scale ordering in the stacking of the fibrils. At a fundamental level, the protein sequence can be altered to design the biochemical properties of amyloids. Similarly, chemical functionalization can be added to realize hierarchical structures of molecules that would not naturally form. At larger hierarchical levels, the arrangement of amyloid fibrils in the material developed by Welland and coworkers can be controlled by introducing plasticizing molecules, enabling one to precisely tune the material’s mechanical properties – films produced through protein self-assembly are highly rigid with a Young’s modulus up to 5–7 GPa, comparable to the highest values for proteinaceous materials found in Nature. These structural alterations make it possible to create a broad range of functional properties based on a limited set of elements through manipulation of assembly and control of structural hierarchies, that is, the explicit control of the materiome.
It is noted that such amyloid structures were initially investigated due to the pathological association with Alzheimer’s and Parkinson’s diseases.90,91 Not only are there inherent benefits in understanding amyloid-type materials from a diagnostic perspective, but also the ability to manipulate and engineer the properties such a protein material prompts the natural utilization as a potential tissue scaffold. The presence of amyloid structures, albeit in a disease state, substantiates suitable biochemical and physio-chemical factors, which perhaps can be utilized to improve or replace biological functions through tissue engineering.
Tissue engineering is associated with applications that repair or replace portions of or whole tissues (ie, bone, cartilage, blood vessels, etc.). Often, the tissues involved require certain mechanical and structural properties for proper functioning, which can be realized though a materiomic framework. The development of suitable scaffolds for tissue engineering involves an implicit balance between architecture and specific tissue type by consideration of both microstructure and microenvironment.20 For example, a critical parameter of tissue scaffolds is the relative pore size to facilitate nutrient transport.92 Commonly, there is an inverse relation between pore size and mechanical stability, which can be reconciled through a complete understanding of the material’s SPP relations – the same relations that materiomics aims to elucidate.
A potential candidate for tissue engineering scaffold is the extracellular matrix (ECM), which is a key component in natural regeneration and maintenance of tissues and organs.19 Methods of producing ECM-inspired tissue platforms, including hydrogels93 and electrospun micro- and nanofribrous scaffolds,94 are successful in replicating the required physio-chemical properties and structural features of their natural analogs, but, in most cases, do not match the mechanical properties of the tissue to be regenerated. ECM is typically composed of collagen, elastin, glycosaminoglycan, laminin, fibrin, and other proteins that contribute to the functionalities of the material. For example, the elasticity of the matrix can determine stem cell differentiation: soft matrices are neurogenic, stiffer matrices are myogenic, and rigid matrices are osteogenic.95 The structure–property relations of the constituent protein components are fundamental in the development of tissue scaffolds to accurately mimic the natural composition of ECM and to understand the role of material stiffness changes in disease states.
The multiscale investigations and integration of such protein-based materials are naturally within the realm of materiomics, with recent studies being undertaken for collagen,28,57 elastin,96,97 and fibrin,98 for instance. The integration of the result of each investigation ultimately contributes to the materiomic information, required for successful tissue engineering and biomaterial development.
Concluding remarks
The advent of materiomics is not an attempt to introduce a new field of science. Rather, it is a unifying proposition motivated by the convergence of many fields toward a fundamental integrated description of materials and their functional roles. Materiomics takes a materials science perspective toward complex biological systems, explicitly accounting for feedback loops that link functional requirements (and changes thereof) to altered material components and structure, at different scales in both time and length. The incentive for materiomics is fourfold:
Primarily, to amalgamate the advancing knowledge base of chemistry, biology, materials science, and mechanics to a single, holistic description of a material system from nano to macro. This complete description, the materiome, contains the information required to manipulate mechanical function and properties and development of new, predictive materiomic theories and models, which specifically include the hallmark of living systems – feedback loops that facilitate an autonomous sensing, structural change, and as a result, adaptation to altered environmental conditions and functional requirements.
Investigative methods developed from a multidisciplinary perspective for multiscale analysis can be applied to a vast amount of material systems, both current and future, offering new insights and theoretical formulations unavailable to past biochemists, medical engineers, and materials scientists.
The application of materiomics can unlock fundamental design principles of Nature’s materials based on high-throughput computational, theoretical, and experimental methods and utilize this insight in the development of advanced materials, biological and synthetic, micro-and macroscopic. This harnessing of Nature’s ‘trade secrets’ could usher in new technologies that are currently unattainable without the integrative approaches a materiomics perspective provides. Indeed, such integrative approaches are already being implemented as discussed above, albeit by different research groups with disparate goals.
Materiomics highlights the similarities and promotes a mutually beneficial relation between all researchers and scientists concerned with biomimetic materials, advanced composite design, nanotechnology, medical engineering, tissue engineering, mechanisms of disease, genetic defects, and any field requiring the complete description and understanding of a specific materiome.
As illustrated, materiomics is a powerful tool to enhance the understanding of materials in biology, at multiple scales and in a variety of functional contexts, contributing to the development of a holistic understanding of biological processes and to the potential of exploiting novel concepts in technological innovation for de novo materials design. It is vital to overcome the barrier that currently separates the understanding at different length and timescales, through the development of new experimental synthesis and characterization methods, novel model systems, and an enhanced appreciation for a multiscale view of materials in general, to fully understand multiscale or cross-scale interactions in materiomics.
A materiomics perspective can be taken for rather simple materials and material systems (such as a reinforced concrete member or a steel truss), but it is then a rather trivial matter. More complex systems, such as stimuli-responsive polymer composites,8 can be (and have been) understood through multiscale holistic perspectives that materiomics promotes. Beyond structure–property relations, materiomics further wishes to encompass cross-scale interactions, multifunctionality, feedback, and adaptation (as depicted in Figure 6) that is common to biological systems, and currently being developed in advanced polymer systems and emerging nanoscale devices. Such a unifying approach is only made possible by the concurrent advances in multiscale analysis, chemical characterization, computational methods, and experimental techniques developed across a spectrum of disciplines.
The long-term goal is to develop a new engineering paradigm that encompasses the analysis and design of structures and materials, starting from the molecular level, to create new materials that mimic and exceed the properties of biological ones by utilizing material concepts discovered in biological materials. Such work can lead to the development of a new set of tools that can be applied, together with synthetic biological and self-assembly methods, to select, design, and produce a new class of materials, similar to the approaches used today in computer aided design of buildings, cars, and machines. The availability of multifunctional and changeable materials reduces the necessity for the use of different materials to achieve different properties and, as such, may provide significant savings in weight and cost. The utilization of abundant natural building blocks such as organic (eg, peptides or proteins) or inorganic (eg, minerals) constituents, combined with novel synthesis techniques based on self-assembly, could lead to new lightweight materials for structural applications in cars, airplanes, and buildings that could reduce the overall energy consumption and ecological footprint of materials.
Materiomics provides an exciting opportunity for the analysis and engineering of complex biological systems based on quantitative insight into their fundamental physical and chemical features. A rigorous understanding may enable us eventually to integrate concepts from living systems into engineering materials design, seamlessly. Optical, mechanical, and electrical properties at ultrasmall material scales, their control, synthesis, and analysis, as well as their theoretical description, represent major scientific and engineering opportunities. However, as in the case of conventional ‘engineered’ materials, these breakthroughs may only be possible provided that their fundamental concepts are well understood and appropriate models developed. Characterization of materials found in biology – in particular in the context of living systems – enabled through a rigorous materiomics approach is aimed toward the elucidation of these fundamental principles of assembly, deformation, and possible failure.
The field of materiomics provides a powerful integrated theoretical framework for complex hierarchical materials, which enables us to define future scientific hypotheses in the field of biological and synthetic materials and nanotechnology in a systematic way. Such hypothesis must be proved through a unified approach that combines theory, experiment, and simulation, leading to a detailed understanding of how Nature successfully links structure, processes, properties, and functions simultaneously over many length scales, from nano to macro.
Acknowledgments
This work was supported primarily by a NSF CAREER grant (award number CMMI-0642545) and the NSF-MRSEC Program (award number DMR-0819762). Additional support from ONR, AFOSR, MITEI, and ARO is greatly acknowledged.
Disclosure
The authors report no conflicts of interest in this work.
Indeed, from a different viewpoint, the first hierarchical level can be considered the constituent amino acids, forming polypeptides, which subsequently form the triple-helix. The hierarchies presented here are labeled for illustrative purposes.
References
- 1.Lakes R. Materials with structural hierarchy. Nature. 1993;361:511–515. [Google Scholar]
- 2.Buehler MJ, Yung YC. Deformation and failure of protein materials in physiologically extreme conditions and disease. Nat Mater. 2009;8(3):175–188. doi: 10.1038/nmat2387. [DOI] [PubMed] [Google Scholar]
- 3.Buehler MJ, Ackbarow T. Nanomechanical strength mechanisms of hierarchical biological materials and tissues. Comput Methods Biomech Biomed Eng. 2008;11(6):595–607. doi: 10.1080/10255840802078030. [DOI] [PubMed] [Google Scholar]
- 4.Wegst UGK, Ashby MF. The mechanical efficiency of natural materials. Philos Mag. 2004;84(21):2167–2181. [Google Scholar]
- 5.Buehler MJ, Yung YC. How protein materials balance strength, robustness and adaptability. HFSP J. 2010;4(1):26–40. doi: 10.2976/1.3267779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fratzel P, Weinkamer R. Nature’s hierarchical materials. Prog Mat Sci. 2007;52:1263–1344. [Google Scholar]
- 7.Ober CK, Cheng SZD, Hammond PT, et al. Research in macromolecular science: challenges and opportunities for the next decade. Macromolecules. 2009;42(2):465–471. [Google Scholar]
- 8.Stuart MAC, Huck WT, Genzer J, et al. Emerging applications of stimuli-responsive polymer materials. Nat Mater. 2010;9(2):101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 9.Doktycz MJ, Simpson ML. Nano-enabled synthetic biology. Mol Syst Biol. 2007;(3):125. doi: 10.1038/msb4100165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao G, Suresh S. Cell and molecular mechanics of biological materials. Nat Mater. 2003;2(11):715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 11.Dahl KN, Scaffidi P, Islam MF, Yodh AG, Wilson KL, Misteli T. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc Natl Acad Sci U S A. 2006;103(27):10271–10276. doi: 10.1073/pnas.0601058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 13.Meyers MA, Chen PY, Lin AYM, Seki Y. Biological materials: structure and mechanical properties. Prog Mater Sci. 2008;53(1):201–206. doi: 10.1016/j.jmbbm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Kepper N, Foethke D, Stehr R, Wedemann G, Rippe K. Nucleosome geometry and internucleosomal interactions control the chromatin fiber conformation. Biophys J. 2008;95:3692–3705. doi: 10.1529/biophysj.107.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dao M, Lim CT, Suresh S. Mechanics of the human red blood cell deformed by optical tweezers. J Mech Phys Solids. 2005;53(2):493–494. [Google Scholar]
- 16.Buehler MJ, Keten S, Ackbarow T. Theoretical and computational hierarchical nanomechanics of protein materials: deformation and fracture. Prog Mater Sci. 2008;53:1101–1241. [Google Scholar]
- 17.Akita T, Ueda A, Yamada Y, et al. Analytical TEM observations of combinatorial catalyst libraries for hydrogen production – As a part of ‘MATERIOMICS’. Comb Artif Intell Methods Mater Sci II. 2004;804:211–216. [Google Scholar]
- 18.Buehler MJ, Keten S. Elasticity, strength and resilience: a comparative study on mechanical signatures of α-Helix, β-sheet and tropocollagen domains. Nano Res. 2008;1(1):63–71. [Google Scholar]
- 19.Fernandes H, Moroni L, van Blitterswijk C, de Boer J. Extracellular matrix and tissue engineering applications. J Materials Chem. 2009;19:5474–5484. [Google Scholar]
- 20.Papenburg BJ, Liu J, Higuera GA, et al. Development and analysis of multi-layer scaffolds for tissue engineering. Biomaterials. 2009;30:6228–6239. doi: 10.1016/j.biomaterials.2009.07.057. [DOI] [PubMed] [Google Scholar]
- 21.Kang B, Ceder G. Battery materials for ultrafast charging and discharging. Nature. 2009;458:190–193. doi: 10.1038/nature07853. [DOI] [PubMed] [Google Scholar]
- 22.Hafiner J, Wolverton C, Ceder G. Toward computational materials design: the impact of density functional theory on materials research. MRS Bull. 2006;31(9):659–668. [Google Scholar]
- 23.Ulm FJ, Abousleiman Y. The nanogranular nature of shale. Acta Geotechnica. 2006;1(2):77–88. [Google Scholar]
- 24.Buehler MJ. Tu(r)ning weakness to strength. Nano Today. 2010. [DOI]
- 25.Kamien R. Music: an appreciation. New York: McGraw-Hill Humanities/Social Sciences/Languages; 2007. [Google Scholar]
- 26.Dodge C, Jerse TA. Computer music: synthesis, composition, and performance. Florence, KY: Cengage Learning; 1997. [Google Scholar]
- 27.Greenbaum D, Luscombe NM, Jansen R, Qian J, Gerstein M. Interrelating different types of genomic data, from proteome to secretome: ’Oming in on function. Genome Res. 2001;11(9):1463–1468. doi: 10.1101/gr.207401. [DOI] [PubMed] [Google Scholar]
- 28.Buehler MJ. Atomistic and continuum modeling of mechanical properties of collagen: elasticity, fracture and self-assembly. J Mater Res. 2006;21(8):1947–1961. [Google Scholar]
- 29.Sasaki N, Odajima S. Elongation mechanism of collagen fibrils and force-strain relations of tendon at each level of structural hierarchy. J Biomech. 1996;29(9):1131–1136. doi: 10.1016/0021-9290(96)00024-3. [DOI] [PubMed] [Google Scholar]
- 30.van der Rijt JA, van der Werf KO, Bennink ML, Dijkstra PJ, Feijen J. Micromechanical testing of individual collagen fibrils. Macromol Bio-sci. 2006;6:697–702. doi: 10.1002/mabi.200600063. [DOI] [PubMed] [Google Scholar]
- 31.Eppell SJ, Smith BN, Kahn H, Ballarini R. Nano measurements with micro-devices: mechanical properties of hydrated collagen fibrils. J R Soc Interface. 2006;3(6):117–121. doi: 10.1098/rsif.2005.0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buehler MJ. Nanomechanics of collagen fibrils under varying crosslink densities: atomistic and continuum studies. J Mech Behav Biomed Mater. 2008;1:59–67. doi: 10.1016/j.jmbbm.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 33.Spatz H-C, O’Leary EJ, Vincent JFV. Young’s moduli and shear moduli in cortical bone. Proc Biol Sci. 1996;263(1368):287–294. doi: 10.1098/rspb.1996.0044. [DOI] [PubMed] [Google Scholar]
- 34.Bickmore W. Nuclear organisation of chromatin: links to normal genome function and to disease. Chromosome Res. 2007;15:2–2. [Google Scholar]
- 35.Seeman NC. An overview of structural DNA nanotechnology. Mol Biotechnol. 2007;37(3):246–257. doi: 10.1007/s12033-007-0059-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen ES, Dong M, Nielsen MM, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459(7243):73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 37.Lin C, Liu Y, Yan H. Designer DNA nanoarchitectures. Biochemistry. 2009;48(8):1663–1674. doi: 10.1021/bi802324w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackbarow T, Buehler MJ. Hierarchical coexistence of universality and diversity controls robustness and multi-functionality in protein materials. J Comput Theor Nanosci. 2008;5(7):1193–1204. [Google Scholar]
- 39.Buehler MJ. Nanomaterials: strength in numbers. Nat Nanotechnol. 2010;5:172–174. doi: 10.1038/nnano.2010.28. [DOI] [PubMed] [Google Scholar]
- 40.Eichhorn SJ, Baillie CA, Zafeiropoulos N, et al. Current international research into cellulosic fibres and composites. J Mater Sci. 2001;36:2107–2131. [Google Scholar]
- 41.Hamad W. Cellulosic materials: fibers, networks and composites. Boston: Kluwer Academic Publishers; 2002. [Google Scholar]
- 42.Abe K, Yano H. Comparison of the characteristics of cellulose microfibril aggregates of wood, rice straw and potato tuber. Cellulose. 2009;16(6):1017–1023. [Google Scholar]
- 43.Nishiyama Y. Structure and properties of the cellulose microfibril. J Wood Sci. 2009;55:241–249. [Google Scholar]
- 44.Qin Z, Cranford S, Ackbarow T, Buehler MJ. Robustness-strength performance of hierarchical alpha-helical protein filaments. Int J Appl Mech. 2009;1(1):85–112. [Google Scholar]
- 45.Wang RZ, Suo Z, Evans AG, Yao N, Aksay IA. Deformation mechanisms in nacre. J Mater Res. 2001;16(9):2485–2493. [Google Scholar]
- 46.Smith BL, Schäffer TE, Viani M, et al. Molecular mechanistic origin of the toughness of natural adhesives, fibres and composites. Nature. 1999;399:761–763. [Google Scholar]
- 47.Barthelat F, Li C-M, Comi C, Espinosa HD. Mechanical properties of nacre constituents and their impact on mechanical performance. J Mater Res. 2006;21(8):1977–1986. [Google Scholar]
- 48.Ortiz C, Boyce MC. Bioinspired structural materials. Science. 2008;319(5866):1053–1054. doi: 10.1126/science.1154295. [DOI] [PubMed] [Google Scholar]
- 49.Munch E, Launey ME, Alsem DH, Saiz E, Tomsia AP, Ritchie RO. Tough, bio-inspired hybrid materials. Science. 2008;322:1516–1520. doi: 10.1126/science.1164865. [DOI] [PubMed] [Google Scholar]
- 50.Yao H, Dao M, Imholt T, et al. Protection mechanisms of the iron-plated armor of a deep-sea hydrothermal vent gastropod. Proc Natl Acad Sci U S A. 2010;107(3):987–992. doi: 10.1073/pnas.0912988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyers MA, Lin AY, Chen PY, Muyco J. Mechanical strength of abalone nacre: role of the soft organic layer. J Mech Behav Biomed Mater. 2008;1(1):76–85. doi: 10.1016/j.jmbbm.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 52.Song F, Soh AK, Bai YL. Structural and mechanical properties of the organic matrix layers of nacre. Biomaterials. 2009;24:3623–3631. doi: 10.1016/s0142-9612(03)00215-1. [DOI] [PubMed] [Google Scholar]
- 53.Yao N, Epstein AK, Liu WW, Sauer F, Yang N. Organic-inorganic interfaces and spiral growth in nacre. J R Soc Interface. 2009;6:367–376. doi: 10.1098/rsif.2008.0316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang Z, Kotov NA, Magonov S, Ozturk B. Nanostructured artificial nacre. Nat Mater. 2003;2:413–418. doi: 10.1038/nmat906. [DOI] [PubMed] [Google Scholar]
- 55.Prater CB, Butt HJ, Hansma PK. Atomic force microscopy. Nature. 1990;345(6278):839–840. [Google Scholar]
- 56.Cross SE, Kreth J, Zhu L, et al. Nanomechanical properties of glucans and associated cell-surface adhesion of Streptococcus mutans probed by atomic force microscopy under in situ conditions. Microbiology. 2007;153:3124–3132. doi: 10.1099/mic.0.2007/007625-0. [DOI] [PubMed] [Google Scholar]
- 57.Sun YL, Luo ZP, Fertala A, An KN. Stretching type II collagen with optical tweezers. J Biomech. 2004;37(11):1665–1669. doi: 10.1016/j.jbiomech.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 58.Tai K, Ulm FJ, Ortiz C. Nanogranular origins of the strength of bone. Nano Lett. 2006;6(11):2520–2525. doi: 10.1021/nl061877k. [DOI] [PubMed] [Google Scholar]
- 59.Ebenstein DM, Pruitt LA. Nanoindentation og biological materials. Nano Today. 2006;1(3):26–33. [Google Scholar]
- 60.Lim CT, Zhou EH, Li A, Vedula SRK, Fu HX. Experimental techniques for single cell and single molecule biomechanics. Mater Sci Eng C Biomim Supramol Syst. 2006;26(8):1278–1288. [Google Scholar]
- 61.van Vliet KJ, Bao G, Suresh S. The biomechanics toolbox: experimental approaches for living cells and biomolecules. Acta Mater. 2003;51(19):5881–5905. [Google Scholar]
- 62.Goddard WA. A perspective of materials modeling. In: Yip S, editor. Handbook of Materials Modeling. New York: Springer; 2006. [Google Scholar]
- 63.Suresh S, Spatz J, Mills JP, et al. Connections between single-cell biomechanics and human disease states: gastrointestinal cancer and malaria. Acta Biomater. 2005;1:15–30. doi: 10.1016/j.actbio.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 64.Cooke BM, Mohandas N, Coppel RL. Malaria and the red blood cell membrane. Semin Hematol. 2004;41(2):173–188. doi: 10.1053/j.seminhematol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 65.Cooke BM, Mohandas N, Coppell RL. The malaria-infected red blood cell: structural and functional changes. Adv Parasitol. 2001;50:1–86. doi: 10.1016/S0065-308X(01)50029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Suwanarusk R, Cooke BM, Dondrop AM, et al. The deformability of red blood cells parasitized by Plasmodium falciparum and P-vivax. J Infect Dis. 2004;189(2):190–194. doi: 10.1086/380468. [DOI] [PubMed] [Google Scholar]
- 67.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature. 2002;415(6872):673–679. doi: 10.1038/415673a. [DOI] [PubMed] [Google Scholar]
- 68.Evans EA. Bending elastic-modulus of red-blood-cell membrane derived from buckling instability in micropipet aspiration tests. Biophy J. 1983;43(1):27–30. doi: 10.1016/S0006-3495(83)84319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mills JP, Qie L, Dao M, Lim CT, Suresh S. Nonlinear elastic and viscoelastic deformation of the human red bllod cell with optical tweezers. Mech Chem Biosyst. 2004;1(3):169–180. [PubMed] [Google Scholar]
- 70.Shelby JP, White J, Ganesan K, Rathod PK, Chiu DT. A microfluidic model for single-cell capillary obstruction by Plasmodium falciparum infected erythrocytes. Proc Natl Acad Sci U S A. 2003;100(25):14618–14622. doi: 10.1073/pnas.2433968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lim CT, Zhou EH, Quek ST. Mechanical models for living cells – a review. J Biomech. 2006;39(2):195–216. doi: 10.1016/j.jbiomech.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 72.Suresh S. Mechanical response of human red blood cells in health and disease: some structure-property-function relationships. J Mater Res. 2006;21(8):1871–1877. [Google Scholar]
- 73.Rauch F, Glorieux FH. Osteogenesis imperfecta. Lancet. 2004;363(9418):1377–1385. doi: 10.1016/S0140-6736(04)16051-0. [DOI] [PubMed] [Google Scholar]
- 74.Gautieri A, Uzel S, Vesentini S, Redaelli A, Buehler MJ. Molecular and mesoscale mechanisms of osteogenesis imperfecta disease in collagen fibrils. Biophys J. 2009;67:857–865. doi: 10.1016/j.bpj.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gautieri A, Vesentini S, Redaelli A, Buehler MJ. Single molecule effects of osteogenesis imperfecta mutations in tropocollagen protein domains. Prot Sci. 2008;18:161–168. doi: 10.1002/pro.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Buehler MJ. Nature designs tough collagen: explaining the nanostructure of collagen fibrils. Proc Natl Acad Sci U S A. 2006;103:12285–12290. doi: 10.1073/pnas.0603216103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sillence DO, Senn A, Danks DM. Genetic heterogeneity in osteogenesis imperfecta. J Med Genet. 1979;16:101–116. doi: 10.1136/jmg.16.2.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dalgleish R. The human type I collagen mutation database. Nucleic Acids Res. 1997;25(1):181–197. doi: 10.1093/nar/25.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beck K, Chan VC, Shenoy N, Kirkpatrick A, Ramshaw JA, Brodsky B. Destabilization of osteogenesis imperfecta collagen-like model peptides correlates with the identity of the residue replacing glycine. Proc Natl Acad Sci U S A. 2000;97(8):4273–4278. doi: 10.1073/pnas.070050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang SG. Fabrication of novel biomaterials through molecular self-assembly. Nat Biotechnol. 2003;21(10):1171–1178. doi: 10.1038/nbt874. [DOI] [PubMed] [Google Scholar]
- 81.Lu W, Lieber CM. Nanoelectronics from the bottom up. Nat Mater. 2007;6(11):841–850. doi: 10.1038/nmat2028. [DOI] [PubMed] [Google Scholar]
- 82.Kol N, Adler-Abramovich L, Barlam D, Shneck RZ, Gazit E, Rousso I. Self-assembled peptide nanotubes are uniquely rigid bioinspired supramolecular structures. Nano Lett. 2005;5(7):1343–1346. doi: 10.1021/nl0505896. [DOI] [PubMed] [Google Scholar]
- 83.Rothemund PWK. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 84.Goodman RP, Schaap IA, Tardin CF, et al. Rapid chiral assembly of rigid DNA building blocks for molecular nanofabrication. Science. 2005;310(5754):1661–1665. doi: 10.1126/science.1120367. [DOI] [PubMed] [Google Scholar]
- 85.Knowles TP, Oppenheim TW, Buell AK, Chirgadze DY, Welland ME. Nanostructured biofilms from hierarchical self-assembly of amyloidogenic proteins. Nat Nanotechnol. 2010;5:204–207. doi: 10.1038/nnano.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kayed R, Head E, Thompson JL, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 87.Knowles TP, Fitzpatrick AW, Meehan S, et al. Role of intermolecular forces in defining material properties of protein nanofibrils. Science. 2007;318:1900–1903. doi: 10.1126/science.1150057. [DOI] [PubMed] [Google Scholar]
- 88.Cherny I, Gazit E. Amyloids: not only pathological agents but also ordered nanomaterials. Angew Chemie Int Ed. 2008;47(22):4062–4069. doi: 10.1002/anie.200703133. [DOI] [PubMed] [Google Scholar]
- 89.Hamley IW. Peptide fibrillization. Angew Chem Int Ed. 2007;46(43):8128–8147. doi: 10.1002/anie.200700861. [DOI] [PubMed] [Google Scholar]
- 90.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 91.Kelly JW. Towards an understanding of amyloidogenesis. Nat Struct Biol. 2002;9(5):323–325. doi: 10.1038/nsb0502-323. [DOI] [PubMed] [Google Scholar]
- 92.Karande TS, Ong JL, Agrawal CM. Diffusion in muscoskeletal tissue engineering scaffolds: design issues related to porosity, permeability, architecture and nutrient mixing. Ann Biomed Eng. 2004;32(12):1728–1743. doi: 10.1007/s10439-004-7825-2. [DOI] [PubMed] [Google Scholar]
- 93.Lee KY, Mooney DJ. Hydrogels for tissue engineering. Chem Rev. 2001;101(7):1869–1879. doi: 10.1021/cr000108x. [DOI] [PubMed] [Google Scholar]
- 94.Teo WE, He W, Ramakrishna S. Electrospun scaffold tailored for tissue-specific extracellular matrix. Biotechnol J. 2006;1(9):918–929. doi: 10.1002/biot.200600044. [DOI] [PubMed] [Google Scholar]
- 95.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 96.Valiaev A, Lim DW, Schmidler S, Clark RL, Chilkoti A, Zauscher S. Hydration and conformational mechanics of single, end-tethered elastin-like polypeptides. J Am Chem Soc. 2008;130(33):10939–10946. doi: 10.1021/ja800502h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Djajamuliadi J, Kagawa TF, Ohgo K, Kumashiro KK. Insights into a putative hinge region in elastin using molecular dynamics simulations. Matrix Biol. 2009;28(2):92–100. doi: 10.1016/j.matbio.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 98.Brown AEX, Litvinov RI, Discher DE, Purohit PK, Weisel JW. Multiscale mechanics of fibrin polymer: gel stretching with protein unfolding and loss of water. Science. 2009;325(5941):741–744. doi: 10.1126/science.1172484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ward DC, White DC. The new ’omics era. Curr Opin Biotechnol. 2002;13(1):11–13. [Google Scholar]
- 100.Schattner P. Genomics made easier: an introductory tutorial to genome datamining. Genomics. 2009;93(3):187–195. doi: 10.1016/j.ygeno.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 101.Wilkins M. Proteomics data mining. Expert Rev Proteomics. 2009;6(6):599–603. doi: 10.1586/epr.09.81. [DOI] [PubMed] [Google Scholar]
- 102.Wilkins MR, Appel RD, van Eyk JE, et al. Guidelines for the next 10 years of proteomics. Proteomics. 2006;6(1):4–8. doi: 10.1002/pmic.200500856. [DOI] [PubMed] [Google Scholar]
- 103.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stumpf MPH, Thorne T, de Silva E, et al. Estimating the size of the human interactome. Proc Natl Acad Sci U S A. 2008;105(19):6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lang M. Lighting up the mechanome. Bridge. 2007;37(4):11–16. [Google Scholar]
- 106.Sem DS, Yu L, Coutts SM, Jack R. Object-oriented approach to drug design enabled by NMR SOLVE: first real-time structural tool for characterizing protein-ligand interactions. J Cell Biochem Suppl. 2001;37:99–105. doi: 10.1002/jcb.10070. [DOI] [PubMed] [Google Scholar]