Abstract
There is a great deal of interest in the possibility that complex nanoscale devices can be designed and engineered. Such devices will lead to the development of new materials, electronics and smart drugs. Producing complex nanoscale devices, however will present many challenges and the components of such devices will require a number of special features. Devices will be engineered to incorporate desired functionalities but, because of the difficulties of controlling matter precisely at the nanoscale with current technology, the nanodevice components must self-assemble. In addition, nanocomponents that are to have wide applicability in various devices must have enough flexibility to integrate into a large number of potentially very different environments. These challenges are daunting and complex, and artificial nanodevices have not yet been constructed. However, the existence of nanomachines in nature in the form of proteins (eg, enzymes) suggests that they will be possible to produce. As the material from which nature’s nanomachines are made, proteins seem ideal to form the basis of engineered components of such nanodevices. Initially, engineering projects may focus on building blocks such as rings, cages and tubes, examples of which exist in nature and may act as a useful start point for modification and further development. This review focuses on the recent research and possible future development of such protein building blocks.
Keywords: bionanotechnology, protein engineering, nanomachine, building-blocks, synthetic biology
Introduction
Since Richard Feynman’s famous 1959 talk at Caltech; “There’s Plenty of Room at the Bottom” (Feynman), it has been one of science’s most ambitious dreams to build complex, multi-functional, nanoscale devices, also known as nanomachines. There is much controversy in the scientific community as to whether complex nanodevices such as universal assemblers are feasible (Smalley 2001; Baum 2003). To date no one has succeeded in manufacturing a working, complex, artificial nanodevice. Obstacles to successful production include the materials used and the difficulty in manipulating nanometric components. Amongst non-biological approaches, carbon structures such as carbon nanotubes (CNTs) offer exciting possibilities (Baughman et al 2002). However, a number of difficulties are associated with such nanotubes. It is worth considering these in some detail as many of the problems typify the general difficulties inherent in non-biological methods and highlight potential areas in which biological molecules, particularly proteins may be able to offer significant advantages.
Production of CNTs can be difficult and/or expensive and generally relies on one of three methods: arc discharge (Iijima 1991; Ebbesen and Ajayan 1992), laser ablation (Guo et al 1995) or chemical vapor deposition (Jose-Yacaman et al 1993). While much progress has been made in the industrialization of multi-walled CNT production, production of single wall CNTs (SWNTs) whose diameters typically lie in the 0.4 to 3 nm range (Baughman et al 2002) have proved much more difficult to scale up, and remain expensive. Other problems associated with CNT production are controlling precisely the length of the produced tubes and, in the case of SWNTs, avoiding contamination (Baughman et al 2002). Defects in carbon nanotubes are common and ironically these are often necessary to allow modification of the tubes, providing convenient “hooks” for the addition of required molecules (Hirsch 2002). Problems in controlling exactly the length of produced CNTs are obstacles to their use in nanodevices where, as with components of macro-scale machines, precise dimensions engineered within fine tolerances may be required. Furthermore, in order to integrate such components into larger scale devices, modification to incorporate extra functionalities may be necessary. The small size of the basic component of the carbon nanotube (a single carbon atom) in which all the valence bonds are accounted for, has several implications: The basic component has no structure in itself and overall structure is determined by the covalent bonds with its neighbors. Thus, modification of carbon nanotubes without disrupting structure is difficult. Although progress has been made in this areas using, for example, non-covalent modification (Hirsch 2002; Simmons et al 2007), the structure of the CNTs themselves places limits on the extent of modification available. Without the ability to modify CNTs to engineer in desired binding specificities it may be difficult for them, when mixed with other nanometric components of a device, to self-assemble to form the complex, final structure.
Biological molecules have long been recognized as potentially useful components of nanodevices because of their structural and sequence flexibility and because of their ability to self-assemble. Widely used biological molecules include peptides, DNA and proteins. There has been wide, longstanding research into the use of peptides to produce nanostructures having various applications (Aggeli et al 1997; Holmes et al 2000; Ryadnov et al 2003). These structures include fibers (Takahashi 2002; Matsumura et al 2004) rings (Ghadiri et al 1993) and tubes (Ghadiri et al 1993; Saviano et al 1994; Seebach et al 1997; Clark et al 1998; Gao and Matsui 2005). A number of notable successes include the use of a modified beta-sheet forming 10-residue peptide that is able to form uniform tubes of 50 to 70 nm in diameter with an internal cavity 20 to 35 nm in diameter and which can reach many hundreds of nanometers in length (Matsumura et al 2005). Peptide tubes can be patterned on surfaces (Reches and Gazit 2006) and coated with metal on the outer (Reches and Gazit 2006) or inner (Reches and Gazit 2003) cavity. The advantages of using peptides to make nanostructures are that they can easily be synthesized and in many cases the final structures can be predicted in advance. Disadvantages are that, as with carbon nanotubes, the basic unit is small and although they can be modified more readily than carbon nanotubes to give useful functionality (Matsui et al 2001; Nuraje et al 2004) and shapes (Ryadnov 2007) they are nevertheless somewhat limited in the amount of modifications that can be tolerated before structure is adversely affected, with changes typically limited to alterations of individual peptide side chains and chemical modification. It is also difficult to precisely control the polymerization of the peptide tube structures, often resulting in polydispersity of length. Although it is possible to construct peptide tubes with small cavity sizes (Hartgerink et al 1998; Horne et al 2003), this is not trivial and many cavities are considerably larger. While useful for some applications, for others such as acting as moulds for biomineralization of quantum wires, sub 10 nm diameters would be preferable.
DNA has also proven a useful tool for constructing nanoscale shapes. Most notably these include regular 2D tiling arrays (Fu and Seeman 1993; Winfree et al 1998; Seeman 2003; Park et al 2006), 2D arrays of any desired pattern (Rothemund 2006), three-dimensional cages (Shih et al 2004) and tubes (Aldaye and Sleiman 2007). Advantages in using DNA are the ease in which it can be synthesized and the relative simplicity of having only 4 bases as the source of structural variability, meaning that predicting the overall shape is easier, allowing precise control.
It is clear that biological molecules offer exciting possibilities for construction of new complex, self assembled nanodevice components whose structures can be finely controlled. They potentially offer many advantages over non-biological systems. Biological molecules will not of course be able to replace non-biological nano-scale components in all cases. For example, while some biological molecules have impressive physical characteristics (the tensile strength of spider dragline silk, one of the strongest biological fibers known, is around 1.1 GPa (Vollrath and Knight 2001)) they cannot compete with those of nonbiological systems (the tensile strength of carbon nanotubes is at least an order of magnitude higher (Sinnott and Andrews 2001)). In electronics, carbon nanotubes and related materials have the potential to be used as semiconductors or superconductors (Kasumov et al 1999; Sinnott and Andrews 2001), something which biological molecules are unlikely to be capable of. Where biological molecules do show great promise is in the ability to self assemble extremely small yet regularly sized (often monodisperse) structures that can act as templates for the build-up of required structures from inorganic materials which, in the main, currently cannot self assemble. A second area where they may have an application is in the production of complex nanoscale devices that consist of numerous interlocking modules, something which has thus far proven difficult with inorganic materials yet which Nature does with ease. Thirdly, biological molecules may be able to act at the interface between inorganic systems and biological systems, for example in coating medical implants, or acting as detectors in a biological-silicon hybrid devices. Finally, many biological molecules can be produced in large (industrial) scale from bacterial expression systems relatively cheaply.
A major hurdle in constructing components of nanodevices is that of controlling self-assembly to give well defined shapes of precise dimensions and utilizing materials that can be easily modified to give new properties without losing overall structure.
Complex nanomachines found in nature, such as enzymes, are proteins and it is reasonable to suppose that this material is a good candidate for producing artificial nanodevices. When considering the design of nanoscale devices, a common problem is that of how to assemble the components, whose dimensions make them extremely difficult to manipulate. This problem is overcome by using a protein “building block” approach. In this system, protein components are used that naturally have, or can be engineered to have, affinities for other building blocks such that nanodevices can spontaneously self-assemble upon mixing of the individual components. Protein systems offer a number of other advantages such as synthesis under ambient conditions with no toxic by-products.
Protein cages, rings and tubes all offer useful starting points for further modification and have been engineered to produce semi-artificial structures which offer promise as basic components that may be built upon in future work. Recent work and future prospects of each of these proteins types is considered in more detail in the following sections.
Protein cages
Protein cages are three-dimensional protein structures, usually roughly spherical in shape, which enclose a central hollow space into which materials can be deposited. Spherical cages have two surfaces: The inner surface and the outer surface. Protein cages self assemble from monomers in solution and one of their great advantages is that the central cavity is completely enclosed. This means that the interior of the cage can provide an isolated environment in which chemical reactions can take place shielded from bulk solution. The protein cage also acts to physically limit the size of particles produced in the cavity, making production of monodisperse nanoparticles relatively trivial. Protein cages of varying sizes are known. Dps (DNA-binding protein from starved cells, Figure 1; (Harrison and Arosio 1996; Bozzi et al 1997)) from Listeria innocua, with an external diameter of 9 nm, is possibly the smallest. Dps is a bacterial protein produced in response to starvation and oxidative stress which has numerous reported functions including a role in protection of DNA from oxidative damage (Mann 1993). It also has a ferritin-like ability to biomineralize iron in its central cavity. Dps is an extremely small cage protein, with a central core approximately 4.5 nm in diameter (Stillman et al 2005). It has been shown that Dps can be used to biomineralize cobalt oxide in its central cavity (Allen et al 2002). Furthermore, it has been used as part of a “ball-and-spike” supramolecule in which the C-terminal of gp5 protein (gp5c), part of the cell puncturing apparatus from bacteriophage T4 (Kanamaru et al 2002) was fused to the N terminus of Dps via a short (22 residue) linker peptide. Gp5c assembles as a trimer into a triangular prism. Because Dps is a dodecamer it has four 3-fold symmetry axes meaning that with a linker of the appropriate length (22 residues), four full gp5c trimers can be assembled with equal spacing around the surface of Dps, at positions corresponding to the vertices of a tetrahedron (Figure 2). Dps can be filled with metal or semiconductor, which can function as a quantum dot. If gp5c can also be modified to act as a template for biomineralization, then it is hoped that biomineralized gp5c “spikes” could act as electrodes around a central Dps quantum dot, forming the basis of extremely small electronic components.
Figure 1.
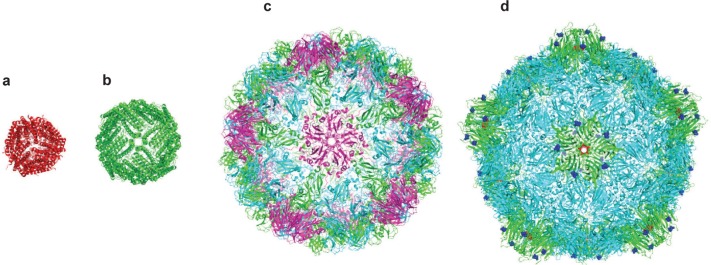
Crystal structures of a range of cage proteins. (a) Dps protein (pdb 2bjy (Ilari et al 2005)), diameter 9 nm; (b) Ferritin (pdb 2za6 (Yoshizawa et al 2007)) diameter 12 nm; (c) Cowpea chlorotic mottle virus (CCMV, pdb 1cwp (Speir et al 1995)), diameter 26 nm; and (d) Cowpea mosaic virus (1NY7 (Lin et al 1999)), diameter 28 nm. In (d), red spheres show the N-terminal glycine that points into the central cavity. Dark blue spheres show the C-terminal lysine on the external surface.
Figure 2.
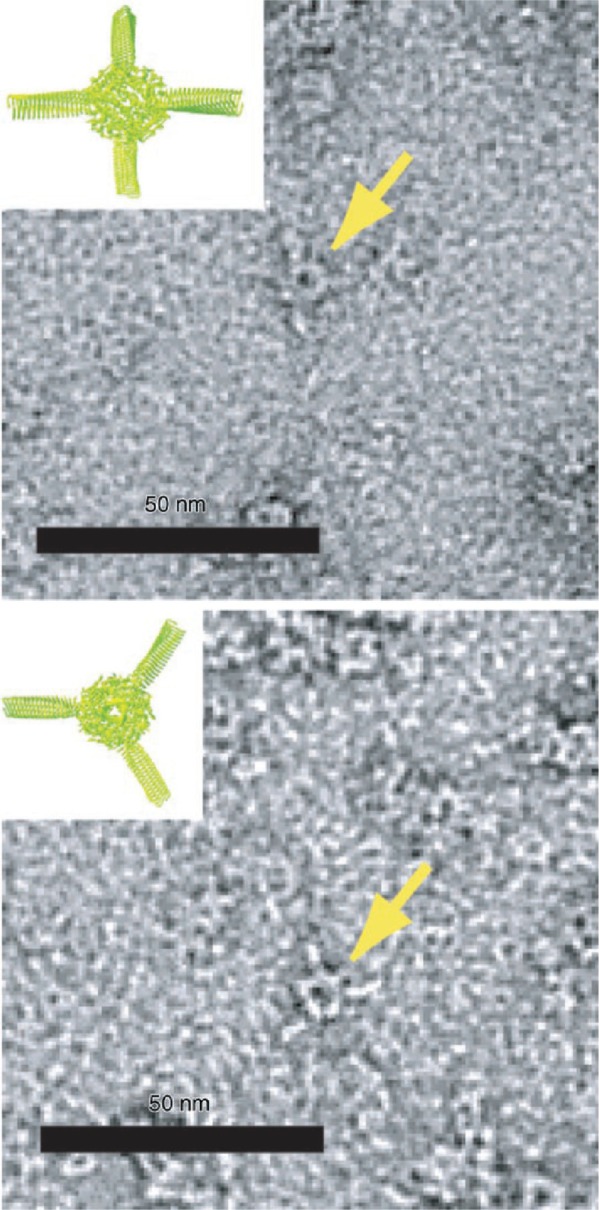
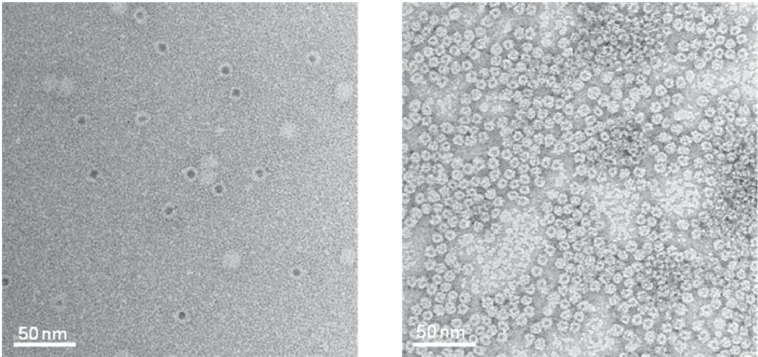
Transmission electron micrographs of “Ball-and-Spike” protein shown in two different orientations. Inset shows an interpretation of the micrograph using existing crystal structures of gp5c and Dps. Reproduced with permission from Sugimoto K, Kanamaru S, et al 2006. Construction of a ball-and-spike protein supramolecule13. Angew Chem Int Ed, 45:2725–8. Copyright ©. Wiley-VCH Verlag GmbH and Co. KGaA.
Dps is a ferritin-like protein and ferritin itself is one of the most widely studied and utilized cage proteins. Found in all domains of life, it is highly conserved, consisting of 24 identical protein subunits (Banyard et al 1978; Yoshizawa et al 2007). In vivo its role is to act as a store of ferric oxide. Ferric iron in the cavity of ferritin is formed from Fe(II) in solution. Fe(II) is able to enter the internal cavity via a channel lined with acidic residues. Once inside, biomineralization begins at specific nucleation sites on the interior surface (Lawson et al 1991).
In higher animals, ferritin is made from two similar proteins; H and L type ferritin with only H type ferritin possessing a true catalytic site, known as a ferroxidase centre (Lawson et al 1989) although both types, when reconstituted as pure H or L type ferritin in vitro, are able to promote biomineralization of ferric iron (Theil et al 2000). Work with ferritin was one of the earliest successes in biomineralization using proteins other than virus capsids. In pioneering work, Mann and colleagues were able to biomineralize a range of materials including manganese and iron oxides in the central cavity of purified ferritins (Meldrum et al 1991, 1992, 1995; Mann 1993; Douglas et al 1995). Ferritin was found to have the useful ability to inhibit biomineralization on its outside surface while catalyzing biomineralization in the cavity (the so-called “Janus effect”). Since then, significant work has been carried out by Yamashita and colleagues to extend the types of materials that ferritin can biomineralize (Figure 3), and they have achieved success with numerous metals and semiconductors including nickel hydroxide (Okuda et al 2003), cadmium selenide (Yamashita et al 2004), zinc selenide (Iwahori et al 2005) and gold sulfide (Yoshizawa et al 2006) while other groups have succeeded in minerlizaing further materials (Douglas and Stark 2000; Kramer et al 2004). The Yamashita group have also developed systems of finely controlling the deposition of ferritin on various surfaces (Kumagai et al 2006; Yamashita et al 2006; Matsui et al 2007) and have used ferritin-based quantum dots as the basis of prototype electronic devices (Yamashita 2001; Miura et al 2006, 2007). It is anticipated that in this way, biomineralized ferritin may become an important component of future electronic devices. Indeed, it has already found a number of other applications, having been used as a catalyst for growth of polycrystalline silicon films (Kirimura et al 2005) and single wall carbon nanotubes (Bonard et al 2002) as well as a reporter protein in MRI (Cohen et al 2005).
Figure 3.
Transmission electron micrographs of ferritin (left) and Dps (right) both filled with CdS cores (Iwahori et al 2007; Iwahori and Yamashita 2007). Figure courtesy of Kenji Iwahori.
The other major class of protein cages used in bionanotechnology research are the virus capsids. Ranging in size from 18 to 500 nm in diameter for icosahedral viruses (Figure 1), they offer the potential for use in a diverse range of applications (Douglas and Young 2006). A particularly useful property of widely utilized capsids such as cowpea chlorotic mottle virus (CCMV) and cowpea mosaic virus (CPMV) is that they have N or C-termini on the outer or inner surfaces of the capsid and have additional external residues that can be addressed genetically or chemically. In a series of experiments by the Evans group, 180 exposed carboxylate groups on surface of CPMV were shown to be addressable and were attached to methyl(aminopropyl)viologen, which resulted in a redox active capsid nanoparticle (Steinmetz et al 2006). Similarly, modification of surface exposed lysines with biotin and surface-exposed cysteines with fluorescent dyes, resulted in fluorescent particles that formed a dense monolayer on a gold surface. If the capsid layer was alternated with layers of streptavidin, multilayers of different fluorescently labeled particles could be achieved (Figure 4; (Steinmetz et al 2006, 2008)). In a similar way surface-exposed amine groups were used for attachment of redox active ferrocenecarboxylate molecules (Steinmetz et al 2006). Plant viruses such as this are particularly attractive for use as potential in vivo therapeutic agents as they are not infectious to animals (although this may be a disadvantage if high efficiency of transport to the interior of target cells is required, see discussion of SV40 below) and they can be made and purified in large quantities from plant hosts.
Figure 4.
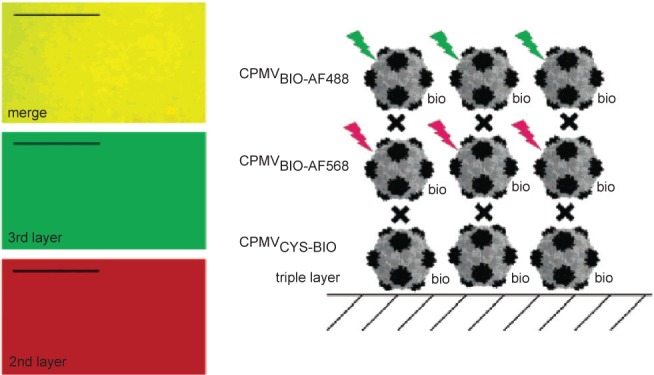
CPMV capsids can be layered on a gold surface with each layer carrying a different modification. In this example, the biotinylated base layer is unlabeled, the second layer is labeled with a red fluorescent dye (AlexaFluor dye AF568) and a third layer with a green fluorescent dye (AF568). Layers are bridged by streptavidin (black cross). Fluorescence imaging microscopy (left) shows that each layer is homogenous. Scale bar is 10 μm. Reproduced with permission from Steinmetz et al 2006a. Plant viral capsids as nanobuilding blocks: construction of arrays on solid supports. Langmuir, 22:10032–37. Copyright © 2006. American Chemical Society.
Like ferritin and Dps, the cavities of plant viruses offer potential uses as nanoreactors or as delivery systems. CCMV has been used to mineralize paratungstate, decavanadate and iron oxide in its cavity, taking advantage of the fact that pores in the capsid can be opened and closed via changes in pH, allowing for fine control of the reaction (Douglas and Young 1998; Douglas et al 2002).
Another virus with potential for in vivo use is Simian virus 40 (Anderer et al 1967; Liddington et al 1991), a spherical virus with a diameter of 40–45 nm. SV40 is a tumorigenic DNA virus and is able to infect a variety of human cells with high efficiency. For this reason it is a potential candidate as a gene delivery system or as a method for transporting a variety of cargos to cellular targets. Furthermore, SV40 represents an interesting example of the flexibility and controllability inherent in many protein nanostructures. In vivo, the virus forms spherical particles constructed from 72 pentamers of VP1 protein, the major capsid protein, and 72 copies of minor proteins VP2 or VP3. VP1 alone is, however, enough to form a capsid and the morphology produced varies significantly depending on experimental conditions (Figure 5). At pH 5.0, for example long, tubular structures are formed whereas at high salt concentrations, small (20 nm) particles predominate. A variety of switching mechanisms are also available to control structure: VP2 switches on assembly of spherical particles with VP1 at neutral pH (Kawano et al 2006) and the capsid can be reversibly disassembled by the addition of EGTA (Kanesashi et al 2003). Another layer of control can be affected by DNA, the presence of which is required for correct assembly of SV40 particles under physiological conditions and which is able to promote switching from tubular to spherical forms at pH5 (Tsukamoto et al 2007a). As well as its potential as a drug or gene delivery nanocapsule SV40 also, like other cage proteins, has potential for biomineralization of inorganic materials.
Figure 5.
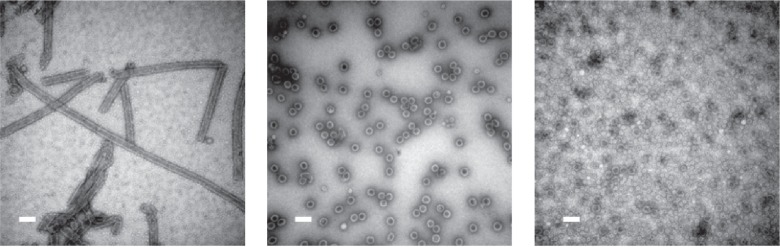
SV40 can form spherical or tubular structures depending on buffer conditions. Left: SV40VP1 pentamers assembled into tubular structure in vitro. Middle: SV40VP1 pentamers assembled into 40 nm spherical particles in vitro in the presence of DNA. Right: VP1 assembled into VLPs (Virus Like Particles) inside insect cells. In all cases, scale bar is 100 nm. Images courtesy of Hiroko Tsukamoto.
Protein rings
Protein rings are squat, three-dimensional tubes whose length is less than their diameter. Ring structures have a central hole and generally, four distinguishable surfaces; the surface lining the central hole, the corresponding outer surface and two “end” surface orthogonal to the hole axis. Ring proteins are common in nature where they perform numerous roles (Figure 6). One well-known example is Rad52, an important protein involved in homologous recombination of DNA where it has a role in promoting annealing of single DNA strands. The protein forms a ring structure of 7 monomers (Stasiak et al 2000) and, if only the N-terminal half of the protein is present, it forms an 11-membered ring (Kagawa et al 2002; Singleton et al 2002). The resulting structure is approximately 12 nm across with a central hole ranging from 2.5 to 5 nm in diameter and it is thought that DNA wraps around the outside of the ring. Another ring protein with similarity to RAD52 is the phage recombinase known as β. β is a 30 kDa protein that achieves single stranded DNA-annealing working in tandem with a partner protein, Exo (Poteete 2001; Court et al 2002). In solution, β forms an approximately 12-membered ring but forms an approximately 12- to 18-membered ring if ssDNA is present (Passy, Yu et al 1999).
Figure 6.
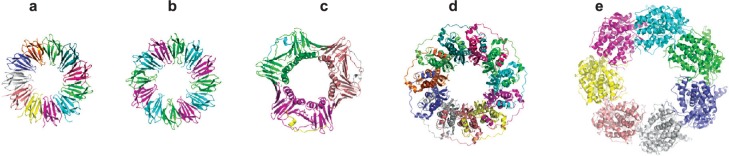
Crystal structures of various ring-shaped proteins. (a) wild-type TRAP protein (pdb 1qaw (Chen et al 1999), diameter approximately 8 nm); (b) mutant 12-membered TRAP protein (pdb 2zd0 (Watanabe et al 2008)); (c) PCNA (pdb 1axc (Gulbis et al 1996)); (d) RAD52 (pdb 1kno (Kagawa et al 2002)), (e) GROEL (pdb 1grl (Braig et al 1994)). All proteins shown approximately to scale.
The holes within ring proteins can be used to capture inorganic materials and array them on surfaces. The beta subunit of the HSP60 chaperonin protein from Sulfolobus shibatae, forms a barrel-like ring structure that has been successfully used to capture gold nanodots over the central core and arrange them into a well ordered 2D array (McMillan et al 2002).
Recently TRAP (trp RNA-binding attenuation protein) has emerged as a potentially useful component for constructing bionano components. The protein, found in species of Bacillus, is involved in regulation of tryptophan synthesis (Babitzke et al 1994, 1995; Babitzke 1997, 2004; Gollnick et al 2005). The structure of the protein shows it to be a small ring consisting of 11 monomers, approximately 8.5 nm in diameter with a central hole approximately 2.5 nm in diameter (Antson et al 1995). A further structure shows that the protein can bind single stranded RNA of a specific sequence around the circumference (Antson et al 1999). TRAP is extremely thermostable (Baumann et al 1997; Heddle et al 2006) and is also tolerant to mutation. In our own work we have begun to use TRAP as an engineerable building block from which to construct more complex structures. For example, we were able to line the central hole with cytseine residues in order to capture gold nanodots (Heddle et al 2007). The gold-binding protein ring was further modified with a titanium binding peptide (Sano and Shiba 2003) such that the gold bound ring was able to be specifically placed on a titanium or silicon oxide surface in a known orientation. Arraying gold nanodots in this way, we were able to construct a prototype MOS capacitor (Figure 7; (Heddle et al 2007)). Furthermore, by fusing together TRAP monomer genes in tandem it was possible to produce a TRAP protein that self-assembled not into the preferred 11mer form but a 12-mer form with 12-fold rotational symmetry which may be most suitable for formation of 2D crystals (Figure 6b; (Heddle et al 2006; Watanabe et al 2008)).
Figure 7.
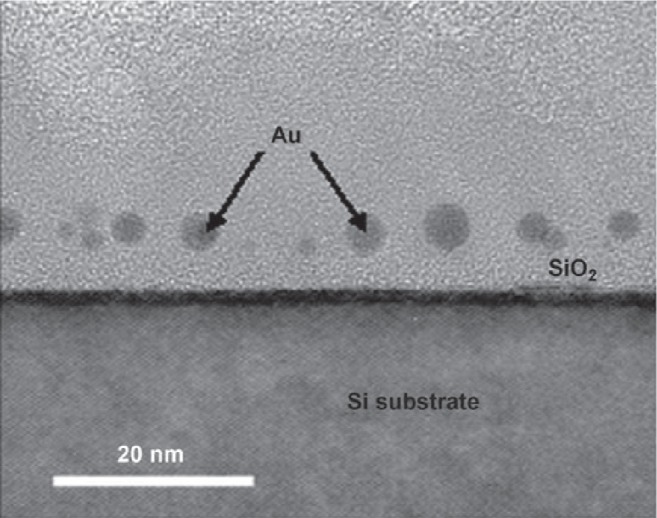
Cross-sectional TEM image of gold nanodots captured by TRAP and embedded in the SiO2 layer of a MOS capacitor. Reproduced with permission from Heddle et al 2007. Using the ring-shaped protein TRAP to capture and confine gold nanodots on a surface. Small, 3:1950–6. Copyright © 2007. Wiley-VCH Verlag GmbH and Co. KGaA.
Proteins that interact with nucleic acids appear to commonly form ring structures: RuvB is another protein involved in homologous recombination, specifically resolution of Holliday junctions, and forms a hexameric ring 12 nm in diameter with a central hole 2–3 nm in diameter (Miyata et al 2000; Yamada et al 2001) through which double-stranded DNA can bind. Other ring proteins, which also bind DNA include helicases such as gene 4 from bacteriophage T7 (Singleton et al 2000) and recombinases such as Dmc1 (Passy et al 1999). Proteins of shapes that may be useful in future nanodevices and also have the ability to bind DNA may be of particular interest as DNA itself is a well established, programmable nano-scaf-fold. It is possible to take advantage of the well-understood nature of Watson-Crick base-pairing in DNA molecules and the fact that DNA can be made with complimentary “sticky ends,” to construct “DNA tiles” that can be assembled into large-scale arrays (Seeman 1999). More recently, a different approach, known as “DNA origami” has proved that it is possible to construct arbitrary two dimensional shapes using DNA (Rothemund 2006). It should be possible to construct DNA patterns containing specific areas for attachment of proteins. This offers the promise of three dimensional protein devices that can be templated onto surfaces in any given pattern using a DNA scaffold. Indeed, such experiments have already been carried out and include construction of a grid of RuvA protein arranged on a lattice of DNA Holliday junctions (Malo et al 2005) and use of DNA nanoarrays containing aptamers specific for certain proteins to arrange those proteins into arbitrary patterns (Chhabra et al 2007).
There are many other protein rings of different sizes and characteristics which may provide useful building blocks for nanostructures. Naturally occurring protein rings may still offer surprising structures such as the catenated rings recently reported in crystals of mitochondrial peroxiredoxin III (Cao et al 2005).
Protein tubes
Tubes resemble elongated rings, where length is significantly greater than width. Like rings they have 4 addressable surfaces. Nanotubes have a number of potential uses, for example as containers for controlled release drug formulations, materials and electronics (Bong et al 2001; Son et al 2006).
Many of these functions have of course already been suggested as possible uses for CNTs. We have seen that, while CNTs offer vast potential in a wide variety of fields, they may not be able to satisfy all the needs of future nanodevices. Nanometric protein structures may be able to fill many of the niches not available to CNTs and the two materials may complement each other.
The idea of using peptide rings stacked to form tubes has a long history, with a system for using cyclic peptides in this way being proposed as long ago as 1974 (De Santis et al 1974). More recently the use of proteins and protein rings to form tubes has been investigated. The use of larger proteins consisting of many amino acids gives a measure of redundancy to the system as these larger building blocks can be modified more extensively without a significant change in overall structure.
Naturally occurring protein tubes include microtubules, flagella and pili. While these may be useful they have a number of features which are not ideal. In some cases, naturally occurring tubes are made from multiple proteins, lack ease of modification, cannot be assembled easily in vitro and may have structural features that make addressing all 4 surfaces difficult. In addition, they may have a relatively short persistence length. For use in nanotechnological applications it would be preferable to be able to make a small diameter protein tube from a single subunit that can be expressed in large quantities and can be easily modified and assembled in solution in vitro. Such a tube should have all four surfaces accessible and it should be possible to program its length.
A protein that fulfills some of these requirements is tobacco mosaic virus (TMV). TMV consists of a tubular shaped ribonucleoprotein made from 2130 copies of the coat protein that assemble around a single-stranded RNA core. The resulting virus particle is approximately 300 nm in length (Shenton et al 1999). Longer tubes can be obtained by assembling the coat protein in the absence of RNA although in this instance, exact control of length is lost. The inner cavity has been used as a template for mineralization with nickel, cobalt, cobalt-platinum (Figure 8) and iron-platinum nanowires (Knez et al 2003; Tsukamoto et al 2007b). The external surface has also been used as a template for the biomineralization of metals (Dujardin et al 2003; Górzny et al 2008) and the insertion of cysteine residues on the surface via genetic engineering allowed the chemical attachment of fluorescent chromophores to form a self-assembling light-harvesting system which was able to collect light over a wide spectrum with high efficiency (Miller et al 2007) and may be the basis for future components of optical devices.
Figure 8.
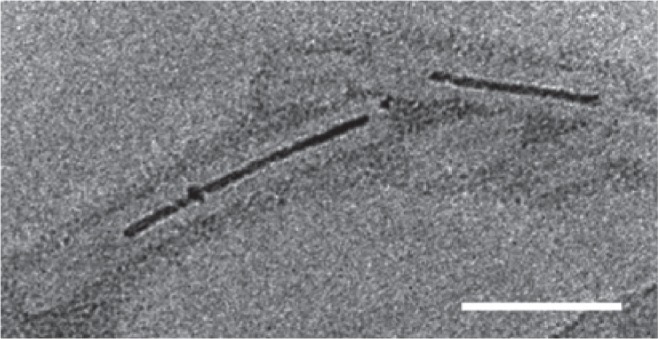
Aurothioglucose-stained TEM image showing TMV with biomineralized Co-Pt forming a nanowire in its cavity. Scale bar is 50 nm. Reproduced with permission from Tsukamoto et al 2007b. Synthesis of CoPt and FePt3 nanowires using the central channel of tobacco mosaic virus as a biotemplate. Chem Mater, 19:2389–91. Copyright © 2007. American Chemical Society.
While the TMV tube consists of a building block of a single coat protein arranged in a helical pattern, other tubes of similar dimensions can be made by using stacked protein rings as building blocks. Recently, a number of groups have succeeded in using synthetic biology approaches to produce modified ring proteins that form self-assembled nanotubes, this exciting new area of research may result in new ways of producing nanowires, biosensors, drug delivery systems and structural components of future complex nanodevices.
In one example, Stable Protein 1 (SP1) was used (Medalsy et al 2008). This is a nano-ring made from 12 identical proteins, found in aspen plants (Wang et al 2002). The 12 proteins form a double-layered 6-membered ring which is 11 nm in diameter with a central hole 2 to 3 nm across (Dgany et al 2004). The central cavity was modified by addition of 6 copies of histidine to the N-terminus of each copy of the protein, lining the central hole. The histidine residues bound strongly to two 1.8 nm diameter Ni-NTA modified gold nanodots giving a total of two dots bound to SP1, one at each end of the double ring. The gold dots did not completely enter the hole; part of the dot protruded from each end of the double ring and so provided attachment points for further rings. In this way, the rings polymerized into tube-like chains with each double ring bridged by a gold nanodot (see Figure 9). The presence of the gold in the rings offers the potential that the tubes could be used as nanowires.
Figure 9.
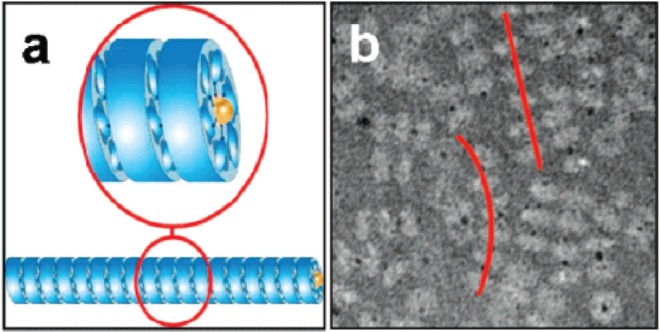
(a) A schematic showing the polymerization of SP1 rings (shown in blue) via interaction with gold nanodots (yellow) placed in the central cavity of the ring; (b) a TEM image of tube-like chains (highlighted by red lines) of SP1 rings (light grey) mediated by gold nanodots (black dots). Reprinted in part with permission from Medalsy et al 2008. SP1 protein-based nanostructures and arrays. Nano Lett, 8:473–7. Copyright © 2008. American Chemical Society.
In a second example, hcp was used. Hcp is a homohexameric protein found in Pseudomonas aeruginosa, a protein ring that forms part of the bacteria’s type VI secretion system; it was modified to self-assemble into a tube (Ballister et al 2008). The protein naturally forms a ring approximately 9 nm in diameter with a central hole approximately 4 nm in diameter (Mougous et al 2006). To promote stacking of the ring into a tube, cysteine residues were engineered into the top and bottom faces so that interring disulfide bonds would form between opposing cysteines (see Figure 10). In this way the protein was able to polymerize into tubes of up to 100 nm. Furthermore, by introducing rings with only one face modified with cysteines, it was possible to cap the end of the growing tubes, a form of statistical length control. The capping rings could further be modified to plug the central hole such that the tubes became nanocapsules.
Figure 10.
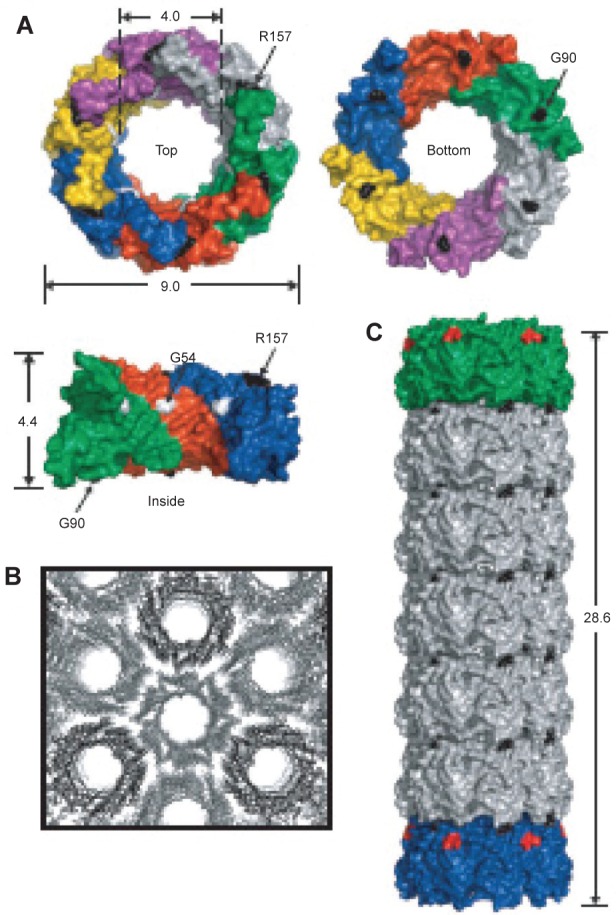
(A) Crystal structure of hcp (pdb 1y12 (Mougous, Cuff et al 2006)). Shown from the top, bottom and side. Dimensions are given in nanometers. Each monomer is shown in a different color and three monomers are removed for the side view. Based on the packing of the rings in the crystal structure into tubes (B) residues R157 and G90 were mutated to cysteine residues to facilitate tube formation via disulfide bond formation (C) Reproduced with permission from Ballister et al 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Nat Acad Sci U S A, 105:3733–8. Copyright © 2008. National Academy of Sciences, USA.
Future prospects
Modifying proteins to produce new nanometric structures endowed with novel structures and properties is an innovative and exciting synthesis of bionanotechnology and synthetic biology and is already being used to make components of prototype nanodevices including nanocapsules (protein cages), nanorings, nanotubes and nanowires. As a catalogue of self-assembled nanoshapes is compiled, we may see the emergence of more complex nanodevices that consist of several such components that assemble together to make a complex, functioning system or “nanomachine”. A recent example demonstrating how this may be achieved is the “ball and spike protein” (Figure 2) a multicomponent device made from a single gene formed by the fusion of genes for Dps and gp5c (described above). This could be used as part of an electronic device with the biomineralized Dps cavity addressed by 4 “spikes” which, if decorated with appropriate materials, could play the roles of the source, drain and gate of a three dimensional, self assembling nanoscale transistor (illustrated in Figure 11). Applying this “building-block” approach to other engineered proteins could result in novel devices with applications as smart drugs and biosensors as well as nanoscale electronics and the field looks set to progress rapidly in coming years.
Figure 11.

Imagined array of ball and spike protein binding to nanodots and electrodes that interface it to a microelectronic device. Image courtesy of Ichiro Yamashita.
Acknowledgments
The author thanks Ichiro Yamashita, Kenji Iwahori and Mime Kobayashi for critical reading of the manuscript. The author is funded by MEXT Special Coordination Funds for Promoting Science and Technology and Grant-in-Aid for Young Scientists (WAKATE B-20710083) from the ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Disclosures
The author has no conflicts of interest to declare.
References
- Aggeli A, Bell M, et al. Responsive gels formed by the spontaneous self-assembly of peptides into polymeric β-sheet tapes. Nature. 1997;386:259–62. doi: 10.1038/386259a0. [DOI] [PubMed] [Google Scholar]
- Aldaye FA, Sleiman HF. Modular access to structurally switchable 3D discrete DNA assemblies. J Am Chem Soc. 2007;129:13376–7. doi: 10.1021/ja075966q. [DOI] [PubMed] [Google Scholar]
- Allen M, Willits D, et al. Protein cage constrained synthesis of ferrimagnetic iron oxide nanoparticles. Adv Mater. 2002;14:1562–5. [Google Scholar]
- Anderer FA, Schlumberger HD, et al. Structure of simian virus 40. II. Symmetry and components of the virus particle. Virology. 1967;32:511–23. doi: 10.1016/0042-6822(67)90303-0. [DOI] [PubMed] [Google Scholar]
- Antson AA, Dodson EJ, et al. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–42. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- Antson AA, Otridge J, et al. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- Babitzke P. Regulation of tryptophan biosynthesis: Trp-ing the TRAP or how Bacillus subtilis reinvented the wheel. Mol Microbiol. 1997;26:1–9. doi: 10.1046/j.1365-2958.1997.5541915.x. [DOI] [PubMed] [Google Scholar]
- Babitzke P. Regulation of transcription attenuation and translation initiation by allosteric control of an RNA-binding protein: the Bacillus subtilis TRAP protein. Curr Opin Microbiol. 2004;7:132–9. doi: 10.1016/j.mib.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Babitzke P, Bear DG, et al. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc Nat Acad Sci U S A. 1995;92:7916–20. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Stults JT, et al. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269:16597–604. [PubMed] [Google Scholar]
- Ballister ER, Lai AH, et al. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Nat Acad Sci U S A. 2008;105:3733–8. doi: 10.1073/pnas.0712247105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banyard SH, Stammers DK, et al. Electron density map of apoferritin at 2.8-A resolution. Nature. 1978;271:282–4. doi: 10.1038/271282a0. [DOI] [PubMed] [Google Scholar]
- Baughman RH, Zakhidov AA, et al. Carbon nanotubes-the route toward applications. Science. 2002;297:787–92. doi: 10.1126/science.1060928. [DOI] [PubMed] [Google Scholar]
- Baum R. Nanotechnology: Drexler and Smalley make the case for and against molecular assemblers. Chem Eng News. 2003;81:37–42. [Google Scholar]
- Baumann C, Xirasagar S, et al. The trp RNA-binding attenuation protein (TRAP) from Bacillus subtilis binds to unstacked trp leader RNA. J Biol Chem. 1997;272:19863–9. doi: 10.1074/jbc.272.32.19863. [DOI] [PubMed] [Google Scholar]
- Bonard JM, Chauvin P, et al. Monodisperse multiwall carbon nanotubes obtained with ferritin as catalyst. Nano Lett. 2002;2:665–7. [Google Scholar]
- Bong DT, Clark TD, et al. Self-assembling organic nanotubes. Angew Chem Int Ed. 2001;40:988–1011. [PubMed] [Google Scholar]
- Bozzi M, Mignogna G, et al. A novel non-heme iron-binding ferritin related to the dna-binding proteins of the Dps family in Listeria innocua. J Biol Chem. 1997;272:3259–65. doi: 10.1074/jbc.272.6.3259. [DOI] [PubMed] [Google Scholar]
- Braig K, Otwinowski Z, et al. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994;371:578–86. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Cao Z, Roszak AW, et al. Bovine mitochondrial peroxiredoxin III forms a two-ring catenane. Structure. 2005;13:1661–64. doi: 10.1016/j.str.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Chen X, Antson AA, et al. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus. J Mol Biol. 1999;289:1003–16. doi: 10.1006/jmbi.1999.2834. [DOI] [PubMed] [Google Scholar]
- Chhabra R, Sharma J, et al. Spatially addressable multiprotein nanoarrays templated by aptamer-tagged DNA nanoarchitectures. J Am Chem Soc. 2007;129:10304–5. doi: 10.1021/ja072410u. [DOI] [PubMed] [Google Scholar]
- Clark TD, Buehler LK, et al. Self-assembling Cyclic β-peptide nanotubes as artificial transmembrane ion channels. J Am Chem Soc. 1998;120:651–6. [Google Scholar]
- Cohen B, Dafni H, et al. Ferritin as an endogenous MRI reporter for noninvasive imaging of gene expression in C6 glioma tumors. Neoplasia. 2005;7:109–17. doi: 10.1593/neo.04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court DL, Sawitzke JA, et al. Genetic engineering using homologous recombination. Annu Rev Genet. 2002;36:361–88. doi: 10.1146/annurev.genet.36.061102.093104. [DOI] [PubMed] [Google Scholar]
- De Santis P, Morosetti S, et al. Conformational analysis of regular enantiomeric sequences. Macromolecules. 1974;7:52–8. [Google Scholar]
- Dgany O, Gonzalez A, et al. The structural basis of the thermostability of SP1, a novel plant (Populus tremula) boiling stable protein. J Biol Chem. 2004;279:51516–23. doi: 10.1074/jbc.M409952200. [DOI] [PubMed] [Google Scholar]
- Douglas T, Dickson DP, et al. Synthesis and structure of an iron(III) sulfide-ferritin bioinorganic nanocomposite. Science. 1995;269:54–7. doi: 10.1126/science.269.5220.54. [DOI] [PubMed] [Google Scholar]
- Douglas T, Stark VT. Nanophase cobalt oxyhydroxide mineral synthesized within the protein cage of ferritin. Inorg Chem. 2000;39:1828–30. doi: 10.1021/ic991269q. [DOI] [PubMed] [Google Scholar]
- Douglas T, Strable E, et al. Protein engineering of a viral cage for constrained nano-materials synthesis. Adv Mater. 2002;14:415–8. [Google Scholar]
- Douglas T, Young M. Host-guest encapsulation of materials by assembled virus protein cages. Nature. 1998;393:152–5. [Google Scholar]
- Douglas T, Young M. Viruses: making friends with old foes. Science. 2006;312:873–5. doi: 10.1126/science.1123223. [DOI] [PubMed] [Google Scholar]
- Dujardin E, Peet C, et al. Organization of metallic nanoparticles using tobacco mosaic virus templates. Nano Lett. 2003;3:413–7. [Google Scholar]
- Ebbesen TW, Ajayan PM. Large-scale synthesis of carbon nanotubes. Nature. 1992;358:220–2. [Google Scholar]
- Feynman RP. A transcript of this talk is available online ( www.zyvex.com/nanotech/feynman.html)
- Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–20. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- Gao X, Matsui H. Peptide-based nanotubes and their applications in bionanotechnology. Adv Mater. 2005;17:2037–50. doi: 10.1002/adma.200401849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadiri MR, Granja JR, et al. Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature. 1993;366:324–7. doi: 10.1038/366324a0. [DOI] [PubMed] [Google Scholar]
- Gollnick P, Babitzke P, et al. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis. Annu Rev Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- Górzny M, Walton AS, et al. Four-probe electrical characterization of Pt-coated TMV-based nanostructures. Nanotechnology. 2008;165704 doi: 10.1088/0957-4484/19/16/165704. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, et al. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Guo T, Nikolaev P, et al. Self-assembly of tubular fullerenes. J Phys Chem. 1995;99:10694–7. [Google Scholar]
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophy Acta Bioenerg. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Hartgerink JD, Clark TD, et al. Peptide nanotubes and beyond. Chem Eur J. 1998;4:1367–72. [Google Scholar]
- Heddle JG, Fujiwara I, et al. Using the ring-shaped protein TRAP to capture and confine gold nanodots on a surface. Small. 2007;3:1950–6. doi: 10.1002/smll.200700400. [DOI] [PubMed] [Google Scholar]
- Heddle JG, Yokoyama T, et al. Rounding up: engineering 12-membered rings from the cyclic 11-Mer TRAP. Structure. 2006;14:925–33. doi: 10.1016/j.str.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Hirsch A. Functionalization of single-walled carbon nanotubes. Angew Chem Int Ed. 2002;41:1853–9. doi: 10.1002/1521-3773(20020603)41:11<1853::aid-anie1853>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Holmes TC, de Lacalle S, et al. Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc Nat Acad Sci U S A. 2000;97:6728–33. doi: 10.1073/pnas.97.12.6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne WS, Stout CD, et al. A heterocyclic peptide nanotube. J Am Chem Soc. 2003;125:9372–6. doi: 10.1021/ja034358h. [DOI] [PubMed] [Google Scholar]
- Iijima S. Helical microtubules of graphitic carbon. Nature. 1991;354:56–8. [Google Scholar]
- Ilari A, Latella MC, et al. The unusual intersubunit ferroxidase center of Listeria innocua Dps is required for hydrogen peroxide detoxification but not for iron uptake. A study with site-specific mutants. Biochemistry. 2005;44:5579–87. doi: 10.1021/bi050005e. [DOI] [PubMed] [Google Scholar]
- Iwahori K, Enomoto T, et al. Cadmium sulfide nanoparticle synthesis in Dps protein from Listeria innocua. Chem Mater. 2007;19:3105–11. [Google Scholar]
- Iwahori K, Yamashita I. Fabrication of CdS nanoparticles in the bio-template, apoferritin cavity by a slow chemical reaction system. Journal of Physics: Conference Series. 2007:492. [Google Scholar]
- Iwahori K, Yoshizawa K, et al. Fabrication of ZnSe nanoparticles in the apoferritin cavity by designing a slow chemical reaction system. Inorg Chem. 2005;44:6393–400. doi: 10.1021/ic0502426. [DOI] [PubMed] [Google Scholar]
- Jose-Yacaman M, Miki-Yoshida M, et al. Catalytic growth of carbon microtubules with fullerene structure. Appl Phys Lett. 1993;62:657–9. [Google Scholar]
- Kagawa W, Kurumizaka H, et al. Crystal structure of the homologous-pairing domain from the human Rad52 recombinase in the undecameric form. Mol Cell. 2002;10:359–71. doi: 10.1016/s1097-2765(02)00587-7. [DOI] [PubMed] [Google Scholar]
- Kanamaru S, Leiman PG, et al. Structure of the cell-puncturing device of bacteriophage T4. Nature. 2002;415:553–7. doi: 10.1038/415553a. [DOI] [PubMed] [Google Scholar]
- Kanesashi SN, Ishizu K, et al. Simian virus 40 VP1 capsid protein forms polymorphic assemblies in vitro. J Gen Virol. 2003;84:1899–905. doi: 10.1099/vir.0.19067-0. [DOI] [PubMed] [Google Scholar]
- Kasumov AY, Deblock R, et al. Supercurrents through single-walled carbon nanotubes. Science. 1999;284:1508–11. doi: 10.1126/science.284.5419.1508. [DOI] [PubMed] [Google Scholar]
- Kawano M-A, Inoue T, et al. The VP2/VP3 minor capsid protein of simian virus 40 promotes the in vitro assembly of the major capsid protein VP1 into particles. J Biol Chem. 2006;281:10164–73. doi: 10.1074/jbc.M511261200. [DOI] [PubMed] [Google Scholar]
- Kirimura K, Uraoka Y, et al. Study of low-temperature crystallization of amorphous Si films obtained using ferritin with Ni nanoparticles. Appl Phys Lett. 2005;86:262106. [Google Scholar]
- Knez M, Bittner AM, et al. Biotemplate synthesis of 3–nm nickel and cobalt nanowires. Nano Lett. 2003;3:1079–82. [Google Scholar]
- Kramer RM, Li C, et al. Engineered protein cages for nanomaterial synthesis. J Am Chem Soc. 2004;126:13282–6. doi: 10.1021/ja046735b. [DOI] [PubMed] [Google Scholar]
- Kumagai S, Yoshii S, et al. Electrostatic placement of single ferritin molecules. Appl Phys Lett. 2006;88:153103. [Google Scholar]
- Lawson DM, Artymiuk PJ, et al. Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature. 1991;349:541–4. doi: 10.1038/349541a0. [DOI] [PubMed] [Google Scholar]
- Lawson DM, Treffry A, et al. Identification of the ferroxidase centre in ferritin. FEBS Lett. 1989;254:207–10. doi: 10.1016/0014-5793(89)81040-3. [DOI] [PubMed] [Google Scholar]
- Liddington RC, Yan Y, et al. Structure of simian virus 40 at 3.8 resolution. Nature. 1991;354:278–84. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- Lin T, Chen Z, et al. The refined crystal structure of cowpea mosaic virus at 2.8 A resolution. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- Malo J, Mitchell JC, et al. Engineering a 2D protein-DNA crystal. Angew Chem Int Ed. 2005;44:3057–61. doi: 10.1002/anie.200463027. [DOI] [PubMed] [Google Scholar]
- Mann S. Molecular tectonics in biomineralization and biomimetic materials chemistry. Nature. 1993;365:499–505. [Google Scholar]
- Matsui H, Porrata P, et al. Protein tubule immobilization on self-assembled monolayers on Au substrates. Nano Lett. 2001;1:461–4. [Google Scholar]
- Matsui T, Matsukawa N, et al. Direct production of a two-dimensional ordered array of ferritin-nanoparticles on a silicon substrate. Jpn J Appl Phys. 2007;46:L713–L15. [Google Scholar]
- Matsumura S, Uemura S, et al. Fabrication of nanofibers with uniform morphology by self-assembly of designed peptides. Chem Eur J. 2004;10:2789–94. doi: 10.1002/chem.200305735. [DOI] [PubMed] [Google Scholar]
- Matsumura S, Uemura S, et al. Construction of biotinylated peptide nanotubes for arranging proteins. Mol Biosyst. 2005;1:146–8. doi: 10.1039/b504516a. [DOI] [PubMed] [Google Scholar]
- McMillan RA, Paavola CD, et al. Ordered nanoparticle arrays formed on engineered chaperonin protein templates. Nat Mater. 2002;1:247–52. doi: 10.1038/nmat775. [DOI] [PubMed] [Google Scholar]
- Medalsy I, Dgany O, et al. SP1 protein-based nanostructures and arrays. Nano Lett. 2008;8:473–7. doi: 10.1021/nl072455t. [DOI] [PubMed] [Google Scholar]
- Meldrum FC, Douglas T, et al. Reconstitution of manganese oxide cores in horse spleen and recombinant ferritins. J Inorg Biochem. 1995;58:59–68. doi: 10.1016/0162-0134(94)00037-b. [DOI] [PubMed] [Google Scholar]
- Meldrum FC, Heywood BR, et al. Magnetoferritin: in vitro synthesis of a novel magnetic protein. Science. 1992;257:522–3. doi: 10.1126/science.1636086. [DOI] [PubMed] [Google Scholar]
- Meldrum FC, Wade VJ, et al. Synthesis of inorganic nanophase materials in supramolecular protein cages. Nature. 1991;349:684–7. [Google Scholar]
- Miller RA, Presley AD, et al. Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J Am Chem Soc. 2007;129:3104–9. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- Miura A, Hikono T, et al. Floating nanodot gate memory devices based on biominerlized inorganic nanodot array as a storage node. Jpn J Appl Phys. 2006;45:L1–L3. [Google Scholar]
- Miura A, Uraoka Y, et al. Bionanodot monolayer array fabrication for nonvolatile memory application. Surf Sci. 2007;601:L81–L5. [Google Scholar]
- Miyata T, Yamada K, et al. Two different oligomeric states of the ruvb branch migration motor protein as revealed by electron microscopy. J Struct Biol. 2000;131:83–9. doi: 10.1006/jsbi.2000.4290. [DOI] [PubMed] [Google Scholar]
- Mougous JD, Cuff ME, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312:1526–30. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuraje N, Banerjee IA, et al. Biological bottom-up assembly of antibody nanotubes on patterned antigen arrays. J Am Chem Soc. 2004;126:8088–9. doi: 10.1021/ja048617u. [DOI] [PubMed] [Google Scholar]
- Okuda M, Iwahori K, et al. Fabrication of nickel and chromium nanoparticles using the protein cage of apoferritin. Biotechnol Bioeng. 2003;84:187–94. doi: 10.1002/bit.10748. [DOI] [PubMed] [Google Scholar]
- Park SH, Pistol C, et al. Finite-Size, fully addressable DNA tile lattices formed by hierarchical assembly procedures. Angew Chem Int Ed. 2006;45:735–9. doi: 10.1002/anie.200503797. [DOI] [PubMed] [Google Scholar]
- Passy SI, Yu X, et al. Rings and filaments of beta protein from bacteriophage lambda suggest a superfamily of recombination proteins. Proc Nat Acad Sci U S A. 1999;96:4279–84. doi: 10.1073/pnas.96.8.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passy SI, Yu X, et al. Human Dmc1 protein binds DNA as an octameric ring. Proc Nat Acad Sci U S A. 1999;96:10684–8. doi: 10.1073/pnas.96.19.10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteete AR. What makes the bacteriophage lambda Red system useful for genetic engineering: molecular mechanism and biological function. FEMS Microbiol Lett. 2001;201:9–14. doi: 10.1111/j.1574-6968.2001.tb10725.x. [DOI] [PubMed] [Google Scholar]
- Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–7. doi: 10.1126/science.1082387. [DOI] [PubMed] [Google Scholar]
- Reches M, Gazit E. Controlled patterning of aligned self-assembled peptide nanotubes. Nat Nanotech. 2006;1:195–200. doi: 10.1038/nnano.2006.139. [DOI] [PubMed] [Google Scholar]
- Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Ryadnov MG. A self-assembling peptide polynanoreactor. Angew Chem Int Ed. 2007;46:969–72. doi: 10.1002/anie.200604014. [DOI] [PubMed] [Google Scholar]
- Ryadnov MG, Ceyhan B, et al. Belt and braces: a peptide-based linker system of de novo design. J Am Chem Soc. 2003;125:9388–94. doi: 10.1021/ja0352045. [DOI] [PubMed] [Google Scholar]
- Sano K, Shiba K. A hexapeptide motif that electrostatically binds to the surface of titanium. J Am Chem Soc. 2003;125:14234–5. doi: 10.1021/ja038414q. [DOI] [PubMed] [Google Scholar]
- Saviano M, Lombardi A, et al. A structural two-ring version of a tubular stack of β-Rings in crystals of a cyclic D, L-hexapeptide. J Inclusion Phenom. 1994;18:27–36. [Google Scholar]
- Seebach D, Matthews JL, et al. Cyclo-β-peptides: structure and tubular stacking of cyclic tetramers of 3-aminobutanoic acid as determined from powder diffraction data-peptides. Helv Chim Acta. 1997;80:173–82. [Google Scholar]
- Seeman NC. DNA engineering and its application to nanotechnology. Trends Biotechnol. 1999;17:437–43. doi: 10.1016/s0167-7799(99)01360-8. [DOI] [PubMed] [Google Scholar]
- Seeman NC. DNA in a material world. Nature. 2003;421:427–31. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- Shenton W, Douglas T, et al. Inorganic-organic nanotube composites from template mineralization of tobacco mosaic virus. Adv Mater. 1999;11:253–6. [Google Scholar]
- Shih WM, Quispe JD, et al. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–21. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- Simmons JM, In I, et al. Optically modulated conduction in chromophore-functionalized single-wall carbon nanotubes. Phys Rev Lett. 2007;98:086802. doi: 10.1103/PhysRevLett.98.086802. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Sawaya MR, et al. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- Singleton MR, Wentzell LM, et al. Structure of the single-strand annealing domain of human RAD52 protein. Proc Nat Acad Sci U S A. 2002;99:13492–13497. doi: 10.1073/pnas.212449899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott SB Andrews R. Carbon nanotubes: synthesis, properties, and applications. Crit Rev Solid State Mater, Sci. 2001;26:145–249. [Google Scholar]
- Smalley RE. Of chemistry, love and nanobots. Sci Am. 2001;285:76–7. doi: 10.1038/scientificamerican0901-76. [DOI] [PubMed] [Google Scholar]
- Son SJ, Bai X, et al. Template synthesis of multifunctional nanotubes for controlled release. J Control Release. 2006;114:143–52. doi: 10.1016/j.jconrel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- Speir JA, Munshi S, et al. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak AZ, Larquet E, et al. The human Rad52 protein exists as a heptameric ring. Curr Biol. 2000;10:337–40. doi: 10.1016/s0960-9822(00)00385-7. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Bock E, et al. Assembly of multilayer arrays of viral nanoparticles via biospecific recognition: a quartz crystal microbalance with dissipation monitoring study. Biomacromolecules. 2008;9:456–62. doi: 10.1021/bm700797b. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Calder G, et al. Plant viral capsids as nanobuild-ing blocks: construction of arrays on solid supports. Langmuir. 2006a;22:10032–37. doi: 10.1021/la0621362. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Lomonossoff GP, et al. Cowpea mosaic virus for material fabrication: addressable carboxylate groups on a programmable nanoscaffold. Langmuir. 2006b;22:3488–90. doi: 10.1021/la060078e. [DOI] [PubMed] [Google Scholar]
- Steinmetz NF, Lomonossoff GP, et al. Decoration of cowpea mosaic virus with multiple, redox-active, organometallic complexes13. Small. 2006c;2:530–3. doi: 10.1002/smll.200500453. [DOI] [PubMed] [Google Scholar]
- Stillman TJ, Upadhyay M, et al. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol. 2005;57:1101–12. doi: 10.1111/j.1365-2958.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Kanamaru S, et al. Construction of a ball-and-spike protein supramolecule13. Angew Chem Int Ed. 2006;45:2725–8. doi: 10.1002/anie.200504018. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Ueno A, Mihara H. Amyloid architecture: complementary assembly of heterogeneous combinations of three or four peptides into amyloid fibrils. Chembiochem. 2002;3:637–42. doi: 10.1002/1439-7633(20020703)3:7<637::AID-CBIC637>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Theil EC, Takagi H, et al. The ferritin iron entry and exit problem. Inorg Chim Acta. 2000;297:242–51. [Google Scholar]
- Tsukamoto H, Kawano, et al. Evidence that SV40 VP1 DNA interactions contribute to the assembly of 40-nm spherical viral particles. Genes to Cells. 2007a;12:1267–79. doi: 10.1111/j.1365-2443.2007.01134.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto R, Muraoka M, et al. Synthesis of CoPt and FePt3 nanowires using the central channel of tobacco mosaic virus as a biotemplate. Chem Mater. 2007b;19:2389–91. [Google Scholar]
- Vollrath F, Knight DP. Liquid crystalline spinning of spider silk. Nature. 2001;410:541–8. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- Wang W-X, Pelah D, et al. Characterization of SP1, a stress-responsive, boiling-soluble, homo-oligomeric protein from aspen. Plant Physiol. 2002;130:865–75. doi: 10.1104/pp.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Mishima Y, et al. Intersubunit linker length as a modifier of protein stability: Crystal structures and thermostability of mutant TRAP. Protein Sci. 2008;17:518–526. doi: 10.1110/ps.073059308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfree E, Liu F, et al. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–44. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- Yamada K, Kunishima N, et al. Crystal structure of the Holliday junction migration motor protein RuvB from Thermus thermophilus HB8. Proc Nat Acad. Sci U S A. 2001;98:1442–7. doi: 10.1073/pnas.031470598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita I. Fabrication of a two-dimensional array of nano-particles using ferritin molecule. Thin Solid Films. 2001;393:12–18. [Google Scholar]
- Yamashita I, Hayashi J, et al. Bio-template synthesis of uniform CdSe nanoparticles using cage-shaped protein, apoferritin. Chem Lett. 2004;33:1158–9. [Google Scholar]
- Yamashita I, Kirimura H, et al. Selective nanoscale positioning of ferritin and nanoparticles by means of target-specific peptides13. Small. 2006;2:1148–52. doi: 10.1002/smll.200600220. [DOI] [PubMed] [Google Scholar]
- Yoshizawa K, Iwahori K, et al. Fabrication of gold sulfide nanoparticles using the protein cage of apoferritin. Chem Lett. 2006;35:1192. [Google Scholar]
- Yoshizawa K, Mishima Y, et al. Effect of N-terminal residues on the structural stability of recombinant horse L-chain apoferritin in an acidic environment. J Biochem (Tokyo) 2007;142:707–13. doi: 10.1093/jb/mvm187. [DOI] [PubMed] [Google Scholar]