ABSTRACT
WhiA is a highly unusual transcriptional regulator related to a family of eukaryotic homing endonucleases. WhiA is required for sporulation in the filamentous bacterium Streptomyces, but WhiA homologues of unknown function are also found throughout the Gram-positive bacteria. To better understand the role of WhiA in Streptomyces development and its function as a transcription factor, we identified the WhiA regulon through a combination of chromatin immunoprecipitation-sequencing (ChIP-seq) and microarray transcriptional profiling, exploiting a new model organism for the genus, Streptomyces venezuelae, which sporulates in liquid culture. The regulon encompasses ~240 transcription units, and WhiA appears to function almost equally as an activator and as a repressor. Bioinformatic analysis of the upstream regions of the complete regulon, combined with DNase I footprinting, identified a short but highly conserved asymmetric sequence, GACAC, associated with the majority of WhiA targets. Construction of a null mutant showed that whiA is required for the initiation of sporulation septation and chromosome segregation in S. venezuelae, and several genes encoding key proteins of the Streptomyces cell division machinery, such as ftsZ, ftsW, and ftsK, were found to be directly activated by WhiA during development. Several other genes encoding proteins with important roles in development were also identified as WhiA targets, including the sporulation-specific sigma factor σWhiG and the diguanylate cyclase CdgB. Cell division is tightly coordinated with the orderly arrest of apical growth in the sporogenic cell, and filP, encoding a key component of the polarisome that directs apical growth, is a direct target for WhiA-mediated repression during sporulation.
IMPORTANCE
Since the initial identification of the genetic loci required for Streptomyces development, all of the bld and whi developmental master regulators have been cloned and characterized, and significant progress has been made toward understanding the cell biological processes that drive morphogenesis. A major challenge now is to connect the cell biological processes and the developmental master regulators by dissecting the regulatory networks that link the two. Studies of these regulatory networks have been greatly facilitated by the recent introduction of Streptomyces venezuelae as a new model system for the genus, a species that sporulates in liquid culture. Taking advantage of S. venezuelae, we have characterized the regulon of genes directly under the control of one of these master regulators, WhiA. Our results implicate WhiA in the direct regulation of key steps in sporulation, including the cessation of aerial growth, the initiation of cell division, and chromosome segregation.
Introduction
The filamentous, Gram-positive bacterium Streptomyces has a complex life cycle, culminating in the formation of specialized reproductive structures called aerial hyphae, which extend upwards from the substrate mycelium into the air. Subsequently, the orderly arrest of aerial growth is coordinated with a remarkable synchronous septation event with concomitant segregation of chromosomes that leads to the formation of long chains of 50 to 100 unigenomic spores (1–4).
Genetic analysis of differentiation in the classical model organism, Streptomyces coelicolor, defined two classes of developmental mutant, blocked at distinct stages of the life cycle. bld mutants are unable to form aerial hyphae and therefore lack the characteristic “fuzzy” appearance of the wild type (WT), exhibiting a shiny, “bald” phenotype (1, 3). In contrast, whi mutants are able to erect aerial hyphae but are unable to complete the life cycle to form mature chains of spores. Such mutants fail to synthesize the characteristic polyketide pigment associated with mature wild-type spores and therefore appear white on plates (1, 3).
The principal focus of this work is the WhiA protein. S. coelicolor whiA mutants fail to halt aerial growth or to initiate sporulation septation and the partitioning of chromosomes. Instead, they keep growing, forming extremely long, undivided aerial hyphae containing uncondensed DNA that appears continuous on staining (5, 6). This phenotype is indistinguishable from that of whiB mutants, leading to suggestions that WhiA and WhiB function together in the same regulatory pathway (5, 6).
WhiA is of great interest, not only because of its critical biological role in controlling the initiation of sporulation but also because of the highly unusual nature of the protein itself, first predicted bioinformatically (7) and dramatically confirmed when the structure of a WhiA homologue from Thermotoga maritima was determined (8). The N-terminal domain of WhiA, constituting two-thirds of the protein, is homologous to eukaryotic “LAGLIDADG” homing endonucleases, but this domain is degenerate in that it has lost key residues required for metal binding and catalysis, and it also displays an extensively altered DNA-binding surface compared with true homing endonucleases (7, 8). Instead, DNA binding in WhiA is mediated through a smaller, C-terminal helix-turn-helix (HTH) domain that is completely absent from classical homing endonucleases (7–9). The two domains are connected by a flexible linker, and the C-terminal HTH domain binds DNA site-specifically by itself (9). Thus, the function of the much larger N-terminal degenerate homing endonuclease domain remains unknown.
A further intriguing aspect of WhiA biology is that it is not confined to streptomycetes or even sporulating actinomycetes but is present in essentially all Gram-positive bacteria, including, therefore, many nonsporulating genera (for example, Staphylococcus). This observation, combined with its relationship with homing endonucleases, raises very interesting questions about the evolution of WhiA as a transcription factor (10) and about the reassortment of the target genes under its control in sporulating and nonsporulating bacteria.
The regulons of the bld gene regulators BldA, BldD, BldH (also called AdpA), and, most recently, σBldN have all been characterized and reveal much of the network that governs the switch from vegetative to aerial growth (11–16). Thus, for example, we now know that σBldN controls the expression of the chaplin and rodlin proteins, the principal components of the hydrophobic sheath that enables the aerial hyphae to escape surface tension and extend into the air (16). However, the whi gene regulatory network that controls the later stages of the Streptomyces life cycle remains largely uncharacterized. Consequently, our understanding of the regulation of processes such as the orderly cessation of aerial growth, the initiation of sporulation septation, and chromosome segregation remains incomplete.
In-depth studies of the regulatory networks that control sporulation in Streptomyces have been greatly facilitated by the introduction of Streptomyces venezuelae as a new model system for the genus (16). The classical model system, S. coelicolor, poses certain logistical problems when seeking to characterize the regulons of the Bld and Whi master regulators. S. coelicolor sporulates only on solid medium, and the reproductive structures undergoing morphogenesis constitute just ~5% of the total biomass, making the application of global techniques such as chromatin immunoprecipitation-sequencing (ChIP-seq) to development-specific transcription factors problematic. This study takes advantage of S. venezuelae, which, unlike S. coelicolor, sporulates synchronously in liquid culture to near-completion (17), greatly facilitating application of global techniques to the analysis of differentiation (16). We apply in vivo ChIP-seq and microarray transcriptional profiling to confirm the function of WhiA as a transcription factor and to characterize the regulon of genes directly under its control. These results implicate WhiA in the direct control of key steps in sporulation, including the cessation of aerial growth, the initiation of cell division, and chromosome segregation.
RESULTS AND DISCUSSION
WhiA is required for sporulation in S. venezuelae.
Before characterizing the WhiA regulon, a whiA null mutant of S. venezuelae was constructed to determine its phenotype and to compare it to the equivalent S. coelicolor whiA mutant (5). Using the Redirect PCR targeting method (18, 19), a whiA mutant was generated in which the coding region was replaced with an apramycin resistance (apr) cassette. As expected, the S. venezuelae ΔwhiA::apr strain was unable to sporulate on solid MYM (maltose-yeast extract-malt extract) medium or to synthesize the characteristic green spore pigment, instead appearing white (Fig. 1). In the wild type, the orderly arrest of aerial growth is followed by a septation event that divides the long, multigenomic hyphae into chains of unigenomic spores (Fig. 2A). To examine the effect of the whiA deletion in more detail, fluorescence microscopy of DAPI (4′,6-diamidino-2-phenylindole)-stained cells and scanning electron microscopy were conducted. As seen previously for S. coelicolor whiA mutants, the S. venezuelae whiA mutant formed unusually long aerial hyphae that fail to initiate septation or chromosome segregation (Fig. 2B). It should be noted that wild-type S. venezuelae forms straight aerial hyphae, in contrast to the coiled aerial hyphae made by wild-type S. coelicolor, and this is reflected in the phenotypes of the corresponding whiA mutants (Fig. 2B) (5, 6). All aspects of the S. venezuelae whiA null mutant were fully complemented in trans by a wild-type copy of whiA introduced into the ΦBT1 integration site (Fig. 1 and data not shown). Based on the phenotypes of the whiA mutants, we concluded that WhiA is likely to play similar roles in S. venezuelae and S. coelicolor.
FIG 1 .

Phenotypes of wild-type S. venezuelae (WT), the constructed ΔwhiA::apr null mutant SV11 (ΔwhiA::apr), SV11 carrying the empty vector pMS82 (ΔwhiA::apr attBΦBT1::pMS82), and the complemented strain (ΔwhiA::apr attBΦBT1::whiA). Strains were grown on MYM solid medium and photographed after 4 days.
FIG 2 .
Phenotypes of wild-type S. venezuelae (A) and the constructed ΔwhiA::apr null mutant SV11 (B), examined by scanning electron microscopy (i), transmission electron microscopy (ii), and fluorescence microscopy after staining membranes and DNA with 10 µg/ml Nile red (iii) and 10 µg/ml DAPI (iv), respectively. Strains were grown on MYM solid medium for 2 days before fluorescence microscopy and 4 days before electron microscopy.
Defining the WhiA regulon.
In order to determine which genes are directly regulated by WhiA in S. venezuelae, ChIP-seq experiments were conducted. We constructed a strain of S. venezuelae that lacked whiA at its native locus but expressed an N-terminal, triple FLAG-tagged version of WhiA from the ΦBT1 integration site. This strain sporulated on solid MYM medium (see Fig. S1 in the supplemental material), although it displayed a slight delay in spore pigment production and was slower to sporulate in liquid culture. The best results were obtained when an additional [Gly4Ser]3 linker was engineered between the tag and WhiA. The resulting S. venezuelae strain sporulated to near-completion in liquid culture, albeit slightly slower than the wild type (Fig. S1 and data not shown). Furthermore, 3×FLAG-[Gly4Ser]3-WhiA was readily detected using anti-FLAG antibody in Western blot assays and there was no visible cross-reaction with any proteins in wild-type S. venezuelae (Fig. S2), suggesting optimal conditions for clean ChIP-seq experiments.
ChIP-seq was conducted with M2 anti-FLAG antibody at time points representing three distinct developmental stages: vegetative growth, the onset of sporulation (fragmentation), and midsporulation. The wild-type strain (expressing nontagged WhiA from the native locus) was used as a negative control to eliminate any signals that might arise from cross-reaction of the antibody with other DNA-binding proteins. In addition, total (nonimmunoprecipitated) input DNA from each time point was also subjected to sequencing. This very useful additional control enables nonuniform shearing of the chromosome to be taken into account. More relaxed regions of DNA shear more readily than compact regions, and so the former can give rise to higher background signals in ChIP-seq (20). Using P < 10−4 as the threshold for significance, a total of 284 peaks were detected in the FLAG-tagged WhiA strain over the three time points selected (see Fig. S3 and Table S1 in the supplemental material). Forty-eight of these were located more than 300 bp upstream of the nearest start codon (Table S1) and were not analyzed further. Using the same threshold for significance, only one peak was found in the wild-type (non-FLAG-tagged strain) control experiment. This peak (upstream of sven5092) also appeared in the FLAG-tagged WhiA strain and was subsequently removed from the data set.
Next, in order to determine how WhiA influences the expression of its target genes, wild-type S. venezuelae and the congenic ΔwhiA mutant were subjected to time-resolved, genome-wide transcriptional profiling during vegetative growth and sporulation. Strains were grown under the same conditions used for the ChIP-seq experiments. RNA samples were prepared at 2-hour intervals from 8 to 20 h, by which time sporulation was nearing completion, and following cDNA synthesis and labeling, samples were hybridized to Affymetrix DNA microarrays. Three independent biological replicates were performed for each strain, and the transcriptional profiling data were examined for each of the genes potentially under WhiA control. Almost all the peaks identified by ChIP-seq were found in intergenic regions. Of these, 40% were found in intergenic regions between divergent genes. In the case of these divergent regulatory regions, the transcriptional profiling data set was used to assess which gene was a target for regulation by WhiA.
Strikingly, 40% of WhiA target genes are downregulated in a whiA mutant and 35% are upregulated in a whiA mutant (see Table S1 in the supplemental material). This strongly suggests that WhiA is bifunctional, working almost equally as an activator and as a repressor to control differentiation in S. venezuelae.
Identification of a WhiA consensus binding site.
To confirm and extend the ChIP-seq data and to help establish a consensus WhiA-binding sequence, DNase I footprinting analysis using purified N-terminally histidine-tagged WhiA was carried out on radiolabeled probes derived from the upstream regions of six of the strongest WhiA targets, as judged by ChIP-seq. In each case, addition of WhiA protein resulted in protection of a clearly defined region, consistent with high-affinity binding of monomeric WhiA (Fig. 3A). The sequences of these six confirmed WhiA target promoters were then examined using the MEME algorithm (21). This identified a short but perfectly conserved sequence (GACAC) in each of the submitted intergenic regions that correlated precisely with the position of the DNase I-protected region (Fig. 3B). This finding is in agreement with analysis conducted by Kaiser and Stoddard (9), in which WhiA binding was demonstrated at its own promoter. Following DNase I footprinting analysis, these authors tested the specificity of WhiA binding using a fluorescence competition assay and were able to determine those bases most important for the affinity of binding. Importantly, the GACAC motif independently identified here corresponds exactly with the sequence identified by Kaiser and Stoddard (9).
FIG 3 .
DNase I footprinting analysis of WhiA binding to the promoter regions of nrdR, ftsW, filP, cslA, and sven1406 and the intergenic region between sven4724 and sven4725. 5′-end-labeled probes were incubated with increasing concentrations of WhiA (indicated in μM above the lanes) and subjected to DNase I footprinting analysis as described in Materials and Methods. Footprints are flanked by Maxam and Gilbert sequence ladders (AG). Black brackets indicate the position of the WhiA-protected region. (B) Summary of DNase I footprinting results presented in panel A. Brackets indicate the protected sequences with the consensus GACAC motif shown in bold. The number indicates the distance (bp) between the consensus motif and the putative start codon of the downstream gene. (C) Overrepresented motif associated with ~70% of WhiA target genes, identified using the MEME algorithm (21). The height of the letters in the sequence logo, in bits, is proportional to the frequency of the A, C, T, or G nucleotides at each position of the motif. Both the forward (i) and the reverse complement (ii) versions of the consensus are shown.
Following the identification of this short sequence, the remaining ChIP-seq targets were manually examined for the presence of the overrepresented WhiA-binding motif. Approximately 70% of the potential gene targets had a GACAC motif correlating closely with the position of the ChIP-seq peak. These target promoters were reexamined using the MEME algorithm, which failed to identify any additional overrepresented sequences associated with the GACAC motif (Fig. 3C). Within this subset, approximately 50% of genes showed greater than a 2-fold change in expression when comparing wild-type S. venezuelae to the whiA mutant. However, WhiA ChIP-seq targets that lacked a GACAC motif were just as likely to exhibit clear whiA dependence.
Because WhiA binds DNA as a monomer and it contains a large N-terminal homing endonuclease-like domain of unknown function, in addition to its small C-terminal HTH DNA-binding domain (8, 9), the orientation of the asymmetric GACAC motif relative to the target gene might well have regulatory significance. However, our analysis showed that there was no clear consistency in the directionality of the conserved motif, when present. We found that approximately twice the number of ChIP-seq-identified targets had upstream GACAC motifs compared to targets with the reverse complement GTGTC sequence upstream. Out of those ChIP-seq targets that show clear whiA dependence in microarray analysis, ~50% have upstream GACAC motifs, ~25% have upstream GTGTC motifs, and ~25% have no such sequence. Furthermore, this pattern does not change significantly when activated and repressed genes are analyzed separately. These data suggest that WhiA’s ability to function as an activator or as a repressor is not tied to its orientation on the DNA.
The identification of WhiA ChIP targets that depend on whiA for their expression, yet lack the GACAC motif, suggests that in some cases other sequences are sufficient to recruit WhiA. Three ChIP targets lacking the GACAC motif (sven3535, sven3229, and treZ, Table 1) were therefore tested for WhiA binding in vitro by DNase I footprinting. WhiA footprinted on only one of these three targets, treZ (sven5890). treZ is a clear WhiA target in ChIP-seq (Fig. 4A) that shows decreased expression in a whiA mutant, suggesting that WhiA activates transcription of this gene (see Table S1 in the supplemental material). DNase I footprinting of WhiA on the treZ promoter revealed two protected regions, both falling within the in vivo ChIP-seq peak (Fig. 4B). Although the determinants of this binding were not investigated, both protected regions contain the sequence GACGC, differing from the GACAC consensus sequence by 1 nucleotide (nt) (Fig. 4C). Thus, it is clear that WhiA can bind in vivo and in vitro to sites lacking the GACAC consensus sequence. For those WhiA ChIP-seq targets that did not bind WhiA in vitro, we speculate that additional transcription factors or accessory proteins might be required.
TABLE 1 .
Selected ChIP-identified WhiA targets
| Flanking genesa |
Distanceb (bp) | Productc | Adjusted P valued |
|||
|---|---|---|---|---|---|---|
| Left (−1) | Right (+1) | V | F | S | ||
| ftsZ | 158 | Cell division protein FtsZ | 2.97E−04 | 2.42E−21 | ||
| ftsK | 11 | Cell division protein FtsK | 1.98E−18 | |||
| sepF3 | 22 | Cell division protein SepF3 | 1.50E−31 | |||
| ftsW | 269 | Cell division protein FtsW | 1.01E−06 | 6.49E−05 | ||
| filP | 58 | Coiled-coil protein FilP | 2.75E−32 | 1.89E−39 | 7.12E−55 | |
| cslA | 54 | Putative cellulose synthase CslA | 8.68E−29 | 5.97E−51 | 1.41E−43 | |
| whiG | 298 | RNA polymerase sigma factor WhiG | 4.23E−16 | |||
| pyrG | sven1411 | 203; 447 | CTP synthase; hypothetical protein | 2.49E−72 | 7.72E−49 | |
| sven4033 | cdgB | 271; 55 | Putative reductase; diguanylate cyclase CdgB | 2.55E−06 | ||
| glpF | 130 | Glycerol uptake facilitator protein GlpF | 1.64E−10 | |||
| sven3676 | sven3677 | 130; 649 | Transcriptional regulator, PadR family; putative secreted penicillin binding protein |
1.73E−13 | ||
| sven4755 | sven4756 | 170; 79 | Hypothetical protein; putative lipoprotein | 5.48E−04 | ||
| sven3340 | crp | −73; 259 | Putative endonuclease; cyclic AMP-binding protein Crp | 7.76E−07 | 5.18E−12 | |
| sven3229 | 14 | Hypothetical protein | 4.65E−10 | |||
| sven3534 | sven3535 | 21; 167 | Putative ABC transporter ATP-binding component; hypothetical protein |
1.34E−09 | 7.88E−58 | 5.77E−50 |
| treZ | 47 | Malto-oligosyltrehalose trehalohydrolase TreZ | 1.54E−17 | |||
| whiA | −65 | Sporulation transcription regulator WhiA | 4.50E−07 | |||
| slzA | 133 | Putative coiled-coil protein SlzA | 3.73E−14 | |||
| ftsE | sven2728 | 175; 61 | Cell division transporter, ATP-binding protein FtsE; putative membrane protein |
1.33E−08 | 2.43E−10 | |
| sven1406 | 91 | Chromosome (plasmid) partitioning ParA-like protein | 9.45E−23 | |||
| sven5480 | nrdR | −76; 94 | Hypothetical protein; ribonucleotide reductase transcriptional regulator NrdR |
1.21E−06 | 5.11E−05 | 1.32E−21 |
| pyrR | bldD | 78; 169 | Pyrimidine operon regulatory protein PyrR; regulator of morphogenesis and antibiotic production BldD |
6.98E−04 | 5.04E−12 | 4.66E−15 |
| sigN | 271 | RNA polymerase sigma factor SigN | 3.97E−20 | 1.05E−10 | ||
| sven6396 | wblH | 132; 63 | Transcriptional regulator, IclR family; WhiB-type transcriptional regulator WblH |
1.67E−27 | 4.98E−08 | |
| infA | 173 | Translation initiation factor 1 | 6.70E−27 | 5.30E−65 | 2.25E−48 | |
| efp | 191 | Translation elongation factor P | 5.64E−11 | |||
| rpsH | 139 | SSU ribosomal protein S8p | 8.25E−14 | 1.99E−16 | ||
| rpsJ | 75 | SSU ribosomal protein S10p | 4.50E−11 | 8.03E−06 | ||
| rpsA | sven1624 | 132; 216 | SSU ribosomal protein S1p; hypothetical protein | 1.03E−06 | ||
Genes flanking the ChIP peak are listed.
Distance to the predicted start codon of the flanking genes.
(Possible) gene function based on annotation in StrepDB (http://strepdb.streptomyces.org.uk).
Maximum significance values (P < E−04) for the 3 ChIP samples; V, vegetative growth; F, onset of sporulation/fragmentation; S, midsporulation.
FIG 4 .
treZ, a WhiA target lacking a perfect GACAC motif. (A) ChIP-seq data for the WhiA target gene treZ. Color coding of the ChIP samples is as follows: 3×FLAG-[Gly4Ser]3-WhiA strain during vegetative growth (V, red), at the onset of sporulation/fragmentation (F, purple), and at midsporulation (S, blue) and the S. venezuelae negative-control strain (C, green). The plot spans approximately 3 kb of DNA sequence. Genes running left to right are shown in green, and genes running right to left are shown in red. The black arrow indicates the gene subject to WhiA regulation. (B) DNase I footprinting analysis of WhiA binding to the promoter region of treZ. 5′-end-labeled probes were incubated with increasing concentrations of WhiA (indicated in μM above the lanes) and subjected to DNase I footprinting analysis as described in Materials and Methods. The footprint is flanked by a Maxam and Gilbert sequence ladder (AG). Black and red brackets indicate the positions of the WhiA-protected regions. (C) Sequence summary of DNase I footprinting results presented in panel B. Black and red brackets indicate the protected sequences, with the near-consensus motif shown in bold. The number indicates the distance (bp) between the nearest consensus motif and the putative start codon of the treZ gene.
In Fig. 3, additional sites of weak protection were observed in the DNase I footprints on nrdR, ftsW, cslA, and the parA-like gene sven1406. It is not clear what these sites represent. Some contain GACAC motifs, and some contain the related sequence GACAG, but others do not show any obvious relationship to the consensus binding site.
WhiA functions as an autoregulator.
whiA appears to be the last gene in a four-gene operon, and previous work in S. coelicolor had suggested that whiA is transcribed throughout growth from a constitutive promoter located well upstream of the whiA gene itself (5). Consistent with this suggestion, our microarray transcriptional profiling data showed that S. venezuelae whiA is transcribed throughout vegetative growth and sporulation, and Western blot analysis of WhiA expressed from its native locus using polyclonal WhiA antiserum showed that the protein is similarly present throughout vegetative growth and sporulation (Fig. 5). In S. coelicolor, whiA expression is upregulated during development from a sporulation-specific promoter, whiAp2, lying just inside the upstream gene (5), and expression from whiAp2 is reduced to a low constitutive level in a whiA null mutant, suggesting positive autoregulation (22). Furthermore, the ability of WhiA to bind to the S. coelicolor whiAp2 promoter has been demonstrated in vitro (9). In line with these findings, our ChIP-seq experiments showed that whiA is a direct in vivo target of WhiA in S. venezuelae (Fig. 6A and Table 1). DNase I footprinting using purified N-terminally histidine-tagged S. venezuelae WhiA showed that WhiA binds to the S. venezuelae whiAp2 promoter region (Fig. 6B and C), in agreement with equivalent experiments carried out in S. coelicolor (9). Overall, it appears that WhiA binds to the region immediately upstream of the whiA gene in vivo to increase its expression prior to sporulation in Streptomyces. These and other experiments reported here suggest that WhiA activity is not a simple reflection of WhiA abundance, likely implying that WhiA activity is regulated posttranslationally (see “Conclusions”).
FIG 5 .

Western blot analysis of WhiA abundance during submerged sporulation in wild-type S. venezuelae (+) and in the congenic whiA mutant control strain, SV11 (Δ). Strains were grown in MYM liquid sporulation medium, 10 µg total protein was loaded per lane, and the blot was developed using a polyclonal antiserum raised against S. venezuelae WhiA. Bars indicate vegetative growth, fragmentation, and sporulation in the wild type, as judged by microscopic examination.
FIG 6 .
WhiA functions as an autoregulator. (A) ChIP-seq data for WhiA binding to the whiAp2 promoter. Color coding of the ChIP samples is as follows: 3×FLAG-[Gly4Ser]3-WhiA strain during vegetative growth (V, red), at the onset of sporulation/fragmentation (F, purple), and at midsporulation (S, blue) and the S. venezuelae negative-control strain (C, green). The plot spans approximately 3 kb of DNA sequence. The black arrow indicates the gene subject to WhiA regulation. (B) DNase I footprinting analysis of WhiA binding to the whiAp2 promoter. 5′-end-labeled probes were incubated with increasing concentrations of WhiA (indicated in μM above the lanes) and subjected to DNase I footprinting analysis as described in Materials and Methods. The footprint is flanked by a Maxam and Gilbert sequence ladder (AG). The black bracket indicates the position of the WhiA-protected region. (C) Sequence summary of DNase I footprinting results presented in panel B. The bracket indicates the protected sequence, and the consensus motif in shown in bold. The number indicates the distance (bp) between the consensus motif and the putative start codon of the whiA gene.
WhiA targets involved in polar growth.
Streptomyces hyphae grow by tip extension, and this polar growth is directed by a complex of coiled-coil cytoskeletal proteins called the polarisome (also called the tip organizing center or TIPOC). The three known polarisome proteins are DivIVA, which is essential for viability of the organism, and two nonessential cytoskeletal proteins that nevertheless play vital roles in tip growth, Scy and FilP (23–26). All three members of the polarisome interact directly with each other, and Scy has been suggested to function as a molecular scaffold for the other components (26). The intermediate filament-like protein FilP is recruited by DivIVA and localizes just behind DivIVA at the hyphal tip (25). The reduced rigidity of the cell wall at hyphal tips in a filP mutant (revealed by atomic force microscopy) and its distorted morphology during growth suggest that FilP plays a mechanical role in controlling cell shape (27). Consistent with this suggestion, FilP can form potentially load-bearing net-like structures in vitro (25). These data suggest that FilP plays a true cytoskeletal role supporting tip growth. During differentiation, aerial hyphae arrest tip extension and initiate a massive, synchronous septation event, and during this switch the polarisome disappears (28–30). The orderly arrest of aerial growth requires whiA (5, 6), but the mechanisms underlying this dependence are unknown. One of the strongest WhiA targets, as judged by ChIP-seq, is filP (sven5047) (Fig. 7 and Table 1). Expression of filP is increased in a whiA mutant (Fig. 8), suggesting that WhiA represses filP transcription. Repression of filP by WhiA during development may therefore contribute to the orderly cessation of aerial growth prior to sporulation and thus help to explain why whiA mutants form abnormally long aerial hyphae. In the context of coiled-coil proteins like FilP, it is interesting that we also identified slzA (small leucine zipper protein A; sven5271) as a member of the WhiA regulon (Table 1). slzA encodes a coiled-coil protein of the leucine zipper class that interacts strongly with itself in bacterial two-hybrid assays, and slzA mutants show a mild developmental phenotype (A. Kotun and J. McCormick, personal communication).
FIG 7 .
ChIP-seq data for 12 selected WhiA target genes: ftsZ, ftsK, sepF3, ftsW, filP, whiG, cdgB, crp, sven3676, glpF, sven3229, and sven4756. Color coding of the ChIP samples is as follows: 3×FLAG-[Gly4Ser]3-WhiA strain during vegetative growth (V, red), at the onset of sporulation/fragmentation (F, purple), and at midsporulation (S, blue) and the S. venezuelae negative-control strain (C, green). Plots span approximately 3 kb of DNA sequence. Genes running left to right are shown in green, and genes running right to left are shown in red. The black arrow indicates the gene subject to WhiA regulation. The arrangement of the 12 panels mirrors that in Fig. 8.
FIG 8 .
Microarray transcriptional profiling data for 12 selected WhiA target genes (ftsZ, ftsK, sepF3, ftsW, filP, whiG, cdgB, crp, sven3676, glpF, sven3229, and sven4756) during submerged sporulation in wild-type S. venezuelae (purple circles) and in the congenic whiA mutant, SV11 (red triangles). In each panel, the x axis indicates the age of the culture in hours, and the y axis indicates the per-gene normalized transcript abundance (log2). For the wild type, 10 to 14 h corresponds to vegetative growth, 14 to 16 h corresponds to the onset of sporulation (fragmentation), and 16 h onwards corresponds to sporulation. The arrangement of the 12 panels mirrors that in Fig. 7.
Another WhiA target identified here is cslA (sven5061) (Table 1), which encodes a putative cellulose synthase that localizes to hyphal tips (31, 32). CslA is proposed to reinforce growing tips through cellulose production, and disruption of cslA leads to a severe delay in the formation of aerial hyphae (31, 32). Although the expression of cslA is largely unchanged in a whiA mutant, there is a 2-fold increase at a late stage in development (20 h), suggesting that WhiA represses cslA (see Table S1 in the supplemental material). cslA appears to be the first gene of a bicistronic operon that also includes glxA, encoding a galactose oxidase required for proper aerial development during osmotic stress. Although the two genes are cotranscribed, glxA is also expressed from an independent promoter within the clsA coding sequence (33).
WhiA targets involved in cell division and chromosome segregation.
Strong signals for WhiA binding were found upstream of several key genes involved in cell division in Streptomyces, including ftsZ, ftsW, and ftsK (Fig. 7 and Table 1). Cell division is not essential for viability in Streptomyces, but it is required for sporulation, which involves the synchronous formation of a ladder of 50 or more cytokinetic rings (2). ftsZ transcription is under developmental control, being substantially upregulated from the ftsZp2 promoter during sporulation (34). Further, in S. coelicolor at least, constitutive overexpression of ftsZ restores sporulation septation to whi mutants, suggesting that appropriate regulation of FtsZ levels is a vital function of the whi gene products (35). BldD was the first transcription factor to be identified as a direct regulator of ftsZ transcription, functioning to repress ftsZ expression during vegetative growth (14), when FtsZ is required only for the formation of occasional vegetative crosswalls, which divide hyphae but do not lead to constriction and cell-cell separation. Microarray transcriptional profiling conducted here shows that ftsZ expression is significantly downregulated in a whiA mutant, consistent with WhiA acting as a direct activator of ftsZ expression (Fig. 8).
Similarly, our data suggest that WhiA functions as an activator of ftsW and ftsK transcription via direct binding to their respective promoters (Fig. 7 and 8; Table 1). (Note that ftsW lies in the middle of the dcw operon, suggesting that the WhiA-binding site identifies an ftsW-specific promoter.) In Escherichia coli, FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site (36), and FtsW is also likely to function as the lipid II “flippase” (37), transporting bacterial cell wall precursors across the membrane to FtsI and other transpeptidases for incorporation into newly synthesized septal peptidoglycan. S. coelicolor ftsW null mutants grown under most conditions have a white phenotype and make aerial hyphae that are unable to assemble FtsZ rings, form sporulation septa, or segregate their chromosomes (38). However, cell division occurs independently of ftsW and ftsI when S. coelicolor is grown on low-osmolarity media (39). Since the S. coelicolor genome encodes four SEDS (shape, elongation, division, and sporulation)/penicillin binding protein (PBP) pairs, it is likely that another pair is able to support cell division under certain growth conditions. Indeed, FtsZ is the only known component of the division machinery absolutely required for cell division under all conditions in Streptomyces, although it is not essential for viability (2, 40).
Sporulation septa constrict over unsegregated nucleoids, and FtsK functions as a DNA translocase, removing trapped DNA from the closing septa (1–3, 41). FtsK localizes to sporulation septa, but it is not required for Z-ring assembly or sporulation septation. However, ftsK mutants have irregular DNA content in their spores and show greatly increased genetic instability, most often associated with large deletions at the ends of the linear chromosome, observations consistent with FtsK’s role as a septal DNA pump (41).
Our ChIP-seq data also reveal binding of WhiA upstream of the ftsE gene (Table 1). In E. coli, FtsE is required for constriction; it resembles the ATP-binding cassette of an ABC transporter, and it interacts with its membrane component, FtsX, as well as with FtsZ (42–45). Despite the presence of a WhiA ChIP peak upstream of the gene, ftsE does not show strong dependence on whiA for its expression (see Table S1 in the supplemental material).
Chromosome segregation during sporulation also requires the ParAB system. ParA is a Walker A ATPase that assists ParB in the formation of foci formed through the binding of ParB to ~20 parS sites that surround oriC (4, 46, 47). The observation that ParA forms filaments that originate from the tip and extend down the sporogenic cell suggests that it provides the force to organize the ParB-bound chromosomes prior to septation (4, 48). More recently, this process has been shown to be coordinated by a direct interaction between ParA and Scy, one of the components of the polarisome that directs tip extension in vegetative and aerial hyphae (30). A previous study showed that the sporulation-specific upregulation of parAB transcription in S. coelicolor depends, directly or indirectly, on several of the whi genes, including whiA (22), and S. coelicolor WhiA was shown to bind to the parAB promoter in vitro (9). Our microarray transcriptional profiling shows that the sporulation-specific upregulation of parAB transcription also depends on whiA and several other whi genes in S. venezuelae, but we did not detect binding of WhiA to the parAB promoter in our ChIP-seq experiments at any of the three time points examined. sven1406 is annotated as parA-like, although its function has yet to be investigated. Our experiments identify sven1406 as a direct WhiA target and show that its expression depends on whiA (Table 1; see also Table S1 in the supplemental material).
SepF functions as a component of the cell division machinery in Bacillus, where it is suggested to play an early role in the assembly of the FtsZ ring (49), in addition to a later role in constriction (50). In S. venezuelae, there are three SepF-like proteins (SepF1, SepF2, and SepF3), but the individual (and presumably related) functions of these homologues are unknown. Our data show that the sepF3 (sven5776) promoter is a direct target of WhiA (Fig. 7 and Table 1) and that expression of sepF3 is significantly downregulated in a whiA mutant, suggesting that WhiA activates transcription of sepF3 (Fig. 8).
Overall, these results suggest that WhiA plays a central role in the initiation of sporulation in Streptomyces through the activation of multiple components of the cell division machinery.
Other WhiA targets involved in morphological differentiation.
cdgB is a target that appears to be moderately repressed by WhiA during the later stages of differentiation (Fig. 7 and 8; Table 1). cdgB encodes a diguanylate cyclase that synthesizes c-di-GMP, and both overexpression and deletion of cdgB inhibit the formation of aerial hyphae (51), the first experiments to establish a role for c-di-GMP signaling in Streptomyces development. Strikingly, in S. coelicolor, BldD directly regulates cdgB, in addition to two other genes encoding predicted diguanylate cyclases, cdgA and sco5511 (14, 51). The fact that cdgB is controlled by two key developmental regulators, WhiA and BldD, reinforces the emerging notion that c-di-GMP signaling plays an important role in the regulatory network controlling differentiation in Streptomyces (14, 51, 52).
ChIP-seq identified whiG (sven5300), encoding the sporulation-specific sigma factor σWhiG, as a WhiA target (Fig. 7 and Table 1). Although the full σWhiG regulon has not been defined, σWhiG has been shown to directly activate transcription of whiH and whiI, encoding two further key sporulation transcription factors (Fig. 9) (53, 54). Our transcriptional profiling shows decreased whiG expression in an S. venezuelae whiA mutant, suggesting that WhiA activates whiG transcription (Fig. 8). Previous ChIP-seq studies showed that whiG is a target for regulation by BldD (14). Therefore, both the release of BldD-mediated repression of whiG and WhiA-mediated activation appear to contribute to the regulation of whiG expression during the transition from aerial growth to sporulation (Fig. 9).
FIG 9 .
Model of the whi gene regulatory network. Flat-headed arrows indicate repression, and pointed arrows indicate activation.
It has been suggested, based on in vitro transcription, that σWhiG directs transcription of the p2 promoter just upstream of whiA (9). However, Aínsa et al. (5) failed to detect EσWhiG-directed in vitro transcription from the whiAp2 promoter under conditions in which the same preparation of σWhiG holoenzyme actively transcribed the well-validated σWhiG target promoter, whiIp. Further, Aínsa et al. (5) found, using S1 nuclease protection assays, that the whiAp2 promoter is active in an S. coelicolor whiG deletion mutant, thus ruling out the in vivo dependence of whiAp2 on σWhiG. It has also been suggested that WhiA and σWhiG directly interact and that WhiA might function as a σWhiG-specific anti-sigma factor (9). We have been unable to detect interaction between S. venezuelae WhiA and σWhiG in an E. coli bacterial two-hybrid system. Resolution of these issues will require further experimental work.
In Streptomyces, the nrdR gene encodes a repressor of the nrdAB and nrdJ genes, which encode class Ia and class II ribonucleotide reductases, respectively (55–57). Ribonucleotide reductases catalyze the reduction of nucleoside diphosphates (NDPs) into deoxynucleoside diphosphates (dNDPs), which are then converted into deoxynucleoside triphosphates (dNTPs), the immediate biosynthetic precursors of DNA, and ribonucleotide reductases play a critical role in regulating the total rate of DNA synthesis in vivo (58). Visualization of active replisomes using enhanced green fluorescent protein (EGFP) fused to a subunit of DNA polymerase III (DnaN-EGFP) indicates that, during the development of aerial hyphae, DNA replication is dramatically upregulated in the long, unseptated sporogenic cell, generating the 50 to 100 chromosomes that will eventually segregate into spores (59). This substantial enhancement of DNA replication presumably necessitates an increased supply of dNTPs. Here we show that WhiA directly binds to a region upstream of the nrdR gene and that nrdR expression is significantly upregulated in a whiA mutant (Table 1; see also Table S1 in the supplemental material), suggesting that WhiA represses the transcription of nrdR. Decreased levels of NrdR would, in turn, lead to derepression of nrdAB and nrdJ transcription and increased levels of the ribonucleotide reductases, thereby enhancing the levels of the dNTPs required for DNA synthesis in the sporogenic cell. Consistent with this logic, our microarray analysis also reveals a corresponding indirect dependence of nrdAB and nrdJ expression on WhiA prior to sporulation.
The largest ChIP-seq peak identified in our analysis lies upstream of the pyrG gene, encoding CTP synthase (Table 1), an enzyme that generates CTP from UTP, ATP, and glutamine. The expression of this gene is downregulated in a whiA mutant, suggesting that WhiA activates pyrG (see Table S1 in the supplemental material). Furthermore, we also identified pyrR (sven1088) as a WhiA target in ChIP-seq, and transcriptional profiling suggests that WhiA represses transcription of pyrR (Table 1; see also Table S1). In other bacteria, PyrR blocks pyrimidine biosynthesis by binding to mRNA derived from the pyr operon in the presence of uridine nucleotides (60). pyrG is not part of the pyr operon and therefore not regulated by PyrR. Activation of pyrG expression by WhiA might lead to an increase in CTP levels, but this could feed DNA replication in sporogenic hyphae only if it were converted to dCTP via three steps (CTP to CDP to dCDP to dCTP), and, apart from the ribonucleotide reductases, no other genes involved in dNTP synthesis were identified as WhiA targets. A speculative alternative explanation for the increase in pyrG expression during Streptomyces morphogenesis might be drawn from the observation that, in several organisms, CTP synthases have the ability to form cytoskeletal-like filaments (reviewed in references 61 and 62). In Caulobacter crescentus, these CTP synthase filaments appear to have been coopted for a cytoskeletal role in determining cell shape, regulating the curvature of cells independently of the enzyme’s catalytic function (63). The mechanism by which the CTP synthase filaments regulate cell shape involves cross talk with another cytoskeletal element, the intermediate filament crescentin (63). It will be interesting to see whether the developmental regulation of pyrG by WhiA reflects a similar cytoskeletal role for CTP synthase in Streptomyces.
Other direct WhiA targets identified here that are linked to morphological development include four additional genes encoding transcription factors: the cyclic AMP receptor protein (Crp), a PadR-family transcriptional regulator (sven3676), the SigB-like RNA polymerase sigma factor SigN (sven3785), and WblH (sven6397) (Fig. 7 and Table 1). Crp appears to function principally as a regulator of secondary metabolism and antibiotic production in S. coelicolor, but several Crp targets are associated with morphogenesis (64). WhiA appears to repress transcription of crp both during vegetative growth and following differentiation (Fig. 8). The PadR-like regulator is likely to function as a repressor to influence the expression of its downstream genes in S. coelicolor, and both deletion and overexpression of the corresponding gene lead to a bald phenotype with sparse aerial hyphae (65). Transcriptional profiling in S. venezuelae reveals clear upregulation in a whiA mutant (Fig. 8), suggesting that WhiA represses transcription of the PadR-family repressor gene. σSigN activity appears to be restricted to the “subapical stem,” the cell lying immediately below the sporogenic cell in aerial hyphae, where it plays a poorly defined role in morphological development and the general stress response (66). Our ChIP-seq analysis shows that sigN is a WhiA target in S. venezuelae, although our transcriptomic data suggest that WhiA does not exert a very substantial effect on sigN expression (see Table S1 in the supplemental material). WblH is a member of the Wbl (WhiB-like) family of Actinobacteria-specific proteins that also includes WhiB and WhiD (67–69). Wbl proteins contain four perfectly conserved cysteine residues that bind an oxygen-sensitive, nitric oxide (NO)-sensitive [4Fe-4S] cluster (70–72). The biochemical role of Wbl proteins has been a matter of dispute (reviewed in reference 68), but recent evidence strongly suggests that Wbl proteins function as transcription factors (73). The regulation of σWhiG, σSigN, Crp, WblH, and other transcription factors by WhiA emphasizes the pleiotropic potential of WhiA control.
treZ, the WhiA target investigated above that lacks the GACAC consensus sequence but carries two copies of a GACGC motif (Fig. 4), encodes the metabolic enzyme TreZ, which functions in a pathway that breaks down the storage compound glycogen to yield trehalose (74). The accumulation and degradation of glycogen, stored as insoluble granules, are under developmental control in Streptomyces, and glycogen reserves are actively degraded during sporulation (75, 76), leading to an accumulation of trehalose that contributes substantially to the heat and desiccation resistance of mature spores (77). Thus, the activation of treZ transcription by WhiA (see Table S1 in the supplemental material) provides a direct link between the regulatory cascade that controls differentiation in Streptomyces and the accumulation of trehalose in spores.
WhiA targets involved in translation.
Unexpectedly, several of the WhiA targets that we identified through ChIP-seq are involved in general translation, including the genes encoding translation initiation factor 1 (IF-1), translation elongation factor P, small-subunit (SSU) ribosomal protein S8, SSU ribosomal protein S1, and SSU ribosomal protein S10 (Table 1; see also Table S1 in the supplemental material). Expression of each of these targets is significantly downregulated in a whiA mutant, suggesting that an increased abundance of these components of the translation machinery may be a feature of morphological development in Streptomyces. It should be noted that S1 is an RNA-binding protein that has also been coopted as a transcription termination factor (78), and S10 (NusE) is involved in antitermination in E. coli (79). It is therefore possible that these proteins are being used for something other than translation in S. venezuelae.
Conclusions.
The work reported here confirms, on a genome-wide scale, that WhiA functions as a transcription factor and shows that it behaves almost equally as an activator and as a repressor. Further, supporting previous in vitro analysis (9), we show that most in vivo binding sites are associated with a highly conserved GACAC motif, but we find that there is no clear relationship between the orientation of this asymmetric sequence and whether the corresponding target gene is activated or repressed by WhiA.
WhiA consists of an N-terminal homing endonuclease domain and a C-terminal HTH domain, connected by a flexible linker (8), and the C-terminal HTH domain (which is completely absent from true homing endonucleases) binds DNA site-specifically by itself (9). It is therefore possible that the HTH domain of WhiA has entirely replaced the DNA-binding function of the homing endonuclease domain. However, although the homing endonuclease domain cannot footprint on DNA in isolation, it does appear to contribute to the affinity of DNA binding by full-length WhiA (9). Therefore, it is conceivable that the homing endonuclease domain could also contribute to the specificity of DNA binding. It may also mediate protein-protein interaction or be a target for posttranslational regulation controlling the activity of WhiA. Extensive further research will be required to understand how the evolutionarily adapted structural and biochemical features of WhiA enable it to fulfill its bifunctional role as repressor and activator.
Based on Western blot analysis, WhiA is constitutively present at all time points examined (Fig. 5; see also Fig. S2 in the supplemental material). In contrast, based on ChIP-seq analysis, WhiA binds to its target promoters predominantly at the onset of sporulation during fragmentation phase (Fig. 4, 6, and 7; see also Fig. S3). These results suggest that WhiA activity is subject to posttranslational regulation.
Counting both DNA strands, there are 15,116 GACAC sequences in the S. venezuelae chromosome, but only a fraction of these sequences are bound by WhiA in vivo (as judged by ChIP-seq). In addition, there are clear targets (such as treZ) that bind WhiA in vivo and in vitro but do not possess this exact motif. Additional specificity determinants must presumably exist in addition to the short (albeit highly conserved) GACAC motif. The shortness of this sequence and the consequent difficulties in bioinformatic prediction of true in vivo binding sites are reminiscent of eukaryotic transcription factors, something that could conceivably reflect the evolutionary history of WhiA.
Comparing the sequences of the S. venezuelae WhiA-binding sites defined by DNase I footprinting with the equivalent promoter regions in S. coelicolor, the GACAC motifs within the footprints are conserved in S. coelicolor for the whiA, filP, ftsW, nrdR, and parA-like targets but not for cslA and sven4724/4725. These results suggest that the WhiA regulon is likely to be broadly conserved between these two model species. However, because the GACAC sequence occurs so frequently in streptomycete genomes, it is not possible to make a valid global comparative analysis for WhiA-binding sites defined only by ChIP-seq.
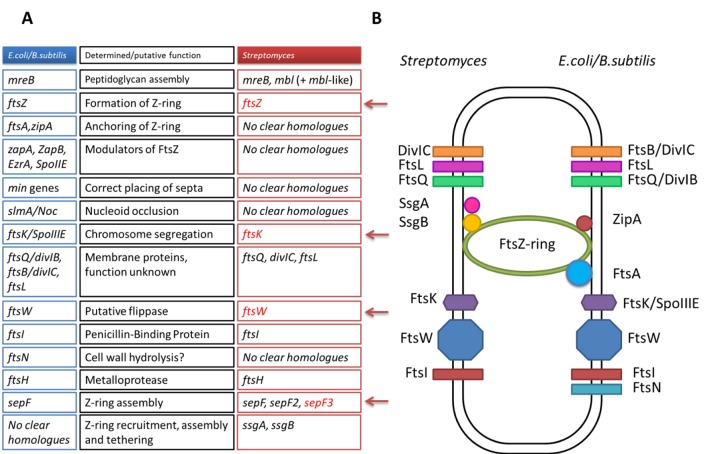
Our constructed S. venezuelae null mutant shows that WhiA is required for the initiation of sporulation septation and chromosome segregation, as it is in S. coelicolor. It is therefore striking that several pivotal proteins of the Streptomyces cell division machinery, such as FtsZ, FtsW, and FtsK (Fig. 10), are encoded by genes that are direct targets for activation by WhiA during development. The initiation of the massive cell division event is tightly coordinated with the orderly arrest of apical growth in the sporogenic cell. It may therefore be significant that filP becomes repressed by WhiA at approximately the same time that ftsZ, ftsW, and ftsK are activated, given that FilP is a key component of the polarisome that directs polar growth at the tips of hyphae (25, 27). Further, recent evidence suggests that the direct interaction of another crucial component of the polarisome, Scy, with the partition protein ParA coordinates the arrest of tip extension in the sporogenic cell with the initiation of chromosome segregation prior to septation (30). The sporulation-specific upregulation of parAB transcription depends on whiA in S. venezuelae, but we did not detect binding of WhiA to the parAB promoter in ChIP-seq, suggesting that this dependence is indirect.
FIG 10 .
Comparison of the cell division machinery of E. coli/B. subtilis and Streptomyces. (A) Genes encoding key components of the divisome in E. coli and B. subtilis, their putative or determined function, and their presence or absence in Streptomyces. Components of the Streptomyces cell division machinery identified in this work as direct targets for WhiA regulation are highlighted in red and marked with an arrow. (B) Differences between the known components of the cell division machinery in E. coli/B. subtilis and Streptomyces.
As illustrated in Fig. 10, many important components of the divisome in E. coli and Bacillus subtilis have no clear homologues in Streptomyces. For example, the FtsZ ring has to be anchored to the membrane, and in E. coli, this role is fulfilled by FtsA and ZipA (80), proteins absent in Streptomyces (2). This suggests that in Streptomyces there exist other proteins that perform functions central to the ability of the organism to divide and sporulate. The Streptomyces proteins SsgA and SsgB that function to recruit FtsZ during sporulation perfectly illustrate this point (81). Further cell division proteins unique to streptomycetes may well be under the control of one or more of the key developmental regulators, perhaps especially WhiA and WhiB, which are the only Whi regulators absolutely required for sporulation septation in S. venezuelae (M. J. Bibb and M. J. Buttner, unpublished data). In seeking out these proteins, it might be particularly fruitful to explore the overlap between the regulons of WhiA and WhiB, given their suggested close interplay and the near-identical phenotypes of the whiA and whiB mutants in both S. coelicolor and S. venezuelae (Fig. 9) (5, 6; Bibb and Buttner, unpublished).
WhiA-like proteins are found in virtually all Gram-positive bacteria, including many that do not sporulate, raising interesting questions about the evolution of WhiA and the reassortment of the genes under its control in different bacteria. Outside the streptomycetes, the only published genetic analysis of whiA function is in Bacillus subtilis. In this endospore-forming bacterium, deletion of whiA (yvcL) has no obvious phenotypic consequences (82), suggesting that WhiA does not play an important role in the control of sporulation or cell division in this organism. It will be interesting to confirm that WhiA functions as a transcription factor in Bacillus and, if so, to identify the genes it regulates.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and growth conditions.
Strains, plasmids, and oligonucleotides used in this study are described in Table S2 in the supplemental material. Escherichia coli K-12 strain DH5α was used for plasmid and cosmid propagation. BW25113 (83) containing a λ RED plasmid, pIJ790, was used to create the disruption cosmid. Cosmids were conjugated from the dam dcm hsdS E. coli strain ET12567 containing pUZ8002 (84) as described by Gust et al. (18, 19). Although ET12567 was used for this purpose, it should be noted that S. venezuelae does not restrict methylated DNA, and so cosmids can be conjugated from methylating E. coli strains. S. venezuelae strains were cultured in MYM-TAP (85) made with 50% tap water and supplemented with 200 µl of trace element solution (86) per 100 ml. Conjugations between E. coli and S. venezuelae were essentially carried out as described for S. coelicolor (86). However, because S. venezuelae grows much more quickly than S. coelicolor, the plates were incubated either at room temperature overnight or at 30°C for only 7 h before overlaying with the selective antibiotics (incubation at 30°C for longer periods before overlaying results in confluent lawns of S. venezuelae). Additionally, there is no need to heat shock S. venezuelae spores prior to the mating. Where conjugation was carried out using the nonsporulating ΔwhiA::apr strain, MYM liquid cultures were set up using mycelium as an inoculum and actively growing cultures were concentrated and used in the conjugation.
Spores of wild-type S. venezuelae were harvested from MYM plates using sterile cotton pads. Three milliliters of 20% glycerol was gently applied across the plate using sterile forceps and the cotton pad. The spores were collected through the pad using a 2-ml syringe. For the nonsporulating whiA mutant, mycelium in 20% glycerol was homogenized manually 10 times in a Dounce homogenizer and appropriate (and reproducible) inocula were determined in trial experiments, as judged by optical density (OD) and microscopic inspection.
Construction and complementation of an S. venezuelae whiA null mutant.
Using the Redirect PCR targeting method of Gust et al. (18, 19), whiA mutants were generated in which the coding region was replaced with a single apramycin resistance (apr) cassette. A cosmid library that covers >98% of the S. venezuelae genome (Bibb and Buttner, unpublished) is fully documented at http://strepdb.streptomyces.org.uk/. Cosmid Sv-2-B11 was introduced into E. coli BW25113 containing pIJ790, and the relevant genes were replaced with the apr-oriT cassette amplified from pIJ773 using the primer pairs whiAdisfor and whiAdisrev (see Table S2 in the supplemental material). The resulting disrupted cosmids were confirmed by restriction digestion and by PCR analysis using the flanking primers whiAconfor and whiAconrev and introduced into S. venezuelae by conjugation. Null mutant derivatives, generated by double crossing-over, were identified by their apramycin-resistant, kanamycin-sensitive, and morphological phenotypes, and their chromosomal structures were confirmed by PCR analysis using the flanking primers whiAconfor and whiAconrev and by Southern hybridization using the entire cosmid Sv-2-B11, partially digested with Sau3AI, as a probe. A representative whiA null mutant was designated SV11. For complementation, whiA was amplified with the primers whiAcompfor and whiAcomprev, generating a fragment carrying the coding sequence and the whiA promoter, and cloned into HindIII-cut pMS82 (87) to create pIJ6760. The plasmid was introduced into the whiA mutant by conjugation and fully complemented all aspects of the mutant phenotype.
Construction of a 3×FLAG-WhiA-complemented S. venezuelae strain.
In order to engineer an S. venezuelae strain expressing a form of WhiA with an N-terminal, triple-FLAG tag (DYKDHDGDYKDHDIDYKDDDDK), a pMS82-derived construct, pIJ10600, was created via a two-step fusion-PCR approach. In the first step, the Sv-2-B11 cosmid was used as a template for two separate PCRs. The first reaction amplified the promoter region of the whiA gene using the primer pair whiAFLAGextfor and whiAFLAGfusrev. The second reaction amplified the coding region of the whiA gene using the primer pair whiAFLAGfusfor and whiAFLAGextrev. Together, the whiAFLAGfusrev and whiAFLAGfusfor primers contain the sequence encoding the triple-FLAG tag via a 24-bp overlapping section. In the second step, a PCR using the nested primers whiAFLAGnesfor and WhiAFLAGnesrev was used to amplify the entire whiA gene and its promoter, fusing the two products from step 1 together and incorporating the 3×FLAG tag sequence between them. The nested primers whiAFLAGnesfor and WhiAFLAGnesrev additionally contain the HindIII and KpnI sites, respectively, to enable cloning into HindIII-, KpnI-cut pMS82. The resulting vector was named pIJ10600. In order to insert a [Gly4Ser]3 linker between the 3×FLAG tag and the coding region of whiA, a second fusion experiment was conducted using pIJ10600 as a template. Once again, two PCRs were carried out in a first step using the primer pairs whiAFLAGnesfor/whiALINKfusrev and whiAFLAGnesrev/whiALINKfusfor. Together, the whiALINKfusrev and whiALINKfusfor primers contain the sequence encoding the [Gly4Ser]3 linker (GGTGGAGGCGGTTCAGGCGGAGGTGGCTCTGGCGGTGGCGGTAGT) via a 24-bp overlapping section. The second step, employing the same nested primers, resulted in fusion of the two products in the first step and incorporated the linker sequence between them. Restriction digestion using HindIII and KpnI, followed by ligation into HindIII-, KpnI-cut pMS82, created pIJ10601. The plasmids pIJ10600 and pIJ10601 were introduced individually into the ΔwhiA mutant by conjugation, and their ability to restore sporulation was assessed both on solid and in liquid MYM.
RNA isolation and DNA microarray analysis.
RNA isolation from S. venezuelae and DNA microarray analysis were performed as described previously (16, 88). The CEL files received from the scanner were read into the R package for statistical computing (89) using the ReadAffy function of the affy package (90). The rma function of the affy package was used to compute an ExpressionSet object from the CEL files. This ExpressionSet object contains the expression values (log2) for each gene in each CEL file. The function lmFit of the limma package (91), along with a suitable design matrix, was used to combine replicate arrays into single coefficients of expressions for each gene at each time point or strain into an MArrayLM object. Expression values were retrieved from the MArrayLM object and subjected to a per-gene normalization to the median before being used to generate the graphs shown in this paper.
Chromatin immunoprecipitation.
S. venezuelae ATCC 10712 and S. venezuelae SV11-pIJ10601 (ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA) were grown in four 30-ml volumes of MYM-TAP for the appropriate length of time. Formaldehyde was added to cultures at a final concentration of 1% (vol/vol), and incubation was continued for 30 min. Glycine was then added to a final concentration of 125 mM to stop the cross-linking. The samples were left at room temperature for 5 min and washed twice in phosphate-buffered saline (PBS) buffer (pH 7.4). The pellets were resuspended in 0.5 ml of lysis buffer (10 mM Tris-Cl, pH 8.0, 50 mM NaCl), containing 15 mg ml−1 of lysozyme and 1× protease inhibitor (Roche Applied Science) and incubated at 37°C for 25 min or until lysed. Subsequently, 0.5 ml immunoprecipitation (IP) buffer (50 mM Tris-Cl, pH 8.0, 250 mM NaCl, 0.8% [vol/vol] Triton X-100), containing 1× protease inhibitor, was added, and the samples were chilled on ice. The samples were sonicated for six cycles of 20 s each at 10 µm to shear chromosomal DNA into fragments ranging from 300 to 1,000 bp on average. The samples were centrifuged twice at 16,000 × g at 4°C for 10 min to clear the cell extracts, after which 50 µl of each was set aside for total-DNA extraction. The remaining lysates were added to M2 (Sigma-Aldrich A2220) gel suspension (40 µl per ml of lysate), prepared and washed as described in the manufacturer’s instructions. The mixtures were then incubated on a rotating wheel at 4°C overnight. The samples were centrifuged for 30 s at 4°C and 5,500 × g, and the pellets were washed twice with 0.5× IP buffer and then twice with 1× IP buffer and transferred to new Eppendorf tubes after the first washing step. The pellets and 50 µl of total cell extracts (set aside earlier) were eluted overnight at 65°C in 100 µl of IP elution buffer (50 mM Tris-Cl, pH 7.6, 10 mM EDTA, 1% SDS) to reverse the cross-links. The samples were centrifuged at 16,000 × g for 5 min to remove the beads. The pellets were reextracted with 50 µl of TE buffer (10 mM Tris-Cl, pH 7.4, 1 mM EDTA) and incubated with 0.2 mg ml−1 proteinase K (Roche) for 2 h at 55°C. The samples were extracted twice with phenol-chloroform and once with chloroform and further purified using QiaQuick columns (Qiagen). DNA was eluted in 50 µl EB buffer (10 mM Tris-Cl, pH 8.5) and quantified using a NanoDrop spectrophotometer (Thermo Scientific).
Library construction and sequencing.
Library construction and sequencing were performed by The Genome Analysis Centre (TGAC), Norwich Research Park, Norwich, United Kingdom. The TruSeq ChIP sample preparation kit from Illumina Inc. (part 15023092, revision A, August 2012) was used according to the recommended protocol. In brief, 5 to 10 ng ChIP DNA is blunt ended and phosphorylated, and a single “A” nucleotide is added to the 3′ ends of the fragments in preparation for ligation to an adapter with a single-base “T” overhang. The ligation products are purified and accurately size selected by agarose gel electrophoresis. Size-selected DNA is then purified and PCR amplified to enrich for fragments that have adapters on both ends. The final purified product is then quantified using a combination of Bioanalyzer DNA HS Chip from Agilent Technology 2100 Bioanalyzer and Qubit 2.0 from Invitrogen. The libraries were normalized to 10 nM and pooled, and quantitative PCR (q-PCR) was performed prior to cluster generation.
Each library pool was diluted to 2 nM with NaOH, and 5 µl was transferred into 995 µl HT-1 to give a final concentration of 10 pM. The normalized library (120 µl) was then transferred into a 200-µl strip tube and placed on ice before loading onto the Illumina cBot. Flow cells were clustered using TruSeq Single-Read Cluster Generation Kit v3, using the SR_Amp_Lin_Block_Hyb_v8.0 recipe. Following the clustering procedure, the flow cell was loaded onto the Illumina HiSeq2000 instrument according to the manufacturer’s instructions. The sequencing chemistry used was TruSeq SBS version 3 using Illumina software HCS 1.5.15.1 and RTA 1/13/48. Each library pool was run on the Illumina HiSeq2000 instrument with 100-bp single-read run metrics.
ChIP-seq data analysis.
The reads in the fastq files received from the sequencing contractor were aligned to the S. venezuelae genome (GenBank accession number NC_018750) using the bowtie2 software (92), which resulted in one SAM (.sam) file for each fastq file. All further operations described below were carried out using a combination of Perl scripts dependent on the BioPerl toolkit (93) and R (89) scripts. From each SAM file, coverage at (number of reads mapping to) each nucleotide position of the S. venezuelae genome was calculated and the output was saved in files here referred to as coverage files. For each coverage file, a local enrichment was calculated in a moving window of 51 nucleotides (nt) moving in steps of 25 nucleotides as (the sum of coverage at each nucleotide position in the 51-nt window) divided by (the sum of coverage at each nucleotide position in a 2,001-nucleotide window centered around the 51-nucleotide window). This results in an enrichment ratio value for every 25 nucleotides along the genome. The enrichment ratios thus calculated were stored in files referred to as enrichment files.
In each difference file, the P value of each difference was calculated assuming that the differences were normally distributed using the pnorm function of R.
Now, for each pair of total DNA and IP DNA enrichment ratios, the values of total DNA were subtracted from the values of IP DNA and the differences stored in files here referred to as the difference files. Thus, we arrived at a difference file for each of the three time points for the FLAG-tagged pairs and one file for the wild-type (WT) pair. In each difference file, the P value of each difference was calculated assuming that the differences were normally distributed using the pnorm function of R. Differences with adjusted P values (using the pnorm function of R with the Hochberg method) less than or equal to 1e−4 were saved to new files referred to as sigDiff files. All the four types of files, i.e., coverage, enrichment, difference, and sigDiff, are two-column files where the first column is always position on the genome and the second column contains the relevant value.
The gene coordinates in the GenBank file and the genomic positions in the sigDiff files were combined to arrive at lists of genes on the left and right of the significantly different genomic positions. The relationships of the difference values and these genes were manually verified by viewing the difference files and the features in the S. venezuelae GenBank file as tracks in the Integrated Genome Browser (94). Significant different genomic positions located more than 300 bp upstream of the nearest start codon were separated from the data set. The most significant genomic position for each transcription unit was then selected. Finally, the information above was combined with Affymetrix microarray data to generate Table S1 in the supplemental material.
Western blotting, DNase I footprinting, and microscopy.
For details on Western blotting, DNase I footprinting, and microscopy, see Text S1 in the supplemental material.
SUPPLEMENTAL MATERIAL
Supplemental materials and methods. Download
Phenotypes of wild-type S. venezuelae (WT), the constructed ΔwhiA::apr null mutant SV11 (ΔwhiA::apr), SV11 carrying the empty vector pMS82 (ΔwhiA::apr attBΦBT1::pMS82), and SV11 expressing WhiA (ΔwhiA::apr attBΦBT1::whiA) or N-terminal, triple FLAG-tagged versions of WhiA with or without a [Gly4Ser]3 linker (ΔwhiA::apr attBΦBT1::3×FLAGlink-whiA and ΔwhiA::apr attBΦBT1::3×FLAG-whiA). (A) Appearance on MYM solid medium after 4 days. (B) Phase-contrast images of coverslip impressions taken from MYM plates after 4 days. Download
Western blot analysis of wild-type S. venezuelae (−) and ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA (+) grown in MYM liquid cultures and sampled from 14 to 40 h. Five micrograms total protein was loaded per lane, and 3×FLAG-[Gly4Ser]3-WhiA was detected using M2 anti-FLAG antibody. Protein size markers (NEB) are shown with molecular masses in kDa. Download
Chromosome-wide distribution of WhiA-binding sites in S. venezuelae identified by ChIP-seq analysis. ChIP-seq was conducted using M2 anti-FLAG antibody on the ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA strain at time points representing three distinct developmental stages: vegetative growth, the onset of sporulation (fragmentation), and midsporulation. The wild-type strain (expressing nontagged WhiA from the native locus) analyzed at the onset of sporulation (fragmentation) was used as a negative control. Download
Complete ChIP-seq data set for ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA. Each row represents a ChIP “peak” based on the analysis of 25-bp segments of the S. venezuelae genome. Only those peaks with significance of P < E−04 for at least one of the ChIP samples are included in the analysis. A single entry that had significance of <E−04 in the negative control was discarded from the analysis. Pos, position of peak in the S. venezuelae genome in bases. Diff, the difference between the local normalized (ln) values of the immunoprecipitated (ChIP) samples and the total (non-ChIP) DNA samples, for each of the ChIP samples, i.e., V (vegetative growth), F (onset of sporulation/fragmentation), S (midsporulation), and C (wild-type negative control, analyzed at the onset of sporulation/fragmentation). Adjusted P values (apv), significance values for each of the ChIP samples, i.e., V, F, S, and WT negative control (C) after adjusting for multiple testing by the Hochberg method as implemented in the p.adjust function of R. Peak (pk), each ChIP sample is qualified as being significant (P < E−04) by the identifier TRUE and insignificant (P > E−04) by the identifier FALSE. Left Gene, the identifier (SVEN no.) for the gene on the left of the identified ChIP peak. Right Gene, the identifier (SVEN no.) for the gene on the right of the identified ChIP peak. Distance, the distance (in bases) between the ChIP peak and the predicted start codon of the downstream gene. Start, the start position on the S. venezuelae genome (in bases) of the gene downstream of the ChIP peak. End, the end position on the S. venezuelae genome (in bases) of the gene downstream of the ChIP peak. Strand, the strand on which the gene is found (forward = 1; reverse = −1). Product, (possible) gene function based on annotation in StrepDB (http://strepdb.streptomyces.org.uk). Affy LogFC, the log fold change (log2 scale) in expression of the whiA mutant compared to wild-type S. venezuelae at the 10-, 12-, 14-, 16-, 18-, and 20-h time points. − indicates a decrease in expression of the gene in a whiA mutant compared to the wild type; + indicates an increase in expression of the gene in a whiA mutant compared to the wild type. Cells highlighted in red represent greater than a 2-fold increase in expression in the whiA mutant. Cells highlighted in yellow represent greater than a 2-fold decrease in expression in the whiA mutant.
Strains, plasmids, and oligonucleotide primers used in this study.
ACKNOWLEDGMENTS
We are grateful to The Genome Analysis Centre (TGAC) for performing the library construction and sequencing of the ChIP samples, to Ágota Domonkos for complementing the whiA null mutant, and to Klas Flärdh for helpful discussion.
This work was funded by BBSRC grant BB/H006125/1 (to M. J. Buttner) and by the MET Institute Strategic Program Grant to the John Innes Centre from the BBSRC.
Footnotes
Citation Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ. 2013. Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. mBio 4(5):e00684-13. doi:10.1128/mBio.00684-13.
REFERENCES
- 1. Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 2. McCormick JR. 2009. Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Microbiol. 12:689–698 [DOI] [PubMed] [Google Scholar]
- 3. McCormick JR, Flärdh K. 2012. Signals and regulators that govern Streptomyces development. FEMS Microbiol. Rev. 36:206–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jakimowicz D, van Wezel GP. 2012. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85:393–404 [DOI] [PubMed] [Google Scholar]
- 5. Aínsa JA, Ryding NJ, Hartley N, Findlay KC, Bruton CJ, Chater KF. 2000. WhiA, a protein of unknown function conserved among Gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3. J. Bacteriol. 182:5470–5478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Flärdh K, Findlay KC, Chater KF. 1999. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3. Microbiology 145:2229–2243 [DOI] [PubMed] [Google Scholar]
- 7. Knizewski L, Ginalski K. 2007. Bacterial DUF.199/COG1481 proteins including sporulation regulator WhiA are distant homologs of LAGLIDADG homing endonucleases that retained only DNA binding. Cell Cycle 6:1666–1670 [DOI] [PubMed] [Google Scholar]
- 8. Kaiser BK, Clifton MC, Shen BW, Stoddard BL. 2009. The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease. Structure 17:1368–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kaiser BK, Stoddard BL. 2011. DNA recognition and transcriptional regulation by the WhiA sporulation factor. Sci. Rep. 1:156. 10.1038/srep00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor GK, Stoddard BL. 2012. Structural, functional and evolutionary relationships between homing endonucleases and proteins from their host organisms. Nucleic Acids Res. 40:5189–5200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. 2001. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol. Microbiol. 40:257-269 [DOI] [PubMed] [Google Scholar]
- 12. Chater KF, Chandra G. 2008. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J. Microbiol. 46:1–11 [DOI] [PubMed] [Google Scholar]
- 13. Chandra G, Chater KF. 2008. Evolutionary flux of potentially bldA-dependent Streptomyces genes containing the rare leucine codon TTA. Antonie Van Leeuwenhoek 94:111–126 [DOI] [PubMed] [Google Scholar]
- 14. den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. 2010. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Mol. Microbiol. 78:361–379 [DOI] [PubMed] [Google Scholar]
- 15. Higo A, Hara H, Horinouchi S, Ohnishi Y. 2012. Genome-wide distribution of AdpA, a global regulator for secondary metabolism and morphological differentiation in Streptomyces, revealed the extent and complexity of the AdpA regulatory network. DNA Res. 19:259–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bibb MJ, Domonkos A, Chandra G, Buttner MJ. 2012. Expression of the chaplin and rodlin hydrophobic sheath proteins in Streptomyces venezuelae is controlled by σBldN and a cognate anti-sigma factor, RsbN. Mol. Microbiol. 84:1033–1049 [DOI] [PubMed] [Google Scholar]
- 17. Glazebrook MA, Doull JL, Stuttard C, Vining LC. 1990. Sporulation of Streptomyces venezuelae in submerged cultures. J. Gen. Microbiol. 136:581–588 [DOI] [PubMed] [Google Scholar]
- 18. Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gust B, Chandra G, Jakimowicz D, Yuqing T, Bruton CJ, Chater KF. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54:107–128 [DOI] [PubMed] [Google Scholar]
- 20. Teytelman L, Ozaydin B, Zill O, Lefrançois P, Snyder M, Rine J, Eisen MB. 2009. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One 4:e6700. 10.1371/journal.pone.0006700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p 28–36 In Altman R, Brutlag D, Karp P, Lathrop R, Searls D. (ed), Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA: [PubMed] [Google Scholar]
- 22. Jakimowicz D, Mouz S, Zakrzewska-Czerwinska J, Chater KF. 2006. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. J. Bacteriol. 188:1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hempel AM, Cantlay S, Molle V, Wang SB, Naldrett MJ, Parker JL, Richards DM, Jung YG, Buttner MJ, Flärdh K. 2012. The Ser/Thr protein kinase AfsK regulates polar growth and hyphal branching in the filamentous bacteria Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 109:E2371–E2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Flärdh K, Richards DM, Hempel AM, Howard M, Buttner MJ. 2012. Regulation of apical growth and hyphal branching in Streptomyces. Curr. Opin. Microbiol. 15:737–743 [DOI] [PubMed] [Google Scholar]
- 25. Fuchino K, Bagchi S, Cantlay S, Sandblad L, Wu D, Bergman J, Kamali-Moghaddam M, Flärdh K, Ausmees N. 2013. Dynamic gradients of an intermediate filament-like cytoskeleton are recruited by a polarity landmark during apical growth. Proc. Natl. Acad. Sci. U. S. A. 110:E1889–E1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Holmes NA, Walshaw J, Leggett RM, Thibessard A, Dalton KA, Gillespie MD, Hemmings AM, Gust B, Kelemen GH. 2013. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc. Natl. Acad. Sci. U. S. A. 110:E397–E406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bagchi S, Tomenius H, Belova LM, Ausmees N. 2008. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 70:1037–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Flärdh K. 2003. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2). Mol. Microbiol. 49:1523-1536 [DOI] [PubMed] [Google Scholar]
- 29. Flärdh K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6:564–571 [DOI] [PubMed] [Google Scholar]
- 30. Ditkowski B, Holmes N, Rydzak J, Donczew M, Bezulska M, Ginda K, Kedzierski P, Zakrzewska-Czerwinska J, Kelemen GH, Jakimowicz D. 2013. Dynamic interplay of ParA with the polarity protein, Scy, coordinates the growth with chromosome segregation in Streptomyces coelicolor. Open Biol. 3:130006. 10.1098/rsob.130006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. de Jong W, Wösten HA, Dijkhuizen L, Claessen D. 2009. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol. Microbiol. 73:1128–1140 [DOI] [PubMed] [Google Scholar]
- 32. Xu H, Chater KF, Deng Z, Tao M. 2008. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. J. Bacteriol. 190:4971–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liman R, Facey PD, van Keulen G, Dyson PJ, Del Sol R. 2013. A laterally acquired galactose oxidase-like gene is required for aerial development during osmotic stress in Streptomyces coelicolor. PLoS One 8:e54112. 10.1371/journal.pone.0054112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flärdh K, Leibovitz E, Buttner MJ, Chater KF. 2000. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Mol. Microbiol. 38:737–749 [DOI] [PubMed] [Google Scholar]
- 35. Willemse J, Mommaas AM, van Wezel GP. 2012. Constitutive expression of ftsZ overrides the whi developmental genes to initiate sporulation of Streptomyces coelicolor. Antonie Van Leeuwenhoek 101:619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mercer KL, Weiss DS. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, Diepeveen-de Bruin M, Nguyen-Distèche M, de Kruijff B, Breukink E. 2011. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 30:1425–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mistry BV, Del Sol R, Wright C, Findlay K, Dyson P. 2008. FtsW is a dispensable cell division protein required for Z-ring stabilization during sporulation septation in Streptomyces coelicolor. J. Bacteriol. 190:5555–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bennett JA, Yarnall J, Cadwallader AB, Kuennen R, Bidey P, Stadelmaier B, McCormick JR. 2009. Medium-dependent phenotypes of Streptomyces coelicolor with mutations in ftsI or ftsW. J. Bacteriol. 191:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14:243–254 [DOI] [PubMed] [Google Scholar]
- 41. Wang L, Yu Y, He X, Zhou X, Deng Z, Chater KF, Tao M. 2007. Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2). J. Bacteriol. 189:2310-2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Arends SJ, Kustusch RJ, Weiss DS. 2009. ATP-binding site lesions in FtsE impair cell division. J. Bacteriol. 191:3772–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schmidt KL, Peterson ND, Kustusch RJ, Wissel MC, Graham B, Phillips GJ, Weiss DS. 2004. A predicted ABC transporter, FtsEX, is needed for cell division in Escherichia coli. J. Bacteriol. 186:785–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. de Leeuw E, Graham B, Phillips GJ, ten Hagen-Jongman CM, Oudega B, Luirink J. 1999. Molecular characterization of Escherichia coli FtsE and FtsX. Mol. Microbiol. 31:983–993 [DOI] [PubMed] [Google Scholar]
- 45. Corbin BD, Wang Y, Beuria TK, Margolin W. 2007. Interaction between cell division proteins FtsE and FtsZ. J. Bacteriol. 189:3026–3035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jakimowicz D, Chater K, Zakrzewska-Czerwínska J. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45:1365–1377 [DOI] [PubMed] [Google Scholar]
- 47. Jakimowicz D, Gust B, Zakrzewska-Czerwinska J, Chater KF. 2005. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 187:3572–3580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65:625–641 [DOI] [PubMed] [Google Scholar]
- 49. Ishikawa S, Kawai Y, Hiramatsu K, Kuwano M, Ogasawara N. 2006. A new FtsZ-interacting protein, YlmF, complements the activity of FtsA during progression of cell division in Bacillus subtilis. Mol. Microbiol. 60:1364–1380 [DOI] [PubMed] [Google Scholar]
- 50. Hamoen LW, Meile JC, de Jong W, Noirot P, Errington J. 2006. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 59:989–999 [DOI] [PubMed] [Google Scholar]
- 51. Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. 2011. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. J. Bacteriol. 193:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hull TD, Ryu MH, Sullivan MJ, Johnson RC, Klena NT, Geiger RM, Gomelsky M, Bennett JA. 2012. Cyclic di-GMP phosphodiesterases RmdA and RmdB are involved in regulating colony morphology and development in Streptomyces coelicolor. J. Bacteriol. 194:4642–4651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF. 1998. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3. Mol. Microbiol. 29:343–357 [DOI] [PubMed] [Google Scholar]
- 54. Aínsa JA, Parry HD, Chater KF. 1999. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3. Mol. Microbiol. 34:607–619 [DOI] [PubMed] [Google Scholar]
- 55. Borovok I, Gorovitz B, Yanku M, Schreiber R, Gust B, Chater K, Aharonowitz Y, Cohen G. 2004. Alternative oxygen-dependent and oxygen-independent ribonucleotide reductases in Streptomyces: cross-regulation and physiological role in response to oxygen limitation. Mol. Microbiol. 54:1022–1035 [DOI] [PubMed] [Google Scholar]
- 56. Grinberg I, Shteinberg T, Gorovitz B, Aharonowitz Y, Cohen G, Borovok I. 2006. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 188:7635–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Grinberg I, Shteinberg T, Hassan AQ, Aharonowitz Y, Borovok I, Cohen G. 2009. Functional analysis of the Streptomyces coelicolor NrdR ATP-cone domain: role in nucleotide binding, oligomerization, and DNA interactions. J. Bacteriol. 191:1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herrick J, Sclavi B. 2007. Ribonucleotide reductase and the regulation of DNA replication: an old story and an ancient heritage. Mol. Microbiol. 63:22–34 [DOI] [PubMed] [Google Scholar]
- 59. Ruban-Ośmiałowska B, Jakimowicz D, Smulczyk-Krawczyszyn A, Chater KF, Zakrzewska-Czerwinska J. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 188:7311–7316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Turnbough CL, Switzer RL. 2008. Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors. Microbiol. Mol. Biol. Rev. 72:266–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ingerson-Mahar M, Gitai Z. 2012. A growing family: the expanding universe of the bacterial cytoskeleton. FEMS Microbiol. Rev. 36:256–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Barry RM, Gitai Z. 2011. Self-assembling enzymes and the origins of the cytoskeleton. Curr. Opin. Microbiol. 14:704–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gao C, Hindra, Mulder D, Yin C, Elliot MA. 2012. Crp is a global regulator of antibiotic production in Streptomyces. mBio 3(6):e00407-12. 10.1128/mBio.00407-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang G, Tian Y, Hu K, Feng C, Tan H. 2010. SCO3900, co-transcripted with three downstream genes, is involved in the differentiation of Streptomyces coelicolor. Curr. Microbiol. 60:268–273 [DOI] [PubMed] [Google Scholar]
- 66. Dalton KA, Thibessard A, Hunter JI, Kelemen GH. 2007. A novel compartment, the “subapical stem” of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Mol. Microbiol. 64:719–737 [DOI] [PubMed] [Google Scholar]
- 67. Molle V, Palframan WJ, Findlay KC, Buttner MJ. 2000. WhiD and WhiB, homologous proteins required for different stages of sporulation in Streptomyces coelicolor A3. J. Bacteriol. 182:1286–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. den Hengst CD, Buttner MJ. 2008. Redox control in Actinobacteria. Biochim. Biophys. Acta 1780:1201–1216 [DOI] [PubMed] [Google Scholar]
- 69. Gao B, Paramanathan R, Gupta RS. 2006. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek 90:69–91 [DOI] [PubMed] [Google Scholar]
- 70. Jakimowicz P, Cheesman MR, Bishai WR, Chater KF, Thomson AJ, Buttner MJ. 2005. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J. Biol. Chem. 280:8309–8315 [DOI] [PubMed] [Google Scholar]
- 71. Singh A, Guidry L, Narasimhulu KV, Mai D, Trombley J, Redding KE, Giles GI, Lancaster JR, Steyn AJ. 2007. Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U. S. A. 104:11562–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Crack JC, Smith LJ, Stapleton MR, Peck J, Watmough NJ, Buttner MJ, Buxton RS, Green J, Oganesyan VS, Thomson AJ, Le Brun NE. 2011. Mechanistic insight into the nitrosylation of the [4Fe-4S] cluster of WhiB-like proteins. J. Am. Chem. Soc. 133:1112–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Smith LJ, Stapleton MR, Fullstone GJ, Crack JC, Thomson AJ, Le Brun NE, Hunt DM, Harvey E, Adinolfi S, Buxton RS, Green J. 2010. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochem. J. 432:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chandra G, Chater KF, Bornemann S. 2011. Unexpected and widespread connections between bacterial glycogen and trehalose metabolism. Microbiology 157:1565–1572 [DOI] [PubMed] [Google Scholar]
- 75. Bruton CJ, Plaskitt KA, Chater KF. 1995. Tissue-specific glycogen branching isoenzymes in a multicellular prokaryote, Streptomyces coelicolor A3. Mol. Microbiol. 18:89–99 [DOI] [PubMed] [Google Scholar]
- 76. Yeo M, Chater K. 2005. The interplay of glycogen metabolism and differentiation provides an insight into the developmental biology of Streptomyces coelicolor. Microbiology 151:855–861 [DOI] [PubMed] [Google Scholar]
- 77. Rueda B, Miguélez EM, Hardisson C, Manzanal MB. 2001. Changes in glycogen and trehalose content of Streptomyces brasiliensis hyphae during growth in liquid cultures under sporulating and non-sporulating conditions. FEMS Microbiol. Lett. 194:181–185 [DOI] [PubMed] [Google Scholar]
- 78. Vasilyev NN, Kutlubaeva ZS, Ugarov VI, Chetverina HV, Chetverin AB. 2013. Ribosomal protein S1 functions as a termination factor in RNA synthesis by Qβ phage replicase. Nat. Commun. 4:1781. 10.1038/ncomms2807 [DOI] [PubMed] [Google Scholar]
- 79. Greive SJ, Lins AF, von Hippel PH. 2005. Assembly of an RNA-protein complex. Binding of NusB and NusE (S10) proteins to boxA RNA nucleates the formation of the antitermination complex involved in controlling rRNA transcription in Escherichia coli. J. Biol. Chem. 280:36397–36408 [DOI] [PubMed] [Google Scholar]
- 80. Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 [DOI] [PubMed] [Google Scholar]
- 81. Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25:89–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Görke B, Foulquier E, Galinier A. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology 151:3777–3791 [DOI] [PubMed] [Google Scholar]
- 83. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. 1999. Evidence that the extracytoplasmic function sigma factor, σE, is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Stuttard C. 1982. Temperate phages of Streptomyces venezuelae: lysogeny and host specificity shown by phages SV1 and SV2. Microbiology 128:115–121 [Google Scholar]
- 86. Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, United Kingdom [Google Scholar]
- 87. Gregory MA, Till R, Smith MC. 2003. Integration site for Streptomyces phage ΦBT1 and development of site-specific integrating vectors. J. Bacteriol. 185:5320–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Hesketh A, Kock H, Mootien S, Bibb M. 2009. The role of absC, a novel regulatory gene for secondary metabolism, in zinc-dependent antibiotic production in Streptomyces coelicolor A3. Mol. Microbiol. 74:1427–1444 [DOI] [PubMed] [Google Scholar]
- 89. Core R. Team 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/ [Google Scholar]
- 90. Gautier L, Cope L, Bolstad BM, Irizarry RA. 2004. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315 [DOI] [PubMed] [Google Scholar]
- 91. Smyth GK. 2005. Limma: linear models for microarray data, p 397–420 In Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. (ed), Bioinformatics and computational biology solutions using R and Bioconductor. Springer Verlag, New York, NY. [Google Scholar]
- 92. Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Fuellen G, Gilbert JG, Korf I, Lapp H, Lehväslaiho H, Matsalla C, Mungall CJ, Osborne BI, Pocock MR, Schattner P, Senger M, Stein LD, Stupka E, Wilkinson MD, Birney E. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12:1611–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nicol JW, Helt GA, Blanchard SG, Raja A, Loraine AE. 2009. The integrated genome browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25:2730–2731 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental materials and methods. Download
Phenotypes of wild-type S. venezuelae (WT), the constructed ΔwhiA::apr null mutant SV11 (ΔwhiA::apr), SV11 carrying the empty vector pMS82 (ΔwhiA::apr attBΦBT1::pMS82), and SV11 expressing WhiA (ΔwhiA::apr attBΦBT1::whiA) or N-terminal, triple FLAG-tagged versions of WhiA with or without a [Gly4Ser]3 linker (ΔwhiA::apr attBΦBT1::3×FLAGlink-whiA and ΔwhiA::apr attBΦBT1::3×FLAG-whiA). (A) Appearance on MYM solid medium after 4 days. (B) Phase-contrast images of coverslip impressions taken from MYM plates after 4 days. Download
Western blot analysis of wild-type S. venezuelae (−) and ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA (+) grown in MYM liquid cultures and sampled from 14 to 40 h. Five micrograms total protein was loaded per lane, and 3×FLAG-[Gly4Ser]3-WhiA was detected using M2 anti-FLAG antibody. Protein size markers (NEB) are shown with molecular masses in kDa. Download
Chromosome-wide distribution of WhiA-binding sites in S. venezuelae identified by ChIP-seq analysis. ChIP-seq was conducted using M2 anti-FLAG antibody on the ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA strain at time points representing three distinct developmental stages: vegetative growth, the onset of sporulation (fragmentation), and midsporulation. The wild-type strain (expressing nontagged WhiA from the native locus) analyzed at the onset of sporulation (fragmentation) was used as a negative control. Download
Complete ChIP-seq data set for ΔwhiA::apr attBΦBT1::3×FLAG-[Gly4Ser]3-whiA. Each row represents a ChIP “peak” based on the analysis of 25-bp segments of the S. venezuelae genome. Only those peaks with significance of P < E−04 for at least one of the ChIP samples are included in the analysis. A single entry that had significance of <E−04 in the negative control was discarded from the analysis. Pos, position of peak in the S. venezuelae genome in bases. Diff, the difference between the local normalized (ln) values of the immunoprecipitated (ChIP) samples and the total (non-ChIP) DNA samples, for each of the ChIP samples, i.e., V (vegetative growth), F (onset of sporulation/fragmentation), S (midsporulation), and C (wild-type negative control, analyzed at the onset of sporulation/fragmentation). Adjusted P values (apv), significance values for each of the ChIP samples, i.e., V, F, S, and WT negative control (C) after adjusting for multiple testing by the Hochberg method as implemented in the p.adjust function of R. Peak (pk), each ChIP sample is qualified as being significant (P < E−04) by the identifier TRUE and insignificant (P > E−04) by the identifier FALSE. Left Gene, the identifier (SVEN no.) for the gene on the left of the identified ChIP peak. Right Gene, the identifier (SVEN no.) for the gene on the right of the identified ChIP peak. Distance, the distance (in bases) between the ChIP peak and the predicted start codon of the downstream gene. Start, the start position on the S. venezuelae genome (in bases) of the gene downstream of the ChIP peak. End, the end position on the S. venezuelae genome (in bases) of the gene downstream of the ChIP peak. Strand, the strand on which the gene is found (forward = 1; reverse = −1). Product, (possible) gene function based on annotation in StrepDB (http://strepdb.streptomyces.org.uk). Affy LogFC, the log fold change (log2 scale) in expression of the whiA mutant compared to wild-type S. venezuelae at the 10-, 12-, 14-, 16-, 18-, and 20-h time points. − indicates a decrease in expression of the gene in a whiA mutant compared to the wild type; + indicates an increase in expression of the gene in a whiA mutant compared to the wild type. Cells highlighted in red represent greater than a 2-fold increase in expression in the whiA mutant. Cells highlighted in yellow represent greater than a 2-fold decrease in expression in the whiA mutant.
Strains, plasmids, and oligonucleotide primers used in this study.