Abstract
BACKGROUND & AIMS
Identification of intestinal stem cells (ISCs) has relied heavily on the use of transgenic reporters in mice, but this approach is limited by mosaic expression patterns and difficult to directly apply to human tissues. We sought to identify reliable surface markers of ISCs and establish a robust functional assay to characterize ISCs from mouse and human tissues.
METHODS
We used immunohistochemistry, real-time reverse-transcription polymerase chain reaction, and fluorescence-activated cell sorting (FACS) to analyze intestinal epithelial cells isolated from mouse and human intestinal tissues. We compared different combinations of surface markers among ISCs isolated based on expression of Lgr5–green fluorescent protein. We developed a culture protocol to facilitate the identification of functional ISCs from mice and then tested the assay with human intestinal crypts and putative ISCs.
RESULTS
CD44+CD24loCD166+ cells, isolated by FACS from mouse small intestine and colon, expressed high levels of stem cell–associated genes. Transit-amplifying cells and progenitor cells were then excluded based on expression of GRP78 or c-Kit. CD44+CD24loCD166+ GRP78lo/− putative stem cells from mouse small intestine included Lgr5-GFPhi and Lgr5-GFPmed/lo cells. Incubation of these cells with the GSK inhibitor CHIR99021 and the E-cadherin stabilizer Thiazovivin resulted in colony formation by 25% to 30% of single-sorted ISCs.
CONCLUSIONS
We developed a culture protocol to identify putative ISCs from mouse and human tissues based on cell surface markers. CD44+CD24loCD166+, GRP78lo/−, and c-Kit− facilitated identification of putative stem cells from the mouse small intestine and colon, respectively. CD44+CD24−/loCD166+ also identified putative human ISCs. These findings will facilitate functional studies of mouse and human ISCs.
Keywords: Stemness, Differentiation, Single-Cell Sorting, Flow Cytometry Analysis
The intestinal epithelium offers an elegant model for studying mammalian adult stem cells because of its simple structure and high turnover rate. In vivo genetic marking, coupled with lineage tracing, is a reliable approach for characterizing mouse intestinal stem cells (ISCs); however, this method depends on transgenic or knock-in animal models such as Lgr5–green fluorescent protein (GFP) to identify crypt base columnar (CBC) cells and reporter constructs for Bmi-1, mTert, Hopx, and Lrig1 to mark +4 ISCs.1–6 In addition, these transgenic reporter mice often display mosaic expression; more importantly, their use in isolating these populations cannot be translated directly to human studies.
Several groups have reported sorting ISCs using single surface markers, including EphB2, CD24, and CD166.7–9 However, the common caveat is that a single surface marker is unable to effectively exclude progenitor and differentiated cells. For example, EphB2 is broadly expressed in >60% of human crypt cells at variable levels, but even EphBhi cells contain both ISCs and Muc2+ progenitor cells.7 The expression of Sox9-GFP or CD24+ was used to isolate ISCs but revealed only 0.2% to 0.6% colony-forming efficiency (CFE)10; therefore, neither of these molecules alone can serve as a robust ISC biomarker.
Functional analysis is required to verify candidate markers for sorted ISCs. However, robust survival of ISCs using the reported in vitro culture method is dependent on Paneth cells (PCs).11,12 Although Wnt3a was reported to replace PCs in this in vitro culture system,12 there are controversial reports that single Lgr5+ CBCs can efficiently grow only when mixed with PCs, not with the addition of Wnt3a.13 Furthermore, the colon does not have PCs, and c-Kit+ goblet cells may support the growth of colonic stem cells (CoSCs) but with very low efficiency.14 These data highlight the need to develop a robust in vitro culture method for ISC studies.
Currently, 2 subpopulations of ISCs have been reported to regulate tissue homeostasis: CBCs with rapidly cycling properties intermingled among PCs and quiescent ISCs located above the PC zone near the +4 position.1–3,15–17 Although CBCs were reported to also express all the +4 ISC markers,18 recent in vivo studies have shown that after genetic ablation or radiation damage, a reserve ISC population located above the PC zone is activated to regenerate the damaged epithelium.4,6,19,20 Despite this important finding, no surface markers are available to efficiently isolate either of the ISC populations.
In this study, we identified a combination of surface markers for isolating mouse ISCs with different Lgr5 expression levels, which were functionally characterized using a newly developed culture method. This method also improves culture of either human crypt or fluorescence-activated cell sorting (FACS)-isolated single human ISCs.
Materials and Methods
Mice and Human Tissues
The Lgr5–enhanced GFP mouse model was provided by Hans Clevers. Adult mice (6–12 weeks old) were used in all experiments. All mice were housed in the animal facility at Stowers Institute for Medical Research and handled according to Stowers Institute for Medical Research and National Institutes of Health guidelines. All procedures were approved by the Institutional Animal Care and Use Committee of Stowers Institute for Medical Research. Use of human tissues was approved by the institutional review board at Cincinnati Children’s Hospital Medical Research Center and by an institutional review board–exempt protocol at University of California, Los Angeles.
Histological Analysis of Tissue
Mouse intestinal tissue was fixed in 1× zinc formalin. The fixed tissues were embedded in paraffin and processed as previously described.21
Mouse Intestinal Crypt Isolation and Cell Dissociation
For information on mouse intestinal crypt isolation and cell dissociation, see Supplementary Materials and Methods.
Cell Culture
Sorted cells were collected at low temperature, pelleted, and embedded in Growth Factor Reduced Matrigel (BD Biosciences, San Jose, CA), followed by seeding on a prewarmed 96-well plate (~200 to 300 single cells in 8.5–10 μL Matrigel per well). Matrigel contained 750 ng/mL epidermal growth factor (Peprotech), 1.5 μg/mL Noggin (Peprotech, Rocky Hill, NJ), and 15 μmol/L Jagged-1 (Anaspec, Fremont, CA). Prewarmed seeding media (100 μL) was added after full polymerization of Matrigel. Seeding medium was Advanced Dulbecco’s modified Eagle medium/F12, supplemented with penicillin (100 U/mL)/streptomycin (100 μg/mL), Glutamax, 10 mmol/L HEPES, 1× N2, 1× B27 (all from Invitrogen, Carlsbad, CA), 1 mmol/L N-acetylcysteine (Sigma St. Louis, MO), Thiazovivin (2.5 μmol/L; Sheng Ding Lab, San Francisco, CA), and CHIR99021 (2.5 μmol/L; Stemgent, Cambridge, MA). Growth factors containing 1 μg/mL R-Spondin 1 (R&D Systems, Minneapolis, MN), 1 μmol/L Jagged-1 peptide (Anaspec), 50 ng/mL epidermal growth factor (Invitrogen), and 100 ng/mL Noggin at final concentration were added on day 2 after cell seeding.11 The first media change was processed on day 4 by removing 50% and adding 50% freshly prepared Advanced Dulbecco’s modified Eagle medium/F12, supplemented with penicillin/streptomycin, Glutamax, HEPES, 2× N2, 2× B27, 2 μg/mL R-Spondin 1, 100 ng/mL epidermal growth factor, and 200 ng/mL Noggin. The culture was then maintained regularly by adding growth factors without Jagged-1 every other day and then changing the entire medium every 4 days. The same strategy was used for single colonic ISC culture. FGF4 (200 ng/mL; R&D Systems) can be added during the first 8 days but is not essential for colony formation.
Flow Cytometric Analysis
Cells (2–6 × 106/mL) were stained with antibodies (Supplementary Materials and Methods) for 30 minutes at 4°C followed by secondary antibody staining for another 30 minutes. After washing, cells were analyzed by MoFlo (DakoCytomation, Carpinteria, CA). Cell size gate, based on forward scatter and side scatter, was set up first to exclude cell debris and clumps; the other gates were set as shown in Figure 1. Flow cytometry data collection and cell sorting were performed with a MoFlo Legacy Cell Sorter (Beckman Coulter, Indianapolis, IN) (see details in Supplementary Materials and Methods). Postsort analysis of cells confirmed successful sorting of desired populations at high purity (Supplementary Figure 6). Data analysis was performed using FlowJo (Tree Star Inc, Ashland, OR).
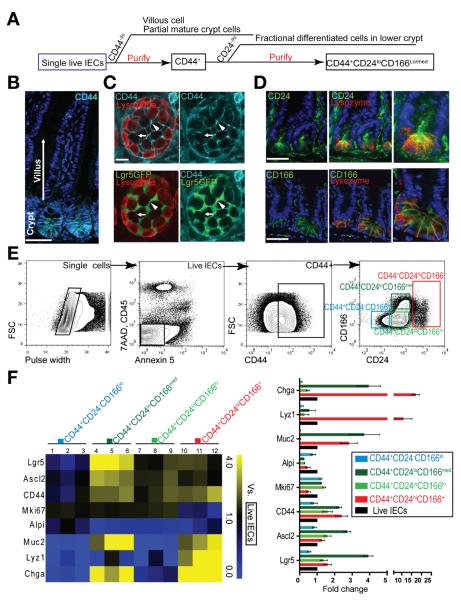
Figure 1.
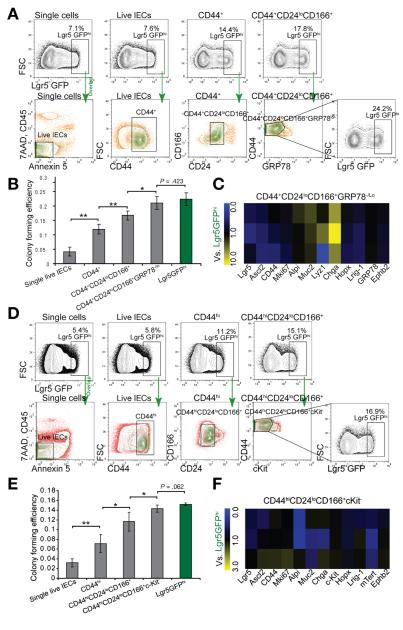
Combination of CD44, CD24, and CD166 preliminarily excludes small intestinal differentiated cells. (A) Experimental scheme of excluding villus and crypt differentiated cells. (B) Immunohistochemistry (IHC) staining shows gradient expression of CD44 in crypts but not in villi. (C) Confocal cross section of crypt bottom, showing strong staining of CD44 (bright blue) in the junctions of Lgr5-GFPhi stem cells (green, arrowheads) and weak staining between PCs (red, arrows). (D) IHC staining of CD24 and CD166 (green) shows higher expression at the crypt base. (E) FACS strategy for sequentially gating single cells, live IECs, CD44+ cells, and cells with different CD24 and CD166 expression. (F) qRT-PCR analysis showing the stem cell and differentiation marker expression in 4 subpopulations gated from CD44+ cells versus total live IECs by heat map (left) and column chart (right). Data are presented as mean ± SEM (n = 3). Scale bars = 50 μm (B and D); 20 μm (C).
Measurement of CFE
When seeding the sorted cells in Matrigel for culture, we fixed and stained 3 to 4 wells with 4′,6-diamidino-2-phenylindole to count the initial number of seeding cells as the denominator. The average number of enteroids formed from the remaining 4 to 7 wells served as the numerator.
Quantitative Reverse-Transcription Polymerase Chain Reaction
TaqMan gene expression assays (Applied Biosystems, Foster City, CA) were performed on triplicate samples (Supplementary Materials and Methods). Data were normalized relative to 3 reference genes: GUSB, TBP, and Hprt1.22 Ct values were calculated to obtain fold changes for sample comparison (DataAssist v3.0, Applied Biosystems, Foster City, CA).
Statistical Analysis
Data are represented as mean ± SEM. Two-tailed Student t test was used to determine statistical significance for bar charts or pairwise comparisons with a significance cutoff of P < .05.
Results
A Combination of Antibodies to CD44, CD24, and CD166 Purified Putative ISCs by Excluding Differentiated Cells From the Villus and Crypt
We used a general strategy to enrich ISCs and to exclude differentiated cells with a combination of positive and negative markers, respectively (Figure 1A). Besides confirming a broad crypt-restricted CD44 expression (Figure 1B), we further found that CD44 expression was higher at the interface between 2 adjacent Lgr5-GFPhi CBCs than that between 2 adjacent PCs (Figure 1C and Supplementary Figures 1 and 3). Furthermore, 2 additional ISC surface markers, CD24 and CD166 (ALCAM),8–10 were highly expressed in the lower region of crypts where ISCs are located (Figure 1D). Together, these data suggest that CD44 can be used to select intestinal epithelial crypt cells, including ISCs, and that CD24 and CD166 can be further used to exclude transient-amplifying and some progenitor cells.
Using FACS, we performed sequential doublets via pulse-width gating, dead/apoptotic cells using 7-aminoactinomycin D plus annexin V, and blood cells with CD45 staining (Figure 1E). We also found that more than 97% of live CD45− cells were EpCAM+ cells, an epithelial cell marker, and thereby eliminated the need to incorporate EpCAM into our procedure (Supplementary Figure 2). After the purification of CD44+ crypt cells, we further applied CD24 and CD166 gating to cosegregate the cells into 4 subpopulations based on isotype staining (Figure 1E and Supplementary Figure 1). To determine the cellular composition of these subpopulations, we then used reverse-transcription polymerase chain reaction (RT-PCR) to measure expression levels of stem cells and differentiation markers (Figure 1F) using the total unfractioned live intestinal epithelial cells (IECs) as the reference population.
Among these 4 groups (Figure 1F), the high expression of stem cell–associated genes in the CD44+CD24loCD166med population and the low expression of differentiation genes in the CD44+CD24loCD166lo population indicated the identification of putative ISCs with different Lgr5 expression levels in these 2 gates. The combination of CD166 and CD24 staining revealed a CD24lo subpopulation with a more distinctive boundary compared with CD24 alone (Supplementary Figure 4). Thus, we combined CD44+CD24loCD166lo and CD44+CD24loCD166med populations to define the CD44+CD24loCD166+(lo+med) population that was used for further purifying ISCs.
The CD44+CD24−CD166lo population revealed the lowest expression of stem cell–related genes (Figure 1F, columns 1–3). On the other hand, the CD44+CD24hiCD166+ subpopulation expressed a high level of secretory lineage markers but a low level of Ki67 expression (Figure 1F, columns 10–12), indicating that this population was enriched for secretory cells. Using immunostaining of Ki67, we confirmed the quantitative RT-PCR (qRT-PCR) results (Supplementary Figures 5 and 6). In addition, a previous study reported that a combination of side scatter and CD24 can separate CD24hi cells into 2 subpopulations that respectively select endocrine cells and PCs.12,13 Interestingly, the CD24hiCD166hi cells derived from the CD44− population also fell into the “PC” gate (Supplementary Figures 1 and 3), providing a potential method to analyze and sort heterogeneous secretory lineage cells.
GRP78+ and GRP78−/lo Distinguished Progenitor Cells and Stem Cells, Respectively
To further exclude remaining differentiated cells from the CD44+CD24loCD166+ population, we tested other candidate surface markers, including the 78-kilodalton glucose-regulated protein (GRP78) (Figure 2A).23 We found that GRP78 was expressed at the highest level in the villus (differentiated cell zone), at an intermediate level in the upper crypt (transient-amplifying cell zone), and at the lowest level in the lower crypt, thus negatively selecting for the majority of ISCs (Figure 2B). A combination of GRP78 and CD44 formed 3 subpopulations of cells defined as CD44−GRP78+, CD44loGRP78+, and CD44hiGRP78−/lo (Figure 2C). qRT-PCR gene expression analysis showed that the CD44hiGRP78−/lo and CD44loGRP78+ populations, respectively, expressed the highest level and lowest level of ISC-related genes, indicating that the combination of GRP78 and CD44 can largely distinguish ISCs from progenitor cells (Figure 2D).
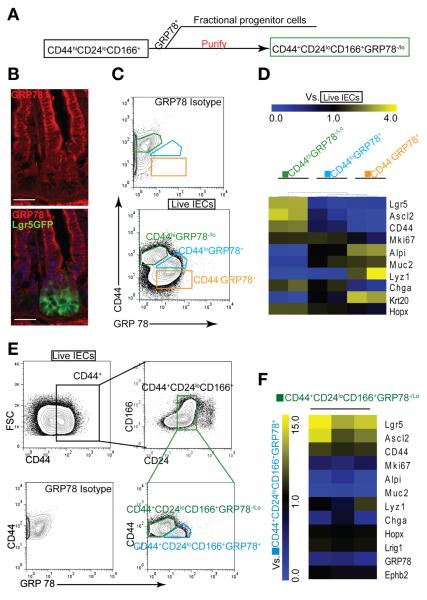
Figure 2.
GRP78 excludes progenitors in the CD44+ CD24loCD166+ population. (A) Experimental scheme of excluding progenitor and differentiated cells from CD44, CD24, and CD166 cells using GRP78+. (B) IHC staining shows high expression of GRP78 (red) in the villi and upper crypt but low expression in the stem cell zone at the crypt base. (C) Single live IECs divided into 3 subgates of CD44hiGRP78−/lo, CD44lo GRP78+, and CD44− GRP78+. (D) Heat map of qRT-PCR results shows the gene expression relative to single live IECs in 3 subgates (C) (n = 2). (E) Two subgates of CD44+CD24loCD166+GRP78−/lo and CD44+CD24loCD166+GRP78+. (F) Heat map of qRT-PCR results shows the gene expression profiling in CD44+CD24loCD166+GRP78−/lo cells relative to the CD44+CD24loCD166+GRP78+ population (n = 3). Scale bars = 50 μm (B).
However, CD44+GRP78−/lo could not efficiently exclude CD24hi differentiated cells (Supplementary Figure 7), so we combined GRP78 with the previously identified CD44+CD24loCD166+ population (Figure 2E). qRT-PCR assay showed that CD44+CD24loCD166+GRP78−/lo cells expressed higher levels (10-fold) of active ISC-related genes (Lgr5 and Ascl2) but lower levels of differentiation markers (ChgA, Alpi, and Muc2) than did CD44+CD24loCD166+ GRP78+ cells (Figure 2F). Taken together, these data indicate that inclusion of GRP78−/lo expression facilitates further purifying ISCs from the CD44+CD24loCD166+ subpopulation. Interestingly, these 2 subpopulations express Lrig1 and Hopx, 2 reserve ISC-related genes, at similar levels, consistent with a recent report.18
CD44hiCD24loCD166+c-Kit− Combinatorial Markers Identified Putative CoSCs
To purify CoSCs, we applied a strategy similar to that used for ISCs (Figure 3A). CD44 immunostaining revealed a gradient expression in the colonic crypt, with the highest level in the putative CoSC zone of the crypt base1 (Figure 3B). Consistently, a clear separation of CD44hi, CD44med, and CD44−/lo subpopulations could be attained by flow cytometry (Figure 3C). Similar to a reported ISC marker EphB2hi,24 CD44hi can identify cells at the crypt base, including Lgr5+ stem cells, adjacent goblet cells, and Lgr5GFPlo cells (Figure 3B, arrows). Therefore, additional markers are required to exclude progenitor cells from CD44hi cells.
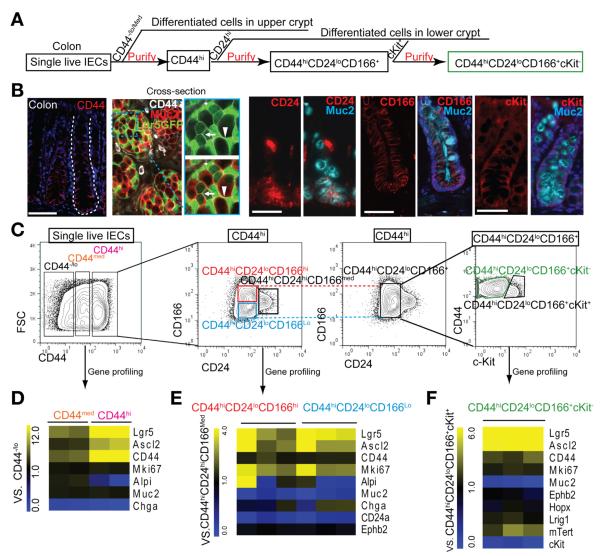
Figure 3.
CD44, CD24, CD166, c-Kit combination identifies CoSCs. (A) Experimental scheme of gradually purifying colonic ISCs. (B) IHC staining shows expression pattern of CD44, CD24, CD166, and c-Kit in colon. The cross section shows the expression of CD44 (white) in Muc2+ (red) cells adjacent to Lgr5-GFPhi (green) cells (arrowheads) and in the conjunction of Lgr5-GFPlo cells (arrows). (C) Sequential FACS plots and gates as indicated. (D) qRT-PCR results show gene expression in the CD44med and CD44hi cells versus CD44lo/− cells (n = 2). (E) Gene expression in CD44hiCD24loCD166lo and CD44hiCD24loCD166hi populations relative to the CD44hiCD24hiCD166med population (E)and further separation with c-Kit (F) (n = 3). Scale bars = 50 μm (B); 10 μm (B, magnification).
Immunostaining of colonic tissues showed that CD24hi cells localized mainly in the lower crypt, where they overlapped with some Muc2+ goblet cells (Figure 3B), whereas CD166 demonstrated a broader expression along the entire crypt axis (Figure 3B). With the addition of CD24 and CD166, the CD44hi population could be further separated into 3 subpopulations by flow cytometry: CD24loCD166lo, CD24loCD166hi, and CD24hiCD166med (Figure 3C). Compared with CD24hiCD166med cells, CD24loCD166lo/hi populations expressed a high level of stem cell–related genes and a low level of the goblet cell gene Muc2 (Figure 3E). We also found that EphB2 was expressed at similar levels in these populations (Figure 3E), indicating its inability to distinguish CoSCs from neighboring Muc2+ goblet cells.
Because we failed to detect GRP78 expression in the colon, we used c-Kit14 to negatively select Muc2+ cells (Figure 3B). Thus, CD44hiCD24loCD166+ cells could be further divided into c-Kit+ and c-Kit− populations by flow cytometry (Figure 3C). RT-PCR analysis revealed that CD44hiCD24loCD166+c-Kit− cells expressed a 5-fold higher level of Lgr5 and Ascl2 and a much lower level of Muc2 (Figure 3F). In addition, the combination of CD44hic-Kit− could exclude largely differentiated cells; however, it still contained a small portion of CD24hi cells (recognizing secretory cells) (Supplementary Figure 8). Taken together, these data show that, although the combined markers CD44hic-Kit− can largely purify for CoSCs by FACS, a more pure population can be defined by CD44hiCD24loCD166+cKit−.
Inhibition of GSK3β and Stabilization of E-cadherin Enable Robust Colony Formation for Testing Single Stem Cells
Characterizing single cells isolated using cell surface markers requires a robust functional assay. However, the current assay achieved only very low CFE even when supplementing the current medium with Wnt3a and other reported factors11 (Figure 4A and Supplementary Figure 9). This is consistent with a recent report that Wnt3a cannot replace PCs to support efficient in vitro colony formation from single ISCs.13 We showed that Wnt3a conditional medium, containing myriad unknown factors, could only modestly increase the CFE of single ISCs (Supplementary Figure 10). However, a high frequency of cell death occurred during the first 4 days of culture, indicating that additional signaling pathways besides Wnt are essential for survival of ISCs.
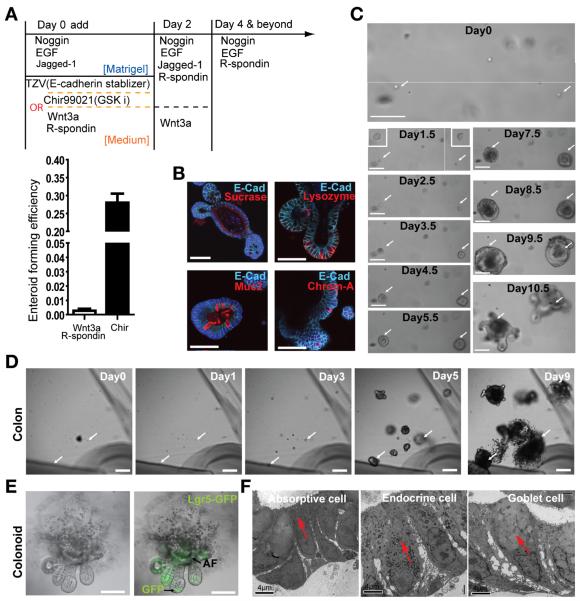
Figure 4.
Small molecules enable robust culturing of bona fide single ISCs or CoSCs. (A) Strategies to culture single Lgr5-GFPhi cells and comparison of Wnt3a and CHIR99021 have an impact on CFE. Data are presented as mean ± SEM (P = .0004, t test, n = 3). (B) IHC staining shows the differentiated cell type in the enteroid derived from single ISCs. (C) Representative images recording the process of single Lgr5-GFPhi cell–forming enteroids. (D) A growth process from a single Lgr5-GFPhi CoSC into colonoid. (E) An open colonoid structure derived from a single Lgr5-GFPhi CSC. AF, autofluorescence. (F) Colonoids containing differentiated cell types identified by electron microscopic analysis. Scale bars = 50 μm (B); 100 μm (C, D, E); 4 μm (F).
CHIR99021, a GSK3β inhibitor, can efficiently activate β-catenin and potential other survival pathways. It has been effectively used for in vitro culture of embryonic25 and hematopoietic stem cells.26 We found that only a low dose of CHIR99021 (2.5 μmol/L) during the first 2 days of culture (Figure 4A) enabled the survival of and subsequent robust growth of single Lgr5-GFPhi ISCs (Figure 4C and Supplementary Figures 9 and 10), whereas a high dose (5–10 μmol/L) inhibited colony growth (Supplementary Figure 11).
Thiazovivin has been reported to increase the stability of E-cadherin and inhibit Rock activity,27 both of which are essential to reduce apoptosis and support survival of single cells during the first 2 days of culture. In our hands, Thiazovivin was more effective than the Rock inhibitor Y27632, as shown with either bigger colony sizes (Supplementary Figure 10) or higher CFE (see Materials and Methods for accurate measurement of CFE). With use of both CHIR99021 and Thiazovivin, we could increase the CFE of single Lgr5GFPhi ISCs up to 25% to 30% (Figure 4A).
We next used the same strategy for CoSC culture and observed formation of colonoids from single CoSCs28 (Figure 4D). We observed 2 types of colonoid structures during the growth of Lgr5GFPhi colonies: a closed form similar to enteroids and an open form with a typical colon-like polarized morphology (Figure 4E). We also observed that the closed form could transform into an open form as the colony grew (Supplementary Figure 12). These colonoids contained both Lgr5-GFP+ stem cells (Figure 4E) and differentiated cells, as revealed by electron microscopy analysis and immunostaining (Figure 4F). Taken together, our work supports the establishment of a robust culture system to evaluate stemness of sorted ISCs and CoSCs.
Combinatorial Cell Surface Markers Show Purification of Functional ISCs and CoSCs
Using the robust single-cell culture method, we measured the extent of ISC enrichment by our surface marker combination. We first used Lgr5-GFPhi ISCs as the reference to evaluate sorted cells for ISC identification. Given the mosaic expression of Lgr5-GFP, an increase in the percentage of Lgr5-GFPhi cells from 7.1% of the unfractioned live cells to 24.2% of the CD44+CD24loCD166+GRP78−/lo population is significant (Figure 5A). Consistently, with each additional antibody applied to our population (from CD44+ to CD44+CD24loCD166+ and to CD44+CD24loCD166+ GRP78−/lo cells), we observed an increase in CFE (Figure 5B). The latter population revealed a CFE similar to that of FACS-isolated Lgr5-GFPhi cells from the transgenic reporter mouse (P = .42, t test, 2 tailed, Figure 5B); thus, we showed that the combination of CD44+CD24loCD166+ GRP78−/lo surface markers enriched for ISCs to a similar degree as the best available genetic reporter line. Although the CD44+CD24loCD166+ GRP78+ population could cover more than 82% of total Lgr5-GFPhi cells, as measured by backgating analysis (Supplementary Figure 13), the RT-PCR result revealed that it still contained a minor portion of ChgA-expressing endocrine cells (Figure 5C).
Figure 5.
The surface marker combination identifies the ISC population with CFE comparable to that of Lgr5-GFPhi cells. (A) Lgr5-GFPhi cells in parent gates were largely confined in daughter gates, showing the gradual selection by surface marker combination. (B) CFE analysis of different subpopulations. Data are presented as mean ± SEM, (*P < .05, **P < .01, n = 4, t test). (C) The gene profiling of CD44+CD24loCD166+ GRP78−/lo cells, taking Lgr5-GFPhi ISCs as control (n = 3). (D) Colonic Lgr5-GFPhi cells in parent gates were largely confined in daughter gates, showing the gradual purification by surface marker combination. (E) CFE analysis of series of gates. Data are presented as mean ± SEM (*P < .05, **P < .01, n = 4, t test). (F) The gene profiling of CD44hiCD24loCD166+cKit− cells, taking Lgr5-GFPhi CoSCs as control (n = 3).
We next measured the enrichment of CoSCs isolated using our surface marker combination. A similar increase in the percentage of Lgr5-GFPhi cells was observed in colon cells and reached about 17% in CD44+CD24loCD166+c-Kit− cells (Figure 5D). Accordingly, the CFE and gene profiling of CD44+CD24loCD166+c-Kit– cells was similar to that of Lgr5-GFPhi cells (Figure 5E and F).
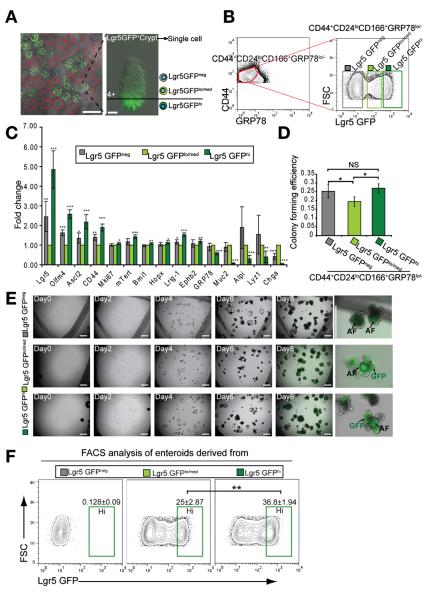
CD44+CD24loCD166+ GRP78−/lo Identifies Putative ISCs Independent of Lgr5-GFPhi Cells
One significant limitation of using the Lgr5-GFP reporter mouse line to label stem cells is the mosaic pattern of GFP+ crypts (Figure 6A). Consistently, we found that the Lgr5 expression level in the CD44+CD24loCD166+ GRP78− population isolated from Lgr5-GFPneg crypts was 50% that of Lgr5-GFPhi cells (Figure 6C), indicating that this population included the genuine Lgr5hiCBC cells. Also, the Lgr5-GFPneg cells had a CFE (>90%) similar to that of Lgr5-GFPhi cells (Figure 6D), indicating that the CD44+CD24loCD166+ GRP78−/lo markers enabled sorting putative ISCs from Lgr5-GFPneg crypts, independent of Lgr5-GFP reporter.
Figure 6.
CD44+CD24loCD166+ GRP78−/lo identifies ISCs independent of Lgr5-GFPhi cells. (A) The mosaic distribution of Lgr5-GFP+ crypt in jejunum. Gradient GFP expression in Lgr5-GFP+ crypt. (B) Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi gates in the CD44+CD24loCD166+ GRP78−/lo population. (C) qRT-PCR results show the gene expression of Lgr5-GFPneg and Lgr5-GFPhi cells relative to Lgr5-GFPlo/med cells from the CD44+CD24loCD166+ GRP78−/lo population. Data are presented as mean ± SEM (n = 4, *P < .05, **P < .01, ***P < .001). (D) CFE analysis of the Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi cells in the gates of B. Data are represented as mean ± SEM (*P < .05, n = 3, t test). (E) A growth process of single cells sorted from Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi gates in B, showing that Lgr5-GFPlo/med cells can generate Lgr5-GFPhi enteroid. (F) FACS analysis of enteroids derived from single Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi cells, showing that single Lgr5-GFPlo/med cells can generate enteroids containing Lgr5-GFPhi cells as single Lgr5-GFPhi cells did. Data are presented as mean ± SEM (n = 3, **P < .01).
Different GFP expression levels in the Lgr5-GFP+ crypts (Figure 6A) were reported to faithfully reflect the endogenous Lgr5 expression levels and to distinguish CBC cells and progenitor cells.18,29 Derived from the CD44+CD24loCD166+GRP78−/lo population, Lgr5-GFPlo/med and Lgr5-GFPhi subpopulations had an average CFE of 19% and 27%, respectively (Figure 6C and D). In contrast, Lgr5-GFPlo/med cells within the CD44+CD24loCD166+GRP78+ gate barely formed colonies (data not shown). Recent studies confined the high level of Lgr5 expression to the PC zone, while Olfm4 and Ascl2 showed broader expression in both CBC and +4 position.18,30 Similarly, we also detected that expression levels of Olfm4 and Ascl2 in the Lgr5-GFPlo/med population were approximately 40% that of the Lgr5-GFPhi population, consistent with the notion that Lgr5-GFPlo/med cells are most likely located at the +4 position above the CBC zone. Thus, based on the gene expression signature and CFE tests of these 2 different subpopulations, we concluded that within the Lgr5-GFP–positive crypts, not all colony-forming cells were derived from Lgr5-GFPhi cells. Some Lgr5-GFPlo/med cells located mainly above the CBC-PC zone (Figure 6A) were capable of forming enteroids.
We then asked whether Lgr5-GFPlo/med and Lgr5-GFPneg cells identified by surface markers could give rise to Lgr5-GFPhi enteroids. We showed that while the Lgr5-GFPneg cells gave rise mainly to Lgr5-GFPneg enteroids, the Lgr5-GFPlo/med cells could generate new enteroids containing Lgr5-GFPhi cells (Figure 6E and F). Compared with Lgr5-GFPhi ISC-derived enteroids, Lgr5-GFPlo/med ISC-derived enteroids shared similar expression levels of Lgr5, Ascl2, and other differentiated cell markers. Both groups can be maintained long-term in vitro (Supplementary Figure 14). Taken together, these results provide new evidence for putative ISCs located above the PC zone, consistent with the recently reported interconversion between ISC populations residing in distinct zones of the crypt.4,6,31 It was recently reported that Dll1+CD24med cells could generate enteroids with 1% CFE under high doses of Wnt ligand32; however, our CD24lo sorted cells could largely exclude Dll1+ cells and the corresponding CFE was much higher (20%–30%).
The New Culture Strategy Could Be Translated to Human Intestinal Tissue
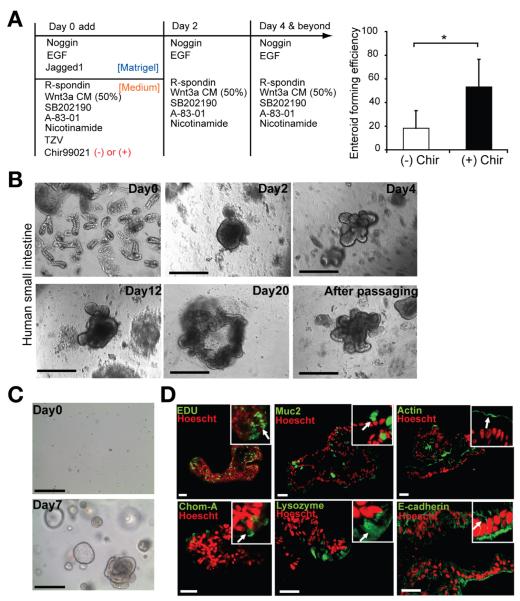
The current barrier in translating mouse studies into human tissue has been the low efficiency of culture of human small intestinal crypts and the difficulty of long-term in vitro maintenance. We tested the culture method that we developed in mice on human small intestinal tissue. Adding CHIR99021 (2.5 μmol/L) and other reported factors33 during the first 2 days increased the efficiency of enterosphere formation (Figure 7A and B). The enterospheres underwent extensive budding to give rise to enteroids after 4 days and could be maintained over 3 months (Figure 7C). After withdrawing Wnt3a factor,33 the major differentiated cell types could be detected in the human enteroids (Figure 7D).
Figure 7.
The new culture strategy enables highly efficient human crypt culture and maintenance. (A) Crypt culture strategy. CHIR99021 significantly increased the efficiency of culturing human small intestinal crypts. Data are presented as mean ± SEM (n = 6, *P .011, t test). (B) Representative growth process of human small intestinal crypt culture with addition of a single dose of GSK inhibitor CHIR99021 and other factors.33 (C) Human enteroid can be passaged up to 10 times over a 3-month period, as shown by their growth after passaging from day 0 to day 7. (D) IHC staining show the cellular proliferation and differentiated cell types in cultured enteroid. Positive staining is indicated by the arrow in the inset. Scale bars = 200 μm (B and C); 50 μm (D).
We further applied our culture method to surface marker–mediated FACS-isolated single human intestinal cells. We found that CD44+CD166+CD24lo cells could identify IECs expressing stem cell markers (Supplementary Figure 15). However, we also noted that adding CD166 and GRP78 did not distinguish distinctive subpopulations as observed in mouse intestinal tissue. Furthermore, we found that the level of CD24 expression varied from proximal (high) to distal (low) regions of human intestine (data not shown). Finally, our data indicated that the addition of the GSK inhibitor, instead of other reported factors,33 enhanced colony formation from single CD44+CD166+CD24lo cells but that the efficiency was not more than 1% (Supplementary Figure 15). Taken together, our proposed surface marker paradigm and culture method provide a strong foundation for continued optimization for isolation and characterization of human ISCs.
Discussion
Identification of highly purified functional ISCs is required for understanding their properties and for translational studies using human ISCs. Using transgenic reporters to isolate ISCs has several limitations. (1) The mosaic expression pattern of the reporter gene results in an inability to distinguish a truly negative population. (2) Direct application of these studies to the human system is difficult. (3) A single reporter could exclude important ISC subpopulations. Progress in other stem cell systems, particularly the hematopoietic system, which depended on the use of multiple, combinatorial cell surface markers to identify different hematopoietic stem cell subsets, is being applied to basic and clinical research. The study presented here will begin alleviating these deficiencies in the ISC field.
Our study showed that differentiated cells and progenitor cells share many reported single markers such as CD24, CD166, and EphB2hi with ISCs (Figures 1 and 3).9,12 Similar to the hematopoietic system, multiple cell surface marker combinations were shown to be essential for the effective exclusion of differentiated cells. We systemically tested combinations of new as well as previously reported cell surface markers by gene profiling and functional assays. We also tested a new strategy to exclude villous cells using antibody B6A6, which recognizes most villous cells except goblet cells (Supplementary Figure 16). The new marker GRP78 showed the ability to separate ISCs from progenitor cells when combined with markers CD44, CD24, and CD166. For human tissue, however, additional markers will be required to further exclude differentiated cells from the CD44+CD24loCD166+ population. Upon completion of this work, a new anti-Lgr5 antibody recognizing overexpressed Lgr5 in cancer cell lines was reported.34,35 This antibody should solve the problem of mosaic expression patterns in the transgenic reporter line; however, whether Lgr5hi versus Lgr5med/lo can be distinguished using this antibody is unknown.
Previously reported high CFE of ISCs depended on coculturing with niche cells, which cannot be replaced by exogenous Wnt3a.13 Similarly, a recent report indicated that constitutive activation of the Wnt pathway by deletion of Apc in Lgr5-GFPhi cells still led to only a 0.6% CFE.36 Here, we showed that transient application of the GSK-3β inhibitor CHIR99021 instead of Wnt3a and R-Spondin promoted the survival of single ISCs during the first 2 days, suggesting that antiapoptotic pathways are activated downstream of GSK-3β inhibition. Moreover, compared with Y27632, the E-cadherin stabilizer Thiazovivin also supported higher CFE, and this may be due to decreasing anoikis by improved E-cadherin stabilization. This new culture protocol is also applicable to human ISCs, emphasizing its clinical relevance.
Despite being a valuable tool, overreliance on single-gene reporters may result in failure to recognize the complexity of different ISC subsets. Benefiting from the Lgr5-GFP mice, we unexpectedly found that the Lgr5-GFPlo/med cells within the Lgr5-GFP–positive crypts isolated from GRP78−/lo but not from GRP78+ of the CD44+CD24loCD166+ subpopulation could also efficiently form enteroids. This observation indicates that our surface markers can select enteroid-forming Lgr5lo/med cells outside the PC zone, supporting the existence of a non-Lgr5hi ISC population. In addition, the low CD24 expression and high CFE of these cells indicate that they are different from recently reported Dll1+CD24med progenitors, which have very low CFE.32
In summary, we showed that CD44+CD24loCD166+ plus GRP78lo/− or c-Kit− identifies putative ISCs and CoSCs in mice. These surface marker combinations and the clonal assay, in principle, can be applied to the human system, which will be critical for future clinical application.
Supplementary Material
Supplementary Figure 1. CD44 expression in the small intestine and the new strategy to enrich PCs. (A) IHC staining shows Muc2-positive (red) and CD44-positive cells in crypts. (B) IHC staining shows chromogranin A–positive (red) and CD44-positive cells in crypts. (C) Setting up CD44+ gate based on isotype control. (D) CD24 isotype control. (E) CD166 isotype control. (F) CD44− and CD44+ cells were distributed into an FACS plot of CD24 versus CD166. CD24hi cells in CD44− and CD44+ populations were divided further, depending on the CD166 expression. (G) The reported gates relatively enriched endocrine cells (blue) and PCs (red) in the FACS plot of CD24 versus SSC. (H) The subgates in F were analyzed in the FACS plot of CD24 versus SSC. It is noteworthy that only CD44–CD24hiCD166hi (red polygon) cells fell mainly into the PC gate (red).
Supplementary Figure 2. ESA (EpCAM) is not required to distinguish IECs in the present FACS analysis. (A) Based on the isotype staining, the majority of cells in the original live IEC gate are ESA+. Upper panel, small intestine. Lower panel, colon. (B) ESA staining can affect the other surface marker staining including CD44. The CD44+ percentage and the FACS plots of CD24 versus CD166 derived from CD44+ or CD44− gates were changed by adding ESA antibody.
Supplementary Figure 3. Lgr5GFPhi cells express a much higher level of CD44 than CD24hiCD166hi PCs. (A) Confocal image of cross sections of the crypt bottom from Figure 1, showing strong staining of CD44 (bright blue) in the junctions of Lgr5-GFPhi stem cells (green, arrowheads) and weak staining between PCs (red, arrows). (B) CD44+ gate drawn out of total live cells. (C) CD24hiCD166hi PC gate drawn out of total live cells (see Supplementary Figure 3). (D) Lgr5 GFPhi cell gate drawn out of total live cells. (E) Backgating CD24hiCD166hi PCs into CD44 gate, showing that the low percentage of PCs is CD44+. (F) Backgating Lgr5 GFPhi Cells into CD44 gate, showing that almost all of Lgr5 GFPhi cells are CD44+.
Supplementary Figure 4. CD24 versus CD166 generates a more natural FACS pattern than CD24 alone. Two representative FACS tests show the cell populations with different CD24 levels in CD24 versus FSC plots or CD24 versus CD166 plots. CD166med cells help define the boundary between CD24neg and CD24lo.
Supplementary Figure 5. Testing the Ki67 expression of 4 subpopulations in the CD24 versus CD166 plot derived from CD44+ cells. (A) Representative FACS plot of CD24 versus CD166 derived from CD44+ cells. Four subpopulations were labeled with different colors (bright blue, deep green, bright green, red), which is consistent with Figure 1. (A) Cells sorted from 4 subpopulations were stained with Ki67 (red) and counterstained by DAPI. Bar = 100 μm. (B) The percentage of Ki67+ cells in different subpopulations (n = 3).
Supplementary Figure 6. Post sorting confirms the sorting purity. (A) The sorted CD44+ CD24loCD166+ GRP78−/lo cells from the small intestine were verified by flow cytometry, showing a high percentage (>90%) overlapping with the gates used for primary sorting. (B) The sorted CD44+ CD24loCD166+ cKit− cells from colon were verified by flow cytometry, showing a high percentage overlapping with the gates used for primary sorting.
Supplementary Figure 7. CD44hiGRP78−/lo cannot exclude the CD44+CD24hi population. (A) CD44 and positive gates from a single live IEC. (B) CD44+ cells distributed in an FACS plot of CD24 versus CD166. (C) Overlap CD44+CD24hi cells into an FACS plot of GRP78 versus CD44 from a single live IEC.
Supplementary Figure 8. The enrichment of clonogenic colonic ISCs by CD44 and c-Kit combination. (A) c-Kit isotype staining used to set the criterion for the c-Kit+ population. (B) Representative FACS analysis shows that the CD44hi colonic IECs can be clearly separated into a c-Kit+ and c-Kit− population. (C) CD44hi c-Kit− cells have heterogeneities for CD24 and CD166 expression and CD44hi c-Kit−CD24loCD166+ gate was drawn based on contour plot. (D) The CFE of CD44hi c-Kit−, CD44hi c-Kit+, CD44hi c-Kit−CD24loCD166+ cells (n = 3). ***P < .001.
Supplementary Figure 9. Comparison of ISC colony formation induced by different combinations of Wnt3a, R-spondin, Chir99021, and Thiazovivin (TZV) on the first 2 days.Different combinations of factors and inhibitors (including Wnt3a, 100 ng/mL; R-Spondin, 1 μg/mL, Chir99021, 2.5 μmol/L; TZV, 2.5 μmol/L) were tested by adding them into culture medium on day 0. Wnt3a was added on day 2 if it was added on day 0. The high efficiency was achieved by adding Chir99021 and TZV on day 0. Adding Wnt3a, R-Spondin, and Chir99021 together inhibited colony growth, partially mimicking the effect of the high concentration of Chir99021 in Supplementary Figure 11. Y-27632 combined with Wnt3a and R-Spondin was also tested and no high CFE was observed. Other growth factors including epidermal growth factor, Noggin, and Jagged-1 were used as shown in Figure 5 in every group. n = 3 wells. Similar results were obtained in 2 independent experiments. Bars = 100 μm.
Supplementary Figure 10. Wnt3a conditional medium and R-spondin 1 cannot replace GSK inhibitor. Compared with Wnt3a, Wnt3a conditional medium has complicated components. Combination of Wnt3a conditional medium (50%), R-Spondin 1 μg/mL, Chir99021 2.5 μmol/L, and TZV 2.5 μmol/L only partially inhibits colony growth. In addition, wnt3a conditional medium combined with R-spondin, TZV, or Y27632 can lead to a CFE of <5%, which is higher than commercial Wnt3a (Supplementary Figure 9). However, Chir99021 combined with TZV led to even higher CFE and much bigger colony size. Other growth factors including epidermal growth factor, Noggin, and Jagged-1 were used as shown inFigure 4 in every group. n = 3 wells. Similar results were obtained in 2 independent experiments. Bar = 100 μm.
Supplementary Figure 11. The effect of Chir99021, TZV, and Y-27632 on single ISC culture. (A) Representative process of colony forming from single Lgr5-GFPhi small intestinal cells with different concentration of Chir99021 in the medium for the first 2 days of culture. Note that the high concentration (5 μmol/L and 10 μmol/L) can cause the spreading of the colony. (B) Adding Y27632 (10 μmol/L) or TZV (2.5 μmol/L) into the medium on the first 2 days leads to a different size of colony derived from single Lgr5-GFPhi small intestinal cells. Chir99021 and other growth factors were used in the same way in both groups. (C) Adding Y27632(10 μmol/L) or TZV (2.5 μmol/L) into the medium on the first 2 days led to a different size of colony derived from single Lgr5-GFPhi colonic intestinal cells. Chir99021 and other growth factors were used in the same way in both groups. Bars = 200 μm.
Supplementary Figure 12. The heterogeneities of morphology of colonoid derived from single Lgr5GFPhi ISCs. (A) Representative growth process of single ISC-derived colonoid from day 6 to day 12. White arrows label the same colony on different days. It is noteworthy that most “closed” colonoids can become “open” along with the cell proliferation and differentiation. (B) High magnification of colonoids on day 10 shows different morphology (sphere, closed enteroid, open enteroid). (C) Representative Muc2 (red) and E-cadherin (bright blue) IHC staining shows the differentiation in colonoids with different morphology.
Supplementary Figure 13. CD44+ CD24loCD166+ GRP78−/lo enrich high percentage of Lgr5GFPhi cells. (A) The distribution of Lgr5GFPhi cells in CD44+, CD44+ CD24loCD166+, and CD44+ CD24loCD166+ GRP78−/lo gates. (B) The percentage of total Lgr5GFPhi ISCs covered by the gate of CD44+ CD24loCD166+ GRP78−/lo (n = 3).
Supplementary Figure 14. The enteroids derived from different Lgr5GFP levels share similarity in gene profiling and long-term culture maintenance. (A) Lgr5GFPhi cells or Lgr5GFPmed/lo cells derived from CD44+ CD24lo CD166+ GRP78−/lo population were cultured and passaged. qPCR test shows the similar expression levels of ISC related genes and differentiation associated genes in these 2 groups (n = 3). (B) Representative passaging image (from day 0 to day 2) showing the robust colony forming of these 2 groups, even after about 2.5 months of culture. Bar = 500 μm.
Supplementary Figure 15. Single human ISC isolation and culture analysis. (A) Experimental scheme of enriched single live ISCs. (B) qRT-PCR results show the gene expression in the CD44+CD166+CD24lo population relative to unsorted live IECs, indicating the enrichment of ISCs, although some differentiated cells were remained (n = 2). (C) Strategies to culture single CD44+CD166+CD24lo cells. Cells grew only in the group with a single dose of CHIR99021 on the first 2 days. Data are represented as mean ± SEM (n = 2). (D) Representative growth process of single CD44+CD166+CD24lo cells with addition of a single dose of CHIR99021. (E) IHC staining of CD44+CD166+CD24lo cells after 14 days of culture shows differentiation into intestinal epithelial lineages. Arrows in the inset indicate positive staining. Scale bars = 100 μm (D); 50 μm (E).
Supplementary Figure 16. Rat mAB-B6A6, CD24, CD166 combination to analyze ISCs and differentiated cells. (A) FACS plots show the exclusion of cell aggregates (left panel) and blood cells and dead cells (right panel). It is noteworthy that doublet cells cannot be efficiently removed without using Pulse width. (B) IHC staining shows that rat monoclonal B6A6 antibody facilitates exclusion of villus enterocytes but not some secretory cells, including goblet cells (arrow). (C) FACS pattern with FSC-W versus FSC-A to exclude cell aggregates. (D) FACS pattern with B6A6 versus FSC-W to show the B6A6− cells. FACS pattern with B6A6 versus FSC-W to show the enrichment of B6A6− cells by using anti-rat immunoglobulin G beads to deplete the B6A6+ cells. FACS pattern with CD24 versus CD166 to show different cell populations in B6A6− cells. It is noteworthy that B6A6− contains some CD44− cells. (E) qRT-PCR to test ISC and lineage related gene expression in different populations (G1–G4) from G c compared with unsorted cells. The G4 population shows the highest lysozyme and Lgr5 expression indicating PC and CBC-PC doublets. G1 shows the highest level of Muc2 expression, indicating the enrichment of goblet cells. G3 shows the high level of Lgr5 and Ascl2, indicating Lgr5hi and Lgr5lo/med cells may be included in this population.
Supplementary Table 1. Antibodies Used in Flow Cytometry for Mouse Tissue
Supplementary Table 2. TaqMan Assay Used in qPCR for Mouse Tissue
Supplementary Table 3. Antibodies Used in Flow Cytometry for Human Tissue
Supplementary Table 4. TaqMan Assay Used in qPCR for Human Tissue
Acknowledgments
The authors thank H. Clevers for providing Lgr5-GFP mice; M. Hembree, M. McClain, R. T. Ross, T. Johnson, H. Marshall, B. Lewis, D. Dukes, C. Semerad, J. Park, S. Beckham, F. Guo, and W. McDowell for technical support; A. Venkatraman, R. Sugimura, M. Zhao, and F. Tao for scientific discussion; K. Tannen for editing; and I. Schmid and the FACS facilities at University of California, Los Angeles Jonsson Comprehensive Cancer Center and AIDS Research Center for extra sorting support.
Funding This research was performed as a project of the Intestinal Stem Cell Consortium, L. Li (U01DK085507), M. H. Wong (U01DK85525), M. Martin (U01DK85535), S. T. Magness (U01DK085547: SJH/STM), and the Coordinating Center J. Niland (U01DK85532). L. Li is supported in part by Stowers Institute for Medical Research.
Abbreviations used in this paper
- CBC
crypt base columnar
- CFE
colony-forming efficiency
- CoSC
colonic stem cell
- FACS
fluorescence-activated cell sorting
- GRP
green fluorescent protein
- IEC
intestinal epithelial cell
- IHC
immunohistochemistry
- ISC
intestinal stem cell
- PC
Paneth cell
- qRT-PCR
quantitative reverse-transcription polymerase chain reaction
Footnotes
Supplementary Material Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at http://dx.doi.org/10.1053/j.gastro.2013.04.050.
Author names in bold designate shared co-first authorship.
Conflicts of interest The authors disclose no conflicts.
References
- 1.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 2.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Montgomery RK, Carlone DL, Richmond CA, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian H, Biehs B, Warming S, et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell AE, Wang Y, Li Y, et al. The Pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takeda N, Jain R, LeBoeuf MR, et al. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Merlos-Suarez A, Barriga FM, Jung P, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 8.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298:G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levin TG, Powell AE, Davies PS, et al. Characterization of the intestinal cancer stem cell marker CD166 in the human and mouse gastrointestinal tract. Gastroenterology. 2010;139:2072–2082. e5. doi: 10.1053/j.gastro.2010.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Furstenberg RJ, Gulati AS, Baxi A, et al. Sorting mouse jejunal epithelial cells with CD24 yields a population with characteristics of intestinal stem cells. Am J Physiol Gastrointest Liver Physiol. 2011;300:G409–G417. doi: 10.1152/ajpgi.00453.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato T, Vries RG, Snippert HJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 12.Sato T, van Es JH, Snippert HJ, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yilmaz OH, Katajisto P, Lamming DW, et al. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature. 2012;486:490–495. doi: 10.1038/nature11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rothenberg ME, Nusse Y, Kalisky T, et al. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology. 2012;142:1195–1205. doi: 10.1053/j.gastro.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: the mucosal governor. Int J Exp Pathol. 1997;78:219–243. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He XC, Zhang J, Tong WG, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am J Anat. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 18.Munoz J, Stange DE, Schepers AG, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan KS, Chia LA, Li X, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Landeghem L, Santoro MA, Krebs AE, et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1111–G1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang F, Zou Z, Liu D, et al. Active deformation of apoptotic intestinal epithelial cells with adhesion-restricted polarity contributes to apoptotic clearance. Lab Invest. 2011;91:462–471. doi: 10.1038/labinvest.2010.182. [DOI] [PubMed] [Google Scholar]
- 22.Wang F, Wang J, Liu D, et al. Normalizing genes for real-time polymerase chain reaction in epithelial and nonepithelial cells of mouse small intestine. Anal Biochem. 2010;399:211–217. doi: 10.1016/j.ab.2009.12.029. [DOI] [PubMed] [Google Scholar]
- 23.Miharada K, Karlsson G, Rehn M, et al. Cripto regulates hematopoietic stem cells as a hypoxic-niche-related factor through cell surface receptor GRP78. Cell Stem Cell. 2011;9:330–344. doi: 10.1016/j.stem.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 24.Jung P, Sato T, Merlos-Suarez A, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 25.Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry JM, He XC, Sugimura R, et al. Cooperation between both Wnt/{beta}-catenin and PTEN/PI3K/Akt signaling promotes primitive hematopoietic stem cell self-renewal and expansion. Genes Dev. 2011;25:1928–1942. doi: 10.1101/gad.17421911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Zhu X, Hahm HS, et al. Revealing a core signaling regulatory mechanism for pluripotent stem cell survival and self-renewal by small molecules. Proc Natl Acad Sci U S A. 2010;107:8129–8134. doi: 10.1073/pnas.1002024107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stelzner M, Helmrath M, Dunn JC, et al. A nomenclature for intestinal in vitro cultures. Am J Physiol Gastrointest Liver Physiol. 2012;302:G1359–G1363. doi: 10.1152/ajpgi.00493.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snippert HJ, van der Flier LG, Sato T, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 30.Itzkovitz S, Lyubimova A, Blat IC, et al. Single-molecule transcript counting of stem-cell markers in the mouse intestine. Nat Cell Biol. 2012;14:106–114. doi: 10.1038/ncb2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Es JH, Sato T, van de Wetering M, et al. Dll1(+) secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14:1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sato T, Stange DE, Ferrante M, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 34.Kemper K, Prasetyanti PR, De Lau W, et al. Monoclonal antibodies against lgr5 identify human colorectal cancer stem cells. Stem Cells. 2012;30:2378–2386. doi: 10.1002/stem.1233. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi S, Yamada-Okabe H, Suzuki M, et al. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30:2631–2644. doi: 10.1002/stem.1257. [DOI] [PubMed] [Google Scholar]
- 36.Schepers AG, Snippert HJ, Stange DE, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. CD44 expression in the small intestine and the new strategy to enrich PCs. (A) IHC staining shows Muc2-positive (red) and CD44-positive cells in crypts. (B) IHC staining shows chromogranin A–positive (red) and CD44-positive cells in crypts. (C) Setting up CD44+ gate based on isotype control. (D) CD24 isotype control. (E) CD166 isotype control. (F) CD44− and CD44+ cells were distributed into an FACS plot of CD24 versus CD166. CD24hi cells in CD44− and CD44+ populations were divided further, depending on the CD166 expression. (G) The reported gates relatively enriched endocrine cells (blue) and PCs (red) in the FACS plot of CD24 versus SSC. (H) The subgates in F were analyzed in the FACS plot of CD24 versus SSC. It is noteworthy that only CD44–CD24hiCD166hi (red polygon) cells fell mainly into the PC gate (red).
Supplementary Figure 2. ESA (EpCAM) is not required to distinguish IECs in the present FACS analysis. (A) Based on the isotype staining, the majority of cells in the original live IEC gate are ESA+. Upper panel, small intestine. Lower panel, colon. (B) ESA staining can affect the other surface marker staining including CD44. The CD44+ percentage and the FACS plots of CD24 versus CD166 derived from CD44+ or CD44− gates were changed by adding ESA antibody.
Supplementary Figure 3. Lgr5GFPhi cells express a much higher level of CD44 than CD24hiCD166hi PCs. (A) Confocal image of cross sections of the crypt bottom from Figure 1, showing strong staining of CD44 (bright blue) in the junctions of Lgr5-GFPhi stem cells (green, arrowheads) and weak staining between PCs (red, arrows). (B) CD44+ gate drawn out of total live cells. (C) CD24hiCD166hi PC gate drawn out of total live cells (see Supplementary Figure 3). (D) Lgr5 GFPhi cell gate drawn out of total live cells. (E) Backgating CD24hiCD166hi PCs into CD44 gate, showing that the low percentage of PCs is CD44+. (F) Backgating Lgr5 GFPhi Cells into CD44 gate, showing that almost all of Lgr5 GFPhi cells are CD44+.
Supplementary Figure 4. CD24 versus CD166 generates a more natural FACS pattern than CD24 alone. Two representative FACS tests show the cell populations with different CD24 levels in CD24 versus FSC plots or CD24 versus CD166 plots. CD166med cells help define the boundary between CD24neg and CD24lo.
Supplementary Figure 5. Testing the Ki67 expression of 4 subpopulations in the CD24 versus CD166 plot derived from CD44+ cells. (A) Representative FACS plot of CD24 versus CD166 derived from CD44+ cells. Four subpopulations were labeled with different colors (bright blue, deep green, bright green, red), which is consistent with Figure 1. (A) Cells sorted from 4 subpopulations were stained with Ki67 (red) and counterstained by DAPI. Bar = 100 μm. (B) The percentage of Ki67+ cells in different subpopulations (n = 3).
Supplementary Figure 6. Post sorting confirms the sorting purity. (A) The sorted CD44+ CD24loCD166+ GRP78−/lo cells from the small intestine were verified by flow cytometry, showing a high percentage (>90%) overlapping with the gates used for primary sorting. (B) The sorted CD44+ CD24loCD166+ cKit− cells from colon were verified by flow cytometry, showing a high percentage overlapping with the gates used for primary sorting.
Supplementary Figure 7. CD44hiGRP78−/lo cannot exclude the CD44+CD24hi population. (A) CD44 and positive gates from a single live IEC. (B) CD44+ cells distributed in an FACS plot of CD24 versus CD166. (C) Overlap CD44+CD24hi cells into an FACS plot of GRP78 versus CD44 from a single live IEC.
Supplementary Figure 8. The enrichment of clonogenic colonic ISCs by CD44 and c-Kit combination. (A) c-Kit isotype staining used to set the criterion for the c-Kit+ population. (B) Representative FACS analysis shows that the CD44hi colonic IECs can be clearly separated into a c-Kit+ and c-Kit− population. (C) CD44hi c-Kit− cells have heterogeneities for CD24 and CD166 expression and CD44hi c-Kit−CD24loCD166+ gate was drawn based on contour plot. (D) The CFE of CD44hi c-Kit−, CD44hi c-Kit+, CD44hi c-Kit−CD24loCD166+ cells (n = 3). ***P < .001.
Supplementary Figure 9. Comparison of ISC colony formation induced by different combinations of Wnt3a, R-spondin, Chir99021, and Thiazovivin (TZV) on the first 2 days.Different combinations of factors and inhibitors (including Wnt3a, 100 ng/mL; R-Spondin, 1 μg/mL, Chir99021, 2.5 μmol/L; TZV, 2.5 μmol/L) were tested by adding them into culture medium on day 0. Wnt3a was added on day 2 if it was added on day 0. The high efficiency was achieved by adding Chir99021 and TZV on day 0. Adding Wnt3a, R-Spondin, and Chir99021 together inhibited colony growth, partially mimicking the effect of the high concentration of Chir99021 in Supplementary Figure 11. Y-27632 combined with Wnt3a and R-Spondin was also tested and no high CFE was observed. Other growth factors including epidermal growth factor, Noggin, and Jagged-1 were used as shown in Figure 5 in every group. n = 3 wells. Similar results were obtained in 2 independent experiments. Bars = 100 μm.
Supplementary Figure 10. Wnt3a conditional medium and R-spondin 1 cannot replace GSK inhibitor. Compared with Wnt3a, Wnt3a conditional medium has complicated components. Combination of Wnt3a conditional medium (50%), R-Spondin 1 μg/mL, Chir99021 2.5 μmol/L, and TZV 2.5 μmol/L only partially inhibits colony growth. In addition, wnt3a conditional medium combined with R-spondin, TZV, or Y27632 can lead to a CFE of <5%, which is higher than commercial Wnt3a (Supplementary Figure 9). However, Chir99021 combined with TZV led to even higher CFE and much bigger colony size. Other growth factors including epidermal growth factor, Noggin, and Jagged-1 were used as shown inFigure 4 in every group. n = 3 wells. Similar results were obtained in 2 independent experiments. Bar = 100 μm.
Supplementary Figure 11. The effect of Chir99021, TZV, and Y-27632 on single ISC culture. (A) Representative process of colony forming from single Lgr5-GFPhi small intestinal cells with different concentration of Chir99021 in the medium for the first 2 days of culture. Note that the high concentration (5 μmol/L and 10 μmol/L) can cause the spreading of the colony. (B) Adding Y27632 (10 μmol/L) or TZV (2.5 μmol/L) into the medium on the first 2 days leads to a different size of colony derived from single Lgr5-GFPhi small intestinal cells. Chir99021 and other growth factors were used in the same way in both groups. (C) Adding Y27632(10 μmol/L) or TZV (2.5 μmol/L) into the medium on the first 2 days led to a different size of colony derived from single Lgr5-GFPhi colonic intestinal cells. Chir99021 and other growth factors were used in the same way in both groups. Bars = 200 μm.
Supplementary Figure 12. The heterogeneities of morphology of colonoid derived from single Lgr5GFPhi ISCs. (A) Representative growth process of single ISC-derived colonoid from day 6 to day 12. White arrows label the same colony on different days. It is noteworthy that most “closed” colonoids can become “open” along with the cell proliferation and differentiation. (B) High magnification of colonoids on day 10 shows different morphology (sphere, closed enteroid, open enteroid). (C) Representative Muc2 (red) and E-cadherin (bright blue) IHC staining shows the differentiation in colonoids with different morphology.
Supplementary Figure 13. CD44+ CD24loCD166+ GRP78−/lo enrich high percentage of Lgr5GFPhi cells. (A) The distribution of Lgr5GFPhi cells in CD44+, CD44+ CD24loCD166+, and CD44+ CD24loCD166+ GRP78−/lo gates. (B) The percentage of total Lgr5GFPhi ISCs covered by the gate of CD44+ CD24loCD166+ GRP78−/lo (n = 3).
Supplementary Figure 14. The enteroids derived from different Lgr5GFP levels share similarity in gene profiling and long-term culture maintenance. (A) Lgr5GFPhi cells or Lgr5GFPmed/lo cells derived from CD44+ CD24lo CD166+ GRP78−/lo population were cultured and passaged. qPCR test shows the similar expression levels of ISC related genes and differentiation associated genes in these 2 groups (n = 3). (B) Representative passaging image (from day 0 to day 2) showing the robust colony forming of these 2 groups, even after about 2.5 months of culture. Bar = 500 μm.
Supplementary Figure 15. Single human ISC isolation and culture analysis. (A) Experimental scheme of enriched single live ISCs. (B) qRT-PCR results show the gene expression in the CD44+CD166+CD24lo population relative to unsorted live IECs, indicating the enrichment of ISCs, although some differentiated cells were remained (n = 2). (C) Strategies to culture single CD44+CD166+CD24lo cells. Cells grew only in the group with a single dose of CHIR99021 on the first 2 days. Data are represented as mean ± SEM (n = 2). (D) Representative growth process of single CD44+CD166+CD24lo cells with addition of a single dose of CHIR99021. (E) IHC staining of CD44+CD166+CD24lo cells after 14 days of culture shows differentiation into intestinal epithelial lineages. Arrows in the inset indicate positive staining. Scale bars = 100 μm (D); 50 μm (E).
Supplementary Figure 16. Rat mAB-B6A6, CD24, CD166 combination to analyze ISCs and differentiated cells. (A) FACS plots show the exclusion of cell aggregates (left panel) and blood cells and dead cells (right panel). It is noteworthy that doublet cells cannot be efficiently removed without using Pulse width. (B) IHC staining shows that rat monoclonal B6A6 antibody facilitates exclusion of villus enterocytes but not some secretory cells, including goblet cells (arrow). (C) FACS pattern with FSC-W versus FSC-A to exclude cell aggregates. (D) FACS pattern with B6A6 versus FSC-W to show the B6A6− cells. FACS pattern with B6A6 versus FSC-W to show the enrichment of B6A6− cells by using anti-rat immunoglobulin G beads to deplete the B6A6+ cells. FACS pattern with CD24 versus CD166 to show different cell populations in B6A6− cells. It is noteworthy that B6A6− contains some CD44− cells. (E) qRT-PCR to test ISC and lineage related gene expression in different populations (G1–G4) from G c compared with unsorted cells. The G4 population shows the highest lysozyme and Lgr5 expression indicating PC and CBC-PC doublets. G1 shows the highest level of Muc2 expression, indicating the enrichment of goblet cells. G3 shows the high level of Lgr5 and Ascl2, indicating Lgr5hi and Lgr5lo/med cells may be included in this population.
Supplementary Table 1. Antibodies Used in Flow Cytometry for Mouse Tissue
Supplementary Table 2. TaqMan Assay Used in qPCR for Mouse Tissue
Supplementary Table 3. Antibodies Used in Flow Cytometry for Human Tissue
Supplementary Table 4. TaqMan Assay Used in qPCR for Human Tissue