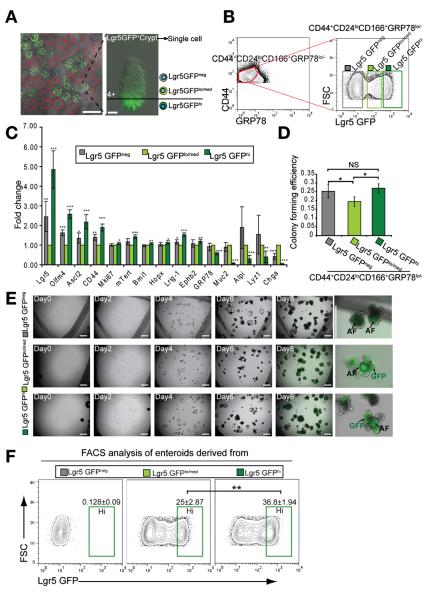
Figure 6.
CD44+CD24loCD166+ GRP78−/lo identifies ISCs independent of Lgr5-GFPhi cells. (A) The mosaic distribution of Lgr5-GFP+ crypt in jejunum. Gradient GFP expression in Lgr5-GFP+ crypt. (B) Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi gates in the CD44+CD24loCD166+ GRP78−/lo population. (C) qRT-PCR results show the gene expression of Lgr5-GFPneg and Lgr5-GFPhi cells relative to Lgr5-GFPlo/med cells from the CD44+CD24loCD166+ GRP78−/lo population. Data are presented as mean ± SEM (n = 4, *P < .05, **P < .01, ***P < .001). (D) CFE analysis of the Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi cells in the gates of B. Data are represented as mean ± SEM (*P < .05, n = 3, t test). (E) A growth process of single cells sorted from Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi gates in B, showing that Lgr5-GFPlo/med cells can generate Lgr5-GFPhi enteroid. (F) FACS analysis of enteroids derived from single Lgr5-GFPneg, Lgr5-GFPlo/med, and Lgr5-GFPhi cells, showing that single Lgr5-GFPlo/med cells can generate enteroids containing Lgr5-GFPhi cells as single Lgr5-GFPhi cells did. Data are presented as mean ± SEM (n = 3, **P < .01).