Abstract
Gene expression profiling of transplant recipient blood and urine can potentially be used to monitor graft function, but the multitude of protocols in use make sharing data and comparing results from different laboratories difficult. The goal of this study was to evaluate the performance of current methods of RNA isolation, reverse transcription, and quantitative polymerase chain reaction (qPCR) and to test whether multiple centers using a standardized protocol can obtain the same results. Samples, reagents, and detailed instructions were distributed to six participating sites that performed RNA isolation, reverse transcription and qPCR for 18S, PRF, GZB, IL8, CXCL9 and CXCL10 as instructed. All data were analyzed at a single site. All sites demonstrated proficiency in RNA isolation and qPCR analysis. Gene expression measurements for all targets and samples had correlations >0.938. The coefficient of variation of fold-changes between pairs of samples was less than 40%. All sites were able to accurately quantify a control sample of known concentration within a factor of 1.5. Collectively, we have formulated and validated detailed methods for measuring gene expression in blood and urine that can yield consistent results in multiple laboratories.
Introduction
Current diagnosis of acute and chronic renal allograft injury depends on evaluation of a biopsy performed either at a protocol-defined time point or after rising serum creatinine levels indicate decreased graft function, by which time injury is already in progress. Gene expression analysis of biopsy tissue by quantitative polymerase chain reaction (qPCR) and microarray has provided a wealth of information about the molecular changes in a graft that accompany ongoing injury (1–4). A problem with this approach is the continued need for an invasive procedure, the biopsy. There has been a great deal of interest and effort in the identification of biomarkers in the blood or urine of transplant patients that are generated as the result of graft tissue injury. Ideally such biomarkers should provide a rapid, noninvasive approach for diagnosis, and possibly prediction, of graft injury that could lead to more immediate treatment and improved outcomes.
The critical role of donor-reactive T cells in acute and chronic graft injury has raised the hypothesis that expression of T cell effector molecules involved in tissue injury might be detectable in the blood or urine of renal transplant patients during rejection. Previous studies of peripheral blood have found upregulated mRNA expression of the T cell-derived cytolytic mediators perforin, granzyme B, and Fas ligand during acute rejection of renal grafts (5–10).
For renal transplant patients, the urine potentially provides a more proximate source of immune events occurring in the graft. By profiling RNA isolated from urine sediment, several investigators have observed gene expression changes that correlate with acute rejection, chronic allograft nephropathy, and interstitial fibrosis of renal grafts. These changes include altered expression of mRNA encoding T cell transcription and effector molecules (11–16); mediators of inflammation, tissue repair, and fibrogenesis (17, 18); and, cytokines, chemokines and their receptors (19, 20). These distinct expression profiles generated from individual laboratory studies indicate that, as predicted, injury to kidney grafts is reflected by molecular changes in the urine.
One of the major goals of the Clinical Trials in Organ Transplantation (CTOT) consortium has been to establish standardized methodology to monitor transplant outcomes in large, multicenter studies. The CTOT core laboratories have developed and validated a comprehensive set of protocols for gene expression profiling of blood and urine that has potential use as a diagnostic and/or monitoring tool. The purpose of this study was to evaluate the reproducibility of results obtained using this method from the core laboratory in five separate sites.
Materials and Methods
Study sites, reagents and samples
The following six sites participated in the study: Cleveland Clinic, Mt. Sinai School of Medicine, University of Alabama at Birmingham, Beth Israel-Deaconess Hospital, Stanford University, and University of California Los Angeles. Each site was assigned an identification number for anonymity. All reagents and samples were prepared at Cleveland Clinic and distributed to the other sites for analysis. Unless otherwise indicated, all reagents are from Life Technologies, Carlsbad, California. Detailed protocols are available in the methods supplement.
Patients and samples for this study were from the CTOT-1 study in which blood and urine were collected from renal transplant patients at the time of transplant and at various time points for 2 years post-transplant for immune monitoring assays. Urine and blood were also collected from normal volunteers. All sample collection and studies were conducted under approval of the Institutional Review Boards of all of the participating sites.
RNA isolation
Urine samples were processed as originally described by Li et al (12). In brief, urine was centrifuged at 2,000 x g for 30 minutes at 4°C, and the sediment washed with PBS and stored at −80°C in RNAlater. RNA was isolated from urine samples with the PureLink Mini Kit using a modified protocol. Blood samples were collected in Tempus Blood RNA Tubes and RNA isolated with the Tempus Spin RNA Isolation Kit according to the manufacturer’s protocol.
Reverse transcription and preamplification
RNA was reverse transcribed with random hexamers using the High Capacity cDNA Reverse Transcription Kit. Urine cDNA samples were assayed using the preamplification enhanced real time qPCR assay developed by the Cornell group (13). In the first step, 30 ng of cDNA was amplified in a 10-cycle mutiplex PCR with all test primers (excluding 18S) and TaqMan PreAmp Master Mix. The resulting amplified cDNA was diluted 1:5 with Tris-EDTA (TE) buffer and used for qPCR.
qPCR
Real-time PCR instruments and the amplification protocol used by each lab are summarized in Supplementary Table 1. The primers and probes were all inventoried TaqMan assays, listed in Supplementary Table 2. PCR reactions were run in duplicate, with each reaction containing 25 ng cDNA, 10 μl 2X PCR master mix, and 1 μl TaqMan assay in a 20 μl total reaction volume. Run data was analyzed with the automatic baseline and threshold settings determined by the respective PCR instruments. Samples having CT values >35 or CT replicate standard deviations >0.5 were excluded from analysis. Results were exported as text files and all subsequent analysis was done at Cleveland Clinic.
FRZ1 PCR standard
Quantities of test genes were derived from a calibration curve using an amplicon of the mouse FZR1 gene (NCBI accession #NM_019757.1) as the standard. TaqMan assay primers and probes were designed using Applied Biosystems Primer Express 3.0 software. The sequences were Forward, 5′ CCCTGTCTTCATACCCCATCTC 3′; Reverse, 5′ CAGCCACCACACTGGGAATC 3′; and, Probe, 5′ FAM-CTGCCCCTTGCCTGG-MGB 3′. Sixteen identical PCR reactions were set up containing 200 ng B6 mouse cDNA, 10 μl TaqMan Fast Universal Master Mix, and 150 nM forward and reverse primers in a 20 μl reaction volume. The product size (60 bp) was verified by gel electrophoresis. The PCR products were pooled and purified using a Qiaex II Gel Extraction Kit (Qiagen, Valencia, CA), quantified by Nanodrop, and diluted with TE buffer to 107 copies/μl. Aliquots were stored at −80°C. Calibration curves for PCR were made from 10-fold dilutions from 106 to 10 copies/μl of this stock solution. Linear regression was used to calculate slope and intercept for the calibration curve and only data from curves with slopes between −3.30 and −3.36 was used for analysis. Representative standard curve plots from each site are shown in Supplementary Figure 1.
Statistical evaluation of the data
For each sample and target, the inter-lab mean, standard deviation, and percent coefficient of variation (CV: standard deviation/mean x 100) were calculated. The mean of all of the CVs is the overall inter-lab variation for that experiment. Pairwise Pearson correlations among laboratories were calculated for all samples and targets.
Results
Variation of PCR alone
To determine the level of variation among the sites when performing qPCR alone, each of five centers was sent aliquots of samples generated from RNA isolated from the blood or urine of CTOT-01 subjects: five blood cDNA and five pre-amplified urine cDNA, along with TaqMan assays for 18S rRNA, perforin, granzyme B, CXCL9/Mig and CXCL10/IP-10. Each group used their own standard qPCR master mix, instrument, and cycling conditions to measure expression of the five genes in each sample. Results for GZMB in the urine samples and all genes in blood sample 217 were excluded because abnormal amplification plots were obtained in more than one site. Six data points (out of 750 total) were omitted for having replicate standard deviations >0.5. Mean, standard deviation and CV were calculated on the quantity reported by each site for each gene and target. The overall inter-lab CVs were identical for blood and urine samples (62% urine, 63% blood) and were unchanged by normalizing the quantities to 18S.
PCR data are commonly expressed as fold-changes between samples or groups rather than raw quantities. We designated one blood and one urine sample as a calibrator to calculate the fold-change (Qtysample/Qtycalibrator) for all samples and targets. Data for CXCL9 mRNA levels expressed as fold-change versus quantity in urine samples is shown in Table 1. The CVs for quantity (A) ranged from 52–63% while those for fold-change (B) ranged from 7–18%. The same pattern (lower cvs for fold-change measurements than for quantity) was observed in the other target genes and samples.
Table 1.
Interlab variability of quantity versus fold-change measurements in the same samples
| A Quantity | |||||||
|---|---|---|---|---|---|---|---|
| sample | site
|
mean | stdev | cv | |||
| 1 | 2 | 3 | 4 | ||||
| 67 | 116.6 | 337.4 | 276.5 | 120.9 | 213 | 111 | 52 |
| 285 | 641.6 | 2,635.5 | 2,103.6 | 869.2 | 1562 | 961 | 62 |
| 50 | 7,812.3 | 34,844.6 | 24,100.8 | 11,655.7 | 19603 | 12311 | 63 |
| 139 | 12,670.9 | 45,624.4 | 32,626.6 | 14,999.1 | 26480 | 15565 | 59 |
| 886 | 13,819.6 | 42,753.6 | 37,017.4 | 17,014.2 | 27651 | 14379 | 52 |
| B Fold-change | |||||||
|---|---|---|---|---|---|---|---|
| sample | site
|
mean | stdev | cv | |||
| 1 | 2 | 3 | 4 | ||||
| 67 | 1.0 | 1.0 | 1.0 | 1.0 | |||
| 285 | 5.5 | 7.8 | 7.6 | 7.2 | 7.0 | 1.0 | 14.9 |
| 50 | 67.0 | 103.3 | 87.2 | 96.4 | 88.5 | 15.7 | 17.8 |
| 139 | 108.7 | 135.2 | 118.0 | 124.1 | 121.5 | 11.1 | 9.1 |
| 886 | 118.6 | 126.7 | 133.9 | 140.7 | 130.0 | 9.5 | 7.3 |
Expression of CXCL9 in 5 urine cDNA samples isolated from CTOT-01 study subjects was measured by qPCR. Samples and TaqMan assays were prepared and distributed from site 1. Master mix and real-time instrument varied by site (table S1). Four sites submitted results for this experiment and no data points were omitted. A) CXCL9 quantity determined by each site with mean, standard deviation and percent CV. B) Fold-change of each sample relative to sample 67 (QtysampleX/Qtysample67). The inter-laboratory variation (CV) for fold-change measurements was lower (7–18%) than for quantity measurements (52–63%). The same pattern was seen for the other target genes in urine and blood samples.
As many published studies employ the ddCT method (21) rather than standard curves to measure relative gene expression between samples, we wanted to compare the inter-lab variability of the two methods (Table 2). Gene expression was measured using a standard curve to calculate quantities (A) or by the ddCT method (B). The columns from left to right indicate the CV calculated for raw quantity (or CT), 18S-normalized quantity (dCT), and fold-change (ddCT) with one blood sample and one urine sample chosen as calibrator. The variability between sites was equivalent for blood and urine samples and for quantification strategy. Regardless of which strategy was used to analyze the data, measurements of fold-change were much more reproducible than analysis of single samples alone. Table 3 shows pair-wise Pearson correlations calculated on the CTs determined for all gene targets in blood (A) and urine (B) specimens. All correlations are greater than 0.97 and are significantly different from zero at p < 0.0001, indicating that there is relatively close agreement between the labs.
Table 2.
Comparison of standard curve to Ct measurements across all samples
| A | |||
|---|---|---|---|
| quantity
|
|||
| sample type | qty | 18S-norm | fold-change |
| blood | 63 | 62 | 41 |
| urine | 63 | 66 | 23 |
| B | ||
|---|---|---|
| CT
| ||
| CT | dCT | ddCT |
| 81 | 63 | 24 |
| 74 | 54 | 18 |
Overall interlab CVs for qPCR analysis of 5 target genes in 10 cDNA samples. Gene expression was measured using a standard curve to calculate quantities (A) or by the ddCT method (B). The columns from left to right indicate the CV calculated for raw quantity (or CT), 18S-normalized quantity (dCT), and fold-change (ddCT) with one blood sample and one urine sample chosen as calibrator. The variability between sites was equivalent for blood and urine samples and for quantification strategy. In all cases, fold-change measurements were more similar between sites than sample-specific measurements.
Table 3.
Multivariate analysis of qPCR results from multiple sites
| A | blood | ||||
|---|---|---|---|---|---|
| site 1 | site 2 | site 3 | site 4 | site 5 | |
|
|
|||||
| site 1 | 1 | 0.9946 | 0.994 | 0.9794 | 0.9967 |
| site 2 | 0.9946 | 1 | 0.9978 | 0.982 | 0.9983 |
| site 3 | 0.994 | 0.9978 | 1 | 0.981 | 0.9984 |
| site 4 | 0.9794 | 0.982 | 0.981 | 1 | 0.9804 |
| site 5 | 0.9967 | 0.9983 | 0.9984 | 0.9804 | 1 |
| B | urine | ||||
|---|---|---|---|---|---|
| site 1 | site 2 | site 3 | site 4 | site 5 | |
|
|
|||||
| site 1 | 1 | 0.9983 | 0.9982 | 0.9913 | 0.9997 |
| site 2 | 0.9983 | 1 | 0.9971 | 0.9908 | 0.9984 |
| site 3 | 0.9982 | 0.9971 | 1 | 0.9929 | 0.9989 |
| site 4 | 0.9913 | 0.9908 | 0.9929 | 1 | 0.9911 |
| site 5 | 0.9997 | 0.9984 | 0.9989 | 0.9911 | 1 |
Expression of 18S, IL8, CXCL9, CXCL10, PRF1, and GZMB were measured in identical blood and urine cDNA samples at 5 clinical sites using different master mixes and PCR instruments Pearson correlations were calculated on CTs.
Pre-analytical steps
To examine the effects of additional processing steps on the inter-lab variation, a series of identical samples was generated from 900 ml of pooled urine collected from a group of healthy volunteers. Each site received identical aliquots of: 1) a frozen urine pellet; 2) isolated pellet RNA; 3) cDNA; and, 4) pre-amplified cDNA. Identical single-use aliquots of all the reagents necessary for RNA isolation, reverse transcription, pre-amplification and qPCR analysis for 18S, CXCL9 and CXCL10 were also provided. All six sites processed the samples using the centrally provided reagents and protocol; only the PCR instruments and cycling conditions were unique to the separate sites. One data point out of 72 was omitted for high replicate standard deviation. The CVs calculated on the quantities reported for each target gene and processing step are shown in Supplemental Table 3. The inter-lab variability of PCR analysis did not increase when more extensive sample processing was done at separate sites. The overall inter-lab CV for all samples and targets was 46%.
Spiked sample
To evaluate our ability to accurately quantify a known amount of template, urine RNA isolated from normal volunteers was equally divided and each aliquot spiked with 1×105 copies of purified CXCL9 amplicon without the knowledge of the participants. This RNA had been previously determined to lack detectable CXCL9 transcripts. The spiked sample was reverse transcribed, pre-amplified, and quantified along with the other test samples in each respective site. The measured quantities by site were: 1) 1.09×105, 2) 1.02×105, 3) 7.42×104, 4) 1.14×105, 5) 9.16×104, and 6) 6.68×104 copies/μl, indicating the ability of each site to quantify the amplicon within a factor of 1.5 when using identical reagents.
Urine RNA quality
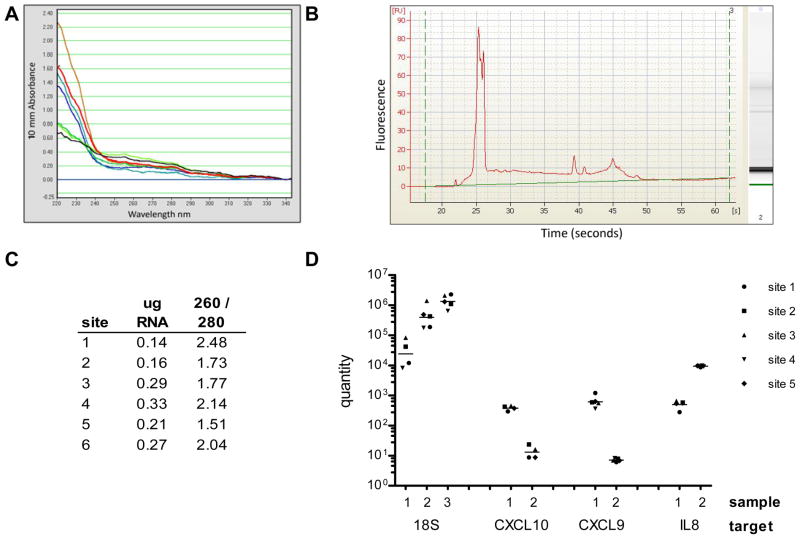
Because only two of the six sites had previous experience isolating RNA from urine samples, we paid particular attention to the RNA isolation step. Each site had been provided with three frozen urine pellets derived from three unique pooled samples, along with all reagents for processing and analysis, as described above. The RNA isolated from the urine pellets was evaluated by Nanodrop and Agilent Bioanalyzer at each site. Representative results from site 1 are shown in Figure 1A and B. The absorbance spectra (A) and electropherogram (B) are normal for urine RNA and illustrate the poor quality and extensive degradation that characterize the samples, even when they have been isolated from freshly collected specimens at highly experienced centers. Yields of RNA isolated from the pellets prepared from 75 ml of urine ranged from 0.14–0.33 μg with OD260/280 ratios between 1.5 and 2.5 Nanodrop readings from urine pellet #1 measured at each of the six sites are shown in Panel C. The three RNA samples were reverse transcribed and analyzed by qPCR in each site. 18S expression was measured in all three samples; CXCL9, CXCL10, and IL8 were measured in two of the samples. One of the six sites does not have PCR data because of a problem with the standards. One value for 18S in sample 1, and for CXCL10 in samples 1 and 2 was omitted for high replicate standard deviation. The gene expression results are shown in Figure 2D.
Figure 1.
Evaluation of urine RNA. Using reagents and samples prepared at site one, each laboratory isolated RNA from three urine pellet samples for qPCR analysis. A) Nanodrop absorbance spectra of 7 urine RNA samples isolated at site 1 illustrates typical appearance of urine RNA. B) Agilent Bioanalyzer electropherogram of a representative urine RNA indicates mostly small fragments of RNA; C) Nanodrop OD readings of the RNA isolated from urine pellet 1 in each site, showing normal yield and purity for urine samples; D) qPCR analysis of 3 urine pellet samples for 18S, and two samples for CXCL9, CXCL10, and IL8. All pre-analytical steps were performed at the respective sites using centrally provided reagents. Quantity and grand mean for each target and sample are plotted on a log scale. 18S sample 1 and CXCL10 samples 1 and 2 have one missing data point; all other samples show data from 5 sites.
Discussion
In order to be able to truly develop gene expression profiling as a diagnostic tool, it is imperative to establish standardized protocols and best practices. As a first step in that direction, we evaluated the performance of the current methods used by the CTOT molecular core laboratory in terms of reproducibility of results in multiple sites. The overall interlab variability (CV) of PCR alone was around 63% regardless of sample type, normalization to 18S, or quantification method, and was not appreciably increased by addition of pre-analytical steps. Our correlations on all samples and targets were >0.93 and we demonstrated consistent measurements of relative gene expression (fold-change) in clinical samples, with CVs of less than 40%. All of the sites were able to accurately quantify the copies of CXCL9 in a spiked RNA sample within a factor of 1.5, which is generally considered the limit of measurements by qPCR (22–24).
Of all the pre-analytical steps involved in this method, we focused on RNA isolation from urine in particular because of the technical challenges involved. All of the sites were able to isolate RNA from urine sediment of sufficient quality and quantity for qPCR. It is important to clarify that quality in this context means RNA that contains quantifiable transcripts. Urine RNA is characterized by extensive degradation that makes traditional approaches to measure quality (i.e. spectrophotometry and capillary electrophoresis) of little value. Spectrophotometry cannot discriminate intact RNA from degraded nucleotides or genomic DNA.
Microfluidic electrophoresis systems such as the Agilent Bioanalyzer characterize the ribosomal fraction of the RNA but do not give information about mRNA or the small fragments that are measured by qPCR. The suitability of an RNA sample for a particular application must be determined empirically. To that end, the CTOT consortium employs a PCR-based method developed by the Cornell group to evaluate urine cDNA samples prior to analysis. Using the standard curve method described in detail in the supplementary material, the copies of 18S rRNA and TGFB1 are measured. Samples that contain a minimum of 5×107 copies/μg 18S and 100 copies/μg TGFB1 will have enough quantifiable transcripts to be included in the analysis (R. Ding, unpublished); samples that do not meet this criteria are excluded. Sites that adhere closely to the collection protocol typically have less than 15% of their samples fail quality control.
In spite of its perceived shortcomings, RNA from urine samples yields quite reproducible results. In this study we found that the inter-laboratory variability of our qPCR data was the same for samples from blood and urine. We found no evidence that the pre-amplification step or the quality of the RNA itself introduce additional variability into the analysis. Previous clinical studies also point to the consistency of results obtained from urine sediment RNA. Between 2001 and 2009, five separate groups using different methods examined the expression of perforin and granzyme B mRNA in the urine sediment of renal transplant patients undergoing acute rejection. The first of those studies (12) employed competitive quantitative PCR, a method that preceded, and has been replaced by, real-time PCR. Two of the groups (14, 16) used purified amplicons of each gene to construct calibration curves, one used an 18S calibration curve (11), and two others the ddCt method for quantification (25, 26). Gene expression was normalized to cyclophilin, HPRT, or nothing at all. Yet in spite of these differences, all found perforin and granzyme B mRNA expression to be upregulated in the urine of acutely rejecting subjects when compared to those with stable graft function. However, the studies reached different conclusions about whether this upregulation is able to distinguish acute rejection from infection.
Quantitative PCR analysis is notoriously difficult to standardize because of the many pre-analytical steps involved and the cyclical magnification of variations along with template. Similar inter-lab comparisons of qPCR to measure breast cancer markers and CMV and EBV viral load reported initial variabilities ranging from 40% to 135% that were markedly improved after implementation of calibrators, international standards, and strategies for data harmonization (27–30). Similarly, we see many opportunities to improve our current method, including optimization of cycling parameters, comparison of different master mix formulations, and the investigation of RNA standards as reaction controls and inter-platform calibrators (31). We anticipate this project will be the beginning of collaborative efforts designed to enhance the further development and identification of non-invasive gene expression tools for use in transplantation.
Supplementary Material
Acknowledgments
This research was performed as part of an American Recovery and Reinvestment (ARRA) funded project under Award Number U0163594 (to PSH), from the National Institute of Allergy and Infectious Diseases. The work was carried out by members of the Clinical Trials in Organ Transplantation (CTOT) and Clinical Trials in Organ Transplantation in Children (CTOT-C) consortia. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. The authors thank the Principal Investigators of CTOT-01 that provided samples used in these studies (D. Hricik, University Hospitals, Case Medical Center, Cleveland, OH; E. Poggio, Cleveland Clinic, Cleveland, OH; K. Newell, Emory University Hospital, Atlanta, GA; R. Formica, Yale University Hospital, New Haven, CT; J. Goebel, Cincinnati Children’s Hospital, Cincinnati, OH; F. Shihab, University of Utah School of Medicine, Salt Lake, UT; D. Rush and P. Nickerson, University of Manitoba, Winnipeg, Manitoba, Canada).
Abbreviations
- CTOT
Clinical Trials in Organ Transplantation
- Ct
threshold cycle
- CV
coefficient of variation
- qPCR
quantitative polymerase chain reaction
- RT
reverse transcriptase
Footnotes
Disclosure
The authors have no financial conflict of interest to declare.
References
- 1.Famulski KS, de Freitas DG, Kreepala C, Chang J, Sellares J, Sis B, et al. Molecular phenotypes of acute kidney injury in kidney transplants. J Am Soc Nephrol. 2012;23(5):948–958. doi: 10.1681/ASN.2011090887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hidalgo LG, Sellares J, Sis B, Mengel M, Chang J, Halloran PF. Interpreting NK cell transcripts versus T cell transcripts in renal transplant biopsies. Am J Transplant. 2012;12(5):1180–1191. doi: 10.1111/j.1600-6143.2011.03970.x. [DOI] [PubMed] [Google Scholar]
- 3.Naesens M, Khatri P, Li L, Sigdel TK, Vitalone MJ, Chen R, et al. Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int. 2011;80(12):1364–1376. doi: 10.1038/ki.2011.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakorchevsky A, Hewel JA, Kurian SM, Mondala TS, Campbell D, Head SR, et al. Molecular mechanisms of chronic kidney transplant rejection via large-scale proteogenomic analysis of tissue biopsies. J Am Soc Nephrol. 2010;21(2):362–373. doi: 10.1681/ASN.2009060628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Netto MV, Fonseca BA, Dantas M, Saber LT, Castro MC, Ferraz AS. Granzyme B, FAS-ligand and perforin expression during acute cellular rejection episodes after kidney transplantation: comparison between blood and renal aspirates. Transplant Proc. 2002;34(2):476–478. doi: 10.1016/s0041-1345(02)02601-5. [DOI] [PubMed] [Google Scholar]
- 6.Sabek O, Dorak MT, Kotb M, Gaber AO, Gaber L. Quantitative detection of T-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation. 2002;74(5):701–707. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- 7.Shin GT, Kim SJ, Lee TS, Oh CK, Kim H. Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract. 2005;100(3):c63–70. doi: 10.1159/000085050. [DOI] [PubMed] [Google Scholar]
- 8.Simon T, Opelz G, Wiesel M, Ott RC, Susal C. Serial peripheral blood perforin and granzyme B gene expression measurements for prediction of acute rejection in kidney graft recipients. Am J Transplant. 2003;3(9):1121–1127. doi: 10.1034/j.1600-6143.2003.00187.x. [DOI] [PubMed] [Google Scholar]
- 9.Vasconcellos LM, Schachter AD, Zheng XX, Vasconcellos LH, Shapiro M, Harmon WE, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66(5):562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 10.Veale JL, Liang LW, Zhang Q, Gjertson DW, Du Z, Bloomquist EW, et al. Noninvasive diagnosis of cellular and antibody-mediated rejection by perforin and granzyme B in renal allografts. Hum Immunol. 2006;67(10):777–786. doi: 10.1016/j.humimm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Dadhania D, Muthukumar T, Ding R, Li B, Hartono C, Serur D, et al. Molecular signatures of urinary cells distinguish acute rejection of renal allografts from urinary tract infection. Transplantation. 2003;75(10):1752–1754. doi: 10.1097/01.TP.0000063931.08861.56. [DOI] [PubMed] [Google Scholar]
- 12.Li B, Hartono C, Ding R, Sharma VK, Ramaswamy R, Qian B, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344(13):947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 13.Muthukumar T, Dadhania D, Ding R, Snopkowski C, Naqvi R, Lee JB, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353(22):2342–2351. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 14.Ozbay A, Torring C, Olsen R, Carstens J. Transcriptional profiles in urine during acute rejection, bacteriuria, CMV infection and stable graft function after renal transplantation. Scand J Immunol. 2009;69(4):357–365. doi: 10.1111/j.1365-3083.2009.02226.x. [DOI] [PubMed] [Google Scholar]
- 15.Seiler M, Brabcova I, Viklicky O, Hribova P, Rosenberger C, Pratschke J, et al. Heightened expression of the cytotoxicity receptor NKG2D correlates with acute and chronic nephropathy after kidney transplantation. Am J Transplant. 2007;7(2):423–433. doi: 10.1111/j.1600-6143.2006.01625.x. [DOI] [PubMed] [Google Scholar]
- 16.Yannaraki M, Rebibou JM, Ducloux D, Saas P, Duperrier A, Felix S, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006;19(9):759–768. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 17.Anglicheau D, Muthukumar T, Hummel A, Ding R, Sharma VK, Dadhania D, et al. Discovery and validation of a molecular signature for the noninvasive diagnosis of human renal allograft fibrosis. Transplantation. 2012;93(11):1136–1146. doi: 10.1097/TP.0b013e31824ef181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mas VR, Mas LA, Archer KJ, Yanek K, King AL, Gibney EM, et al. Evaluation of gene panel mRNAs in urine samples of kidney transplant recipients as a non-invasive tool of graft function. Mol Med. 2007;13(5–6):315–324. doi: 10.2119/2007-00017.Mas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matz M, Beyer J, Wunsch D, Mashreghi MF, Seiler M, Pratschke J, et al. Early post-transplant urinary IP-10 expression after kidney transplantation is predictive of short- and long-term graft function. Kidney Int. 2006;69(9):1683–1690. doi: 10.1038/sj.ki.5000343. [DOI] [PubMed] [Google Scholar]
- 20.Tatapudi RR, Muthukumar T, Dadhania D, Ding R, Li B, Sharma VK, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004;65(6):2390–2397. doi: 10.1111/j.1523-1755.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Bubner B, Gase K, Baldwin IT. Two-fold differences are the detection limit for determining transgene copy numbers in plants by real-time PCR. BMC Biotechnol. 2004;4:14. doi: 10.1186/1472-6750-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo YM, Lun FM, Chan KC, Tsui NB, Chong KC, Lau TK, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc Natl Acad Sci U S A. 2007;104(32):13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmermann B, Holzgreve W, Wenzel F, Hahn S. Novel real-time quantitative PCR test for trisomy 21. Clin Chem. 2002;48(2):362–363. [PubMed] [Google Scholar]
- 25.Aquino-Dias EC, Joelsons G, da Silva DM, Berdichevski RH, Ribeiro AR, Veronese FJ, et al. Non-invasive diagnosis of acute rejection in kidney transplants with delayed graft function. Kidney Int. 2008;73(7):877–884. doi: 10.1038/sj.ki.5002795. [DOI] [PubMed] [Google Scholar]
- 26.Galante NZ, Camara NO, Kallas EG, Salomao R, Pacheco-Silva A, Medina-Pestana JO. Noninvasive immune monitoring assessed by flow cytometry and real time RT-PCR in urine of renal transplantation recipients. Transpl Immunol. 2006;16(2):73–80. doi: 10.1016/j.trim.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch HH, Lautenschlager I, Pinsky BA, Cardeñoso L, Aslam S, Cobb B, et al. An International Multicenter Performance Analysis of Cytomegalovirus Load Tests. Clinical Infectious Diseases. 2013;56(3):367–373. doi: 10.1093/cid/cis900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK, et al. Interlaboratory Comparison of Cytomegalovirus Viral Load Assays. American Journal of Transplantation. 2009;9(2):258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 29.Preiksaitis JK, Pang XL, Fox JD, Fenton JM, Caliendo AM, Miller GG, et al. Interlaboratory Comparison of Epstein-Barr Virus Viral Load Assays. American Journal of Transplantation. 2009;9(2):269–279. doi: 10.1111/j.1600-6143.2008.02514.x. [DOI] [PubMed] [Google Scholar]
- 30.Span PN, Sieuwerts AM, Heuvel JJ, Spyratos F, Duffy MJ, Eppenberger-Castori S, et al. Harmonisation of multi-centre real-time reverse-transcribed PCR results of a candidate prognostic marker in breast cancer: an EU-FP6 supported study of members of the EORTC - PathoBiology Group. Eur J Cancer. 2009;45(1):74–81. doi: 10.1016/j.ejca.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 31.Devonshire AS, Elaswarapu R, Foy CA. Evaluation of external RNA controls for the standardisation of gene expression biomarker measurements. BMC Genomics. 2010;11:662. doi: 10.1186/1471-2164-11-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.