Summary
Bacillus subtilis has adopted a bet-hedging strategy to ensure survival in changing environments. From a clonal population, numerous sub-populations can emerge, expressing different sets of genes that govern the developmental processes of sporulation, competence and biofilm formation. The master transcriptional regulator Spo0A controls the entry into all three fates and the production of the phosphorylated active form of Spo0A is precisely regulated via a phosphorelay, involving at least four proteins. Two proteins, YmcA and YlbF were previously shown to play an unidentified role in the regulation of biofilm formation, and in addition, YlbF was shown to regulate competence and sporulation. Using an unbiased proteomics screen, we demonstrate that YmcA and YlbF interact with a third protein, YaaT to form a tripartite complex. We show that all three proteins are required for proper establishment of the three above-mentioned developmental states. We show that the complex regulates the activity of Spo0A in vivo and, using in vitro reconstitution experiments, determine that they stimulate the phosphorelay, probably by interacting with Spo0F and Spo0B. We propose that the YmcA-YlbF-YaaT ternary complex is required to increase Spo0A~P levels above the thresholds needed to induce development.
Keywords: Spo0A, YmcA, YlbF, YaaT, Phosphorelay, Two-component regulation
Introduction
The well-studied model organism Bacillus subtilis undergoes a variety of developmental fates (reviewed in Lopez et al., 2009) with sub-populations emerging from a genetically homogeneous culture as a bet-hedging strategy to increase the probability of survival (Dubnau and Losick, 2006, Veening et al., 2008). For example, some cells express motility genes, while others enter a sessile state, often as a precursor to forming a multi-cellular biofilm. In addition, different sub-populations express the competence genes, which are required for the uptake of exogenous DNA, while others sporulate. Although the decisions to enter these states are individually regulated (Lopez et al., 2009), all of them rely on the response regulator protein Spo0A. The active form of Spo0A is phosphorylated and Spo0A~P is required for the transcription of genes needed to form a mature spore, as well as for biofilm formation and competence (Branda et al., 2001, Molle et al., 2003, Mirouze et al., 2011). Spo0A is phosphorylated by a phosphorelay (Burbulys et al., 1991), with input from the auto-phosphorylation of a number of histidine kinases (Antoniewski et al., 1990, Trach and Hoch, 1993, Kobayashi et al., 1995, LeDeaux and Grossman, 1995, Jiang et al., 1999, Fabret et al., 1999), followed by transfer of the phosphoryl moiety to the single-domain response regulator Spo0F, then to the phosphotransfer protein Spo0B, and finally to Spo0A.
Each developmental pathway is initiated when the concentration of Spo0A~P reaches a characteristic level (Fujita et al., 2005, Fujita and Losick 2005). A high concentration of Spo0A~P is required for sporulation and lower amounts are needed for biofilm formation or competence. Consequently, for appropriate decision-making the Spo0A~P concentration must be critically regulated in response to environmental signals. Developmental fates differ between cells within a single clone and this choice is made stochastically, with likelihoods that are determined by the amounts of Spo0A~P. Because the relative proportions of different cell-types are adjusted by selection to optimize bet-hedging, the distribution of Spo0A~P concentration among cells must be regulated, as well as the average concentration.
In both Gram-negative and Gram-positive bacteria, auxiliary proteins are known that regulate two-component systems (Buelow and Raivio, 2009, and Mitrophanov and Groisman 2008) and such proteins are important regulators of development in B. subtilis, contributing to the complex mechanisms that control the level of Spo0A~P. Compared to classical two component regulatory systems, the more extended phosphorelay potentially offers enhanced opportunities for signal integration through the use of auxiliary proteins, allowing the timing of production and ultimate amount of Spo0A~P to be precisely regulated. The Rap and Spo0E phosphatase families, which dephosphorylate Spo0F~P and Spo0A~P respectively (Perego et al., 1994, Ohlsen et al., 1994, Perego, 2001, Stephenson and Perego, 2002), are two important classes of auxiliary proteins. The activity of each phosphatase probably responds to specific cues (Jiang et al., 2000, Perego, 2001, Smits et al., 2007). Although to date no regulators of the Spo0E class of phosphatases have been discovered, Rap protein activities are inhibited by cognate Phr peptides, which are often produced late in growth (Perego and Hoch, 1996). Other auxiliary proteins contribute to phosphorelay regulation. KinA, the most important kinase for sporulation when induced by starvation, is regulated by KipI and Sda. KipI, which inhibits KinA auto-phosphorylation, is produced in the presence of sugars, thus presumably inhibiting spore formation in nutrient rich environments, and is further regulated by the anti-anti-kinase KipA (Wang et al., 1997). Sda, produced in response to a replication block, inhibits KinA, thus serving as a DNA replication checkpoint for sporulation (Burkholder et al., 2001). The ClpXP protease degrades Sda, offering yet another potential point of control (Ruvolo et al., 2006). It is likely that additional auxiliary proteins exist that modulate the production of Spo0A~P.
Two genes with unknown function, ymcA and ylbF, were identified in a screen for novel mutants with defects in biofilm formation (Branda et al., 2004). Null mutants of ymcA and ylbF fail to form pellicles at air-liquid interfaces and grow on solid media as smooth, undifferentiated colonies. The ymcA and ylbF mutants are phenotypically indistinguishable, suggesting that their gene products may work in the same pathway, and may even directly interact (Branda et al., 2004). Later, it was discovered that null mutations in sinR, which encodes a master repressor of biofilm formation (Branda et al., 2004, Branda et al., 2006), bypass the defects seen with ymcA and ylbF mutants (Kearns et al., 2005). This suggested that YmcA and YlbF are upstream regulators of SinR. SinR is antagonized by the anti-repressor SinI (Gaur et al., 1986, Bai et al., 1993, Kearns et al., 2005) and sinI transcription is driven by Spo0A~P (Gaur et al., 1988, Shafikhani et al., 2002, Chai et al., 2011). Thus, the expression of biofilm genes is indirectly dependent on Spo0A~P, YlbF and YmcA, with all three acting at least in part via the SinI/SinR pathway. Interestingly, YlbF had been identified in an earlier screen for novel mutants affecting competence development (Tortosa et al., 2000). In ylbF null mutants, both competence and sporulation are severely impaired. Because competence, sporulation and biofilm deficient phenotypes can all be caused by the decreased production of Spo0A~P, YlbF and YmcA may be auxiliary proteins that act positively on the phosphorelay or on its product.
The overall goal of this work was to clarify the mechanism by which YmcA and YlbF function in development. To this end, we used a proteomics approach to identify novel interacting partners of YmcA and YlbF and discovered that both interact with a third protein, YaaT, which had previously been shown to act early during sporulation (Hosoya et al., 2002). YmcA, YlbF and YaaT interact directly with one another, forming a stable ternary complex in vitro. This is consistent with our additional finding that all three proteins are required for competence, sporulation and the formation of biofilms. Analysis with a phosphorelay bypass mutation revealed that the mutational inactivation of ymcA, ylbF or yaaT resulted in a decrease in the amount of Spo0A~P. In vitro experiments with the reconstituted phosphorelay confirmed that the ternary complex accelerates the production of Spo0A~P and bacterial 2-hybrid experiments showed that YmcA interacts with both Spo0F and Spo0B. We propose that the YmcA-YlbF-YaaT complex affects the phosphotransfer between Spo0F and Spo0B, thereby accelerating the production of Spo0A~P.
Results
Identification of YlbF and YmcA binding partners
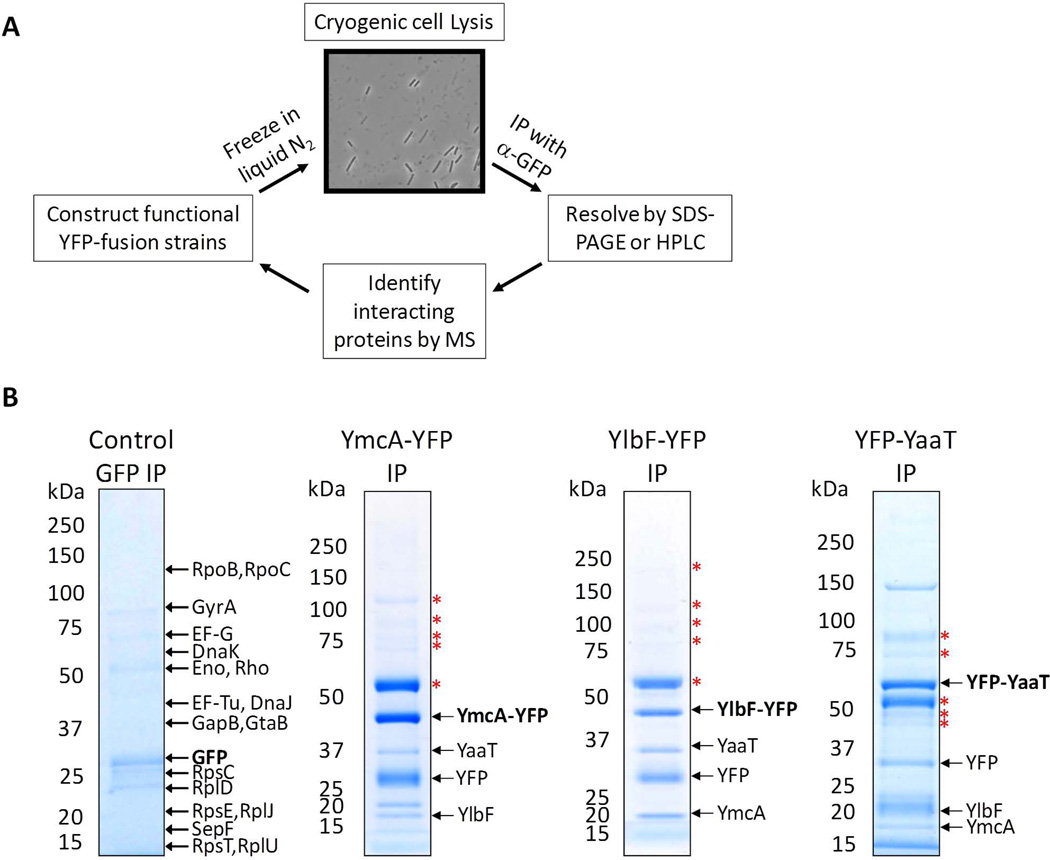
To provide insights into the functions of YlbF and YmcA, we sought to identify their in vivo binding partners using an immunoaffinity purification (IP) strategy as outlined in figure 1A. DNA fragments encoding C-terminal portions of each protein were cloned into the vector pKL184, creating in-frame fusions to the N-terminus of YFP. The resulting plasmids were transformed into BD630 to create fusions of yfp to the C-termini of full length ymcA and ylbF, expressed from their native promoters. Since ylbF null mutants have a defect in comK expression (Tortosa et al., 2000) and ymcA and ylbF null mutants have defects in eps expression (Kearns et al., 2005), we tested the YFP fusion strains for functionality using the lacZ reporter constructs. The YmcA-YFP and YlbF-YFP fusion proteins expressed comK’-lacZ+ and eps’-lacZ+ to the same extents as the wild-type proteins, showing that the fusion proteins were functional (not shown). The strains producing YmcA-YFP or YlbF-YFP were grown in LB, promptly frozen in liquid nitrogen, and the cells were cryogenically disrupted. This technique was previously shown to improve the efficiency of tagged protein isolation, which in turn improves the isolation of interacting partners (Cristea et al., 2005). The number of cycles of cryogenic grinding was optimized, and it was found that after 20 cycles, the majority of cells (>80%) were broken (Fig. 1A). The lysis buffer was optimized for maximum recovery and YmcA-YFP- or YlbF-YFP-containing protein complexes were isolated on magnetic beads coated with purified high-affinity polyclonal anti-GFP antibodies. Proteins were either resolved by SDS-PAGE and digested in-gel, or digested in-solution to help identify lower abundance interacting partners. Peptides were separated by nano liquid chromatography (nLC) and interacting proteins were identified by MS/MS with an LTQ-Orbitrap Velos ETD mass spectrometer. An additional IP was performed, in which GFP was expressed from a xylose-inducible promoter at the amy locus (Fig. 1B, Table S3), to control for the specificity of interactions with the YlbF and YmcA moieties of the fusion proteins. For a summary of the interacting partners detected see Table S3. Two important results emerged from these data. First, a consistently high scoring protein co-isolated with YmcA-YFP was YlbF and conversely, YmcA was reciprocally isolated with YlbF-YFP (Fig. 1B). Because ymcA and ylbF single mutants display phenotypes that are indistinguishable from a double mutant, it had been suggested that they may interact (Branda et al., 2004) and our data present the first experimental evidence for this. Second, in both the YmcA-YFP and YlbF-YFP IPs, another consistently high scoring protein was YaaT (Fig. 1B). YaaT is required for sporulation and its inactivation has been suggested to limit the production of Spo0A~P (Hosoya et al., 2002). The interaction of YaaT with YmcA and YlbF has been independently observed by A. Y. Hsueh and D. Kearns (personal communication) and by DeLoughery and R. Losick (personal communication).
Figure 1.
(A) Proteomics work flow. For each protein of interest, a functional YFP-tagged construct was generated. Cells were frozen rapidly in liquid nitrogen, and lysed by cryogenic grinding. The efficiency of lysis was at least 80%, judged by light microscopy, where lysed cells appear as ghosts, and the dark, opaque cells were those that remained intact. This method allows for more efficient isolation of the tagged protein, and therefore aids in the identification of interacting partners. The YFP-containing protein complexes were isolated on magnetic beads coated with high-affinity anti-GFP antibodies. Proteins were separated by SDS-PAGE, and/or peptides were separated by nano-liquid chromatography (nLC). Co-isolated proteins were identified by tandem MS/MS using an LTQ Orbitrap Velos ETD mass spectrometer. (B) Strains harboring YmcA-YFP (BD5494), YlbF-YFP (BD5402), YFP-YaaT (BD6410), and GFP expressed from Pxly-gfp (BD4319) were grown in DSM media to T2. The gfp construct was induced by the addition of 1% xylose to the media. YFP-containing protein complexes were separated by SDS-PAGE, and bands were visualized by Coomassie blue staining. For the control IP, major contaminants are labeled. The tagged protein is labeled in bold font. The bands labeled with red stars were common to most IP’s, including the control and thus likely represent contaminating proteins.
YmcA, YlbF and YaaT exhibit reciprocal interactions in vivo
From the data described above, it seemed likely that YmcA, YlbF and YaaT interact in vivo perhaps forming a ternary complex. To further investigate this possibility, we generated a functional (not shown) YFP-YaaT fusion at the amyE locus, under Phyperspank control, to avoid interference with the expression of essential genes downstream of yaaT at its native locus (Hosoya et al., 2002). IP of YFP-YaaT revealed that YmcA and YlbF are interacting partners (Fig. 1B, Table S3). The three-way reciprocal interactions detected in these IPs suggested strongly that the proteins are associated in vivo, although any of these associations may be indirect and it remained to be determined if all three proteins exist in a single complex.
YmcA, YlbF and YaaT interact directly and form a stable ternary complex
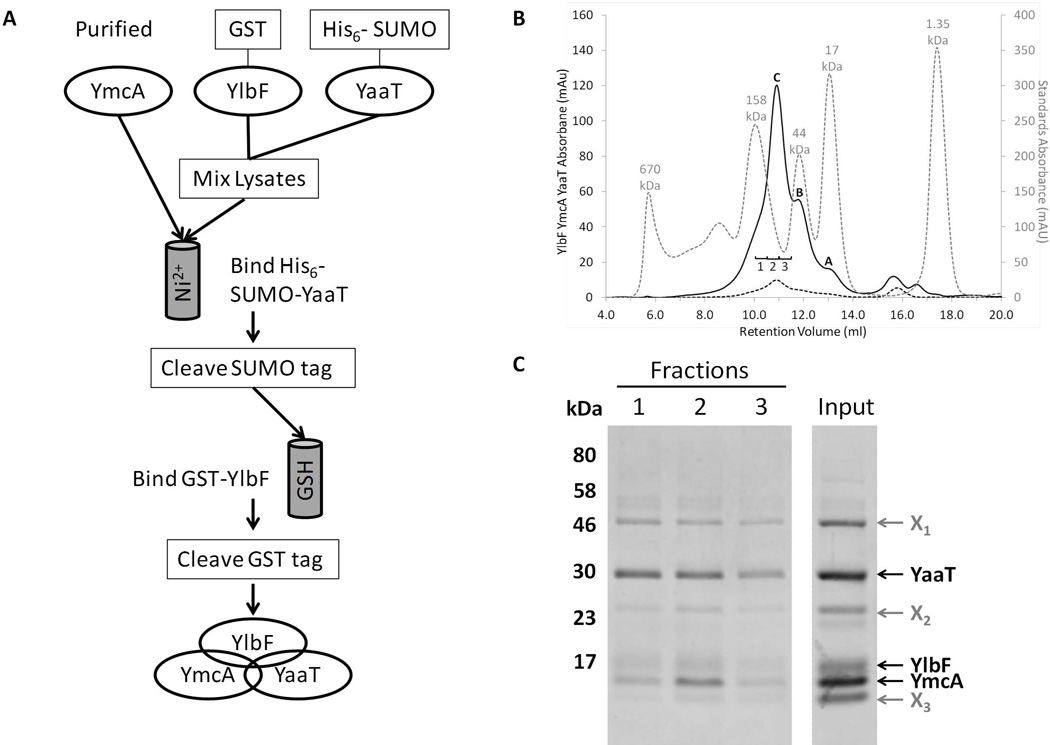
To confirm the direct interactions and the possible formation of a tripartite complex of the three proteins, we first attempted to purify the proteins individually and then combine them to allow in vitro assembly. Although YmcA was soluble, YaaT and particularly YlbF precipitated from solution. We therefore adopted a different strategy (Fig. 2A). A GST-YmcA fusion protein was overproduced in E. coli, purified and its GST tag was removed using PreScission Protease (GE healthcare). Separate cultures expressing His6-SUMO-YaaT and GST-YlbF were centrifuged and their pellets were combined. After cell disruption and centrifugal clarification, the supernatant was combined with the purified YmcA preparation and this mixture was incubated overnight to allow complex formation. This preparation was then loaded onto Ni-NTA resin, permitting binding of the His6-SUMO-YaaT along with its associated proteins. Following elution from the Ni-NTA resin using imidazole, the His6 tag was removed by incubation with SUMO protease. The resulting sample was then loaded onto glutathione (GSH) resin, capturing GST-YlbF and any interacting proteins. The GST tag was then removed using PreScission Protease and the proteins liberated by this cleavage event were recovered from the column flow-through. SDS-PAGE analysis revealed that all three tag-free proteins were present, demonstrating the formation of a ternary complex held together by direct interactions (Fig. 2C, input lane). The fact that the three proteins remained associated throughout this procedure, involving two column steps, provides strong evidence that the mutual interactions are stable. To confirm that the three proteins indeed were part of a complex, and to obtain an approximate molecular weight, the purified protein complex was subjected to gel filtration chromatography followed by SDS-PAGE. The chromatogram (Fig. 2B) showed a significant absorbance at a retention volume of 10.92 ml (Peak C) with two smaller shoulders at 11.81 ml (Peak B) and 13.05 ml (Peak A). YmcA, YlbF and YaaT were all present in the fractions labeled 1, 2 and 3 (Fig. 2B and C) with peak intensity occurring in fraction 2, placing the complex in the center of peak C (Fig. 2B). This corresponds to an estimated size of 80 kDa. The preparation contained three noticeable contaminants (Fig. 2C), two of which have sizes consistent with PreScission protease (X1) and GST (X2). Trace amounts of these two species were commonly observed as contaminants in all of our protein preparations after cleaving GST-tagged proteins. The identity of band X3 is not entirely clear, though its presence seems to be dependent on YlbF. Because a protein of this size is present in other GST-YlbF preparations, it is likely that it is a degradation product of YlbF. Based on deconvolution of our chromatogram into several Gaussian components as described in Experimental procedures, we estimate that the complex comprises about 40% of the total 215 nm-absorbing material in this preparation. Similar results have been obtained in two independent purification experiments. The isolation of the partially pure ternary complex permitted its use in biochemical experiments to be described below.
Figure 2.
YmcA, YlbF and YaaT form a ternary complex. (A) Co-purification scheme of the proteins from E. coli. First, GST-YmcA was expressed, purified, and the GST tag cleaved. Separate cultures of cells expressing GST-YlbF and His6-SUMO-YaaT were mixed and lysed together, and the lysates were incubated with the pure YmcA to allow for complex formation. This mixture was passed over nickel resin to isolate His6-SUMO-YaaT and associated proteins, and after cleavage of the SUMO tag the preparation was passed over glutathione (GSH) resin to isolate the protein complexes containing GST-YlbF. After cleavage of the GST-tag from YlbF, all three untagged proteins were present in the eluate. (B) Co-purified proteins were run on a Superose 12 size exclusion column, as described in Experimental procedures. Depicted are the sample absorbance at 215 nm (A215, solid black line) and absorbance at 280 nm (A280, dashed black line), which has a low intensity due to the presence of a single tryptophan residue in the complex (YaaT). Included for comparison is a mixture of molecular weight sizing standards (A280, dotted grey line). From left to right the standard peaks are: thyroglobulin (670 kDa), γ-globulin (158 kDa), ovalbumin (44 kDa), myoglobin (17 kDa), and vitamin B12 (1.35 kDa). Peaks A and B correspond to contaminating species that are roughly 20 and 45 kDa in size, respectively. Peak C represented the intact YmcA-YlbF-YaaT complex, and corresponds to ~80 kDa in size. (C) Fractions taken from peak C (labeled 1, 2, and 3) were TCA precipitated, resolved by SDS-PAGE on a 12.5% Tris-tricine gel and bands visualized by staining with Coomassie blue. The input lane contained 5 µg of partially purified protein complex. Bands labeled X1-X3 represented major contaminating proteins in the preparation. All samples shown were run on the same gel with irrelevant lanes removed.
YmcA, YlbF and YaaT are required for sporulation, competence and biofilm formation
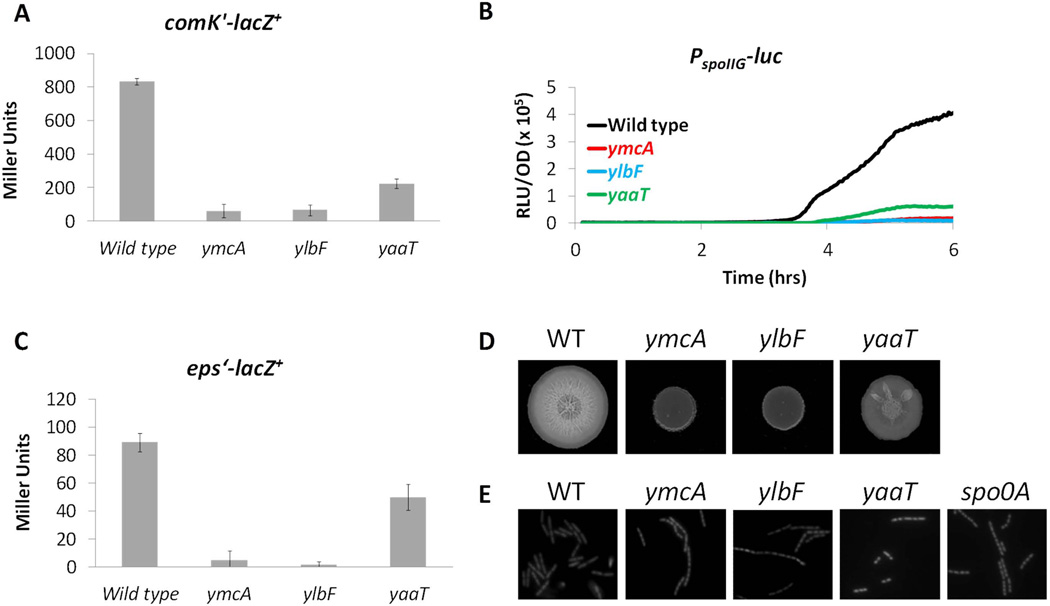
Given the interactions among the three proteins, we thought it likely that they function together. If so, inactivating their genes individually should confer similar phenotypes. It had been shown previously that ylbF mutants are impaired in the expression of competence genes (Tortosa et al., 2000). We wished to determine if ymcA and yaaT were also competence deficient. Null mutations in ymcA, ylbF and yaaT were moved individually into a strain that harbors a lacZ fusion to the promoter of comK and β-galactosidase activity was assayed after growth in competence media to T2 (two hours after the transition to stationary phase, defined as T0). As shown in Fig. 3A, the ylbF mutation caused a 14-fold reduction in β-galactosidase activity in comparison to the wild-type strain, in agreement with previous work (Tortosa et al., 2000). The level measured in the ymcA background was nearly identical to that of the ylbF strain (Fig. 3A), while inactivation of yaaT caused a less severe reduction in β-galactosidase activity. When transformed with leu+ DNA the ymcA and ylbF mutants exhibited 200–250-fold reductions in transformation frequency compared to the wild-type strain (Table 1), while the yaaT mutant again displayed an intermediate phenotype between that of the wild type and those of ymcA and ylbF. These results were consistent with the hypothesis that YmcA and YaaT promote competence development, as shown previously for YlbF.
Figure 3.
YmcA, YlbF and YaaT play regulatory roles in competence (A), sporulation (B), biofilm formation (C + D) and stationary phase chaining (E). (A) Wild type (BD1991), ymcA (BD5410), ylbF (BD5409) and yaaT (BD6411) strains with a comK’-lacZ+ reporter were grown to T2 in CM and β-galactosidase activity was measured. Average determinations of activity from three independent assays are shown together with error bars, which represent standard deviations. (B) Wild type (BD5966, black line), ymcA (BD5669, red line), ylbF (BD5670, blue line) and yaaT (BD5571, green line) strains carrying a PspoIIG-luc construct were grown in LB for 2 hrs, and diluted into fresh DSM in a 96-well plate. After addition of luciferin, OD600 and luminescence were determined at 2 minute intervals during growth. A representative plot of relative luminescence units per OD (RLU/OD) versus time is displayed. (C) Wild type (BD4498), ymcA (BD5563), ylbF (BD5564) and yaaT (BD6412) strains carrying an eps’-lacZ+ reporter were grown in MsGG medium until T2. Samples were taken and β-galactosidase activity was measured. (D) Wild type (WT, NCIB 3610), ymcA (BD5807), ylbF (BD5808) and yaaT (BD5809) were grown in LB liquid media until they reached an OD600 of 1. Five µl of each culture was spotted onto an MsGG plate, and allowed to grow at 30°C. Pictured are colonies after 3 days of growth. (E) Wild type (WT, BD630), ymcA (BD3032), ylbF (BD2741), yaaT (BD5635) and spo0A (BD4576) strains were grown in LB for 6 hours, and then prepared for microscopy as described in Experimental procedures. The preparations were stained with 1 µg/ml propidium iodide to make individual cells easier to distinguish. For all experiments yaaT mutants were grown in the presence of 1 mM IPTG to induce the expression of downstream genes.
Table 1.
ymcA, ylbF, and yaaT mutants have defects in competence, sporulation and cell separation
| Genotype1 | Competence Frequency2 |
Sporulation Frequency |
Chaining Frequency |
|---|---|---|---|
| Wild type | 100% | 68.8% | 34.8% |
| ymcA | 0.4% | 0.006% | 91.7% |
| ylbF | 0.5% | 0.009% | 89.3% |
| yaaT | 0.9% | 0.025% | 48.2% |
| spo0A | 0% | 0% | 82.4% |
Wild type (BD630), ymcA (BD3032), ylbF (BD2741), yaaT (BD5635), spo0A (BD4576).
Percentage of wild-type competence frequency.
ylbF and yaaT mutants were previously shown to have early sporulation defects (Tortosa et al., 2000, Hosoya et al., 2002). To determine if inactivation of ymcA confers a similar deficiency, the three null mutations were transformed into a strain containing the promoter of spoIIG fused to the luciferase reporter gene. The three strains and a wild-type control were grown in sporulation media (DSM) and real-time luciferase assays were carried out. We have shown previously that this assay reflects the rate of transcription rather than gene product accumulation (Mirouze et al., 2011). In the wild-type strain, PspoIIG-luc activity began to increase at 3½ hours, approximately at T0, and continued to increase throughout the experiment (Fig. 3B). However, spoIIG transcription was nearly undetectable in the ymcA and ylbF mutants, consistent with the previously reported effect of ylbF inactivation on spoIIE transcription (Tortosa et al., 2000). The yaaT mutant also exhibited a severe defect in agreement with previous work using a spoIIE reporter (Hosoya et al., 2002), but was less affected than the ymcA and ylbF strains. Consistent with this, the ymcA and ylbF strains were 7,000–12,000-fold deficient in sporulation compared to their wild-type parent while the yaaT mutant was less affected (Table 1). These data confirm that YmcA is an important regulatory protein for sporulation as shown previously for YlbF and YaaT, and that all three act early in this process.
YmcA and YlbF are known to play important positive roles in biofilm formation (Branda et al., 2004), and are likely involved in alleviating SinR-mediated repression of the eps operon (Kearns et al., 2005). To test the role of YaaT in an early step in biofilm formation, three strains carrying a Peps transcriptional fusion to lacZ together with null mutations in either ymcA, ylbF or yaaT were grown to stationary phase in MsGG, a medium that promotes biofilm formation (Branda et al., 2004). In agreement with previous studies (Kearns et al., 2005), the ylbF and ymcA mutations caused a large reduction in eps expression (Fig. 3C). As predicted, the yaaT mutants exhibited a reduction of eps expression, but only about 2-fold compared to the wild-type strain. To further investigate the role of YaaT in biofilm formation, we introduced each of the three mutations into NCIB 3610, a B. subtilis natural isolate that forms more elaborate biofilms than the laboratory strain (Branda et al., 2001). The wild-type NCIB 3610 strain formed a large colony on solid MsGG medium, with an intricate wrinkled pattern clearly visible (Fig. 3D). The ymcA and ylbF mutants formed small, smooth, shiny colonies, in agreement with previous findings (Branda et al., 2004). Once again, the yaaT strain had an intermediate phenotype, with a reduced colony size and less wrinkling compared to the wild-type strain (Fig. 3D). The biofilm phenotype of a yaaT mutant has been observed independently by Y. Hsueh and D. Kearns (personal communication). We conclude that YaaT, like YmcA and YlbF, is required for full biofilm formation. Biofilm formation, sporulation and competence all depend on the production of Spo0A~P and YaaT has been reported to contribute to Spo0A~P formation (Hosoya et al., 2002). These observations suggested that YmcA, YlbF and YaaT work together to increase the levels of Spo0A~P. An accidental observation described in the next section, supported this inference.
ymcA, ylbF, and yaaT mutants form chains in stationary phase
Microscopic examination revealed that the ymcA, ylbF and yaaT strains exhibited excess chaining compared to the wild-type strain, when grown to stationary phase in LB. As seen in figure 3E, the majority of wild-type cells appeared as single cells or as pairs of cells. However, the ymcA and ylbF mutants often appeared as long chains of cells and were noticeably smaller than the wild-type cells. It was determined that ~35% of wild-type cells were in chains of at least 4 cells (Table 1), while about 90% of the ymcA and ylbF mutant cells were present in chains of this length. The mutant chains were typically longer than those seen with the wild-type strain (Fig. 3E). As seen with all the phenotypes discussed thus far, the yaaT mutants displayed an intermediate phenotype, with ~50% of the cells present in chains (Fig. 3E, Table 1). The yaaT mutant chains were a mix of shorter 4-cell chains and longer ones, and again the cell size in general seemed smaller than in the wild type. As expected, the chained cells were non-motile (Chai et al., 2010). In agreement with these data, the ymcA and ylbF mutants could not be transduced by bacteriophage PBS1, which attaches to flagella (Raimundo et al., 1968). yaaT mutants could be transduced, although not as efficiently as wild type (not shown). For all three mutants, these phenotypes were dependent on the medium and the growth stage and excessive chaining was not observed during exponential phase in LB, or in stationary phase in competence or sporulation media.
Because we suspected that the three proteins played a role in the formation or turnover of Spo0A~P we wondered whether a spo0A null mutant might also display a chaining, non-motile phenotype. In fact, spo0A strains behaved nearly identically to the ymcA and ylbF mutants, with ~80% of cells existing in long chains in stationary phase LB cultures (Fig. 3E, Table 1). Although we cannot explain the effect of the spo0A mutation on chaining, this result together with those presented above were consistent with the emerging impression that YmcA, YlbF and YaaT are required for the roles played by Spo0A in a variety of developmental pathways.
The promoters of ymcA and ylbF are regulated by Spo0A
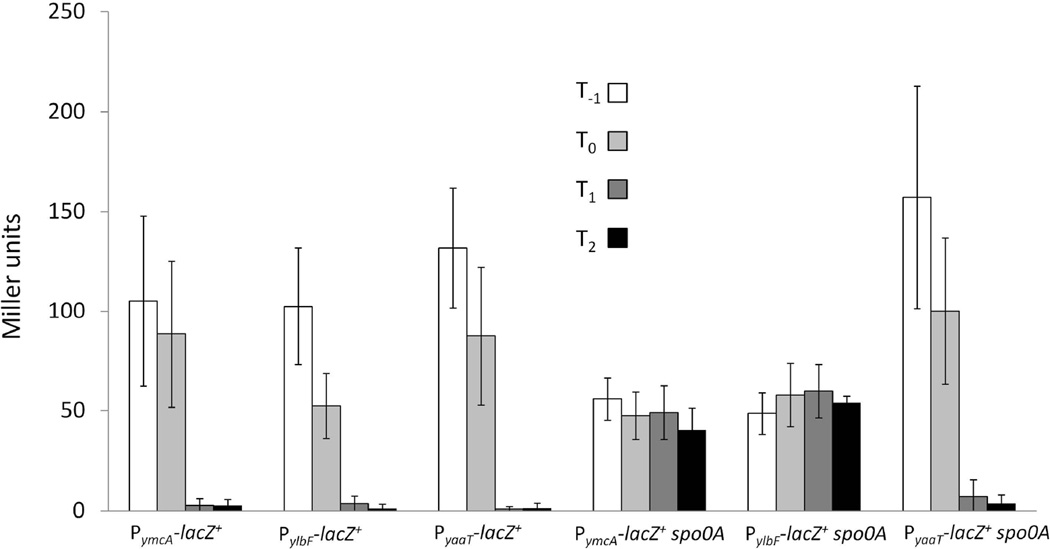
To monitor the expression of the three genes during growth in DSM, we created lacZ transcriptional fusions to each promoter. pMutin4 was used to create the fusions, placing downstream genes under Pspac control. Figure 4 shows that the three promoters are expressed similarly during exponential growth and at the transition to stationary phase. No difference was detected in the expression from PymcA and PylbF in the presence or absence of IPTG (not shown). However, in the case of PyaaT, the expression of downstream genes was required for normal growth, which depended on the addition of the inducer. Expression from all three promoters appeared to turn off as the cells progressed further into stationary phase. Since the stability of LacZ under these conditions is unknown, we cannot definitively conclude that transcription is down-regulated as growth slows, but this is likely to be the case. Since it is known that there is an increase in the amount of Spo0A~P when cells enter stationary phase in DSM, we determined whether the promoters were regulated by spo0A. Inactivation of spo0A had no effect on the yaaT promoter (Fig. 4). However, the ymcA and ylbF promoters displayed constitutive expression that was approximately half of the maximum activity seen in a wild-type background. These data suggest that Spo0A~P directly or indirectly increases expression of ymcA and ylbF during exponential growth and is also responsible for turning off the expression of these genes in stationary phase.
Figure 4.
The promoters of ymcA and ylbF are regulated by Spo0A. The following strains were grown in DSM, and samples were taken at the indicated time points: PymcA-lacZ+ (BD6413), PylbF-lacZ+ (BD6414), PyaaT-lacZ+ (BD5635), PymcA-lacZ+ spo0A (BD6530), PylbF-lacZ+ spo0A (BD6531) and PyaaT-lacZ+ spo0A (BD6532). β-galactosidase activities were determined as described in Experimental procedures. Bars represent an average of 3–4 independent experiments, and error bars represent standard deviations. All strains were grown in the presence of 1 mM IPTG.
sad-67 bypasses the ymcA, ylbF and yaaT spoIIG transcription defects
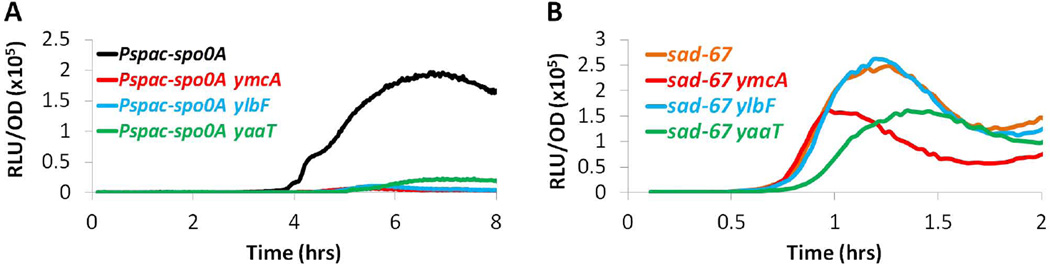
The results presented so far suggest that YmcA, YlbF and YaaT affect either the level of Spo0A protein, or the phosphorylation state of Spo0A in the cell. Western blot analysis revealed that the levels of Spo0A in the ymcA, ylbF, and yaaT mutants were equivalent or slightly increased compared to wild type (Fig. S1), thus ruling out the former possibility. To determine if the phosphorylation of Spo0A was affected, we introduced the sad-67 allele of spo0A into the PspoIIG-luc strains. sad-67 renders Spo0A constitutively active without phosphorylation by introducing a 19 residue deletion in the receiver domain of Spo0A (Ireton et al., 1993). If the phenotypes of the ymcA, ylbF, and yaaT mutants were due to a decrease in Spo0A phosphorylation, the sad-67 allele should bypass these phenotypes. Since the sad-67 mutant gene at the amyE locus is under Pspac control, a Pspac-spo0A construct encoding the wild-type gene was used for comparison. When wild-type spo0A was induced, the ymcA, ylbF, and yaaT mutations caused a severe reduction in luciferase activity, as seen before with spo0A expressed from its native locus (compare Figs. 5A and 3B). This result confirmed that the three mutations do not act by preventing transcription of spo0A, because their effects persisted when spo0A was transcribed from Pspac. In contrast, the spoIIG transcription rate was dramatically increased in the ymcA, ylbF and yaaT strains expressing the sad-67 allele, nearly to the level achieved in the isogenic ymcA+, ylbF+ and yaaT+ strains (Fig. 5B). Because the sad-67 allele is under Pspac control and its product bypasses the phosphorelay, spoIIG expression was induced early in growth in this experiment. The observed lag in expression was likely due to a lag in induction of the IPTG-inducible sad-67 construct. When the wild-type spo0A gene was similarly induced, expression in the ymcA+, ylbF+, and yaaT+ strains increased only as the culture entered stationary phase when the normal signals governing the phosphorelay are in play. These data suggest that YmcA, YlbF and YaaT affect the phosphorylation state of Spo0A. They may do so by accelerating the phosphorelay or by preventing dephosphorylation of a phosphorelay component.
Figure 5.
sad-67 bypasses the spoIIG transcription defects of ymcA, ylbF and yaaT mutants. Luciferase assays for a PspoIIG-luc reporter were carried out as described in Experimental procedures. Strains were grown in the presence of 1 mM IPTG to induce expression of spo0A (A) or sad67 (B) from the Pspac promoter. The graphs are color coded as indicated. (A) Pspac-spo0A (BD6379), Pspac-spo0A ymcA (BD6385), Pspac-spo0A ylbF (BD6386), Pspac-spo0A yaaT (BD6387). (B) sad-67 (BD6380), sad-67 ymcA (BD6381), sad-67 ylbF (BD6382) and sad-67 yaaT (BD6383).
The results presented above, taken together with the phenotypes associated with ymcA, ylbF and yaaT mutants suggested that reduced levels of Spo0A~P are responsible for the observed defects. abrB has a high affinity promoter, which is repressed by a low level of Spo0A~P (Strauch et al., 1990, Fujita et al., 2005). We expected that the effect of our mutations on PabrB would be less than that of a spo0A null mutant. Indeed, we observed an intermediate phenotype for the ylbF strain, when compared to the wild type or spo0A mutant (Fig S2). This result strongly supported the conclusion that the inactivation of ylbF decreases the level of Spo0A~P.
Knockout mutations in spo0E, but not in rapA or kinD bypass the ymcA, ylbF and yaaT defects in spoIIG transcription
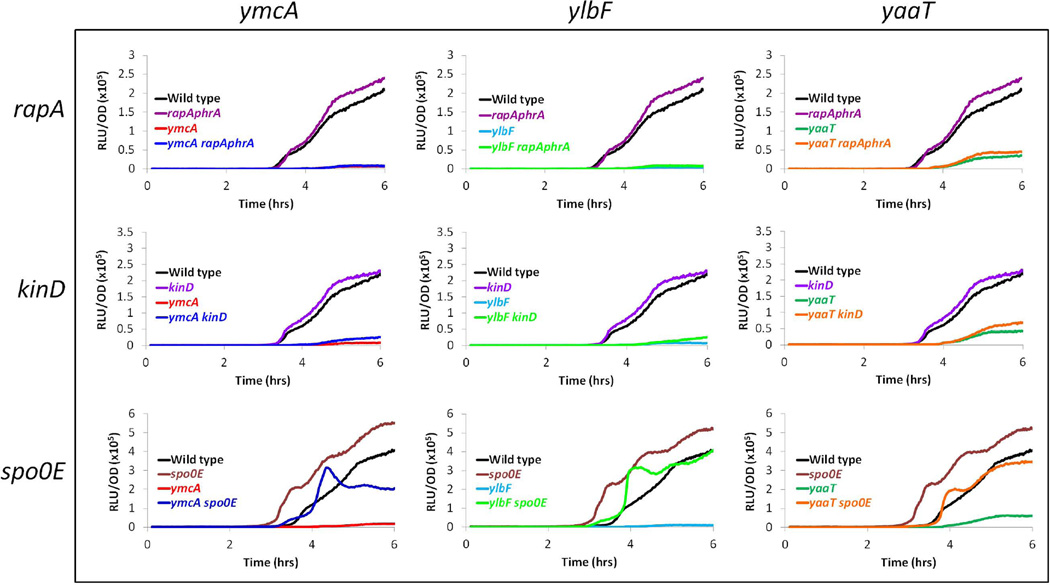
A reasonable explanation for how YmcA, YlbF and YaaT regulate the levels of Spo0A~P is by inhibiting the activity of a phosphatase. This notion is consistent with a report that the mutational inactivation of spo0E bypasses the sporulation defect of a yaaT null mutation (Hosoya et al., 2002) (see below). RapA and Spo0E represent two families of phosphatases that act on phosphorelay proteins (Perego, 1998). As a first test of this possibility, a rapA-phrA null mutation was introduced into PspoIIG-luc strains harboring the ymcA, ylbF and yaaT knockouts. phrA encodes a peptide that down-regulates RapA activity (Perego, 1999). The rapA-phrA mutation did not detectably elevate PspoIIG expression in the mutant strains nor did it affect expression in the wild-type strain (Fig. 6). Although these data are consistent with the conclusion that YmcA, YlbF and YaaT do not work by down regulating the activity of RapA, it is still possible that they regulate other Rap phosphatases, since there are several in B. subtilis.
Figure 6.
Inactivation of spo0E, but not of rapA or kinD bypasses the spoIIG transcription defects of ymcA, ylbF and yaaT mutants. Luciferase assays for expression from the PspoIIG-luc reporter construct were carried out as described in Experimental procedures. The strains carried knockouts of ymcA, ylbF or yaaT, as well as knockout mutations of rapA, kinD or spo0E. All yaaT strains were grown in the presence of 1 mM IPTG. The graphs are color coded as indicated in the figure panels. Strains included are as follows: Wild type (BD5966), ymcA (BD5669), ylbF (BD5670), yaaT (BD5671), rapAphrA (BD6213), ymcA rapAphrA (BD6214), ylbF rapAphrA (BD6215), yaaT rapAphrA (BD6216), kinD (BD6600), ymcA kinD (BD6601), ylbF kinD (BD6602), yaaT kinD (BD6603), spo0E (BD6219), ymcA spo0E (BD6220), ylbF spo0E (BD6221), and yaaT spo0E (BD6222).
It has been reported that the down regulation of KinD by extracellular matrix is needed for sporulation in the context of biofilms (Aguilar et al., 2010). It was postulated that like other histidine kinases, KinD can also act to dephosphorylate response regulators, and that this activity must be inactivated to allow spore formation. To test whether KinD associated phosphatase activity is a target for YmcA, YlbF or YaaT, we used a kinD null mutation with the PspoIIG-luc reporter in an experiment analogous to the one described for rapA-phrA. Only a minor bypass was obtained when kinD was inactivated (Fig. 6). This small increase in luciferase activity probably indicates that KinD associated phosphatase activity limits flux through the phosphorelay to a small extent. Inactivation of kinD thus slightly relieves the effects due to the absence of ymcA, ylbF or yaaT. It does not appear likely that KinD is a major target for these three proteins.
YmcA, YlbF and YaaT could act to promote Spo0A phosphorylation by modulation of Spo0E activity and, as noted above, it has been reported that the mutational inactivation of spo0E bypasses the sporulation defect of a yaaT null mutation (Hosoya et al., 2002). We therefore tested the ability of a spo0E null mutation to bypass the spoIIG transcription defect of the ymcA, ylbF and yaaT knockouts. The PspoIIG-luc activity in the spo0E mutant increased almost an hour earlier than in the spo0E+ strain and increased to a higher extent, suggesting that Spo0E actively limits the amount of Spo0A~P in cells entering the sporulation pathway, as noted previously (Perego and Hoch, 1991). In the ymcA, ylbF and yaaT mutant backgrounds, introduction of the spo0E null mutation restored luciferase activity to near wild-type levels (Fig. 6).Since an spo0E mutation would be expected to increase the amount of Spo0A~P, these data provide further evidence that the absence of ymcA, ylbF or yaaT impedes development by reducing Spo0A~P levels. However, they do not show that the proteins act through inhibition of Spo0E activity. Instead, the inactivation of spo0E might simply compensate for their absence by removing a drain on the Spo0A~P pool. This would be true, for example, if the three proteins acted to increase the flux through the phosphorelay. To test these hypotheses, we determined the effects of the YmcA/YlbF/YaaT ternary complex on the phosphorelay and on Spo0E activity in vitro.
The ternary complex does not regulate Spo0E activity in vitro but does accelerate the phosphorelay
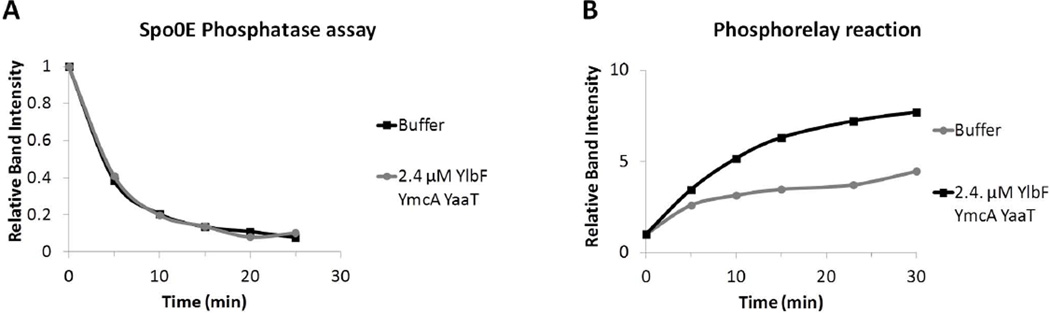
To determine the effect of the YmcA/YlbF/YaaT complex on dephosphorylation of Spo0A~P by Spo0E, the phosphorelay was reconstituted in vitro using purified proteins. The reaction was allowed to run for 60 minutes before Spo0E was added at a final concentration of 50 nM, which was previously determined to result in a rate of dephosphorylation, which would allow the observation of kinetic differences (see Experimental procedures). The time course of the dephosphorylation reaction was then monitored in the presence and absence of added ternary complex. The addition of the ternary complex at a nominal concentration of 2.4 µM did not decrease the rate of dephosphorylation of Spo0A~P, despite its ~50-fold excess over the concentration of added Spo0E (Fig. 7A). In this and subsequent experiments the nominal concentrations of complex were calculated with correction for the 40% purity, determined as described above.
Figure 7.
YmcA-YlbF-YaaT does not regulate Spo0E activity in vitro, but does stimulate the phosphorelay. (A) The phosphorelay was reconstituted in vitro as described in Experimental procedures. The relay was allowed to run for 60 minutes, and then 50 nM of purified Spo0E was added to the reaction in the presence or absence of 2.4 µM complex. Samples were taken at the indicated time points. Because the phosphorelay reactions are reversible, total protein-associated radioactivity was plotted versus time. (B) The phosphorelay proteins were pre-incubated with buffer or with 2.4 µM YmcA-YlbF-YaaT complex to allow for potential interactions. The relay was initiated by the addition of 32P-γ-ATP, and samples were collected at the indicated times. For each experiment, the bands were quantified as described in experimental procedures, and total radioactivity associated with the four phosphorelay proteins is shown. In each panel, a representative graph selected from several replicates is shown.
In an initial attempt to test the complex’s affect on the phosphorelay, we purified each protein separately, and mixed them prior to addition to the phosphorelay. The solubility of YlbF was poor, and to circumvent this issue GST-YlbF was used in place of YlbF, because we have observed enhanced solubility of the fusion protein. Surprisingly, when GST-YlbF was mixed with individually purified YmcA and YaaT, a dramatic inhibition of the phosphorelay was observed (Fig. S3B and C). We suspected that GST-YlbF either alone or after incorporation into a complex inhibited by interaction with one or more phosphorelay components. To test this, we incubated GST-YlbF alone with the phosphorelay, and observed a similar inhibition of the phosphorelay (Fig. S3D). When using purified GST alone, no such inhibition was observed (Fig. S3D). It should be noted that our protein preparations lacked phosphatase or protease contamination (not shown). These data strongly imply that YlbF-GST interacts with one or more of the phosphorelay proteins.
To determine the effect of the ternary complex on the kinetics of the in vitro phosphorelay reaction, we used the partially co-purified preparation, which eliminates the potential complications from the presence of the GST tag. The addition of the complex of YmcA, YlbF and YaaT at a nominal concentration of 2.4 µM increased the rate and yield of Spo0A~P production (not shown) as well as that of the total transfer of phosphoryl groups between KinA, Spo0F, Spo0B, and Spo0A (Fig. 7B). Stimulation of the relay was consistently observed in 4 independent experiments, using two different preparations of the complex. We have observed intermediate rate increases when lower concentrations of the ternary complex were added (Fig. S3A). A mass of bovine serum albumin (BSA) equal to that of the ternary complex was added to control for non-specific effects due to protein addition, and no stimulation of the phosphorelay was observed (not shown). We propose that the in vivo phenotypes associated with the genes encoding the ternary complex proteins are due to this acceleration of the phosphorelay reaction.
YmcA interacts with Spo0B and Spo0F
To detect possible interactions of YmcA, YlbF and YaaT with components of the phosphorelay, the bacterial two-hybrid system (Karimova et al., 1998, Karimova et al., 2000) was used. kinA, spo0F, spo0B, spo0A, ymcA, ylbF and yaaT were cloned as fusions to either the T18 or T25 fragments of adenylate cyclase as described in Supporting material. β-galactosidase activity was used as a readout for reconstitution of adenylate cyclase activity, indicating that the fused proteins had interacted in the Escherichia coli host. β-galactosidase was detected either by inspection of colonies grown in the presence of X-gal (not shown) or by enzyme determination with liquid grown cultures (Table 2). Negative controls using empty T18 and T25 vectors consistently exhibited no blue color on the plates and background levels of β-galactosidase activity in the enzyme determinations. The interaction of both Spo0B and Spo0F with YmcA was ~11-fold higher than the negative control (P < 0.012), and corresponding combinations consistently formed blue colonies on indicator media. The only other strong interaction observed was YmcA with itself. The failure to detect significant interactions of YlbF and YaaT with any of the phosphorelay proteins does not mean they do not interact, because these two proteins may have adopted aberrant conformations in the absence of their binding partners. YmcA, on the other hand forms soluble dimers and may thus present an intact surface for binding to Spo0B and Spo0F.
Table 2.
YmcA interacts with Spo0B, Spo0F, and itself1
| T18-Spo0A | T18-Spo0B | T18-Spo0F | T18-KinA | T18 | T18-YmcA | |
|---|---|---|---|---|---|---|
| YmcA-T25 | 0.83 ± 0.31 | 11.0 ± 3.26 | 11.3 ± 2.79 | 1.06 ± 0.20 | 0.78 ± 0.35 | 48.3 ± 12.2 |
| YlbF-T25 | 1.12 ± 0.27 | 1.57 ± 0.27 | 1.05 ± 0.25 | 0.98 ± 0.19 | 1.16 ± 0.28 | ND |
| YaaT-T25 | 0.77 ± 0.29 | 1.03 ± 0.35 | 0.99 ± 0.56 | 0.83 ± 0.26 | 0.76 ± 0.29 | ND |
| T25 | 1.21 ± 0.18 | 1.19 ± 0.20 | 0.99 ± 0.39 | 1.08 ± 0.49 | 1 | ND |
Pairs of plasmids to be tested by the bacterial 2-hybrid method were co-transformed into the BTH101 reporter strain. Transformants for each pair were grown in liquid media, and assayed for β-galactosidase activity as described in Experimental procedures. Average determinations of the ratio above the T18–T25 negative control are displayed +/− standard deviations. Bold numbers represent positive interactions. ND = Not done.
We performed an ANOVA followed by a Student’s t-test, comparing each combination to the interactions of the protein with the empty vectors T25 and T18 alone or to the T25–T18 pair. The YmcA-Spo0B, YmcA-Spo0F and YmcA-YmcA interactions were all statistically significant, with P-values < 0.012 (n=4), regardless of which negative control was used for comparison.
Discussion
YmcA, YlbF and YaaT form a ternary complex
Using an unbiased proteomics screen (Fig. 1A) we have provided direct evidence that YmcA and YlbF are part of the same protein complex and that they interact directly with one another and with YaaT to form a stable tripartite complex. Using gel filtration, we have found that the molecular weight of the complex is ~80 kDa, consistent with a 2:1:1 stoichiometry with either two YlbF molecules or two YmcA molecules. We suspect that YmcA is present as a dimer in the complex for the following reasons. Quantification of the band intensities from Coomassie blue-stained gels (Fig. 2C) with normalization for protein mass revealed 2.2-fold more YmcA than YlbF or YaaT. Also, YmcA crystallizes as a dimer (Seetharaman et al., 2009), and exhibited strong self-affinity in the bacterial 2-hybrid assay (Table 2).
The YmcA-YlbF-YaaT complex regulates Spo0A-dependent processes
If the three proteins function together in a complex, individual null mutations should show common phenotypes and indeed this appears to be the case, although the yaaT-associated phenotypes are consistently less severe than those caused by inactivation of ylbF or ymcA (Fig. 3 and Table 1). These results confirm and extend published findings (Tortosa et al., 2000, Hosoya et al., 2002, Branda et al., 2004) and reinforce the conclusion that the proteins work on a pathway common to competence, sporulation and biofilm formation, all of which depend on Spo0A~P. The unexpected finding that inactivation of each of the three proteins leads to excessive chaining, predicted that a null mutant of spo0A would do the same. Confirmation of this prediction provided correlative evidence that the proteins act to increase the production of Spo0A~P.
The ternary complex accelerates the phosphorelay
In vivo suppressor analysis with sad-67 (Ireton et al., 1993), as well as in vitro studies with the reconstituted phosphorelay, has demonstrated that the ternary complex accelerates this pathway. These findings, and our observation that the complex does not inhibit purified Spo0E in vitro, argue against a published suggestion that YaaT down regulates the Spo0E phosphatase (Hosoya et al., 2002). Our conclusion that the three proteins positively affect the in vivo level of Spo0A~P is supported by the result of Hosoya et al., who reported that the sof-1 allele of spo0A completely bypasses the sporulation defect of a yaaT mutant (Hosoya et al., 2002). The sof-1 mutation permits the direct phosphorylation of Spo0A by KinC, bypassing the requirement for Spo0F and Spo0B (Kobayashi et al., 1995). Bypass by sof-1 suggests that the phosphorelay itself is defective in the yaaT mutant and presumably in all three mutant backgrounds, consistent with the present results.
What is the mechanism of action of the ternary complex? In its presence, the phosphorelay was reproducibly stimulated in vitro, manifested as an increase in the rate of formation of total phosphorylated protein (Fig. 7). Radiolabeled 32P is expected to become distributed among the various species of phosphorylated proteins, because the phosphorelay steps are reversible (Burbulys et al., 1991). The shape of this distribution is partly a function of the arbitrary amounts of the proteins we have used in the experiment. The ternary complex apparently establishes a new equilibrium among the components by increasing the forward equilibrium constant at one or more steps along the way. Either the complex enhances the rate of the forward reaction at some step, or it inhibits the reverse reaction, thereby driving the overall reaction.
The largest effect on phosphorylation, consistently observed in the presence of the protein complex, was on the formation of Spo0B~P (not shown). This observation suggests that the complex may enhance a step prior to phosphotransfer between Spo0B and Spo0A. YmcA was shown to interact with Spo0B and Spo0F by bacterial 2-hybrid analysis (Table 2). Although our immunopurifications failed to identify interactions with phosphorelay proteins (Table S3), a transient interaction in vivo could easily have been missed. It should be noted that Spo0B has been co-isolated with YaaT-His6 protein (Y. Hsueh and D. Kearns, personal communication). YmcA readily dimerizes (Table 2), and forms a saddle-like structure (Seetharaman et al., 2009) with a large interaction surface potentially available for binding other proteins. Perhaps such a surface on YmcA serves as a docking site for both Spo0B and Spo0F, permitting a more efficient phosphotransfer reaction. In vivo, the levels of Spo0B are at least 10 times lower than those of KinA, Spo0F or Spo0A (Eswaramoorthy et al., 2010), potentially limiting the rate of the allover forward reaction. By bringing Spo0B and Spo0F into close proximity, the complex might serve as a catalyst for the forward reaction, leading to an increased production of Spo0A~P and representing an important point of control. In fact, ever since the phosphorelay was discovered it has been suggested that the addition of Spo0B to the two-component paradigm may provide another control point for Spo0A~P formation (Burbulys et al., 1991). The direct interaction of YmcA with Spo0B may provide the first evidence for such regulation. Clearly the exact mechanism of action and the dynamics of the system will require a more rigorous characterization and will be the subject of future studies, guided by the above speculations.
Spo0A~P regulates the ymcA and ylbF promoters
Feedback loops are common regulatory elements. Although ymcA, ylbF and yaaT are not linked on the chromosome they are transcribed at similar levels during growth and the expression of all three genes is turned off as the culture enters stationary phase (Fig. 4). This coordinated transcription pattern is consistent with the function of the three gene products in a ternary complex. It is also consistent with a requirement for acceleration of the phosphorelay early in sporulation and with a need to avoid excess Spo0A~P production as sporulation proceeds. The downturn in ymcA and ylbF transcription is dependent on spo0A, suggesting the existence of a negative feedback loop operating on these two genes. This is not true of yaaT, which is embedded in a large, multi-gene operon together with genes involved in DNA replication. Also, ymcA and ylbF were constitutively expressed at a decreased level in the spo0A mutant background (Fig. 4), suggesting the existence of a complex regulatory mechanism.
An important outstanding question concerns the possibility that the ternary complex mediates a signal transduction pathway that relays information concerning environmental, metabolic or cell cycle signals to the phosphorelay. Such a pathway may be further modulated by the negative feedback mechanism in which Spo0A~P acts on the promoters of ylbF and ymcA. Different levels of Spo0A~P are required for sporulation, competence and biofilm formation and signaling through the YlbF-YmcA-YaaT ternary complex may participate in decision making among these cell fates.
Experimental procedures
Bacterial Strains, Plasmids, Media and Growth conditions
Bacillus subtilis strains are listed in Table S1. Strain construction was carried out by transformation with selection for appropriate antibiotics (Albano et al., 1987) or by transduction using bacteriophage PBS1 (Takahashi, 1963). Antibiotic concentrations used were as follows: 5 µg/ml chloramphenicol (cm), 5 µg/ml erythromycin (ery), 5 µg/ml kanamycin (kan), 100 µg/ml spectinomycin (spc), and 25 µg/ml tetracycline (tet). Liquid and agar Luria broth (LB), and MsGG media (Kearns et al, 2005), liquid minimal competence media (Albano et al., 1987) and Schaeffer’s sporulation media (DSM) (Schaeffer et al., 1965) were prepared as described previously, with the addition of 50 µg/ml histidine, leucine and methionine to the MsGG and competence media. Bacteria were grown at 37°C with aeration, and growth was monitored either in a Klett colorimeter or in a spectrophotometer by measurement of OD600. Construction of plasmids is described in the Supporting information. E. coli DH5α (Invitrogen) and Stellar (Clontech) competent cells were used for cloning. Plasmids were selected for and maintained in E. coli with the addition of 100 µg/ml ampicillin (amp).
Mass spectrometry
Following is a brief description of the YFP immunopurification-mass spectrometry procedure. For a more detailed description see Supporting information. B. subtilis strains expressing YFP fusions were grown to T1 (one hour after the onset of stationary phase) in DSM, harvested, frozen as small pellets in liquid nitrogen, and subjected to cryogenic cell lysis as described previously (Cristea et al., 2005, Carabetta et al., 2010). The resulting frozen cell powder was resuspended in lysis buffer (20 mM HEPES, pH 7.4, 100 mM potassium acetate, 2 mM MgCl2, 0.1% tween-20 (v/v), 1 µM ZnCl2, 1 µM CaCl2, 0.2% Triton-X, 150 mM NaCl, 10 µg/µl DNaseI, 1:100 protease inhibitor cocktail (Sigma) and 0.1 mg/ml phenylmethylsulphonyl fluoride (PMSF)) and centrifuged. The soluble fraction was mixed with M270 Epoxy Dynabeads (Dynal, Invitrogen) coated with polyclonal anti-GFP antibody, for 1 hour at 4°C. The magnetic beads were recovered, washed and proteins were eluted directly into lithium dodecyl sulfate (LDS, Invitrogen)-PAGE sample buffer if in-gel digestion was to be performed, or into TEL buffer (26 mM Tris-HCl, 35 mM Tris-base, 127 µM EDTA, 0.5% LDS, pH 8.5) for in-solution digestion. All samples were prepared for mass spectrometry analyses as described (Tsai et al., 2012, Greco et al., 2012). Briefly, samples were alkylated with 100 mM iodoacetamide for 30 minutes. Isolated protein complexes were either resolved on a 4–12% NuPAGE Novex Bis-Tris gel (Invitrogen), and digested overnight with 12.5 ng/µl trypsin in-gel, or digested on a filter (Vivacon 500 centrifugal filters (10K cutoff), Sartorius Stedim Biotech, Goettingen, Germany) with 100 µl of 5 ng/µl trypsin by a filter-aided sample preparation method (FASP) as described (Wisniewski et al., 2009). The extracted peptides were pooled and concentrated by vacuum centrifugation to ~12 µl. Half of the elution sample was analyzed by nLC-MS/MS on a Dionex Ultimate 3000 RSLC coupled directly to an LTQ-Orbitrap Velos ETD mass spectrometer (ThermoFisher Scientific, San Jose, CA) as described (Tsai et al., 2012). The MS/MS spectra were acquired by collision induced dissociation (CID) fragmentation, extracted by Proteome Discoverer (ver. 1.3, Thermo Fisher Scientific, San Jose, CA) and then further analyzed by SEQUEST (ver. 1.3.0.339, Thermo Fisher Scientific, San Jose, CA) for peptide database searching against the B. subtilis database, including some common contaminant sequences (4374 entries). Peptide spectrum matches (PSMs) were analyzed and validated by Scaffold (ver. 3.2; Proteome Software, Inc.), as described previously (Tsai et al., 2012).
Microscopy and quantification of chaining
Cells were grown for 6 hrs in 5 ml of LB plus appropriate antibiotics. Then the cells were harvested, resuspended in minimal media, and were allowed to attach to poly-l-lysine-coated slides without fixation (Harry et al., 1995). The samples were stained with propidium iodide (1 mg/ml) for less than 2 min, and were mounted in Slow Fade (Molecular Probes). Microscopy was performed with an upright Nikon Eclipse 90i microscope outfitted with an Orca-ER Digital Camera (Hamamatsu), and a Nikon TIRF 1.45 NA Plan Neo-Fluor 100× oil immersion objective. Semrock Optical filter sets were used for fluorescence detection. The Volocity software package (Version 6.0.1, Improvision) was used for image acquisition and cell counting. Four or more cells attached together were considered a chain. Those cells not present in a chain were either single cells, or two cells that had not separated. Percentage of chaining was determined by dividing the number of cells in chains by the total number of cells in the field. At least 500 cells were counted for each determination, and results were observed more than once.
Sporulation, competence, and biofilm assays
For sporulation assays, strains were streaked on LB plus appropriate antibiotics at 37°C. Single colonies were inoculated into 10 ml of freshly prepared DSM media, and grown for 24 hrs with shaking, at 37°C. Cells were serially diluted, plated for viable counts, and the dilutions were incubated at 80°C for 30 minutes, then plated again onto LB plates and incubated for 16 hrs at 37°C. After colony counting, the percentage of sporulation was calculated as heat-resistant colony forming units divided by the total colony forming units. B. subtilis cells were transformed as described previously (Albano et al., 1987). For the calculation of competence frequency, leu auxotrophs were transformed with genomic DNA prepared from BD170 (trp thr) and plated on minimal media lacking leucine. Transformation frequency was calculated as the number of leu+ colony forming units/ml divided by the total colony forming units/ml, and all values were normalized to the wild-type frequency. For biofilm formation, cells were grown to an OD600 of 1, in LB media, and 5 µl of the suspension was spotted on an MsGG plate. Plates were incubated at 30°C for 2–5 days, with visual observation every day. All experiments were done in triplicate.
β-galactosidase and luciferase assays
β-galactosidase assays were carried out essentially as described previously (Gryczan et al., 1984). Cultures were grown to T2 in either competence or MsGG media, OD600 was measured and 500 µl of each culture was pelleted and frozen at −20°C. Samples were resuspended in an equal volume of buffer Z (0.1M NaPO4, pH 7, 1mM MgSO4, 0.1M β-mercaptoethanol), vortexed for 30 sec with 10 µl of toluene, and incubated on ice for at least 1 hr. For the comK’-lacZ+ and eps’-lacZ+ fusions, 100 µl and 300 µl of toluene-treated lysate was used, respectively. Reactions were initiated by addition of 200 µl of O-nitrophenyl-β-D-galactoside (ONPG, 4 mg/ml in buffer Z) followed by incubation at 30°C. The reactions were stopped by the addition of 500 µl of 1M Na2CO3 and the elapsed time was noted. Miller units were calculated as follows: 1000 * (A420-(1.75 * A550)/(t*v*OD600)), where t is the time (min) and v is the volume (ml) of lysate used. Every experiment was repeated three times.
Luciferase assays were carried out as described previously (Mirouze et al., 2011). Briefly, strains were grown in LB for 2 hrs, 1 ml of culture was harvested and cells were resuspended in freshly supplemented DSM to an OD600 of 2. These suspensions were further diluted 20-fold into DSM, and 200 µl of each culture was added to the well of a 96-well black plate (Corning), in duplicate. 10 µl of luciferin, final concentration 1.5 mg/ml (4.7 mM), was added to each well and plates were incubated at 37°C with agitation in a PerkinElmer Envision 2104 Multilabel Reader equipped with an enhanced sensitivity photomultiplier for luminometry. The plate lids were heated to 38°C to prevent condensation. Relative luminescence units (RLU) and OD600 were measured at 1.5 min intervals.
Bacterial two hybrid experiments
Each target gene was cloned in frame with either the N- (T18) or C-terminal (T25) portions of the cyaA gene. Details of the construction of all plasmids can be found in the Supplemental material. Pairs of interacting proteins to be tested were co-transformed into BHT101, a cya−, lacZ reporter E. coli strain. Following co-transformation, the cultures were allowed to recover for 1h at 37°C with aeration. Dilutions were plated on LB agar containing 100 µg/ml of ampicillin and 25 µg/ml of kanamycin for plasmid selection and grown for 2 days at 25°C. In addition, for each culture, 10 µl was spotted onto selective minimal agar plates containing 100 µg/ml of ampicillin, 25 µg/ml of kanamycin, 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and 0.008% X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside). Colony color was observed daily for 3 days following growth at 30°C. β-galactosidase assays were carried out as previously described (Georgiadou et al., 2012), with modifications. From each co-transformation plate, 2 colonies were inoculated into LB media supplemented with 100 µg/ml of ampicillin, 25 µg/ml of kanamycin, and 0.5 mM IPTG, and grown for 16 hrs at 30°C. The OD600 was recorded, and 1 ml of cells harvested from each culture. Pellets were resuspended in buffer Z (0.1M NaPO4, pH 7, 1mM MgSO4, 0.1M β-mercaptoethanol), 16 µl of chloroform and 16 µl of 0.1% SDS added, and vortexed for 30 seconds. The suspensions were incubated at 37°C for 40 minutes with shaking. For each reaction, 100 µl of permeablized cells was used, and the reaction was carried out as described above for β-galactosidase determination. Statistical significance of each interacting pair was determined using Stata (version 10.1) to perform a Student’s t-test after analysis of variance (ANOVA) applying the Bonferroni correction, as described (Robakiewicz and Ryder, 2000).
Purification of the ternary complex of YmcA, YlbF and YaaT
For expression of His6-SUMO-YaaT, the plasmid pTB146-yaaT (ED1450) was transformed into the T7 polymerase expressing strain, BL21 (DE3). Single colonies of the DH5α strains harboring either the pGEX6P-1-ymcA (ED1333) or the pGEX6P-1-ylbF (ED1334) expression vectors and of the BL21(DE3) strain harboring pTB146-yaaT were each inoculated into 100 ml of LB plus ampicillin and grown overnight at 37°C. These cultures were used to inoculate 4L of fresh LB plus ampicillin (1:50 dilution) for the GST-YmcA producing strains, and 2L for the GST-YlbF and His6-SUMO-YaaT producing strains and allowed to grow at 37°C until they reached an OD600 of 0.6 (~3 hours). After the initial growth at 37°C of the pGEX6P-1-ymcA or pGEX6P-1-ylbF constructs, protein expression was induced with 1 mM IPTG for 2 hours. The strain harboring pTB146-yaaT was induced with 0.5 mM IPTG, moved to 30°C and grown for 5 hours. Cultures were centrifuged in a Beckman Coulter centrifuge (Avanti J-25, rotor JLA 8.1000) at 4500 × g for 15 minutes and the pellets were stored at −80°C.
The frozen pellet of the GST-YmcA-expressing strain was thawed on ice, and resuspended in 20 ml of ice-cold lysis buffer (50 mM Na2HPO4, pH 7.5, 300 mM NaCl, 1:100 protease inhibitor cocktail (Roche Diagnostics) and 0.1 mg/ml PMSF). Cells were lysed by 4–5 passages through an Avestin EmulsiFlex-C5 cell disrupter at an operating pressure of 12,000 psi (Neidtich and Hughson, 2007). The cell lysate was centrifuged (rotor JA 25.50) at 14,000 × g for 1 hr at 4°C. The supernatant was separated from the insoluble pellet, and mixed with 3 ml of pre-equilibrated glutathione-Super flow resin (Clontech) for 1 hr at 4°C, with gentle rotation. The sample was loaded on a column (Biorad), the flow-through was collected, and the column was washed with 20 column volumes of lysis buffer minus the protease inhibitors. To elute free YmcA, the resin was removed from the column and a 50% slurry of resin and cleavage buffer (lysis buffer plus 1 mM dithiothreitol (DTT)) was mixed with 600 µl of a 1:10 dilution of the PreScission Protease (GE healthcare) and incubated overnight at 4°C. The digestion reaction was loaded on a column and the cleaved YmcA was recovered in the flow-through fraction, largely free of protease. The purity of the YmcA protein was estimated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining and the preparation was judged to be about 90% pure.
The pellets of cells expressing GST-YlbF and His-SUMO-YaaT were thawed and mixed together in 80 ml of lysis buffer (50 mM Na2HPO4,pH 7.5, 300 mM NaCl, 10 mM imidazole, 5 mM DTT, 1:100 protease inhibitor cocktail, 0.1 mg/ml PMSF). The cells were lysed, and the supernatant was collected as described above. The total preparation of purified YmcA (~2.8 mg) was added to the supernatant, which was then incubated overnight at 4°C, with gentle rotation, to allow for complex formation. This preparation was incubated for 1 hour at 4°C with gentle rotation with 1 ml of Ni-NTA Superflow resin (Qiagen) that had been equilibrated with 50 mM Na2HPO4, pH 7.5, 300 mM NaCl, 10 mM imidazole, 5 mM DTT. The resin was loaded onto a column (Biorad), and washed with 20 column volumes of lysis buffer, minus protease inhibitors. Twenty fractions were eluted step-wise by increasing concentrations of imidazole, ranging from 10 mM to 1 M. The fractions were analyzed by SDS-PAGE and the presence of His6-SUMO-YaaT, GST-YlbF and YmcA was confirmed in the fractions eluted with 30–90 mM imidazole. Peak fractions were pooled, concentrated and the imidazole was removed by overnight dialysis at 4°C against 50 mM Na2HPO4, pH 7.5, 300 mM NaCl. For cleavage of the SUMO tag, a final concentration of 0.2% NP-40, and 1:20 (w/w) SUMO protease (Invitrogen) were added, followed by digestion overnight at 4°C, with gentle rotation. After digestion, the reaction was mixed with 2 ml glutathione resin pre-equilibrated with 50 mM Na2HPO4 pH 7.5, 300 mM NaCl to bind the GST-YlbF, incubated for 1 hour at 4°C with rotation and loaded onto a column. The flow-through was discarded and the resin was washed with 20 volumes of lysis buffer. The GST-YlbF was cleaved as described for the GST-YmcA. Following digestion, the column flow-through was concentrated and analyzed by SDS-PAGE to determine purity. During protein concentration a filter with a 30K molecular weight cutoff was used to remove excess GST, and potentially uncomplexed YmcA, YlbF and YaaT. The resulting complex was estimated to be ~40% pure (see Gel filtration procedure). Assuming a 2:1:1 (YmcA:YlbF:YaaT) ratio (see Discussion) of the three proteins the concentration of this complex was estimated to be ~5 µM.
Purification of the phosphorelay proteins and of Spo0E
The phosphorelay proteins Spo0A (PP494, Muchová et al., 2004), Spo0F (ED1428), Spo0B (ED1427) and KinA (ED1444) (Fujita and Losick, 2003), were expressed and purified as described previously. GST alone was purified from the vector pQLinkG2 (Scheich et al., 2007). The phosphatase GST-Spo0E (ED1602) was purified in the manner described previously for GST-YmcA, with the following modifications. After growth at 37°C, protein expression was induced with 0.5 mM IPTG, for 3 hours at 30°C. The lysis buffer used was 25 mM Tris, pH 8.5, 500 mM KCl, 1 mM IPTG, 1:100 protease inhibitor cocktail, 0.1 mg/ml PMSF. The resulting protein preparation was analyzed by SDS-PAGE for determination of purity, and was >90% pure.
In vitro phosphorelay reaction
The phosphorelay reaction was carried out as described previously (Seredick et al., 2009). The concentrations of components used were as follows: 0.75 µM KinA, 0.1 µM Spo0B, 0.5 µM Spo0F, 0.5 µM Spo0A in phosphorylation buffer (10 mM HEPES, pH 8.0, 25 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT, 0.1 mg/ml bovine serum albumin (BSA)). The complex of YmcA, YlbF, and YaaT was allowed to pre-incubate with the phosphorelay on ice for 1 hour to allow for potential interactions with the relay components. The reaction was started by the addition of 25 µM ATP + 25 µCi of [γ-32P] ATP [6,000 Ci/mM] (MP Biosciences). The ternary complex was added at a final concentration of 2.4 µM, unless noted otherwise. As a control, an equivalent mass of BSA or volume of buffer was added to the phosphorelay reaction and run in parallel. The reaction was incubated at 23°C for 30 minutes, and at the indicated time points, samples were withdrawn and added to an equal volume of 2× SDS-PAGE sample buffer (100 mM Tris-HCl, pH 6.8, 4% SDS, 150 mM β-mercaptoethanol, 0.2% bromphenol blue and 20% glycerol). Samples were resolved on a 15% gel by SDS-PAGE, radiolabeled bands were identified using a phosphoimager (Typhoon 9410, variable mode imager, Molecular Dynamics) and band intensities were quantified using ImageQuant, version 5.2.
Phosphatase reactions
For assay of Spo0E phosphatase activity, the phosphorelay reaction was allowed to run for 1 hour, to completion. The concentration of Spo0E needed was optimized, and it was determined that 50 nM was sufficient. The co-purified complex of YmcA, YlbF, and YaaT was pre-incubated with Spo0E on ice for 1 hour, and this mixture was added directly to the phosphorelay reaction. An equal volume of buffer was added as a control to a separate reaction. Time points were taken every 5 minutes for 25 minutes, samples were added to equal volumes of 2× SDS sample buffer and then analyzed by SDS-PAGE, as described above.
Size-exclusion chromatography
A 40 µl sample of partially purified YmcA-YlbF-YaaT complex was applied to a Superose 12 FPLC 10/30 (Pharmacia) gel filtration column, which was previously equilibrated with 50 mM Na2HPO4, pH 7.5, 300 mM NaCl. A series of 0.5 ml fractions were collected. The void volume was determined using 2 mg/ml dextran blue (2,000 kDa). Molecular weight estimations were interpolated using gel filtration standards (Bio Rad), graphing on a log-linear plot. Sample purity was estimated by deconvolving the central 10.92 ml peak into a subset of assumed Gaussian curves using a peak fitting algorithm for the MATLAB software package (http://terpconnect.umd.edu/~toh/spectrum/InteractivePeakFitter.htm).
Proteins were precipitated from peak fractions to analyze their content by SDS-PAGE. Fractions of interest were incubated with 0.02% sodium deoxycholate for 30 minutes on ice followed by an overnight incubation with 10% trichloroacetic acid (TCA) at 4°C. Precipitated proteins were collected by centrifugation at 4°C for 15 minutes at 15,000 × g. Pellets were washed with 100% ice cold acetone and dried. All samples were resuspended in an equal volume of SDS loading buffer and resolved on a 12.5% Tris-tricine gel. Gels were stained with Coomassie blue and imaged using the 700 nm channel of the Odyssey CLx Infrared Imaging System (LI-COR). All band intensities were quantified using Image Studio, version 2.0.
Supplementary Material
Acknowledgments
We thank all the members of our lab and those from M. Neiditch’s group for frequent useful discussions and advice. We thank T. Li and A. Guise for technical assistance and advice. We thank D. Kearns for providing us with the NCIB 3610 strain and M. Fujita for providing over-producing strains for purification of the phosphorelay proteins, as well as Spo0A anti-sera. We thank K. Lemon and A. Grossman for providing the pKL184 plasmid. We also wish to express our appreciation to the late Alex Neyfakh for informing us of his results suggesting that YlbF and YmcA work together. This work was supported by NIH grant GM057720 awarded to DD, by a UMDNJ Foundation grant awarded to AT and NIH/NIDA grant DP1DA026192 and HFSPO award RGY0079/2009-C to IMC.
References
- Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio. 2010;1 doi: 10.1128/mBio.00035-10. e00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albano M, Hahn J, Dubnau D. Expression of competence genes in Bacillus subtilis. J Bacteriol. 1987;169:3110–3117. doi: 10.1128/jb.169.7.3110-3117.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniewski C, Savelli B, Stragier P. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J Bacteriol. 1990;172:86–93. doi: 10.1128/jb.172.1.86-93.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai U, Mandic-Mulec I, Smith I. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein–protein interaction. Genes Dev. 1993;7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda SS, Gonźalez-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004;186:3970–3979. doi: 10.1128/JB.186.12.3970-3979.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buelow DR, Raivio TL. Three (and more) component regulatory systems-auxiliary regulators of bacterial histidine kinases. Mol Microbiol. 2009;75:547–566. doi: 10.1111/j.1365-2958.2009.06982.x. [DOI] [PubMed] [Google Scholar]
- Burbulys D, Trach KA, Hoch JA. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Burkholder WF, Kurtser I, Grossman AD. Replication initiation proteins regulate a developmental checkpoint in Bacillus subtilis. Cell. 2001;104:269–279. doi: 10.1016/s0092-8674(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Carabetta VJ, Silhavy TJ, Cristea IM. The response regulator SprE (RssB) is required for maintaining poly(A) polymerase I-degradosome association during stationary phase. J. Bacteriol. 2010;192:3713–3721. doi: 10.1128/JB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. An epigenetic switch governing daughter cell separation in Bacillus subtilis. Genes Dev. 2010;24:754–765. doi: 10.1101/gad.1915010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea IM, Williams R, Chait BT, Rout MP. Fluorescent proteins as proteomic probes. Mol Cell Proteomics. 2005;4:1933–1941. doi: 10.1074/mcp.M500227-MCP200. [DOI] [PubMed] [Google Scholar]
- Dubnau D, Losick R. Bistability in bacteria. Mol Microbiol. 2006;61:564–572. doi: 10.1111/j.1365-2958.2006.05249.x. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy P, Dinh J, Duan D, Igoshin OA, Fujita M. Single-cell measurement of the levels and distributions of the phosphorelay components in a population of sporulating Bacillus subtilis cells. Microbiology. 2010;156:2294–2304. doi: 10.1099/mic.0.038497-0. [DOI] [PubMed] [Google Scholar]
- Fabret C, Feher VA, Hoch JA. Two component signal transduction in Bacillus subtilis: how one organism sees its world. J Bacteriol. 1999;181:1975–1983. doi: 10.1128/jb.181.7.1975-1983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. The master regulator for entry into sporulation in Bacillus subtilis becomes a cell-specific transcription factor after asymmetric division. Genes Dev. 2003;17:1166–1174. doi: 10.1101/gad.1078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, González-Pastor JE, Losick R. High- and low-threshold genes in the Spo0A regulon of Bacillus subtilis. J. Bacteriol. 2005;187:1357–1368. doi: 10.1128/JB.187.4.1357-1368.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005;19:2236–2244. doi: 10.1101/gad.1335705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur NK, Dubnau E, Smith I. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J Bacteriol. 1986;168:860–869. doi: 10.1128/jb.168.2.860-869.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur NK, Cabane K, Smith I. Structure and expression of the Bacillus subtilis sin operon. J Bacteriol. 1988;170:1046–1053. doi: 10.1128/jb.170.3.1046-1053.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiadou M, Castagnini M, Karimova G, Ladant D, Pelicic V. Large-scale study of the interactions between proteins involved in type IV pilus biology in Neisseria meningitidis: characterization of a subcomplex involved in pilus assembly. Mol Microbiol. 2012;84:857–873. doi: 10.1111/j.1365-2958.2012.08062.x. [DOI] [PubMed] [Google Scholar]
- Greco TM, Miteva Y, Conlon FL, Cristea IM. Complementary proteomic analysis of protein complexes. Methods Mol Biol. 2012;917:391–407. doi: 10.1007/978-1-61779-992-1_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan TJ, Israell-Reehes M, Dubnau D. Induction of macrolide-lincosamide streptogramin B resistance requires ribosomes able to bind inducer. Mol Gen Genet. 1984;194:357–361. doi: 10.1007/BF00425544. [DOI] [PubMed] [Google Scholar]
- Harry EJ, Pogliano K, Losick R. Use of immunofluorescence to visualize cell-specific gene expression during sporulation in Bacillus subtilis. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch JA. spo0 genes, the phosphorelay, and the initiation of sporulation. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and other Gram-positive bacteria: biochemistry, physiology and molecular genetics. Washington, DC: American Society for Microbiology; 1993. pp. 747–755. [Google Scholar]
- Hosoya S, Asai K, Ogasawara N, Takeuchi M, Sato T. Mutation in yaaT leads to significant inhibition of phosphorelay during sporulation in Bacillus subtilis. J Bacteriol. 2002;184:5545–5553. doi: 10.1128/JB.184.20.5545-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K, Rudner DZ, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Jiang M, Tzeng YL, Feher VA, Perego M, Hoch JA. Alanine mutants of the Spo0F response regulator modifying specificity for sensor kinases in sporulation initiation. Mol Microbiol. 1999;32:389–395. doi: 10.1046/j.1365-2958.1999.01481.x. [DOI] [PubMed] [Google Scholar]
- Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of the Rap phosphatases in Bacillus subtilis. J. Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Ullmann A, Ladant D. A bacterial two-hybrid system that exploits a cAMP signaling cascade in Escherichia coli. Methods Enzymol. 2000;328:59–73. doi: 10.1016/s0076-6879(00)28390-0. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator of biofilm formation by Bacillus subtilis. Mol Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Shoji K, Shimizu T, Nakano K, Sato T, Kobayaski Y. Analysis of a suppressor mutation ssb (kinC) of sur0B20 (spo0A) mutation in Bacillus subtilis reveals that kinC encodes a histidine protein kinase. J Bacteriol. 1995;177:176–182. doi: 10.1128/jb.177.1.176-182.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDeaux JR, Grossman AD. Isolation and characterization of kinC, a gene that encodes a sensor kinase homologous to the sporulation sensor kinases KinA and KinB in Bacillus subtilis. J Bacteriol. 1995;177:166–175. doi: 10.1128/jb.177.1.166-175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez D, Vlamakis H, Kolter R. Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol Rev. 2009;33:152–163. doi: 10.1111/j.1574-6976.2008.00148.x. [DOI] [PubMed] [Google Scholar]
- Mirouze N, Prepiak P, Dubnau D. Fluctuations in spo0A transcription control rare developmental transitions in Bacillus subtilis. PloS Genet. 2011;7:e1002048. doi: 10.1371/journal.pgen.1002048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrophanov AY, Groisman EA. Signal integration in bacterial two-component regulatory systems. Genes Dev. 2008;22:2601–2611. doi: 10.1101/gad.1700308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchová K, Lewis RJ, Perečko D, Brannigan JA, Ladds JC, Leech A, et al. Dimer-induced signal propagation in Spo0A. Mol Microbiol. 2004;53:829–842. doi: 10.1111/j.1365-2958.2004.04171.x. [DOI] [PubMed] [Google Scholar]
- Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. The Spo0A regulon of Bacillus subtilis. Mol Microbiol. 2003;50:1683–1701. doi: 10.1046/j.1365-2958.2003.03818.x. [DOI] [PubMed] [Google Scholar]
- Ohlsen KL, Grimsley JK, Hoch JA. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Hanstein CG, Welsh KM, Djavakhishvili T, Glaser P, Hoch JA. Multiple protein aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- Perego M, Hoch JA. Negative regulation of Bacillus subtilis sporulation by the spo0E gene product. J Bacteriol. 1991;173:2514–2520. doi: 10.1128/jb.173.8.2514-2520.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M, Hoch JA. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perego M. Kinase-phosphatase competition regulates Bacillus subtilis development. Trends Microbiol. 1998;6:366–370. doi: 10.1016/s0966-842x(98)01350-x. [DOI] [PubMed] [Google Scholar]
- Perego M. Self-signaling by Phr peptides modulates Bacillus subtilis development. In: Dunny GM, Winans SC, editors. Cell-cell signaling in bacteria. Washington, DC: American Society for Microbiology; 1999. pp. 243–258. [Google Scholar]
- Perego M. A new family of aspartyl phosphate phosphatases targeting the sporulation transcription factor Spo0A of Bacillus subtilis. Mol Microbiol. 2001;42:133–143. doi: 10.1046/j.1365-2958.2001.02611.x. [DOI] [PubMed] [Google Scholar]
- Raimondo LM, Lundh NP, Martinez RJ. Primary adsorption site of phage PBS1: the flagellum of Bacillus subtilis. J Virol. 1968;2:256–264. doi: 10.1128/jvi.2.3.256-264.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robakiewicz P, Ryder EF. Statistics: detecting differences among groups. In: Chandra V, Leondar S, editors. Current protocols in protein science. New York, NY: John Wiley & Sons, Inc.; 2000. pp. A.3G.1–A.3G.22. [DOI] [PubMed] [Google Scholar]
- Ruvolo MV, Mach KE, Burkholder WF. Proteolysis of the replication checkpoint protein Sda is necessary for the efficient initiation of sporulation after transient replication stress in Bacillus subtilis. Mol Microbiol. 2006;60:1490–1508. doi: 10.1111/j.1365-2958.2006.05167.x. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheich C, Kümmel D, Soumailakakis D, Heinemann U, Büssow K. Vectors for co-expression of an unrestricted number of proteins. Nucleic Acids Res. 2007;35:e43. doi: 10.1093/nar/gkm067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seredick SD, Seredick BM, Baker D, Spiegelman GB. An A257V mutation in the Bacillus subtilis response regulator Spo0A prevents regulated expression of promoters with low-consensus binding sites. J Bacteriol. 2009;191:5489–5498. doi: 10.1128/JB.00590-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafikhani SH, Mandic-Mulec I, Strauch MA, Smith I, Leighton T. Postexponential regulation of sin operon expression in Bacillus subtilis. J Bacteriol. 2002;184:564–571. doi: 10.1128/JB.184.2.564-571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman J, Abashidze M, Forouhar F, Wang D, Fang Y, Cunningham K, et al. Crystal structure of protein YmcA from Bacillus subtilis. 2009 [Google Scholar]
- Smits WK, Bongiorni C, Veening JW, Hamoen LW, Kuipers OP, Perego M. Temporal separation of distinct differentiation pathways by a dual specificity Rap-Phr system in Bacillus subtilis. Mol Microbiol. 2007;65:103–120. doi: 10.1111/j.1365-2958.2007.05776.x. [DOI] [PubMed] [Google Scholar]
- Stephenson SJ, Perego M. Interaction surface of the Spo0A response regulator with the Spo0E phosphatase. Mol Microbiol. 2002;44:1455–1467. doi: 10.1046/j.1365-2958.2002.02974.x. [DOI] [PubMed] [Google Scholar]
- Strauch M, Webb V, Spiegelman G, Hoch JA. The Spo0A protein of Bacillus subtilis is a repressor of the abrB gene. Proc Natl Acad Sci USA. 1990;87:1801–1805. doi: 10.1073/pnas.87.5.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi L. Transducing phages for Bacillus subtilis. J Gen Microbiol. 1963;31:211–217. doi: 10.1099/00221287-31-2-211. [DOI] [PubMed] [Google Scholar]
- Tortosa P, Albano M, Dubnau D. Characterization of ylbF, a new gene involved in competence development and sporulation in Bacillus subtilis. Mol Microbiol. 2000;35:1110–1119. doi: 10.1046/j.1365-2958.2000.01779.x. [DOI] [PubMed] [Google Scholar]
- Trach KA, Hoch JA. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM. Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA Pol I transcription. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.015156. M111.015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veening JW, Smits WK, Kuipers OP. Bistability, epigenetics and bet-hedging in bacteria. Annu Rev Microbiol. 2008;62:193–210. doi: 10.1146/annurev.micro.62.081307.163002. [DOI] [PubMed] [Google Scholar]
- Wang L, Grau G, Perego M, Hoch JA. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.