SUMMARY
Alternative intronic polyadenylation (IPA) can generate truncated protein isoforms with significantly altered functions. Here, we describe 31 dominant-negative, secreted variant isoforms of receptor tyrosine kinases (RTKs) that are produced by activation of intronic poly(A) sites. We show that blocking U1-snRNP can activate IPA, indicating a larger role for U1-snRNP in RNA surveillance. Moreover, we report the development of an antisense-based method to effectively and specifically activate expression of individual soluble decoy RTKs (sdRTKs) to alter signaling, with potential therapeutic implications. In particular, a quantitative switch from signal transducing full-length vascular endothelial growth factor receptor-2 (VEGFR2/KDR) to a dominant-negative sKDR results in a strong anti-angiogenic effect both on directly targeted cells and on naïve cells exposed to conditioned media, suggesting a role for this approach in interfering with angiogenic paracrine and autocrine loops.
INTRODUCTION
Polyadenylation is an essential part of mRNA processing and is needed to ensure efficient mRNA export, translation and stability of all transcripts except histones (Licatalosi and Darnell, 2010). At least 50% of human genes contain alternative polyA sites (PAS) (Lutz, 2008). These include common tandem PAS within the 3’-UTR and intronic PAS further upstream in the gene sequence (Tian et al., 2007). Usage of these intronic PAS, which must occur concomitantly (and in competition) with pre-mRNA splicing, leads to shortened mature mRNAs and protein isoforms. In contrast to truncated isoforms generated by the introduction of Premature Termination Codons (PTCs) through nonsense mutations or out-of-frame splicing, these mRNA variants are naturally immune to Nonsense-Mediated Decay (NMD) (Lejeune and Maquat, 2005), and are thus stable and efficiently expressed. As the truncations can result in proteins lacking essential C-terminal domains, the function of these isoforms is often dramatically different from their full-length counterparts. This is evident in the processing of trans-membrane (TM) proteins such as receptor tyrosine kinases (RTKs), where usage of upstream intronic PAS can generate soluble isoforms lacking the anchoring TM domain and the intracellular kinase function (Fig. 1A). Such soluble isoforms can retain high affinity ligand binding and can act as dominant negative regulators (or ‘decoy’ receptors) of the signaling pathway (Lemmon and Schlessinger, 2010).
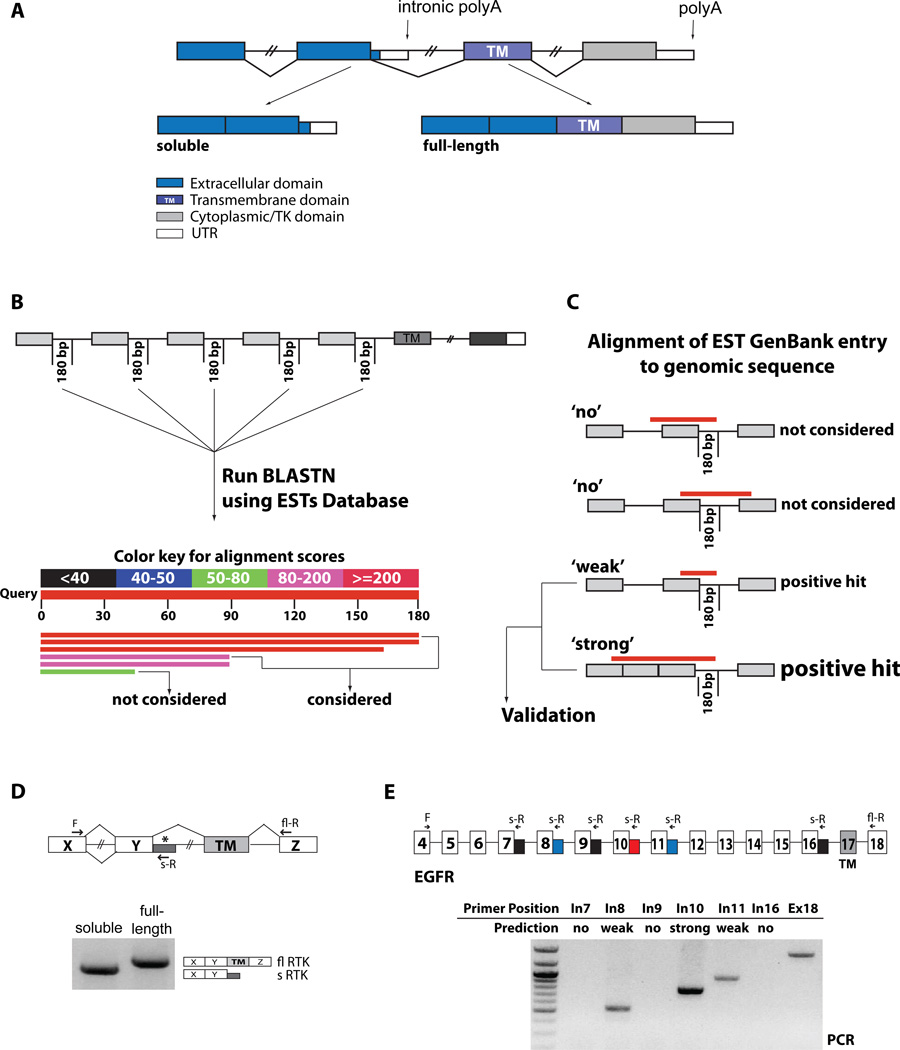
Figure 1. Identification and validation of soluble RTK isoforms.
(A) Alternative polyadenylation in RTKs. Alternative PAS usage upstream of the TM domain leads to soluble isoforms that compete as decoy receptors with the membrane-anchored, full-length isoforms. (B) The first 180 bps of selected introns were used to query EST databases with BLASTN. Positive hits (score>80) were further analyzed. (C) Retrieved GenBank EST hit entries were aligned to their genomic sequences, and hits that crossed the upstream exon-intron boundary were pursued. Hits extending into the upstream intron, reflecting unprocessed RNA or genomic contaminations, were discarded. Hits that reflected full intron retention were also discarded since these variants would still generate soluble variants, but by a different, NMD-sensitive mechanism. Hits that ended within the first upstream exon were considered ‘weak’ since neither upstream processing nor unprocessed RNA can be excluded. Hits that aligned to the exon-intron boundary and spanned one or more upstream exon-exon junctions, indicating evidence of processed RNAs were considered ‘strong’ positive hits. All weak and strong positive hits were further evaluated for validation by oligo-dT RT-PCR. (D) PCR validation was performed with forward primers located on exons upstream of the one preceding the intronic hit, to ensure that the amplified product derives from processed mRNAs. Reverse primers were located within the intronic hit (soluble RTK) or within an exon downstream of the TM domain (full-length) as a measure of expression of the gene. The products of the PCR reaction were analyzed by gel electrophoresis and sequenced. (E) RT-PCR validation of predicted soluble EGFR variants. Reverse primers were designed to amplify the predicted new 3’UTRs (blue and red) from spliced mRNAs. Reverse primers within introns without predicted EST hits were used as negative controls (black). A common forward primer sequence is located in exon 4, the full-length reverse primer is in exon 18. See also Supp. Table s1.
Soluble decoy RTK isoforms (sdRTKs) have been described for isolated RTKs (e.g.: VEGFR1, HER-2, Met) (He et al., 1999; Scott et al., 1993; Tiran et al., 2008), but whether natural ectodomains are a general RTK feature remains unclear. In many cases the mechanism underlying the generation of sdRTK isoforms is unknown, with protease cleavage and ectodomain shedding, alternative splicing, or IPA as proposed mechanisms (Heaney and Golde, 1998; Jin et al., 2008; Swendeman et al., 2008). Enhanced RTK signaling is a driving force in many human malignancies (Lemmon and Schlessinger, 2010), and gaining further insight into regulation of these pathways might result in treatment opportunities.
Pre-mRNA splicing and IPA are closely linked processes, which are mutually exclusive when a common intron is involved (Cartegni et al., 2002; Licatalosi and Darnell, 2010). U1 small nuclear ribonucloparticle (snRNP) (from here on, U1), an essential component of the spliceosome, is primarily responsible for recognition of the 5’ splice site (5’ss) by RNA:RNA base-pairing (Roca and Krainer, 2009). However, it can also strongly inhibit polyadenylation when tethered to the 3’ UTR of a gene (Fortes et al., 2003; Goraczniak et al., 2009). Because U1, by definition, interacts with the 5’ss of virtually all introns, we reasoned that U1 could inhibit intronic PAS in the same manner that it inhibits PAS in the 3’UTR. This inhibition should be particularly evident in cases where the PAS correspond to natural isoforms, where the system may have evolved to respond to variations in U1 binding.
Our investigation led us to the identification of multiple natural sdRTK variants that are individually induced by inhibiting U1 binding to the upstream 5’ss with specific antisense oligonucleotides. Our results are consistent with the recently reported splicing-independent function for U1 snRNP in protecting the transcriptome (Kaida et al., 2010) and support a role for U1 snRNP in mRNA surveillance, distinct and complementary to NMD.
Controlled activation of IPA by modulation of U1 functions could promote sdRTKs with dominant negative properties. The consequent signaling inhibition could act at several levels, recapitulating, with a single compound, the outcome of several therapeutic strategies, like direct receptor knock-down, ligand sequestration and receptor blockade by non-productive heterodimerization.
The specific induction of a soluble isoform of VEGFR2, and the associated anti-angiogenesis effects on target cells and bystanders, showcases how this approach might be useful for modulating the activity of a class of targets that is at the core of multiple therapeutic strategies for cancer and other diseases.
RESULTS
EST-Database Screen and RT-PCR Validation of Soluble Decoy RTKs
RTKs are grouped into 20 families (Lemmon and Schlessinger, 2010). To identify sdRTKs generated by IPA activation, we screened ESTs (http://www.ncbi.nlm.nih.gov/dbEST) for the expression of intronic sequences within processed RTK mRNAs (Fig. 1B) (Tian et al., 2005). We queried the EST database with intronic probes in the 5’ region of ~500 introns around or upstream of the exon encoding the TM domain, from the 39 genes in 16/20 RTK families (Table S1). Seventy-nine of the high-homology EST clusters retrieved were subsequently aligned to their respective genomic sequences. Hits that did not include the exon-intron boundary or that could be attributed to full intron retention, unprocessed pre-mRNAs or DNA contamination (Fig. 1C) were excluded from further analysis, resulting in 42 residual hit clusters. Hits that aligned upstream with spliced products, or where the alignment ended within the first upstream exon, were evaluated as strong and weak candidates, respectively.
To validate these findings, we performed RT-PCR experiments using a forward primer located two or more exons upstream (to ensure that the product is derived from spliced mRNAs) and a reverse primer in the 5’ region of the candidate intron (Fig. 1D). In addition, all non-hit last introns before the TM-encoding exon were also analyzed (Table S1). To exclude that detection of PCR products was simply derived from partially processed pre-mRNAs, we performed control experiments with several reverse primers placed in introns that did not contain PAS, and where EST analysis predicted no intron retention. For example, we predicted three putative soluble variants in the EGFR1 gene, corresponding to PAS in introns 8, 10 and 11. When RT-PCR was performed using a common forward primer in exon 4 and specific reverse primers in introns 7, 9 or 16 (no prediction); 8 or 11 (weak prediction); 10 (strong prediction); and exon 18 (fully spliced product) we could only detect products corresponding to the predicted and full-length variants, with the strongest signal corresponding to the strong prediction (Fig. 1E).
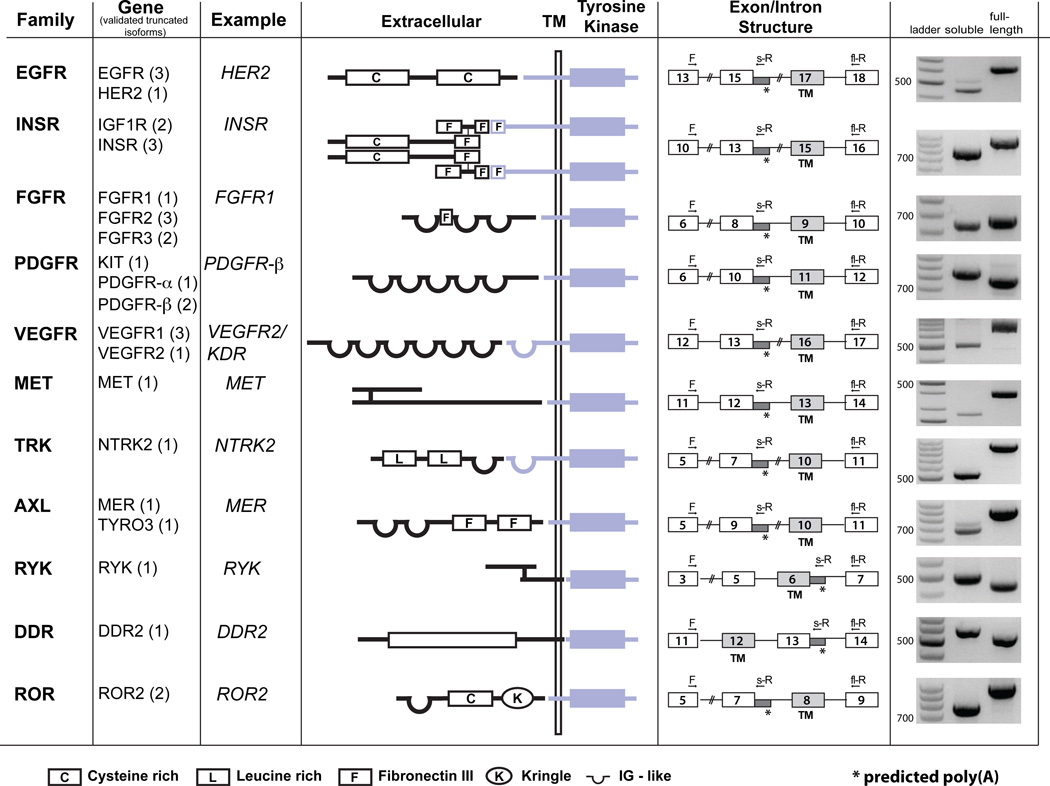
By this method, plus additional evidence that included direct sequencing of all isoforms, previously published data, differential 5’/3’ intronic expression, presence of polyA stretches in ESTs, and 3’ RACE (Table S2, Fig. s1) we identified 31 mRNA variants predicted to encode truncated protein isoforms from 19/24 RTK genes that generated hits, with at least one variant from each of 11 different RTK gene families (Fig. 2). Seven of these had been previously described as IPA variants, whereas 24 correspond to unpublished isoforms (Table S2). In all, the abundance of these isoforms indicates that the expression of secreted, dominant negative RTKs through IPA is a much more general feature than expected given the few examples described in the literature (Table S2). Moreover, this finding can likely be extended to other protein families.
Figure 2. RTK family members and validated truncated isoforms.
Families and respective genes of validated soluble RTKs (number of validated isoform per gene in parenthesis). In the middle, schematic shows domain structure for one example for each RTK family. Intron retention/polyadenylation (represented by asterisks in intron/exon structures) leads to truncated receptors maintaining the extracellular, ligand binding domains (depicted in black), but missing the TM and/or tyrosine kinase domains (blue). RT-PCR of RNA derived from a collection of multiple cell lines (Universal RNA) using the indicated forward (F), reverse (s-R) and full-length reverse (fl-R) primers verified the existence of the soluble RTK isoforms predicted by EST analysis. See also Supp. Table s2 and Supp. Fig s1.
U1-dependent modulation of intronic polyA sites
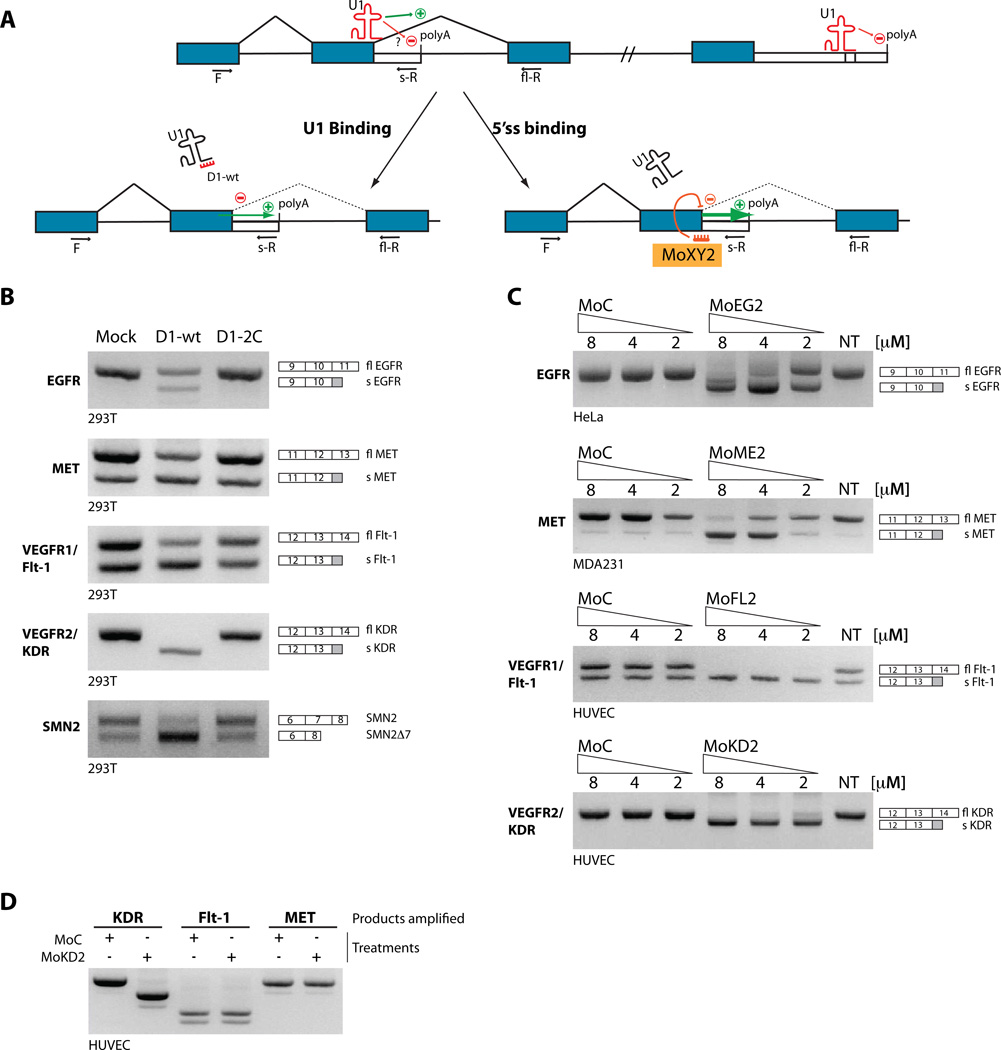
To investigate the relationship between U1, splicing and IPA (Fig. 3A), and to test whether U1 participates in IPA regulation as it does within a 3’UTR context (Fortes et al., 2003), we overexpressed a ‘decoy’ RNA oligonucleotide (D1-wt) which contains a canonical 5’ss sequence (Roca and Krainer, 2009). This short RNA binds to the region of U1 snRNA involved in 5’ss recognition and impedes its recognition of the 5’ss on pre-mRNAs, thus functionally inactivating it. As control, a similar RNA containing a single point mutation (D1-2C) that prevents it from binding efficiently to U1 was used (Fig. s2A).
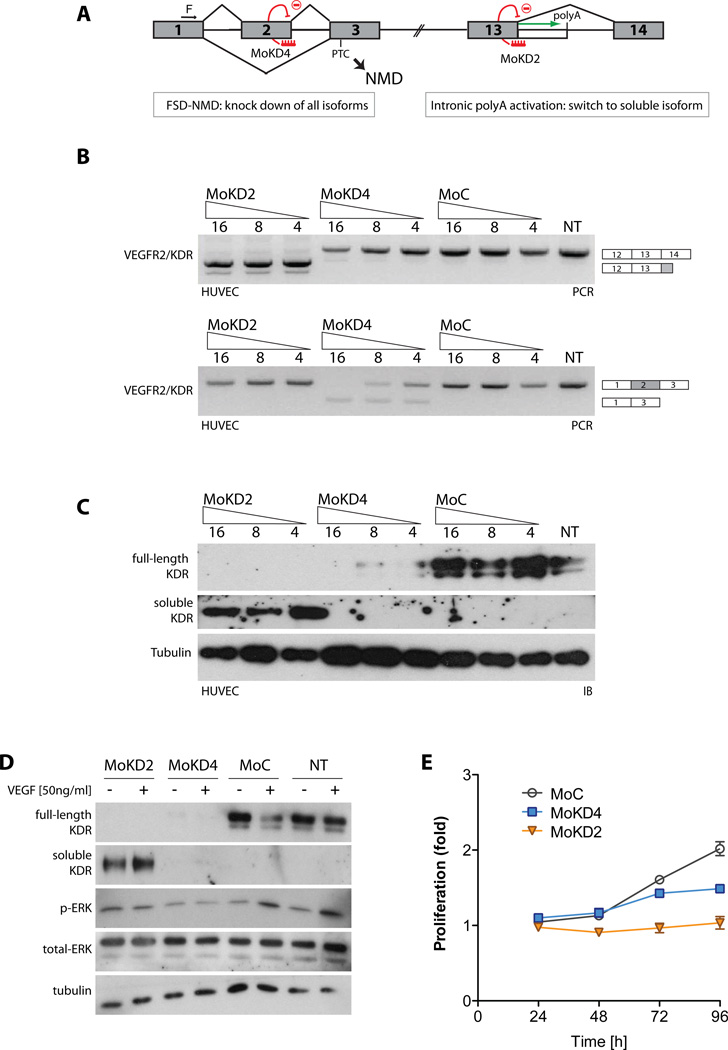
Figure 3. U1-dependent regulation of IPA and target-specific IPA modulation by morpholino.
(A) Binding of U1 to the 5’ss promotes splicing at the expense of IPA, if a usable polyA site is present in the intron. When U1 is targeted to the 3’UTR of the mRNA, strong polyadenylation inhibition ensues. Excess of a ‘decoy’ 5’ss (D1wt) precludes U1 from binding to the 5’ss on the pre-mRNA, decreasing splicing efficiency and/or releasing the polyA site from repression. Similarly, the intronic PolyA site is activated if U1 function is competed out by an antisense compound targeted to the 5’ss. Blue and empty boxes indicate exons and 3’UTRs. (B) 293T cells where mock-transfected or transfected with a plasmid over-expressing either D1-wt (a short RNA complementary to the 5’ end of U1 that mimics a 5’ss) or the D1-2C mutant (with a mutation in the U1-binding region, see Fig. s2A). Activation of intronic PAS in EGFR1, MET, VEGFR1/FLT-1 and VEGFR2/KDR was assessed by 3-oligo PCR, with two exonic (F and fl-R) and one intronic (s-R) primer (quantified in Fig s2B). Top bands corresponds to full-length (fl) products, bottom bands to soluble (s). To verify linearity of amplification, calibration curves using plasmid templates were performed (Fig. s2D). Previously described effect of functional U1 depletion on SMN2 splicing was assessed as a control, after PCR with SMN primers and DdeI digestion. (C) Vivo-morpholinos targeting selected 5’ss block their usage and promote specific IPA. Increasing amounts of compounds or controls were delivered to HUVEC (VEGFR-2/KDR, VEGFR-1/Flt-1), MDA-231 (MET) or HeLa (EGFR) cells. The effect of treatment was analyzed by RT-PCR as in (B). (D) To test for morpholino specificity, MoKD2-treated HUVEC were analyzed as above also for VEGFR-1/Flt-1 and MET expression. See also Supp. Fig. s2 and Supp. Table s3.
Over-expression of D1-wt, but not D1-2C, stimulates IPA from downstream PAS without impairing efficient splicing of upstream introns, and enhances expression of multiple sdRTKs (Fig. 3B and Fig. s2B). As a control of U1 functional downregulation, we confirmed that inclusion of exon 7 of the SMN2 gene was reduced by D1-wt treatment (Fig. 3B), as previously described (Jodelka et al., 2010).
The selective effect on IPA without complete inhibition of upstream splicing likely reflects partial U1 inactivation under our experimental conditions. This might be sufficient to impact regulated events but not constitutive ones, and would affect different events at different rates depending on the PAS context and their dependence on U1, as evidenced from the examples above. Similarly, inhibition of U1 functions by siRNA knock-down of its component U1–70K results in an increase in IPA, albeit to a lesser extent (Fig. s2C), consistent with the described U1–70K role in regulating 3’UTR polyadenylation through direct interactions with polyA polymerase (Gunderson et al., 1998).
The well-characterized dual functions of U1 in splicing and polyadenylation, together with the involvement in modulation of intronic PAS described here, support the intriguing hypothesis that U1 plays a general RNA-surveillance role to silence spurious PAS within introns, therefore limiting the production of potentially damaging truncated isoforms.
Activation of alternative intronic PAS by 5’ss-targeted morpholinos
Any downregulation of general U1 function would likely have broad pleiotropic effects because, in addition to IPA, it would also affects splicing efficiency and splice-site selection (Roca and Krainer, 2009) and would therefore be associated with general RNA processing defects.
To activate individual intronic polyA sites specifically, we developed a modified splicing redirection approach with antisense oligonucleotides (ASOs), which can be used to modulate specific splicing events (Bauman et al., 2010). Typically, ASOs that directly block a splice site cause exon-skipping. However, based on the above observations, when a PAS is present in the downstream intron, an ASO directed to the upstream 5’ss could block U1 binding and promote IPA instead of exon exclusion (Fig. 3A, lower right).
We tested this hypothesis for the human RTKs EGFR (intron 10), MET (intron 12), VEGFR1/Flt-1 (intron 13), and VEGFR2/KDR (intron 13), which we selected because of their direct involvement in multiple pathologies (table S3). We used phosphorodiamidate morpholino oligomers coupled to a dendritic delivery moiety (vivo-morpholinos) (Li and Morcos, 2008), designed to be complementary to the 5’ss upstream of the predicted PAS (Fig. s2E). Treatment of cultured cells (HeLa, MDA-231 or HUVEC, Fig. 3C) with these splice-site blocking morpholinos, but not with nonspecific controls, led in each case to a switch from full-length RTK (flRTK) to sdRTK both at the RNA (Fig. 3C and Fig. s2F) and protein level (Fig. 4A and s2G–I). The effect was specific and treatment with any individual compound did not affect expression of non-targeted RTKs (Fig. 3D).
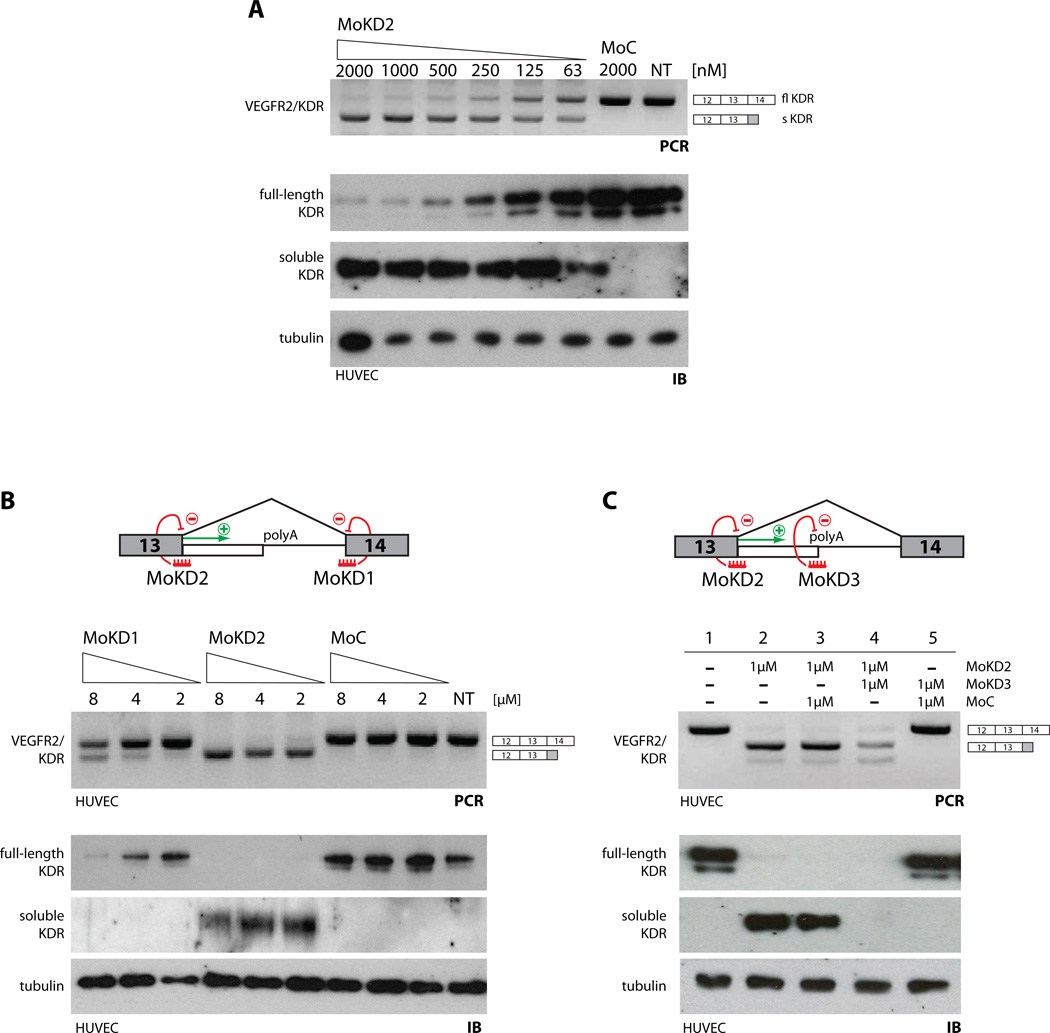
Figure 4. Modulation of VEGFR2/KDR splicing/polyadenylation.
(A) Dose-dependent effect of MoKD2 on VEGFR-2/KDR intron 13 polyadenylation in HUVEC. Increasing amounts of MoKD2 (63 nM to 2 µM) and 2 µM of MoC were delivered to HUVEC. VEGFR-2/KDR RNAs were analyzed as in Fig 3C. Whole-cell lysates (8 µg) were analyzed by immunoblot (IB) with antibodies to KDR or tubulin. Undiluted conditioned media (20 µl) was directly analyzed by IB to measure soluble KDR, with N-terminal directed antibodies. (B) 5’ss are better targets than 3’ss to activate IPA. (Top) MoKD2 and MoKD1 are targeted to the 5’ss and 3’ss of VEGFR2/KDR intron 13. RNAs, lysates and conditioned media from HUVEC treated for 48 h with the indicated amounts of MoKD1, MoKD2 and control vivo-morpholinos were analyzed as in (A). Blocking the 5’ss is much more efficient in activating IPA than blocking the 3’ss, consistent with the proposed non-splicing role of U1 in IPA control. (C) Inhibition of VEGFR-2/KDR polyA signal. (Top) MoKD3 is targeted directly to the mapped proximal polyA site (from Fig. s3). HUVEC were treated with 1 µM of either MoKD2, MoKD3 or MoC in the indicated combinations. RT-PCR and IB analysis (as in (A)) confirmed induction of sKDR and efficient knockdown of flKDR with MoKD2. Adding MoKD3 to MoKD2-treated cells blocks the intronic polyA site and consequently leads to lack of sKDR activation in addition to flKDR knockdown. See also Supp. Fig. s3.
Modulation of endogenous sKDR
Next, we selected VEGFR2/KDR for further analysis. VEGFR2/KDR is the principal mediator of VEGF signaling and controls the angiogenic, lymphangiogenic, mitogenic and survival functions of VEGF (Kerbel, 2008). VEGF-dependent activity is dysregulated in cancer, where it participates in autocrine and paracrine activating loops with VEGF-A as the main activating ligand for both VEGFR2/KDR and VEGFR1/FLT-1. Furthermore, a murine sKDR homolog has been recently described as an essential inhibitor of lymphangiogenesis in mice (Albuquerque et al., 2009) and human sKDR is downregulated in metastatic neuroblastoma, underscoring the physiological relevance and therapeutic potential of this molecule (Becker et al., 2010).
Upon HUVEC treatment with MoKD2, the sKDR isoform is readily detectable in tissue culture medium, whereas the membrane-bound VEGFR2/KDR is ablated from the lysates (Fig. 4A). This dose-dependent shift is potent (low [nM]), rapid and persistent. Suppression of VEGFR2/KDR and concomitant induction of sKDR RNA and protein occurs within 6h of MoKD2 delivery, and lasts for at least 72h post-treatment, when sKDR is still accumulating in the media (Fig. s3A–D).
Competitive inhibition of splicing is not sufficient to drive efficient IPA activation, because targeting the downstream 3’ss of the same intron with a similar compound (MoKD1) is much less effective, both at the RNA and protein levels (Fig. 4B and Fig. s3E). This is not due to intrinsically different ASO efficacies, because both 5’ss- (MoKD2) and 3’ss-targeted (MoKD1) morpholinos block splicing to a similar low degree resulting in some exon skipping (Fig. s3F), indicating that only a small part of IPA activation upon MoKD2 treatment is due to splicing inhibition. Rather, U1 inhibits IPA directly (like in a ‘proper’ UTR setting) and when an activatable polyA site is present, its usage (rather than exon skipping) is the primary consequence of the 5’-ss-blocking treatment.
Overall, these data demonstrate that blocking U1 binding at specific 5’ss activates downstream IPA sites in multiple targets (EGFR, MET, VEGFR1, VEGFR2), leading to appearance of sdRTKs at the expense of the full-length receptors.
Direct Inhibition of IPA sites blocks production of sdRTKs
In the VEGFR2/KDR gene, the soluble isoform is generated by retention of sequences from intron 13, which includes two potential PAS (Fig. s3G–H). Using a primer-walking approach, we show that the upstream PAS is primarily used (Fig. s3I–J). A compound (MoKD3) complementary to this PAS (Fig. 4C, top) inhibited the activation of endogenous sKDR RNA and protein by the 5’ss-blocking morpholino (MoKD2) (Fig. 4C, lanes 4 vs. 3). This result not only demonstrates that IPA is required for expression of sKDR, but also that PAS can be directly targeted when the inhibition of the soluble variant is a desirable outcome.
Uncoupling induction of the sKDR from flKDR knockdown: FSD-NMD
As described, IPA activation results in both the simultaneous reduction of the full-length variant and the induction of the secreted decoy RTK. Given that the simple downregulation of the signaling membrane-bound isoform will have biological consequences by itself, it becomes essential to uncouple these two connected but distinct phenomena, so to distinguish between the biology due to each. To properly compare the effect of MoKD2 treatment with a simple knockdown of KDR, while minimizing non-specific and treatment-derived effects, we applied a strategy that couples the knock-down properties of NMD with the splicing-re-direction mechanics used above, which we termed Forced Splicing-Dependent NMD (FSD-NMD). In brief, we designed a morpholino compound (MoKD4) that induced KDR exon 2 skipping, causing a frameshift that created a premature termination codon (PTC) downstream and triggered NMD (Fig. 5A). Since the approach is the same used for IPA activation (except for the primary sequence of the compound), the resulting data sets are highly comparable and treatments can be done in parallel.
Figure 5. Effects of VEGFR-2/KDR intronic PolyA activation compared to knock-down.
(A) Knock-down of VEGFR2/KDR induced by FSD-NMD compared to splicing re-direction to sKDR. MoKD4 was designed to block VEGFR2/KDR exon 2 splicing and induce its skipping. Since VEGFR2/KDR exon 2 is 94 nt long, its exclusion leads to a frameshift in the VEGFR2/KDR ORFs, which introduces a PTC in exon 3. This in turn triggers NMD and leads to VEGFR2/KDR RNAs degradation. FSD-NMD allows uncoupling of the effects of knockdown (MoKD4) from those of intronic polyA induction (MoKD2), as both outcomes are achieved with directly comparable protocols/chemistries. (B) HUVEC were treated for 48 h with MoKD2, MoKD4 and control (4 µM to 16 µM). Changes in isoform ratios were analyzed by three-oligo RT-PCR as before (upper panel). Primers in exon 1 and 3 were used to assess KDR exon 2 skipping (lower panel). (C) Whole-cell lysates and conditioned media from MoKD2-, MoKD4- or MoC-treated HUVEC were analyzed as in Fig. 4A, resulting in induction of sKDR, ablation of both variants and no effect, respectively. (D) Induction of sKDR or VEGFR2/KDR knock-down inhibits MAPK pathway activation. HUVEC were treated for 48 hours with MoKD2 or MoKD4. 50 ng/ml of VEGF-A (or control) was added for 10 min. flKDR and sKDR were analyzed as above. ERK activation was assessed with phospho-ERK antibody. Total ERK and tubulin are shown as loading control. (E) Induction of sKDR or VEGFR2/KDR knock-down inhibits growth. HUVEC were treated with either 4 µM MoKD2, MoKD4 or MoC and proliferation was measured by a Formazan generation assay (Ab(490)). Each point represents means (±SD, bars) of 3 experiments.
FSD-NMD leads to very efficient inhibition of KDR expression in HUVEC, without the induction of sKDR (Fig. 5B–C, lanes 4–6). Importantly, treatment with either MoKD2 or MoKD4 inhibited VEGF-A-dependent phosphorylation of ERK (Fig. 5D) and cellular proliferation (Fig. 5E).
Induction of sKDR inhibits angiogenesis in primary and secondary target cells
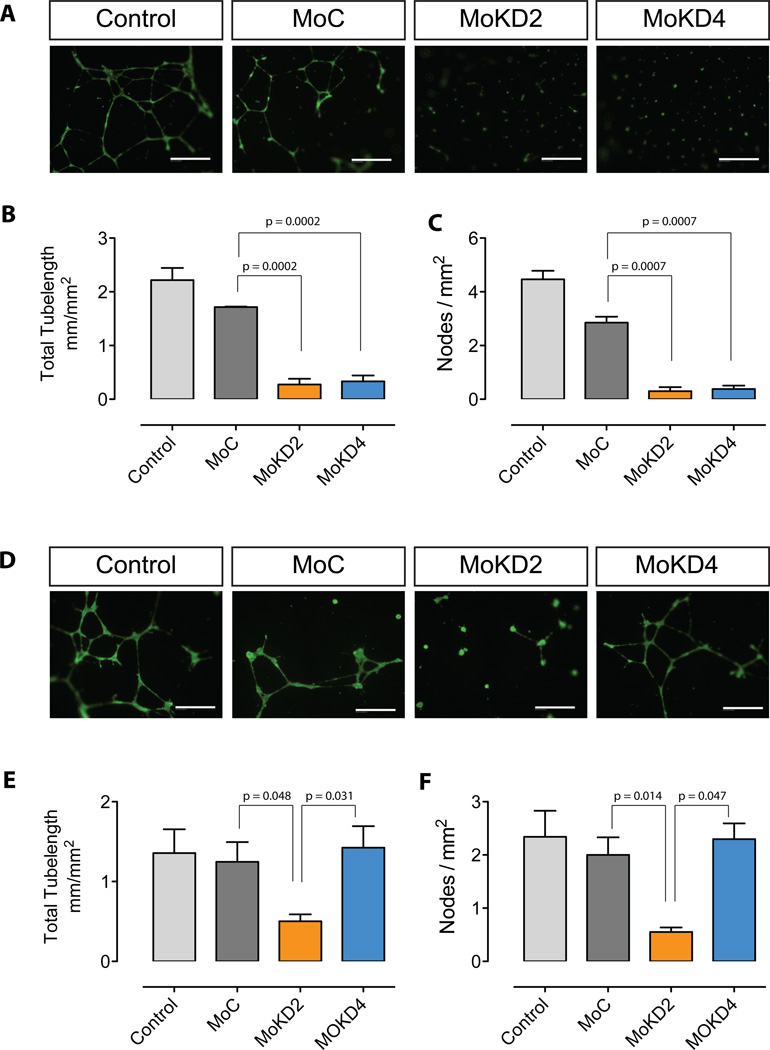
Next, both morpholinos were tested for their ability to inhibit endothelial tube formation on matrigel. To this end, HUVEC were initially treated with MoKD2 or MoKD4 for 48h, then transferred to matrigel-coated culture vessels with their respective medium. Both treatments resulted in reduction of tube formation compared to MoC control oligos (Fig. 6A), both in terms of tube-length (Fig. 6B) and of number of nodes formed (Fig. 6C), consistent with the reduced KDR signaling.
Figure 6. Inhibition of HUVEC tube formation by VEGFR-2/KDR intronic PolyA activation.
(A) Treatment with MoKD2 or MoKD4 decreased ability of HUVEC to form tubes on matrigel. HUVEC seeded at 1.5×105 cells in 6-well plates were grown for 2 days in 2 ml of serum- and VEGF-free EGM-2, containing vivo-morpholinos MoKD2 (2µM), MoKD4 (10µM) and MoC (10µM). Cells were resuspended at 8×104 cells/ml in the respective medium supplemented with 20 ng/ml VEGF-A, seeded at 4×104 cells in matrigel-coated 48-well plates and grown for 20 h. (B–C) Total tube length and number of nodes from experiments described in A) were measured as described in Methods. Mean (±SD, bars) of 3 independent experiments is shown. (D) To test the inhibitory effect of MoKD2-induced sKDR uncoupled from the MoKD2-induced knock-down, the treatment of HUVEC with MoKD2 or MoKD4 was repeated under the same conditions for 72 hours and the respective conditioned medium was used to resuspend untreated HUVEC before seeding them on matrigel as in A). Only conditioned media from cells treated with MoKD2 inhibited tube formation, while the supernatant of cells treated with MoKD4 had no effect on untreated HUVEC. (E–F) Measurements of D) as in B–C) from experiment in A). Size bar: 100 µm
In a second experiment, we collected the medium of HUVEC treated for 72 h with MoKD2 or MoKD4 as described above, added VEGF-A, and used it to stimulate untreated HUVEC seeded on matrigel (Fig. 6D–F). In this setting, conditioned medium from MoKD4-treated cells had no effect, whereas the supernatant from MoKD2 treated cells inhibited tube formation. The effect on the primary target cells could be attributed to downregulation of the full-length isoforms (Fig. 7-1) (consistently, both MoKD2 and MoKD4 treatments work similarly). However, inhibition of angiogenesis in the secondary target cells is due to the inhibitory effect of the sKDR present in the medium, which either binds VEGF (Fig. 7-2) or engages the residual membrane-bound functional VEGFR2 (and/or VEGFR1) on the cell surface into non-functional complexes (Fig. 7-3). The transacting, dominant-negative nature of such interactions underscores how this approach, unlike traditional knock-down strategies, can also affect cells not efficiently targeted by the compounds and could potentiate the biological outcome and therapeutic significance of the treatment via a bystander effect.
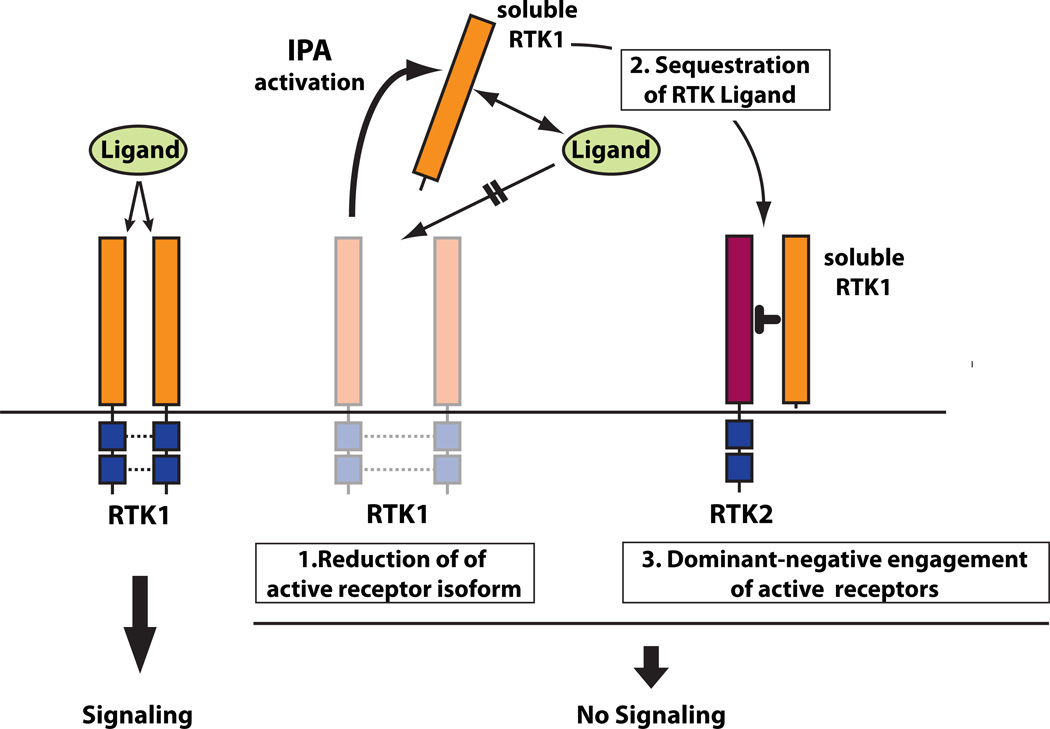
Figure 7. Effects of induction of soluble RTKs by Intronic PolyA Activation.
Left. Binding of RTK ligand to membrane bound FL receptor activates signaling. Right. The generation of soluble RTK isoforms by IPA activation can inhibit signaling in three ways: 1) by reducing the amount of active full-length receptor; 2) by sequestering RTK ligand through direct binding; 3) by inhibiting residual full-length RTK and/or related RTKs by engaging them in non-functional heterodimers, since RTKs can form homo- and hetero-dimers and frequently share common activating ligands with other members of the same RTK family. See also Supp. Table s4.
DISCUSSION
Induction of soluble decoy RTKs, a molecular triple-play
Aberrant RTK signaling is involved in multiple pathological aspects of various human diseases, including tumor development, progression and maintenance (Lemmon and Schlessinger, 2010), and thus RTKs represent preeminent and effective targets for cancer therapy. Current therapeutic approaches include neutralizing antibodies, small drug TK inhibitors, knock-down strategies, or ectopic expression of recombinant dominant-negative variants (Bennasroune et al., 2004; Knight et al., 2010). However, the frequent appearance of resistance to specific RTK inhibitors via neutralizing mutations and/or activation of compensatory RTK pathways (Xu and Huang, 2010) underscores the continuing need for additional approaches.
The activation of ligand-binding sdRTKs, at the expense of the full-length active receptor, recapitulates -within a single compound- the functions of various independent therapeutic strategies by simultaneously working at three levels of inhibition (Fig 7):
Elimination of the active full-length isoform (akin to a canonical knock-down approach, such as RNAi).
Direct binding of the ligand, in a manner similar to ligand-trapping (e.g.: VEGF-Trap) or to anti-ligand antibodies approaches.
Trans-acting blockade of residual full-length receptor (or of other receptors that may dimerize with the target) by non-functional dimerization, both on the same cells and, importantly, on surrounding cells that might not be efficiently targeted by the primary effector.
Such modulation of RTKs expression provides both a useful tool for the general study of receptor biology, and a potential treatment approach in situations where either the upregulation of soluble variants with dominant-negative capabilities is desirable to shut down signaling pathways (e.g., cancer), or where reduction of the secreted variant would be beneficial. For example in preeclampsia, a severe complication of pregnancy affecting ~10% of pregnant women which is associated with excess sFlt-1 (Kita and Mitsushita, 2008).
Indeed, the therapeutic potential of sdRTKs is demonstrated by many studies (Table S4) that ectopically deliver vector-expressed or recombinant sdRTKs, which in some cases are reaching the clinic (e.g.: Aflibercept/VEGF-Trap (Teng et al., 2010)). Compared to such strategies, our approach has the added promising advantage that stimulation of endogenous sdRTK is necessarily linked to flRTK down-regulation, thus IPA activation would result in more efficient inhibition of intracellular signaling.
Importantly, in contrast to sdRTKs generated by ectodomain shedding from the flRTK, IPA activation does not leave behind the transmembrane-intracellular portion, which could lead to constitutive and potentiated signaling, as shown for Met (Matteucci et al., 2009).
Biological role of soluble VEGFR2/KDR
To assess the potential of this three-pronged strategy we selected VEGFR2/KDR, because of its pivotal role in controlling VEGF-dependent functions, which makes it a key target for therapy in cancer and other pathological contexts (Kerbel, 2008).
The induction of IPA within intron 13 of the VEGFR2/KDR gene illustrates the potential of this approach: in cells that are directly targeted by the polyA-inducing compounds, the biological effects of activating the PAS is similar to that of knocking-down VEGFR2/KDR by FSD-NMD. Indeed, early effects are likely due to rapid downregulation of the transmembrane variant (Fig. s3A–B), as it takes time for the soluble variant to accumulate (Fig. s3B,D). However, in cells not directly treated the inhibition of angiogenesis occurs only when sKDR is activated (Fig. 6D–F), pointing to a role in trans (ligand binding or non-productive dimerization, Fig. 7) in the naïve cells and suggesting paracrine/autocrine interference. In vivo, these activities would be further enhanced by the localized production (and maximal concentration) around the cells directly involved in VEGF signaling, eliminating the need to deliver high doses of compound, as is the case for equivalent recombinant compounds administered systemically.
Like other RTKs, both full-length and soluble KDR and FLT-1 (and VEGFR3) can form heterodimers (in any combination), with the essential distinction that dimers that include soluble isoforms are inactive because of the impossibility of reciprocal cross-activation of the kinase domains through phosphorylation (Heidenreich et al., 2004; Kendall et al., 1996). Therefore, on top of blocking activity of VEGFR2 receptors in targeted and surrounding cells, the decoy sKDR would also interfere with signaling mediated by other VEGF-binding receptors.
To be effective, IPA-activating compounds need to be efficiently delivered to the target cells in vivo. In recent years, progress achieved with systemic delivery of antisense compounds, has lead to therapeutic rescue in model systems of molecular dystrophy (Yin et al., 2010) and spinal muscular atrophy (Hua et al., 2010). In addition, we recently showed that systemic delivery of antisense compounds conjugated to neovasculature-homing peptides to tumor endothelial cells in vivo had anti-tumorigenic properties (Henke et al., 2008). This strategy is also applicable to sKDR-activating compounds to optimize their anti-angiogenic and anti-lymphangiogenic potential.
An additional mRNA surveillance mechanism? A role for U1 snRNP in controlling intronic polyadenylation
There are likely more sdRTK variants, beyond the ones we identified, because we only considered IPA deriving from intron retention (and not from the similarly frequent ‘hidden-exon’ mechanism), and because variants expressed at low levels or only in specific contexts would be under-represented in the EST libraries.
The mechanistic details still need to be elucidated, but at least in some cases the level of activation of the intronic polyA sites appears to be physiologically regulated, as in the modulation of VEGFR1/sFlt-1 ratio in pregnancy and its dysregulation in pre-eclampsia (Kita and Mitsushita, 2008) and is at least in part controlled by pre-mRNA cis-elements (Thomas et al., 2010). Regulation of IPA is likely due to a combination of control from dedicated cis-elements/trans-factors and inhibition of U1 binding at the upstream splice site (recapitulated by morpholinos). Reduction of U1 binding to 5’ss could influence the activation of IPA both indirectly, as decrease in splicing efficiency would enhance the competing polyA event, and directly if U1 participates to suppression of the polyA usage within the intron like it does within 3’UTRs (Fortes et al., 2003; Gunderson et al., 1998). In this scenario, blocking U1 would hierarchically release IPA silencing in introns, beginning from the physiologically regulated ones. As the level of functional U1 decreases, a broader usage of weaker IPA sites would be observed, leading to increasingly shorter mRNAs (as PAS in early introns get also used). Since the normal splicing functions of U1 would too be affected, inhibition of U1 would eventually be associated with an overall reduction of mature mRNAs due to the increase of unstable unprocessed RNAs.
A role for U1 snRNP in suppressing IPA would complement NMD functions. NMD is a conserved and essential cellular mRNA surveillance mechanism that protects from potentially dangerous proteins arising from the aberrant introduction of PTCs within ORFs (Lejeune and Maquat, 2005). PTC recognition depends on ongoing translation and on their positioning upstream of the last exon-exon junction. mRNAs harboring PTCs are tagged for degradation, while stop codons occurring beyond the last exon-exon boundary are recognized as ‘normal’ and their mRNAs can proceed to translation.
This scenario leaves a significant gap in the mRNA surveillance network, as equally deleterious products could be generated by the spurious activation of cryptic intronic PAS. Sequence requirements for PAS are not very stringent and such signals are abundant in introns. In addition, given the low selective pressure, intronic sequences can easily mutate into potentially good Poly(A) sequences. An activated intronic PAS generates a ‘last exon’ with its own 3’UTR, where a PTC would, -by definition,- be immune to NMD. If suppression of PTCs in ORFs was sufficient to drive the establishment of the conserved NMD machinery, a complementary mechanism may have also evolved to reduce similar damages from aberrant IPA. From a cell-economy perspective, it would be reasonable to exploit pre-existing machinery, such as U1 snRNP, to carry out a dual function. Whether this additional surveillance U1 function requires dedicated components, like NMD, remains to be elucidated. Consistent with our observations, Kaida and colleagues recently reported that functional knock-down of U1 activity in HeLa cells causes splicing inhibition and the appearance of short mature mRNAs generated by the activation of internal polyA sites from early introns near the start of transcription (Kaida et al., 2010). Our work confirms and extends their findings by providing an important biological context and by identifying technologies that could be exploited for therapeutic applications.
Additionally, these observations have implications for the interpretation of disease-associated mutations. Splicing mutations could also activate intronic polyA sites, with the consequent generation of stable and potentially pathogenic truncated variants. Such events would likely be underestimated, as they would not be predicted based on sequencing, nor would they be detected by routine PCR analysis.
Conclusions
In summary, our systematic analysis of EST data led to the recognition that expression of truncated, antagonistic isoforms via IPA is a common phenomenon among RTKs, suggesting that these variants contribute to the fine modulation of RTK signaling. The use of these intronic PAS, and possibly many others, is controlled by U1 snRNP, in a competition/silencing function that might underscore a broad role in RNA surveillance. Release of U1-dependent silencing by antisense compounds targeted to the 5’ splice site specifically and efficiently induces sdRTK expression.
This mechanism provides both a tool to investigate the role of RTKs in their biological context and, when further developed for in vivo applications, an additional possible approach to tackle dysregulated RTK signaling in disease, especially in cases of resistance to available RTK inhibitors.
MATERIALS AND METHODS
Identification of truncated RTK isoforms
39 Ensembl (http://www.ensembl.org) RTK gene entries were analyzed (Supp. Fig. 1). For all, the first 180 bps of each intron upstream and around the TM domain were selected and a BLASTN analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of EST GenBank database (http://www.ncbi.nlm.nih.gov/projects/dbEST) was performed. Positive hits had alignment scores >80. Hits’ EST GenBank entries were aligned to genomic RTK sequences. Alignement to the upstream exon-intron junction identified hits, weak when the alignment ended in the exon, strong if continued into the upstream exons, without intervening introns. Hits fully included in the intron or aligning to upstream intron-exon-intron junctions were discarded as unprocessed RNAs or genomic contaminations.
RNA Isolation and Reverse Transcription (RT)-PCR
Total RNA isolated using RNAqueous® (Ambion), and RT performed with SuperScript III (Invitrogen). 3-oligo PCR performed with Platinum Taq Polymerase (Invitrogen) (annealing at 60°C, 45s extension at 72°C). Two reverse (R) primers, one for the full-length, one for the soluble isoform were used together with a common forward (F) primer in a PCR reaction (oligos 129–130 in Supp. methods). To ensure that PCR reflected actual isoform ratios, control PCRs were performed using different ratios of cloned soluble and full-length cDNA templates (Supp. Fig. s8). qPCR: qPCR reaction were prepared with Sybr-Green PCR mix (Invitrogen), run on the MJ Research Opticon 2 and quantified by the ddC(t) method. All oligonucleotide sequences are in Supp. Methods.
3’ RACE experiments
3' RACE reactions were performed on RNA purified from MoEG2 treated HeLa cells and the RACE experiments were done according to the manufacture’s instructions (3’ RACE System for Rapid Amplification of cDNA Ends, Invitrogen).
Validation of targets
RT-PCR validation (30 cycles, 60°C annealing, 1min/kb extension) was performed in triplicates from different RNAs (HUVEC, MDA231, Universal Human Ref. RNA, Stratagene), using F primers at least two exons upstream of the positive intron and R primers within the intronic hit, upstream of the first putative PAS.
Cell Culture
Human umbilical vein endothelial cells (HUVEC) (Lonza) were grown in EGM-2 complete medium containing 2% (v/v) fetal bovine serum (FBS), recombinant human growth factors including VEGF, FGF, IGF1, and EGF, as well as hydrocortisone, heparin, and ascorbic acid (cell passage ≤8). Where appropriate, cells were starved in serum-free basal EBM-2 medium for 12 h prior to treatment. MDA231 breast cancer cell line was from ATCC and cultured accordingly.
U1 depletion/U1–70K depletion
siRNAs were synthesized by Integrated DNA Technologies, Inc. (IDT). 40 pmol of siRNAs, mixed with 6 ml of Lipofectamine 2000 in 250 ml of Opti-MEM medium were used to transfected fro 48 h HEK-293Tcells plated in 6-well dish at 1.5 ×105 cells/well. After 48 h cells were split 1:2 and grown for an additional 24. Total RNA was prepared using Trizol (Invitrogen). Transfection of D1-wt and D1-2C decoy plasmids into HEK-293T cells was as described (Roca and Krainer, 2009). After 48h, total RNA was prepared as above.
Morpholino treatment
Morpholinos coupled to a delivery moiety (vivo-morpholino, Gene Tools) were were added directly at indicated concentrations to 80% confluent cells seeded in 200 µl media without FBS in 48 well plates.
Immunoblot Analysis
HUVEC, HeLa or MDA231 cells were lysed in mammalian protein extraction reagent (M-PER, Pierce) with Complete protease inhibitor mixture (Roche). 8 µg of lysate (flRTKs) or 20 µl of conditioned media (sdRTKs) were run 6 or 8% SDS-PAGE and transferred to Immobilon™ PVDF Membranes (Millipore). Primary antibodies were to: KDR C-ter (for flKDR Cell Signaling #55B11) and KDR N-ter (for sKDR, R&D AF357), EGFR (R&D, AF231), MET (R&D, AF276), phospho-ERK (Santa Cruz #sc-7383), total ERK (Santa Cruz #sc-94-G), and β-tubulin (Sigma #T5186). HRP-conjugated secondary antibodies and ECL system was used for visualization.
Soluble KDR quantitation from cell culture supernatant
Production of sKDR and sFlt-1 from HUVEC or of sEGFR from HeLa cells was determined by enzyme-linked immunosorbent assay (ELISA) using ELISA kits (R&D systems) according to the manufacturer’s instructions.
VEGF stimulation and phospho-ERK analysis
HUVEC were plated at 1×104 cells in 48-well plates in EGM-2 complete media. After 24h, media was substituted with 0.2 ml EGM-2 without FBS and VEGF and containing vivo-morpholinos at 4µM. Cells were stimulated with 50 ng/ml VEGF-A for 10 minutes after 48 h of vivo-morpholino treatment. After stimulation, conditioned media was collected and cells were washed and lysed in in M-PER buffer. Conditioned media and lysates were analyzed as described above.
Proliferation Assay
HUVEC were seeded into 48-well plate at 1.2 × 104 cells/well in EGM-2 complete media. After 12 hours, media was substituted with 0.2 ml EGM-2 without FBS and VEGF and containing vivo-morpholinos. After 24h of treatment, cells were stimulated with 10 ng/ml VEGF-A and 2% FBS. Proliferation was assessed by CellTiter 96 Aqueous reagent (Promega).
Tube formation Assay
HUVEC were plated at 1.5×105 cells in 6-well plates in EGM-2 complete media. Media was substituted with 0.2 ml EGM-2 without FBS and VEGF and containing vivo-morpholinos. Cells were harvested after 48 h and resuspended at 8×104/ml in the respective media supernatant supplemented with 20 ng/ml VEGF-A. Cells were seeded at 4×104 cells in 48-well plates coated with matrigel (BD Bioscience, 150 µL matrigel/well, coated for 60 min at 37 °C) and incubated for 20 h at 37 °C. To test the inhibitory effect of sKDR in the conditioned media, cells were incubated for 72 h after morpholino addition. Naive HUVEC were suspended at 8×104/ml in the respective conditioned media supplemented and plated as above. For evaluation, cells on matrigel were carefully washed with PBS and stained with 5 µg/ml CalceinAM (Invitrogen) in PBS for 45 min at 37 °C. Fluorescence images were taken on a Zeiss Axio200 using a fluorescein filter and analyzed with ImageJ (http://rsbweb.nih.gov/ij/). Results were analyzed with Prism 5.0 (GraphPad). P-vals were calculated using t-Test (unpaired, two-tailed).
Supplementary Material
Acknowledgments
We thank R. Benezra, P. Mignatti, E. de Stanchina, M. Myers, X. Roca and R. Karni for helpful comments and suggestions. Work supported by funds from the Brain Tumor Center and the Experimental Therapeutic Center at MSKCC, and by NIH grant NS069759 (MH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Albuquerque RJ, Hayashi T, Cho WG, Kleinman ME, Dridi S, Takeda A, Baffi JZ, Yamada K, Kaneko H, Green MG, et al. Alternatively spliced vascular endothelial growth factor receptor-2 is an essential endogenous inhibitor of lymphatic vessel growth. Nat Med. 2009;15:1023–1030. doi: 10.1038/nm.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38:8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Pavlakovic H, Ludewig F, Wilting F, Weich HA, Albuquerque R, Ambati J, Wilting J. Neuroblastoma progression correlates with downregulation of the lymphangiogenesis inhibitor sVEGFR-2. Clin Cancer Res. 2010;16:1431–1441. doi: 10.1158/1078-0432.CCR-09-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasroune A, Gardin A, Aunis D, Cremel G, Hubert P. Tyrosine kinase receptors as attractive targets of cancer therapy. Crit Rev Oncol Hematol. 2004;50:23–38. doi: 10.1016/j.critrevonc.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Fortes P, Cuevas Y, Guan F, Liu P, Pentlicky S, Jung SP, Martinez-Chantar ML, Prieto J, Rowe D, Gunderson SI. Inhibiting expression of specific genes in mammalian cells with 5' end-mutated U1 small nuclear RNAs targeted to terminal exons of pre-mRNA. Proc Natl Acad Sci U S A. 2003;100:8264–8269. doi: 10.1073/pnas.1332669100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goraczniak R, Behlke MA, Gunderson SI. Gene silencing by synthetic U1 adaptors. Nat Biotechnol. 2009;27:257–263. doi: 10.1038/nbt.1525. [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- He Y, Smith SK, Day KA, Clark DE, Licence DR, Charnock-Jones DS. Alternative splicing of vascular endothelial growth factor (VEGF)-R1 (FLT-1) pre-mRNA is important for the regulation of VEGF activity. Mol Endocrinol. 1999;13:537–545. doi: 10.1210/mend.13.4.0265. [DOI] [PubMed] [Google Scholar]
- Heaney ML, Golde DW. Soluble receptors in human disease. J Leukoc Biol. 1998;64:135–146. doi: 10.1002/jlb.64.2.135. [DOI] [PubMed] [Google Scholar]
- Heidenreich R, Machein M, Nicolaus A, Hilbig A, Wild C, Clauss M, Plate KH, Breier G. Inhibition of solid tumor growth by gene transfer of VEGF receptor-1 mutants. Int J Cancer. 2004;111:348–357. doi: 10.1002/ijc.20260. [DOI] [PubMed] [Google Scholar]
- Henke E, Perk J, Vider J, de Candia P, Chin Y, Solit DB, Ponomarev V, Cartegni L, Manova K, Rosen N, Benezra R. Peptide-conjugated antisense oligonucleotides for targeted inhibition of a transcriptional regulator in vivo. Nat Biotechnol. 2008;26:91–100. doi: 10.1038/nbt1366. [DOI] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010 doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zhang J, Sumariwalla PF, Ni I, Jorgensen B, Crawford D, Phillips S, Feldmann M, Shepard HM, Paleolog EM. Novel splice variants derived from the receptor tyrosine kinase superfamily are potential therapeutics for rheumatoid arthritis. Arthritis Res Ther. 2008;10:R73. doi: 10.1186/ar2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodelka FM, Ebert AD, Duelli DM, Hastings ML. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010 doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall RL, Wang G, Thomas KA. Identification of a natural soluble form of the vascular endothelial growth factor receptor, FLT-1, and its 9 heterodimerization with KDR. Biochem Biophys Res Commun. 1996;226:324–328. doi: 10.1006/bbrc.1996.1355. [DOI] [PubMed] [Google Scholar]
- Kerbel RS. Tumor angiogenesis. N Engl J Med. 2008;358:2039–2049. doi: 10.1056/NEJMra0706596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita N, Mitsushita J. A possible placental factor for preeclampsia: sFlt-1. Curr Med Chem. 2008;15:711–715. doi: 10.2174/092986708783885309. [DOI] [PubMed] [Google Scholar]
- Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer. 2010;10:130–137. doi: 10.1038/nrc2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YF, Morcos PA. Design and synthesis of dendritic molecular transporter that achieves efficient in vivo delivery of morpholino antisense oligo. Bioconjug Chem. 2008;19:1464–1470. doi: 10.1021/bc8001437. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS. Alternative polyadenylation: a twist on mRNA 3' end formation. ACS Chem Biol. 2008;3:609–617. doi: 10.1021/cb800138w. [DOI] [PubMed] [Google Scholar]
- Matteucci E, Bendinelli P, Desiderio MA. Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis. 2009;30:937–945. doi: 10.1093/carcin/bgp080. [DOI] [PubMed] [Google Scholar]
- Roca X, Krainer AR. Recognition of atypical 5' splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol. 2009;16:176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott GK, Robles R, Park JW, Montgomery PA, Daniel J, Holmes WE, Lee J, Keller GA, Li WL, Fendly BM, et al. A truncated intracellular HER2/neu receptor produced by alternative RNA processing affects growth of human carcinoma cells. Mol Cell Biol. 1993;13:2247–2257. doi: 10.1128/mcb.13.4.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swendeman S, Mendelson K, Weskamp G, Horiuchi K, Deutsch U, Scherle P, Hooper A, Rafii S, Blobel CP. VEGF-A stimulates ADAM17-dependent shedding of VEGFR2 and crosstalk between VEGFR2 and ERK signaling. Circ Res. 2008;103:916–918. doi: 10.1161/CIRCRESAHA.108.184416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng LS, Jin KT, He KF, Zhang J, Wang HH, Cao J. Clinical applications of VEGF-trap (aflibercept) in cancer treatment. J Chin Med Assoc. 2010;73:449–456. doi: 10.1016/S1726-4901(10)70097-6. [DOI] [PubMed] [Google Scholar]
- Thomas CP, Raikwar NS, Kelley EA, Liu KZ. Alternate processing of Flt1 transcripts is directed by conserved cis-elements within an intronic region of FLT1 that reciprocally regulates splicing and polyadenylation. Nucleic Acids Res. 2010;38:5130–5140. doi: 10.1093/nar/gkq198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. A largescale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiran Z, Oren A, Hermesh C, Rotman G, Levine Z, Amitai H, Handelsman T, Beiman M, Chen A, Landesman-Milo D, et al. A novel recombinant soluble splice variant of Met is a potent antagonist of the hepatocyte growth factor/scatter factor-Met pathway. Clin Cancer Res. 2008;14:4612–4621. doi: 10.1158/1078-0432.CCR-08-0108. [DOI] [PubMed] [Google Scholar]
- Xu AM, Huang PH. Receptor tyrosine kinase coactivation networks in cancer. Cancer Res. 2010;70:3857–3860. doi: 10.1158/0008-5472.CAN-10-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Moulton HM, Betts C, Merritt T, Seow Y, Ashraf S, Wang Q, Boutilier J, Wood MJ. Functional rescue of dystrophin-deficient mdx mice by a chimeric peptide-PMO. Mol Ther. 2010;18:1822–1829. doi: 10.1038/mt.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.