Abstract
Clustering neurotransmitter receptors at the synapse is crucial for efficient neurotransmission. Here we identify a Caenorhabditis elegans locus, lev-10, required for postsynaptic aggregation of ionotropic acetylcholine receptors (AChRs). lev-10 mutants were identified on the basis of weak resistance to the anthelminthic drug levamisole, a nematode-specific cholinergic agonist that activates AChRs present at neuromuscular junctions (NMJs) resulting in muscle hypercontraction and death at high concentrations1–3. In lev-10 mutants, the density of levamisole-sensitive AChRs at NMJs is markedly reduced, yet the number of functional AChRs present at the muscle cell surface remains unchanged. LEV-10 is a transmembrane protein localized to cholinergic NMJs and required in body-wall muscles for AChR clustering. We also show that the LEV-10 extracellular region, containing five predicted CUB domains and one LDLa domain, is sufficient to rescue AChR aggregation in lev-10 mutants. This suggests a mechanism for AChR clustering that relies on extracellular protein–protein interactions. Such a mechanism is likely to be evolutionarily conserved because CUB/LDL transmembrane proteins similar to LEV-10, but lacking any assigned function, are expressed in the mammalian nervous system and might be used to cluster ionotropic receptors in vertebrates.
Genetic screens for C. elegans mutants that exhibit strong resistance to levamisole have identified four genes encoding AChR subunits and two genes that are required for the biosynthesis of levamisole-sensitive AChRs1–3. However, no genes required for AChR clustering were cloned despite the large size of these screens. We hypothesized that impairing the function of such genes would generate subtle phenotypes for two reasons. First, unclustered levamisole-sensitive AChRs might remain functional if properly inserted into the plasma membrane, thus conferring levamisole-sensitivity. Second, there is an additional class of AChRs present at C. elegans NMJs that are activated by acetylcholine and nicotine but are insensitive to levamisole4. These receptors, of as yet unknown composition, might compensate for a decrease in levamisole-sensitive AChRs at the synapse.
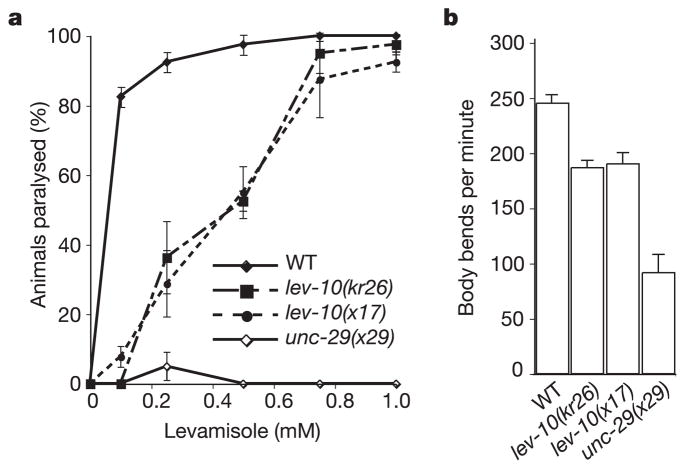
We therefore performed a screen to isolate mutants that exhibited only weak resistance to levamisole. To facilitate the identification of mutated genes we used an insertional mutagenesis based on germ-line mobilization of the Drosophila transposon Mos1 (ref. 5). We isolated a mutant allele, kr26, that resulted from a Mos1 insertion whose interpolated genetic position was in the vicinity of the lev-10 locus. A single mutant allele of lev-10, x17, was isolated previously in a levamisole-resistance screen but was not characterized at the molecular level1. Using a genetic complementation test, we showed that x17 and kr26 are two alleles of the same gene, lev-10. Both lev-10 mutants displayed a slight resistance to levamisole, when assayed by dose–response (Fig. 1a), but after 1 h of exposure to 1 mM levamisole, 100% of the lev-10 mutants became paralysed. Although, in contrast to wild-type animals, lev-10 mutants were able to survive while remaining hypercontracted at this elevated drug concentration. In addition, both lev-10 mutants displayed marginal locomotory defects on plates. When movement was analysed in liquid medium, a subtle but significant movement impairment was detected (Fig. 1b). These phenotypes suggested that mutating lev-10 only partly impairs the function of levamisole-sensitive AChRs.
Figure 1.
Phenotypic characterization of lev-10 mutants. a, The levamisole dose–response curve indicates that lev-10 mutants are only weakly resistant to levamisole when compared with unc-29(x29) mutants, which lack levamisole-sensitive AChRs. Error bars represent s.e.m. (n = 4 independent experiments). WT, wild type. b, lev-10 mutants exhibit weak locomotory defects compared with the wild type in a thrashing assay (ANOVA test; p < 0.01) but are not as impaired as unc-29(x29) mutants (p < 0.01). Error bars represent s.e.m. (n = 6).
To analyse the expression of these receptors, we raised antibodies against UNC-29, a non-α-subunit of the levamisole-sensitive AChR in muscle6. In wild-type animals, UNC-29 was clustered along the ventral and dorsal cords and in the nerve ring where head muscles are innervated (Fig. 2a, c). In lev-10 mutants, no detectable UNC-29 staining was observed along the ventral and dorsal nerve cords, and only weak staining remained in the nerve ring (Fig. 2b, d). To test whether the loss of UNC-29 clusters in lev-10 mutants was due to the absence of cholinergic innervation, we immunostained cholin-ergic varicosities with an antibody against UNC-17, the vesicular acetylcholine transporter in C. elegans7. Staining patterns were similar in wild-type (Fig. 2e) and lev-10 mutant animals (Fig. 2f). In addition to cholinergic innervation, body-wall muscles are also innervated by GABAergic motoneurons. To determine whether lev-10 was globally required for the formation of receptor aggregates or was specifically acting at cholinergic neuromuscular synapses, we immunostained the muscle GABAA receptor UNC-49 (refs 8, 9). In both the wild type and lev-10 mutants, GABA receptors were clustered along the nerve cords (Fig. 2g, h). The inability to detect AChRs by immunostaining could result from decreased receptor expression in lev-10 mutants. Alternatively, a diffuse distribution of a wild-type number of receptors could be below our detection threshold. AChR expression was therefore assessed by western blot analysis of fractionated worm extracts (Fig. 2i). In lev-10(kr26) and lev-10(x17) extracts, the UNC-29 concentrations were similar to that in the wild type (90 ± 12% (n = 4) and 123 ± 17% (n = 4), respectively), suggesting that AChR expression is not reduced in lev-10 mutants.
Figure 2.
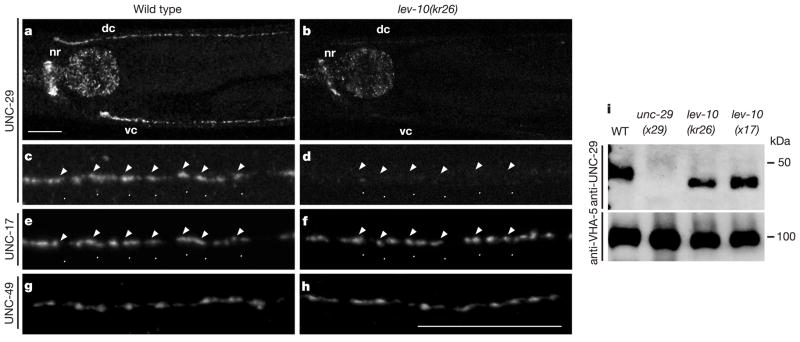
Mutation of lev-10 results in the specific loss of levamisole-sensitive AChR clusters at neuromuscular junctions. a–d, UNC-29 localization detected by immunofluorescence with anti-UNC-29 antibodies. a, Shown are the nerve ring (nr) and the dorsal (dc) and ventral (vc) nerve cords in wild-type animals. c, Individual UNC-29 puncta at high magnification in the dorsal cord from the wild type. b, d, UNC-29 staining in lev-10(kr26) animals at magnifications as in a and c, respectively. The staining in the pharynx is non-specific (data not shown). e, Visualization of cholinergic varicosities by co-immunostaining of the vesicular ACh transporter UNC-17 in wild-type animals shows that UNC-29 clusters are juxtaposed to cholinergic release sites (arrowheads). f, UNC-17 staining in lev-10(kr26) mutants. g, h, Immunostaining of the GABA receptor UNC-49 in wild-type animals (g) and lev-10(kr26) mutants (h). Scale bars, 20 μm. i, Western blot with anti-UNC-29 and anti-VHA-5 antibodies on membrane fractions of C. elegans extracts. The UNC-29 protein has an apparent molecular mass of 47 kDa. VHA-5 detection is used for normalization. WT, wild type.
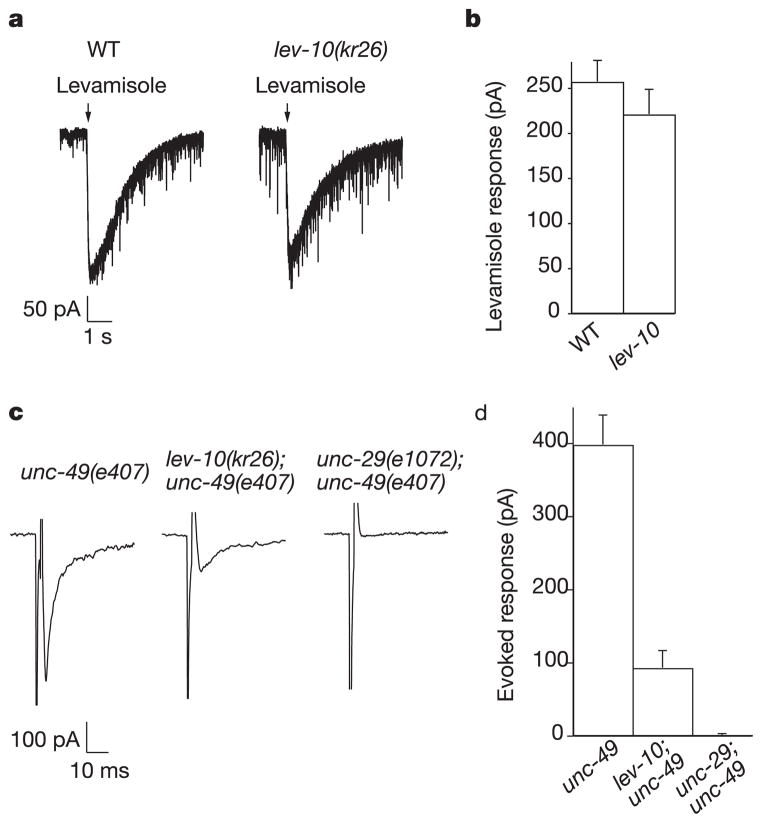
To test whether the UNC-29 protein detected in lev-10 mutants was assembled into functional receptors present at the muscle cell surface, we used electrophysiology4. Pressure-ejection of levamisole onto voltage-clamped body-wall muscles elicited similar currents in the wild type and in lev-10 mutants (Fig. 3a, b), whereas in a unc-29 lev-10 double mutant no levamisole current was detected (data not shown). These data indicate that the overall expression level of functional levamisole-sensitive AChRs in lev-10 mutants is comparable to that in the wild type. To analyse the synaptic population of levamisole-sensitive AChRs, we stimulated motoneurons in the ventral cord and recorded evoked currents in individual muscle cells. To eliminate currents due to activation of the GABA receptor UNC-49, we performed our analysis in an unc-49(e407) null mutant background. Furthermore, we used the nicotinic antagonist dihydro-β-erythroidine (DHβE) to block levamisole-insensitive AChRs present at NMJs4. In lev-10;unc-49 double mutants, the size of the evoked response in the presence of DHβE was decreased by 77% compared with that of unc-49 (Fig. 3c, d). The remaining evoked current was due to the activation of levamisole-sensitive AChRs, because unc-29;unc-49 double mutants, which no longer express levamisole-sensitive AChRs4, exhibited no evoked response in the presence of DHβE. In addition, the time to peak and decay time of the evoked current were increased in lev-10;unc-49 compared with those in unc-49 (6.44 ± 0.41 ms (n = 7) versus 4.64 ± 0.18 ms (n = 7), p < 0.0017, and 16.6 ± 6.9 ms (n = 6) versus 8.4 ± 0.43 ms (n = 7), p < 0.0002, respectively). These kinetic alterations are consistent with a decreased ratio of synaptic versus perisynaptic receptors being activated by synaptic release of acetylcholine in the lev-10 background. Analysis of evoked response amplitudes in the absence of DHβE in lev-10;unc-49 and unc-49 mutants did not reveal any significant difference (3329 ± 221 pA (n = 6) versus 2904 ± 291 pA (n = 7), respectively), thus suggesting that the expression and localization of the levamisole-insensitive AChRs present at the NMJ were not affected in lev-10 mutants. In combination with the immunostaining data, these results indicate that lev-10 is required specifically for the clustering of levamisole-sensitive AChRs at the synapse.
Figure 3.
Levamisole-sensitive AChRs are functional but diffusely distributed in lev-10 body-wall muscle. a, Currents recorded from voltage-clamped body wall muscles in response to pressure-ejection of levamisole (300 μM) in wild-type (WT) and lev-10(kr26) mutants. b, Average amplitude of levamisole-elicited current. c, Evoked currents recorded in a body-wall muscle after eliciting neurotransmitter release by ventral nerve cord depolarization. Experiments were performed in an unc-49(e407) background to eliminate currents due to GABA receptor activation and in the presence of 5 μM DHβE, which blocks the levamisole-insensitive AChRs. d, Average amplitude of evoked response. Error bars in b and d represent s.e.m.
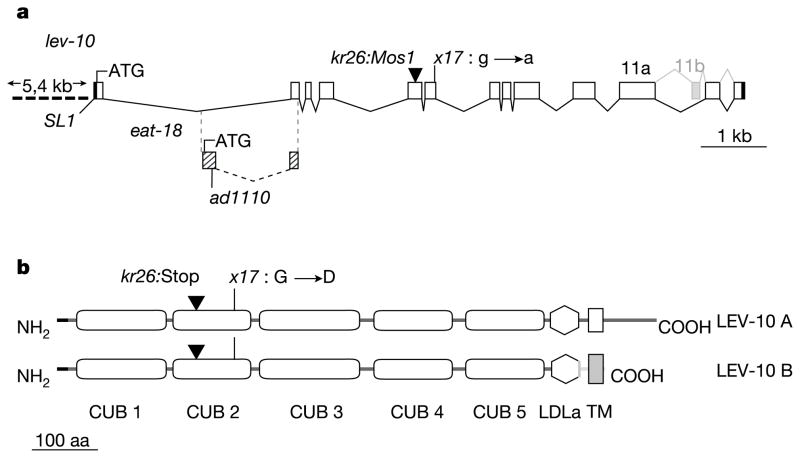
We cloned lev-10 using inverse polymerase chain reaction (PCR) to identify the genomic position of the kr26::Mos1 insertion (Fig. 4a) and confirmed its identity by rescue experiments (Supplementary Table S1). Interestingly, lev-10 overlaps with eat-18, a gene required for the function of AChRs in pharyngeal muscle10,11. Mutation of eat-18 does not confer levamisole resistance. We confirmed that these genes are distinct by genetic complementation (data not shown) and by rescuing lev-10 mutants with a genomic fragment carrying the eat-18(ad1110) nonsense mutation (Supplementary Table S1). lev-10 is predicted to encode a type I transmembrane protein (Fig. 4b). The extracellular part of the protein contains five predicted CUB domains and one LDLa domain. These domains are present in a wide variety of secreted and membrane-bound proteins and mediate protein–protein interactions (for reviews see refs 12–14). Alternative splicing of lev-10 generates two transcripts, lev-10a and lev-10b (Fig. 4b). The lev-10b splice variant represents less than 10% of lev-10 mRNAs (data not shown) and codes for a LEV-10 isoform that differs from LEV-10A in the transmembrane region and contains virtually no intracellular region.
Figure 4.
lev-10 encodes a CUB domain-rich transmembrane protein. a, Genomic organization of lev-10. Open boxes, coding regions; black boxes, 5′ and 3′ untranslated region; ATG, translational start site; SL1, SL1 trans-spliced leader. The first intron of lev-10 contains the first exon of the gene eat-18 (hatched boxes). The eat-18 exon is spliced to the second exon of lev-10 by using a different frame, which ends 16 bp after the splice site. ad1110, nonsense mutation in the first exon of eat-18. b, Predicted structure of the LEV-10 isoforms. Horizontal black line, signal peptide; CUB, complement, urchin epidermal growth factor, and bone morphogenetic protein domain; LDLa, low-density lipoprotein receptor domain class A; TM, transmembrane region; aa, amino acids. Domain predictions were based on SMART (smart.embl-heidelberg.de).
In wild-type animals, LEV-10 is concentrated at cholinergic NMJs (Fig. 5a, c, e). Double labelling experiments with an antibody against the vesicular acetylcholine transporter UNC-17 (ref. 15) demonstrated that 93 ± 3% of LEV-10 puncta were associated with cholinergic varicosities (mean ± SEM, 104 puncta counted in seven worms). However, three-dimensional analysis of confocal image stacks revealed that LEV-10 staining was juxtaposed to, but did not overlap, UNC-17 distribution (Fig. 5f). To determine whether LEV-10 functions postsynaptically, we expressed LEV-10A or LEV-10B under the control of the muscle-specific promoter myo-3 (ref. 16) in lev-10(kr26) animals. Both proteins rescued behavioural defects and UNC-29 synaptic clustering when expressed in muscle (Supplementary Table S1). Genetic mosaic analysis (Supplementary Information) confirmed that LEV-10 is required cell autonomously in postsynaptic muscle cells for AChR clustering at NMJs.
Figure 5.
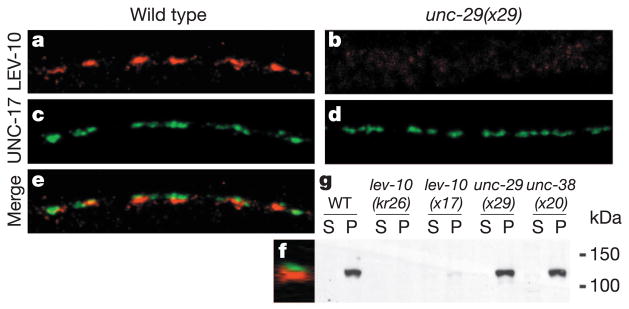
LEV-10 is a synaptic protein that requires levamisole-sensitive AChRs for proper localization but not for expression. a, LEV-10A immunostaining in the dorsal cord of a wild-type animal. c, UNC-17 immunostaining of the same animal labels cholinergic varicosities. e, Merged images. f, Z-optical projection through the entire stack of confocal images at the level of the dashed arrow in e. b, d, LEV-10A (b) and UNC-17 (d) immunostaining in the dorsal cord of an unc-29(x29) mutant. Scale bar, 10 μm. g, Western blot analysis of fractionated C. elegans extracts. P, pellet; S, cytosolic supernatant. The LEV-10 transmembrane protein has an apparent molecular mass of about 120 kDa. Mutants of the levamisole-sensitive AChR subunits unc-29(x29) and unc-38(x20) have LEV-10 concentrations at the membrane comparable to those of the wild type.
Recent results have suggested that proteins involved in neurotransmitter receptor clustering do not accumulate at the synapse in the absence of the receptors17. To test this possibility in our system, we analysed the distribution of LEV-10 in animals lacking levamisole-sensitive AChRs. In unc-29(x29) and unc-38(x20) AChR subunit mutants, no LEV-10 was detected in ventral and dorsal nerve cords by immunofluorescence (Fig. 5b and data not shown) even though presynaptic cholinergic varicosities differentiated normally (Fig. 5d). In parallel, LEV-10 expression level was assessed by western blot analysis (Fig. 5g). LEV-10 was detected as a 120-kDa protein present in the membrane fraction of wild-type worm extracts. This band was absent from lev-10(kr26) extracts and was markedly reduced in lev-10(x17). In unc-29 and unc-38 extracts, LEV-10 concentrations were decreased only slightly in comparison with those in the wild type (72% ± 11 (n = 4) and 81% ± 4 (n = 5), respectively), indicating that a lack of levamisole-sensitive AChRs does not alter LEV-10 expression level. Because LEV-10 is expressed but fails to accumulate at synapses in the absence of levamisole-sensitive AChRs, we cannot exclude the possibility that LEV-10 requires AChRs to reach the plasma membrane, although no intracellular staining of LEV-10 is seen by immunofluorescence in unc-29 and unc-38 mutants. Alternatively, LEV-10 might interact directly or indirectly with AChRs in a complex that is recruited or stabilized at the synapse.
Because most characterized ionotropic receptor clustering proteins are cytoplasmic, relevant interactions are thought to occur on the cytoplasmic side of the postsynaptic membrane18,19. However, complexes formed on the extracellular side of the postsynaptic membrane might also be critical20–23. To test this possibility, we fused the extracellular part of LEV-10 to the human CD4 transmembrane domain. Expression of this chimaeric protein in muscle rescued the defects in levamisole sensitivity, locomotion and AChR clustering of lev-10(kr26) animals. Furthermore, we overexpressed a green fluorescent protein (GFP)-tagged version of LEV-10 truncated before the transmembrane segment. This protein was secreted from muscle cells into the pseudocoelomic cavity (data not shown) but was still able to rescue lev-10(kr26) mutant phenotypes (Supplementary Table S1). The function of LEV-10 in AChR clustering therefore seems to involve only extracellular interactions.
LEV-10 is the first example of a CUB/LDL protein involved in the synaptic clustering of AChRs. The presence of multiple predicted protein–protein interaction domains in the extracellular region indicates that LEV-10 might bind multiple partners. Because we have so far been unable to demonstrate direct interactions between LEV-10 and AChRs, LEV-10 might be indirectly involved in the recruitment of signalling molecules that, in turn, cause AChR clustering. However, the interdependence between LEV-10 and AChR for synaptic localization is consistent with a model that would involve a set of interactions between LEV-10, AChRs or AChR-associated proteins, and a synaptic determinant used to nucleate clustering. Along this line, another C. elegans CUB-domain-rich transmembrane protein, SOL-1, has recently been shown to physically interact with glutamate receptors and to be required for glutamate-gated currents through an as yet unidentified mechanism24.
Of 145 CUB-domain-containing mouse proteins present in non-redundant databases, the first two CUB domains of LEV-10 are most similar to those present in NETO2 (ref. 25) (26% identity, 43% similarity). NETO2 and its paralogue NETO1/BTCL1 (refs 25, 26) are predicted type I transmembrane proteins containing two CUB domains and one LDLa domain in their extracellular region. The two NETO genes are specifically expressed in retina and brain, but their function is unknown. It is therefore tempting to speculate that LEV-10 and NETO proteins are members of a novel class of membrane-spanning proteins engaged in postsynaptic domain organization by means of extracytoplasmic interactions at the synapse.
Methods
Cloning of lev-10
N2 worms were mutagenized by germline mobilization of the Drosophila transposon Mos1 (ref. 5). Young-adult F2 worms were screened for resistance to 1 mM levamisole 3–5 h after transfer to drug-containing plates. In EN 26 [lev-10(kr26::Mos1)], a Mos1 insertion was localized in a predicted exon of the open reading frame Y105E8A.7a, at position 14,403,250 of chromosome I by using inverse PCR (WormBase, www.wormbase.org).
Rescue experiments were performed with a genomic fragment covering the Y105E8A.7a coding region plus 5 kilobases (kb) upstream of the translational start site and 0.21 kb downstream of the lev-10a stop codon. This 15-kb fragment was amplified from N2 or eat-18(ad1110)11 genomic DNA and was injected at 0.85 ng μl−1 with the use of pTG96 (sur-5::GFP)27 as a co-injection marker at 100 ng μl−1. Rescue was scored on the basis of survival on 1 mM levamisole for 16 h.
Tissue-specific rescue
lev-10 complementary DNAs were cloned by PCR after reverse transcription, and sequenced. SL1 splicing was established by PCR with an SL1 primer and a primer in lev-10 exon 13, and by sequencing the expressed sequence tag yk796a04.3′. PCR with rapid amplification of cDNA ends (RACE–PCR) was used to localize the polyadenylation site. lev-10a and lev-10b cDNAs were subcloned into pPD115.62 under the control of the myo-3 promoter. The CD4 transmembrane domain amplified from the human CD4 cDNA (GenBank accession number M12807) was subcloned in frame into Pmyo-3::lev-10a and a stop codon was introduced immediately after the CD4 transmembrane domain. The secreted gfp-lev-10 s was obtained by removing the CD4 transmembrane domain from Pmyo-3::lev-10-CD4 and inserting a GFP cDNA immediately after the lev-10 signal peptide. All constructs were injected at 20 ng μl−1 together with pTG96 (80 ng μl−1) except for Pmyo-3::gfp-lev-10 s (10 ng μl−1).
Levamisole dose–response curve
Young adult worms were scored blind for paralysis after 1 h exposure to levamisole. A distance of one body length of forward movement after mechanical stimulus was required to score a worm as non-paralysed.
Electrophysiological studies
Electrophysiological methods were performed as described previously4. Muscle recordings were made in the whole-cell voltage-clamp configuration (holding potential –60 mV) with an EPC-10 patch-clamp amplifier and digitized at 1 kHz. Data were acquired by Pulse software (HEKA). The bath solution contained 150 mM NaCl, 5 mM KCl, 5 mM CaCl2, 1 mM MgCl2, 10 mM glucose and 15 mM HEPES, pH 7.35, about 340 mOsm. The pipette solution was prepared as described previously4. Subsequent analysis and graphing were performed with Pulsefit (HEKA) and Igor Pro. All statistically derived values are given as means ± s.e.m.
Antibody production and immunocytochemistry
UNC-29: a DNA fragment encoding UNC-29 amino acids 348–431 was inserted into pGEX-3X (Amersham Biosciences). The glutathione-S-transferase (GST)–UNC-29 fusion protein was expressed in Escherichia coli and purified in accordance with the manufacturer’s protocol. Rabbits were injected with 100 μg of fusion protein and boosted three times with 100 μg each.
LEV-10: two synthetic peptides (Eurogentec) corresponding to the LEV-10A amino acids 847–861 and 892–906 were injected into rabbits as described for UNC-29. Both antibodies were purified as described previously28 by using the fusion proteins GST–UNC-29 or GST–LEV-10A (amino acids 836–906 in pGEX-3X) blotted on nitrocellulose. Immunostaining was performed as described9. UNC-29 antibody was used at a dilution of 1:250, and LEV-10 antibody at 1:300. For double-labelling experiments, UNC-17 monoclonal antibody15 was diluted at 1:500 and incubated for 1 h; after 1 h of washing, UNC-29 or LEV-10 antibodies were incubated overnight. The secondary antibody, Cy3-labelled goat anti-rabbit IgG (H +L) (Jackson ImmunoResearch Laboratories), was diluted at 1:900 and the secondary antibody, Alexa488-labelled goat anti-mouse (Molecular Probes) at 1:200.
Protein extraction and western blotting
A mixed staged population of worms (500 μl) was frozen at –80 °C until use. For extraction, worm pellets were ground under liquid nitrogen and thawed on ice. While thawing, 6–10 volumes of ice-cold homogenization buffer (20 mM HEPES pH 7.4, 10 mM KCl, 1 mM EDTA, 400 μM Pefabloc (Roche) and Complete Mini Protease inhibitor cocktail (Roche)) were added and the suspension was further homogenized with ten strokes with the use of a 2-ml tight-fitting glass tissue homogenizer. Afterwards an equal volume of homogenization buffer containing 0.5 M sucrose was added and the suspension was centrifuged twice at 2,000g for 10 min to remove worm debris. The resulting nuclear pellets were pooled and extracted twice with 5 ml of homogenization buffer containing 0.25 M sucrose. The post-nuclear supernatants were pooled and subsequently centrifuged at 150,000g for 1 h. Equal amounts (about 30 μg) of the resulting cytosolic supernatant and membrane pellet were separated by SDS–polyacrylamidegel electrophoresis and blotted onto nitrocellulose membranes. The membranes were subsequently probed with purified anti-LEV-10 serum (dilution 1:1000), anti-UNC-29 (1:600) or anti-VHA-5 (1:3000) (M. Labouesse, unpublished observations) and horseradish-peroxidase-conjugated goat anti-rabbit antibodies (DAKO) and revealed with LumiLight reagents (Roche).
Supplementary Material
Acknowledgments
We thank J Lewis for the lev-10(x17) strain, M. Labouesse for the anti-VAH-5 antibodies, J. Rand for the anti-UNC-17 antibodies, M. Han for the pTG96 plasmid, Y. Kohara for the clone yk796a04, A. Fire for the GFP vectors, the Caenorhabditis Genetic Center for strains, R. Weimer for critical reading of the manuscript, and I. Nuez and H. Gendrot for technical help. C.G. was supported by a fellowship from the Ministère de la Recherche and by the Association pour la Recherche contre le Cancer. S.E. is an EMBO fellow. This work was funded by the Institut National de la Santéet de la Recherche Médicale and the Association Française contre les Myopathies. J.R. was supported by the NIH grant RO1NS41477-03.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare that they have no competing financial interests.
References
- 1.Lewis JA, Wu CH, Berg H, Levine JH. The genetics of levamisole resistance in the nematode Caenorhabditis elegans. Genetics. 1980;95:905–928. doi: 10.1093/genetics/95.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lewis JA, Wu CH, Levine JH, Berg H. Levamisole-resistant mutants of the nematode Caenorhabditis elegans appear to lack pharmacological acetylcholine receptors. Neuroscience. 1980;5:967–989. doi: 10.1016/0306-4522(80)90180-3. [DOI] [PubMed] [Google Scholar]
- 3.Lewis JA, et al. Cholinergic receptor mutants of the nematode Caenorhabditis elegans. J Neurosci. 1987;7:3059–3071. doi: 10.1523/JNEUROSCI.07-10-03059.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richmond JE, Jorgensen EM. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nature Neurosci. 1999;2:791–797. doi: 10.1038/12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bessereau JL, et al. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- 6.Fleming JT, et al. Caenorhabditis elegans levamisole resistance genes lev-1, unc-29, and unc-38 encode functional nicotinic acetylcholine receptor subunits. J Neurosci. 1997;17:5843–5857. doi: 10.1523/JNEUROSCI.17-15-05843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfonso A, Grundahl K, Duerr JS, Han HP, Rand JB. The Caenorhabditis elegans unc-17 gene: a putative vesicular acetylcholine transporter. Science. 1993;261:617–619. doi: 10.1126/science.8342028. [DOI] [PubMed] [Google Scholar]
- 8.Bamber BA, Beg AA, Twyman RE, Jorgensen EM. The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J Neurosci. 1999;19:5348–5359. doi: 10.1523/JNEUROSCI.19-13-05348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gally C, Bessereau JL. GABA is dispensable for the formation of junctional GABA receptor clusters in Caenorhabditis elegans. J Neurosci. 2003;23:2591–2599. doi: 10.1523/JNEUROSCI.23-07-02591.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raizen DM, Lee RY, Avery L. Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans. Genetics. 1995;141:1365–1382. doi: 10.1093/genetics/141.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKay JP, Raizen DM, Gottschalk A, Schafer WR, Avery L. eat-2 and eat-18 are required for nicotinic neurotransmission in the C elegans pharynx. Genetics. 2004;166:161–169. doi: 10.1534/genetics.166.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 13.Christensen EI, Birn H. Megalin and cubilin: multifunctional endocytic receptors. Nature Rev Mol Cell Biol. 2002;3:256–266. doi: 10.1038/nrm778. [DOI] [PubMed] [Google Scholar]
- 14.Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- 15.Duerr JS, Gaskin J, Rand JB. Identified neurons in C. elegans coexpress vesicular transporters for acetylcholine and monoamines. Am J Physiol Cell Physiol. 2001;280:C1616–C1622. doi: 10.1152/ajpcell.2001.280.6.C1616. [DOI] [PubMed] [Google Scholar]
- 16.Okkema PG, Harrison SW, Plunger V, Aryana A, Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono F, Mandel G, Brehm P. Acetylcholine receptors direct rapsyn clusters to the neuromuscular synapse in zebrafish. J Neurosci. 2004;24:5475–5481. doi: 10.1523/JNEUROSCI.0851-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bredt DS, Nicoll RA. AMPA receptor trafficking at excitatory synapses. Neuron. 2003;40:361–379. doi: 10.1016/s0896-6273(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 19.Choquet D, Triller A. The role of receptor diffusion in the organization of the postsynaptic membrane. Nature Rev Neurosci. 2003;4:251–265. doi: 10.1038/nrn1077. [DOI] [PubMed] [Google Scholar]
- 20.O’Brien RJ, et al. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 21.Dalva MB, et al. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 22.Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- 23.Tomita S, Fukata M, Nicoll RA, Bredt DS. Dynamic interaction of stargazin-like TARPs with cycling AMPA receptors at synapses. Science. 2004;303:1508–1511. doi: 10.1126/science.1090262. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Mellem JE, Brockie PJ, Madsen DM, Maricq AV. SOL-1 is a CUB-domain protein required for GLR-1 glutamate receptor function in C. elegans. Nature. 2004;427:451–457. doi: 10.1038/nature02244. [DOI] [PubMed] [Google Scholar]
- 25.Stohr H, Berger C, Frohlich S, Weber BH. A novel gene encoding a putative transmembrane protein with two extracellular CUB domains and a low-density lipoprotein class A module: isolation of alternatively spliced isoforms in retina and brain. Gene. 2002;286:223–231. doi: 10.1016/s0378-1119(02)00438-9. [DOI] [PubMed] [Google Scholar]
- 26.Michishita M, et al. A novel gene, Btcl1, encoding CUB and LDLa domains is expressed in restricted areas of mouse brain. Biochem Biophys Res Commun. 2003;306:680–686. doi: 10.1016/s0006-291x(03)01035-0. [DOI] [PubMed] [Google Scholar]
- 27.Yochem J, Gu T, Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller KG, Emerson MD, McManus JR, Rand JB. RIC-8 (Synembryn): a novel conserved. doi: 10.1016/s0896-6273(00)00037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.