Abstract
Purine riboswitches play an essential role in genetic regulation of bacterial metabolism. This family includes the 2′-deoxyguanosine (dG) riboswitch, involved in feedback control of deoxyguanosine biosynthesis. To understand the principles that define dG selectivity, we determined crystal structures of natural Mesoplasma florum riboswitch bound to cognate dG, as well as non-cognate guanosine, deoxyguanosine monophosphate and guanosine monophosphate. Comparison with related purine riboswitch structures reveals that the dG riboswitch achieves its specificity by modifying key interactions involving the nucleobase and through rearrangement of the ligand-binding pocket, so as to accommodate the additional sugar moiety. In addition, we observe novel conformational changes beyond the junctional binding pocket, extending as far as peripheral loop-loop interactions. It appears that re-engineering riboswitch scaffolds will require consideration of selectivity features dispersed throughout the riboswitch tertiary fold, and that structure-guided drug design efforts targeted to junctional RNA scaffolds need to be addressed within such an expanded framework.
Untranslated mRNA regions, termed riboswitches, provide feedback modulation of gene expression by adopting alternative conformations in the presence or absence of cellular metabolites in all domains of life1,2. Riboswitch selectivity is entirely programmed in the metabolite-sensing domains of riboswitches, which form three-dimensional structures that specifically bind to cognate small molecule ligands and direct the folding of adjacent expression-controlling elements3. Riboswitches typically utilize distinct folds to bind to different metabolites in order to ensure the high specificity required for a precise and fast response4. The requirement for high selectivity causes nucleotides involved in ligand recognition and structure formation to be highly conserved even among distantly related species2. However, ongoing studies keep identifying cases where the same metabolite can be recognized by more than one riboswitch fold, either sharing common elements5 or being structurally distinct6-9.
The crucial role of riboswitches in gene expression circuits in bacterial species, including pathogens, demands an understanding of the molecular mechanisms of riboswitch function. Structural studies constitute the initial step to providing a molecular foundation for the design and implementation of mechanistic experiments. The majority of previous structural studies concentrated on distinct riboswitch classes, while structurally related riboswitches received less attention. Nevertheless, related riboswitches within a distinct class represent an excellent platform for extracting precise information on riboswitch folding, small molecule binding, and mechanisms of genetic control. Structure-function relationships are most intriguing within the purine riboswitch family10, whose representatives, the adenine11, guanine12 and 2′-deoxyguanosine (dG)13 riboswitches, face the serious challenge of discriminating between different purine ligands using related RNA folds. X-ray structures14,15 revealed virtually identical three-dimensional folds for adenine and guanine riboswitches that consist of a regulatory helix P1 connected to hairpins P2-L2 and P3-L3 and stabilized by tertiary loop-loop interactions (Fig. 1a). To discriminate between the binding of adenine or guanine, these riboswitches form Watson-Crick base pairs between the purine ligands and uridine or cytidine residues located within the junctional core14-16. The dG riboswitch carries nucleotide changes in otherwise conserved positions of the core and possesses shortened hairpins expected to change critical tertiary contacts between the terminal loops (Fig. 1b)13. These alterations help the dG riboswitch bind to dG and effectively discriminate against guanine, which lacks the deoxyribose sugar. Since crystal structure determination of the wild type dG riboswitch has turned out to be refractory to date, our understanding of dG riboswitch specificity was instead advanced by a structural study that revealed a modest switch from guanine to dG specificity following the introduction of a limited number of replacements in the ligand-binding pocket of the guanine riboswitch17. Nevertheless, these core mutations need to be supplemented by additional extensive changes in the P2-L2 and P3-L3 hairpins to improve dG binding by ~1,000 fold17 in order to reach the wild-type dG affinity. These data suggest that the ligand-binding pocket is not the sole determinant of dG binding specificity. However, considerable improvement in dG affinity in some mutant constructs, for instance, when the non-conserved P2 helix (Fig. 1a) in the guanine riboswitch is replaced by a corresponding helix from the dG riboswitch17, cannot be easily rationalized with the available structural information, while the remaining specificity determinants for dG recognition cannot not be reliably identified.
Figure 1.
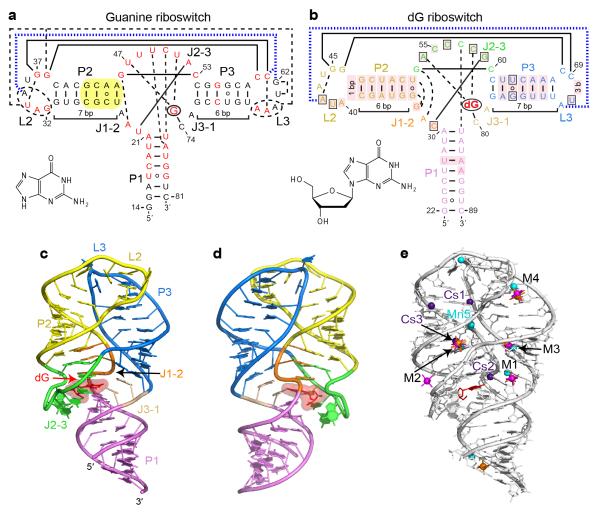
Overall structure and tertiary interactions of the dG-bound M. florum riboswitch. a, Secondary structure and tertiary interactions in the Bacillus subtilis xpt G riboswitch15 (PDB ID 1Y27). Canonical and non-canonical tertiary base pairing is depicted by long solid and dashed lines, respectively. Tertiary stacking interactions are in thick dashed blue lines. Nucleotides in red correspond to the positions that are >90% conserved in the known guanine riboswitch representatives13. Note that some conserved positions preserve chemical signatures (purine or pyrimidine) rather than the identity of nucleotides. Yellow shading shows a helical region whose replacement by the corresponding segment of the dG riboswitch significantly enhances dG binding17. The bound ligand is circled and its chemical formula is shown in inset. b, Secondary structure and tertiary interactions in the dG riboswitch. Pink shading indicates nucleotide differences from the guanine riboswitch. Boxes depict nucleotide variations from the guanine riboswitch that occur at otherwise highly conserved positions. c, Overall riboswitch structure in a ribbon representation, front view. d, Same structure, back view. e, Summary of the metal binding sites located using anomalous scatterers. Cs+ (indicated Cs), Mn2+ (indicated Mn), [Co(NH3)6]3+ and [Ir(NH3)6]3+ cations are shown in purple, cyan, orange and pink colors, respectively. General metal binding sites, where more than one cation type was detected, are designated M1 through M4.
In the present study, we performed structural and biochemical experiments to identify determinants of dG riboswitch selectivity and compared them with determinants of guanine recognition in the guanine riboswitch. Unexpectedly, our studies established a novel conformation of the riboswitch core that explained biochemical data on dG specificity and revealed a previously uncharacterized tertiary loop-loop interaction that critically contributes to dG binding affinity. Therefore, in addition to the residues layering the ligand binding pocket, a representative of the best studied riboswitch family exploits naturally altered structural elements outside of the pocket region to achieve high ligand specificity and affinity.
RESULTS
Recognition of dG in the ligand binding pocket
To discover molecular features essential for specific ligand binding, we have determined the 2.3 Å crystal structure of the complex between dG and the metabolite-sensing domain of the wild type M. florum type I-A dG riboswitch that controls the ribonucleotide reductase gene13 (Figs. 1c,d, Supplementary Results on line, statistics in Supplementary Table 1). The riboswitch structure conforms to the anticipated ‘tuning fork’ global fold previously described for adenine/guanine riboswitches14,15. The structure consists of the regulatory helix P1, the ‘handle’ of the fork, connected via a three-way junction to ‘prongs’, parallel hairpins P2-L2 and P3-L3 held together by interacting loops L2 and L3 (Figs. 1c,d and Supplementary Fig. 1). Since metal cations such as K+ and Mg2+ typically facilitate RNA folding, we identified cation binding sites in the structure by soaking riboswitch crystals in solutions of anomalous scatterers Cs+, Mn2+ and [Co(NH3)6]3+, which mimic K+, Mg2+ and fully hydrated Mg2+ cations, respectively (Fig. 1e, Supplementary Fig. 2, Supplementary Table 1).
The junctional core of the dG riboswitch comprises the top part of helix P1 and bottom sections of helices P2 and P3 joined by J1-2, J2-3 and J3-1 segments (Fig. 2a and Supplementary Fig. 1). Bound dG is positioned in the center of the core where it forms a Watson-Crick base pair with C80, equivalent to the discriminatory C74 in the guanine riboswitch14,15. The major and minor groove edges of the dG base interact with the 2′-OH of C31 and the base of C58, respectively. The O6 atom is proximal to a cesium binding site Cs2 (Fig. 1e). The dG deoxyribose contacts the A30 base and faces a four-nucleotide loop composed of C57 and stacked residues A54-C55-C56 (Fig. 2a). Within this loop segment, the 2′-OH of the C56 sugar and the N4 of C57 form hydrogen bonds with the ligand.
Figure 2.
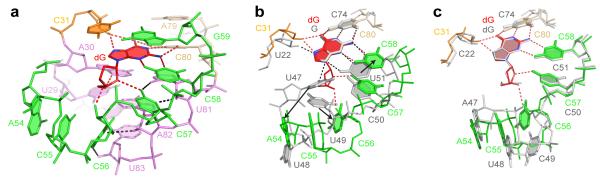
Structural features of the dG binding pocket. a, A view of the junctional core with bound dG. Red and black dashed lines depict putative hydrogen bonds involved in dG binding and tertiary RNA contact formation, respectively. b, Superposition of the ligand binding pockets from the dG (in colors) and guanine15 (in grey) riboswitch structures showing accommodation of the deoxyribose sugar ring. Red and black dashed lines depict putative hydrogen bonds involved in dG and guanine recognition, respectively. Arrow shows major conformational changes. c, Superposition of the ligand binding pockets from dG (in colors) and hybrid guanine-dG 17 (in grey) riboswitch structures. Putative hydrogen bonds between the dG ligand and dG riboswitch are depicted in red dashed lines.
Comparison of dG and guanine riboswitch structures establishes that both riboswitches recognize the Watson-Crick and Hoogsteen edges of the bound purines in a similar manner while several critical changes allow for the accommodation of the additional deoxyribose moiety in dG (Figs. 2b and Supplementary Fig. 3). First, the replacement of conserved U51 by C58 makes room for the sugar since the cytosine base has to shift along the minor groove edge of dG to form a novel set of hydrogen bonds with N3 and N2 of the ligand (Fig. 2b and Supplementary Fig. 3b). In agreement with the structure, a C58U substitution in the context of the dG riboswitch reduced dG binding affinity 83-fold and increased guanine affinity 2.8-fold17. Second, additional space for the deoxyribose is provided by substituting a conserved U47 with A54. In the guanine riboswitch, U47 contacts the ligand and U51, while in the dG riboswitch, A54 is unable to make similar interactions and instead is extruded from the ligand-binding pocket. This substitution modestly contributes to the specificity of dG binding since a U47A mutant in the context of the guanine riboswitch increases dG affinity 4.5-fold and reduces guanine binding 1.4-fold17. Third, C55 is moved slightly inwards in comparison with U48 from the guanine riboswitch, so as to maximize stacking on A54 and C56 (Fig. 2b and Supplementary Fig. 3d). Fourth, the base of C56, equivalent to the conserved U49, is rotated to avoid close juxtaposition with the deoxyribose of the ligand. This rotation prevents the formation of a base triple with the U29-A82 base pair and instead facilitates hydrogen bonding with the 2′-OH groups of A82 and U83, thereby connecting nucleotides of the J2-3 loop to helix P1 (Fig. 2a). Remarkably, the wild-type dG riboswitch structure shares several similarities in the conformation of the dG-recognizing nucleotides with the previously determined hybrid guanine-dG riboswitch structure that carried dG-specific substitutions in the core17. Both structures revealed movements of C58 and A54 to accommodate the deoxyribose moiety (Fig. 2c and Supplementary Fig. 3b) and hydrogen bonding between the dG sugar moiety and A30 and C57 (Supplementary Fig. 3c). The differences include the positioning of A79 further away from the C31-G59 base pair, the presence of a Mg2+ cation in the vicinity of G59 (Supplementary Fig. 3a), an additional water-mediated contact between the deoxysugar and C57 (Supplementary Fig. 3c), and interactions of the J2-3 loop residues with the deoxysugar and A82 (Supplementary Fig. 3d) in the natural dG riboswitch structure. Together, our structure and published structural and biochemical data13,17 suggest that C58 and the nucleotides of the J2-3 loop are important elements of dG riboswitch specificity.
Altered conformation above the ligand binding pocket
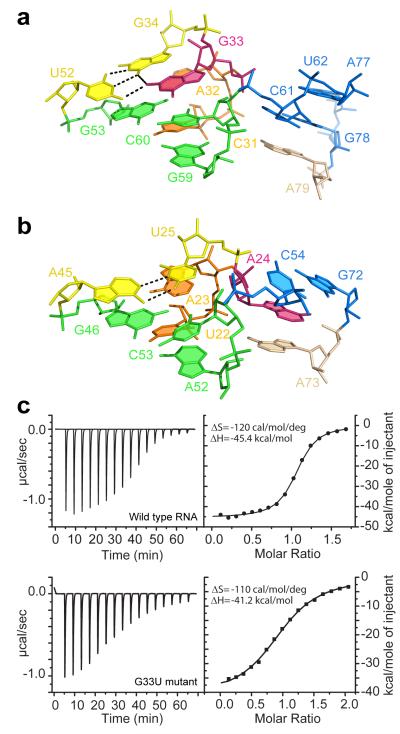
Above the bound ligand, the C31-G59 base pair and A79 form the foundations of two stacks emerging from the junction (Figs. 2a and 3a). The first stack continues with two nucleotide triples that sit on top of the C31-G59 base pair and define the directionality of the adjoining double-stranded P2 helix. In the second stack, A79 is capped by a C61-G78 base pair that begins the 7-bp P3 helix. Comparison with guanine and adenine riboswitches revealed that this junctional conformation is specific to the dG riboswitch (Figs. 3a,b). In the dG riboswitch, the longer helix P3 moves down by a register to accommodate an extra base pair so that the C61-G78 pair occupies the position of the non-paired A24 of the guanine riboswitch. The G33 residue that substitutes for the non-conserved A24 turns its sugar-phosphate backbone around towards the P2-L2 hairpin and forms a G•G platform with G34, which is engaged in base pairing with U52 (Fig. 3a and Supplementary Fig. 4). As a consequence of the swinging movement of G33, the hairpin stems are less interlinked in the junctional region but apparently more stable since more nucleotides are involved in hydrogen bonding and stacking.
Figure 3.
Comparison of junctional regions above the ligand binding pockets in the dG and guanine riboswitches. Purine residues G33 and A24 that adopt alternative conformations are shown in pink. a, Alignment of G33 in the structure of the dG riboswitch. b, Alignment of A24 in the structure of the guanine riboswitch. c, Representative ITC titrations with integrated fitted heat plots of dG binding to the wild type dG riboswitch (top) and the G33U mutant (bottom) at 2 mM Mg2+.
To test the contribution of the G33•(G34•U52) triple to dG binding, we disrupted the alignment of the triple by nucleotide substitutions and measured dG binding affinity by isothermal titration calorimetry (ITC) (Fig. 3c and Supplementary Fig. 5). In buffer containing 2 mM Mg2+, the replacement of the wobble G34•U52 pair with a canonical G-C base pair abolished dG binding (Supplementary Figs. 5a,b), while a G33U substitution that eliminated hydrogen bonding with G34, moderately (~9-fold) decreased dG binding affinity (Fig. 3c). A G33A mutation that introduced an alternative partner for pairing with U52 caused a much stronger (~50-fold) reduction in dG binding affinity. These data establish that the G33•(G34•U52) triple contributes to dG binding and suggest that G33 plays an important role in the formation of the triple. At a higher 20 mM Mg2+ concentration that typically provides extra stability to RNA structures, we observed weakening of the negative effects of these mutations (Supplementary Fig. 5a), thus signifying the involvement of the triple in the formation of the riboswitch conformation. Nevertheless, the U52C mutation still reduces dG binding ~17-fold at this Mg2+ concentration, while binding curve fitting reproducibly suggests that only a quarter of the mutant RNA is capable of binding to dG (N=0.25) despite applied RNA refolding procedures. The metal soaking experiments located three metal binding sites, M2, Cs3 and Mn5, in the vicinity of the triple (Fig. 1e and Supplementary Fig. 2). The M2 and Mn5 cations bridge P2 and P3, and therefore the G33•(G34•U52) triple may also contribute to the spatial arrangement of the hairpins through the coordination of metal cations. Since the riboswitch does not bind to dG in the absence of Mg2+ (Supplementary Fig. 5c), these cations may increase the overall stability of the riboswitch. The formation of the G•(G•U) triple because of the U-A to G•U base pair substitution at the base of the P2 helix reasonably explains the unexpected 270-fold gain in dG binding affinity when the bottom half of P2 helix was converted to the dG riboswitch sequence in the hybrid guanine-dG riboswitch (Fig. 1a)17. Interestingly, among the 1,244 sequences of adenine/guanine riboswitches18, a guanine or a G•U base pair from the triple is present only in several cases and neither sequence has a G•(G•U) triple, while both classes of dG riboswitches possess the triple. Given that nucleotide identity at the position equivalent to G33, typically represented by adenine or uracil, is important for high affinity ligand binding to guanine/adenine riboswitches19,20, we conclude that the G33•(G34•U52) triple is exclusive to dG riboswitches and that position 33 can be considered as one of the ligand specificity determinants in the purine riboswitch family.
Role of peripheral loop-loop contacts in ligand binding
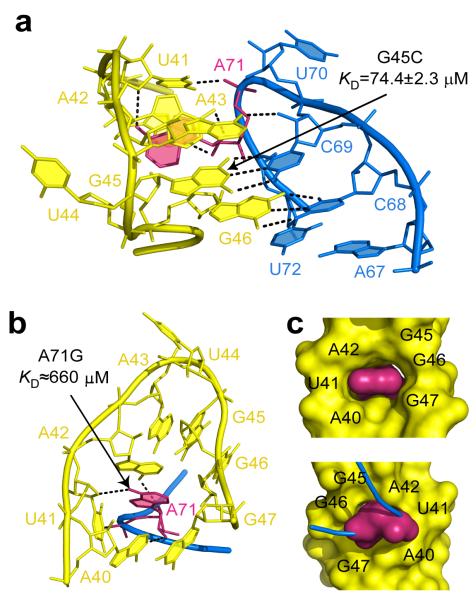
The tertiary contacts between the terminal loops, called kissing loop interactions, represent an additional element that could, to some extent, be tailored for dG binding. As anticipated from chemical probing17, the reduction in the size of the L3 loop and conserved nucleotide substitutions in the L2 loop dramatically changed the conformation and interface of the kissing loops in the dG riboswitch in comparison with the guanine riboswitch. The dG riboswitch preserved two tertiary Watson-Crick G-C base pairs found in the guanine riboswitch but lost three tertiary non-canonical base pairs and stacking interactions (Figs. 1a-b, 4a, and Supplementary Fig. 6), thereby decreasing the number of tertiary hydrogen bonds from 17 to 11. To strengthen loop contacts, the dG riboswitch utilizes an unprecedented21 RNA feature which can be defined as a molecular ‘key-and-lock’ element: A71 from L3, a ‘key’, inserts into the semi-open pocket, a ‘lock’, built by six nucleotides of the P2-L2 hairpin (Figs. 4b,c and Supplementary Fig. 6). In the pocket, A71 stacks on A42 and uses only one atom in the base for tertiary hydrogen bonding, while the other bonds involve the sugar-phosphate moiety of A71 and the bases of A40, U41 and A42. A71 contributes 221 Å2 to the 423 Å2 loop-loop contact surface in the dG riboswitch. This interface value is comparable to the guanine riboswitch inter-loop interface of 410 Å2. However, a strong ~6,500 and ~750-fold decrease in dG binding affinity in the A71G and G45C mutants at 20 mM Mg2+ and abolished binding to A71G RNA at 2 mM Mg2+ (Figs. 4a,b and Supplementary Fig. 5d) indicate that the integrity of tertiary loop interactions is likely more critical for ligand binding in the dG riboswitch than in adenine/guanine riboswitches, where any single nucleotide substitution in the loops resulted in a weaker reduction22,23. The A71G mutation also suggests that the L2 “lock” is attuned to adenine and is not sufficiently flexible to be ‘picked up’ by a guanine ‘key’, which would place the oxygen atoms of guanine and U41 in close proximity. The formation of the inter-loop interactions may depend on metal cations since soaking experiments identified several metal binding sites, including M4 and Cs1, within the inter-loop interface (Fig. 1e and Supplementary Fig. 2).
Figure 4.
Tertiary loop-loop interactions in the dG riboswitch. Dashed lines show putative tertiary hydrogen bonds. Key residue A71 is shown in pink. Binding affinities of dG for mutant RNAs are indicated on the panels. a, Binding interface. b, The key-and-lock element of the interacting loops. c, Surface representation. Front (top) and back (bottom) views.
Riboguanosine binding involves weaker interactions
The specificity of gene expression control demands that the dG riboswitch efficiently discriminate against nucleosides and nucleotides related to dG. The dG riboswitch was postulated to bind guanosine with weaker affinity because of a steric clash between the RNA and the additional 2′-OH group of guanosine13. However, based on the dG-bound structure, it is conceivable that the 2′-OH group could be accommodated in the ligand binding pocket if the guanosine ribose adopted its favorable C3′-endo conformation rather than the C2′-endo conformation, characteristic of deoxyribonucleosides. The crystal structure of the dG riboswitch bound to guanosine (Fig. 5a, Supplementary Table 2 and Supplementary Fig. 7) indeed revealed that the ribose moiety of guanosine is in the C3′-endo conformation. This sugar pucker is incompatible with hydrogen bonding of the 3′-OH to C56, observed in the dG bound structure, and C56, released from ligand binding, extrudes from the pocket and likely destabilizes the J2-3 loop, thereby decreasing guanosine binding by ≈50-fold (Supplementary Fig. 5e).
Figure 5.
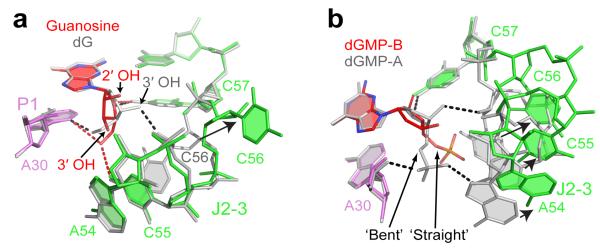
Specificity of dG recognition by the riboswitch. a, Distinct recognition of dG (grey) and guanosine (colored) by the dG riboswitch shown following superposition of the junctional cores from molecules A. Red and black dashed lines depict putative hydrogen bonds between the RNA and ligand’s ribose and deoxyribose, respectively. Arrow shows the conformational change of C56. b, Conformational differences in the J2-3 loops (depicted by arrows) caused by alternative ‘straight’ and ‘bent’ conformations of the sugar-phosphate moiety of dGMP. The view shows overlaid molecules A (grey) and B (colored) from the asymmetric unit of the dGMP bound structure.
Alternative ligand conformation in analog discrimination
In line with published data13, the dG riboswitch binds deoxyguanosine monophosphate (dGMP) ≈3,800-fold (Kd=379±216 μM) weaker than dG; nonetheless, the dG bound structure suggests that a phosphate moiety of dGMP can be accommodated after minimal adjustments in the J2-3 region. The structure of the dG riboswitch bound to dGMP confirmed this expectation. It showed that dGMP binding requires small shifts of the nucleotides in the vicinity of the ligand phosphate (Supplementary Fig. 8a, Supplementary Table 2), thereby accommodating the phosphate moiety in a ‘bent’ conformation and facilitating the formation of two intermolecular hydrogen bonds. Comparison with the structure of the second riboswitch molecule within the asymmetric unit revealed an alternative ‘straight’ conformation of the phosphate moiety (Fig. 5b and Supplementary Figs. 8a,b). In this molecule designated B, the J2-3 loop nucleotides are shifted much further than in molecule A and are not involved in hydrogen bonding with the ligand phosphate. Therefore, a straight phosphate conformation may cause destabilization of the loop, as reflected in the deterioration of electron density around the loop in molecule B (Supplementary Fig. 8c).
Extrusion of C56 and further deterioration of electron density was observed in the guanosine monophosphate (GMP) bound structure, where the phosphate moieties of both GMP molecules adopted straight conformations (Supplementary Fig. 8d,e). Analysis of crystal packing interactions indicated that the bent phosphate conformation observed for the dGMP complex presented above was possibly trapped in the crystal due to the stabilization of the J2-3 loop by the neighboring riboswitch molecule (Supplementary Fig. 9). Nonetheless, these structural data suggest that the ability of the flexible sugar-phosphate moiety to adopt different riboswitch-bound conformations may interfere with the trapping of phosphorylated ligands by the J2-3 loop and preclude the strong and specific binding of nucleotides to the dG riboswitch.
DISCUSSION
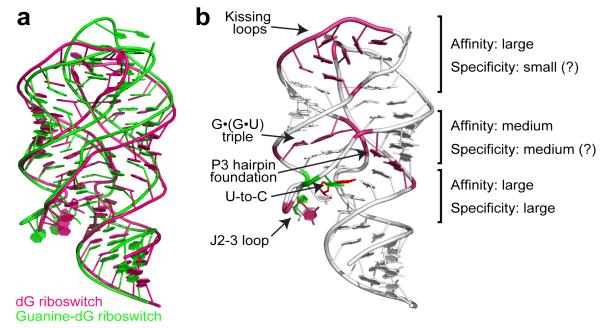
The wealth of adenine/guanine riboswitch structures with bound purine analogs23-27 has established the existence of some plasticity within the purine binding pocket and implied that other purine analogs could potentially be recognized through limited alterations in the pocket. In support of this view, our structural studies showed that the natural dG riboswitch adopts an overall architecture reminiscent of adenine/guanine riboswitches, emphasizing their evolutionary relationship (Fig. 6a). However, multiple nucleotide substitutions cause unexpected conformational rearrangements and reshape the riboswitch for specific binding to dG. Currently, this finding stands out since riboswitches that recognize related metabolites either adopt practically identical structures14,15 or share very little in common28-31.
Figure 6.
Summary of dG recognition. a, Superposition of the natural dG riboswitch structure (this study, pink) and the guanine riboswitch based guanine-dG riboswitch structure17 (green) shows similar global RNA folds. b, Involvement of riboswitch structural features in dG binding to the natural dG riboswitch. Pink and green nucleotides designate areas where the most critical differences from adenine/guanine riboswitches occur. Green nucleotides highlight similarities with the guanine-dG riboswitch structure17. Each element’s contribution to the specificity and affinity of dG binding is roughly estimated based on current and published data13,17.
Our structural results and published data13,17,32 point to nucleotides in the ligand binding pocket as primary dG specificity determinants (Fig. 6b). These determinants include two nucleotides,common to the guanine riboswitch, that distinguish between the purine bases and other nucleotides of the pocket, including C58 and the J2-3 loop, that can be considered specific determinants for dG recognition. Previously, the J2-3 loop was shown to act as a flexible lid that entraps the ligand after its initial docking in the pocket of adenine/guanine riboswitches26,33. In the dG riboswitch, this loop is exploited for both specific dG encapsulation and discrimination against related compounds. Our structural studies showed that destabilization of the J2-3 conformation plays a key role in discrimination against various dG analogs and likely perturbs genetic control in the dG riboswitch system. The tertiary triple interactions between the J2-3 loop and regulatory helix P1 that substantially contribute to helix P1 stabilization in guanine/adenine riboswitches23 are replaced in part by contacts with deoxyribose in dG riboswitches. Since some of these interactions are lost in the analog-bound dG riboswitches, the nucleobases bind to the junction less tightly and cannot provide the hydrogen bonding and stacking surface needed to stabilize the junction and regulatory helix P1. Thus, an alternative riboswitch conformation forms, interfering with transcription termination13. In addition to the J2-3 loop, the regions outside of the binding pocket are likely involved in analog discrimination, possibly by stabilizing a correct junctional scaffold. In-line probing13 and our ITC measurements demonstrated 100- and 50-fold decreases, respectively, in guanosine binding by the wild type dG riboswitch, while the hybrid guanine-dG riboswitch showed only a 3-fold discrimination against guanosine17.
The unexpected conformational rearrangement above the binding pocket, where a non-conserved purine residue (G33 in the dG riboswitch) swung out from the P3-L3 hairpin in the guanine riboswitch to the P2-L2 hairpin in the dG riboswitch, has been validated by recent solution NMR experiments, which supported the close positioning of G33 to G53, C60 and U52 in helix P232. In its new position, G33 participates in the formation of an additional nucleotide triple, thereby likely stabilizing the P2-L2 hairpin and the junction. This is supported by the loss of reactivity in the bottom part of helix P2 in SHAPE experiments performed on the dG riboswitch13,17. Strengthening of the P2-L2 hairpin may be important for genetic regulation by the dG riboswitch, since this hairpin, shown experimentally to fold prior to the P3-L3 hairpin in the related adenine riboswitch34, has been deemed critical for the folding of entire guanine/adenine riboswitches35. The junctional arrangement in the dG riboswitch may prevent the extensive movements of the P2-L2 hairpin observed in single molecule experiments with the guanine riboswitch35. The decreased dynamic properties and increased stability of the P2-L2 hairpin could compensate for the flexibility of helix P3, observed in the NMR study of the free dG riboswitch32, and contribute to the stabilization of the free form, as shown for the guanine riboswitch36. This would decrease the entropic penalty for ligand binding in the context of more complex ligand recognition and less pronounced tertiary inter-loop interactions in the dG riboswitch.
In adenine/guanine riboswitches, tertiary loop-loop interactions are important for ligand binding14,22,23,33. Since the relative position of the P2-L2 and P3-L3 hairpins is slightly different in the dG and guanine riboswitches (Fig. 6a), it is tempting to speculate that the novel core conformation specifically assists the formation of new tertiary loop interactions which, in turn, help the riboswitch maintain the dG-specific junctional conformation. This view is supported by a 2.9-fold reduction in dG binding in the mutant dG riboswitch whose kissing loops are replaced by ones from the guanine riboswitch17. Although this small decrease observed at 20 mM Mg2+ suggests that different sets of kissing loops may be exchanged in a modular fashion, it is conceivable that lower physiological Mg2+ concentrations may negatively impact the inter-loop interactions and reduce dG binding22.
The loop-loop interactions in the adenine/guanine and dG riboswitches bear little resemblance14,15. In the dG riboswitch, the kissing loops adopt a novel ‘key-and-lock’ loop motif, which is less interlinked, lacks base quartets and heavily depends on the insertion of a single adenine from the L3 loop into the L2 loop. Our structural studies generally agree with recent NMR studies32 that showed the formation of two tertiary Watson-Crick G-C base pairs and suggested the close positioning of U41 and A71. Although the crystal structure did not reveal the reverse Hoogsteen U41•A71 base pair suggested by NMR32, these nucleotides do utilize the respective base edges for interactions with each other. Remarkably, guanine riboswitches with loops similar to the dG riboswitch loops13 either show >50-fold reduction in guanine binding (class III riboswitches) or possess other changes, such as shortened P2 and P3 helices (class V-A riboswitch). Variations in loop-loop interactions might serve as a tool for modifying ligand binding affinity20, since the response of riboswitches specific for the same ligand may be attuned to different ligand concentrations.
Numerous studies underlined the importance of Mg2+ cations for the folding of adenine/guanine riboswitches14,15,22,33,36-40, although they are not absolutely critical for guanine binding to the guanine riboswitch15,16. Unlike the guanine riboswitch, the dG riboswitch completely lost dG binding affinity in the absence of Mg2+,reminiscent of the observation that weaker binders like hypoxanthine require Mg2+ for interaction with the guanine riboswitch14. Since the dG riboswitch overcomes the requirement of Mg2+ assisted folding for ligand recognition by the elongation of helix P132, Mg2+ cations likely play a general RNA stabilizing role, as supported by our metal site mapping. Metal binding sites were identified in areas proximate to the loops, where in agreement with NMR data32, cations stabilize the tertiary loop-loop contacts. Sites were also found close to the bottom parts of helices P2 and P3, where as suggested by NMR32, they facilitate the folding of regions adjacent to the junction. Some riboswitches also utilize Mg2+ or K+ cations to compensate for the negative charges of their ligands41,42. However, the structures did not reveal cations in proximity to the phosphate moieties of bound GMP and dGMP, suggesting that the riboswitch does not have a cation-binding surface which would facilitate RNA-phosphate interactions.
Adenine/guanine riboswitches represent one of the best systems for studying RNA folding and RNA-small molecule interactions. Structure determination of the natural dG riboswitch presents an opportunity to expand on these studies so as to decipher how related natural RNA conformations discriminate amongst distinct purine ligands. Our structural results, as well as dG binding17 and folding19,20,43 experiments, demonstrate the multitude of factors and complexity of sequence combinations that may impact on the evolution of new riboswitch specificities. Thus, re-engineering the riboswitch to accommodate even small ligand alterations may necessitate changes in both the ligand binding pocket and remote parts of the RNA. Given the recent successes in the re-engineering of purine riboswitches44 and the development of antibacterials45,46 that target purine riboswitches in a mammalian model of staphylococcal infection46, the dG riboswitch structure provides new frameworks and insights into future structure-guided drug design applications.
METHODS
RNA preparation, crystallization and data collection
We crystallized the dG-riboswitch complex using 68-nt and 70-nt long RNAs, named RNA1 and RNA2, and ammonium sulfate or pentaerythritol propoxylate as precipitants. The RNAs encoding the M. florum riboswitch sequences were transcribed in vitro and purified by denaturing polyacrylamide gel electrophoresis followed by anion-exchange chromatography and ethanol precipitation. Complexes of the dG riboswitch with dG were prepared by mixing 0.5 mM RNA with 0.55 mM dG in a buffer containing 50 mM K-acetate, pH 6.8 and 5 mM MgCl2. Crystals were grown at 20 °C for 3-5 days using the hanging-drop vapor diffusion method after mixing the dG-RNA1 complex at an equimolar ratio with the reservoir solution containing 0.05 M Na-acetate, pH 5.1, ~3.0 M (NH4)2SO4, 0.5 mM spermine and 20 mM MgCl2. Alternatively, crystallization was carried out at 4 °C for 1-2 weeks using the hanging-drop vapor diffusion method after mixing the dG-RNA2 complex at an equimolar ratio with the reservoir solution containing 0.05 M Na-HEPES, pH 7.5, ~40 % (v/v) pentaerythritol propoxylate, 0.2 M KCl and 0.0-2.0 mM spermidine. For data collection, crystals were flash-frozen in liquid nitrogen. For heavy atom and cation soaking, crystals grown in pentaerythritol propoxylate conditions were transferred into the solution containing components of the reservoir solution at 10% higher concentrations in the absence of spermidine, supplemented with 2 mM [Ir(NH3)6]Cl3, 10 mM [Co(NH3)6]Cl3, 10 mM BaCl2 or 10 mM MnCl2 and soaked for 4 h. Data were collected at 100 K at the X29a beamline of the National Synchrotron Light Source (NSLS, Brookhaven) and processed with HKL2000 (HKL Research). Crystals with dG analogs were obtained by co-crystallization in ammonium sulfate conditions.
Structure determination, refinement and analysis
Since molecular replacement trials using adenine, guanine as well as hybrid guanine/dG riboswitches and their fragments as search models were unsuccessful, the structure of the dG-bound RNA2 was determined by single-wavelength anomalous diffraction (SAD) method using 2.9 Å iridium hexamine data, that was processed using SHELXD47 and autoSharp48 (Supplementary Table 1). The initial RNA model was autobuilt in Phenix49 and improved by iterative rounds of building/refinement in Turbo-Frodo (http://afmb.cnrs-mrs.fr/rubrique113.html), Phenix and Refmac50 using a 2.3 Å native data set. dG was added to the model at a later stage based on the experimental and refined electron density maps. Cations and their coordinated waters were added based on the coordination geometry, coordination distances, and analysis of omit and anomalous maps. All other riboswitch structures were determined by molecular replacement using the native riboswitch structure as a starting model (Supplementary Tables 1 and 2) and refined in Refmac and Phenix. The contact surface areas were calculated by using PISA webserver (http://www.ebi.ac.uk/msdsrv/prot_int/cgi-bin/piserver).
ITC measurements
Experiments were performed 2-6 times using the Microcal calorimeter ITC200 at 30 °C. Prior to titration, 0.01-0.3 mM RNA samples were dialyzed overnight at 4 °C against experimental buffer containing 50 mM HEPES-KOH, pH 6.8, 100 mM KCl and 2 or 20 mM MgCl2. The conditions with 20 mM MgCl2 are comparable with the conditions used for KD measurements in published literature13,17. In the conditions without MgCl2, dialysis was performed in the buffer supplemented with 0.5 mM ethylenediaminetetraacetic acid (EDTA). For measurements, the ligands, dissolved in dialysis buffer at concentrations ~3-100 fold higher than the RNA concentration, were typically titrated into RNA in the sample cell (V=207 μL) by 17-20 serial injections of ~2.4 μL each, with 180 s intervals between injections and a reference power of 10 μcal sec−1. The thermograms were integrated and analyzed using Origin 7.0 software (Microcal, Inc) assuming a one-to-one binding model.
Supplementary Material
Acknowledgments
We thank the personnel of beamline X29 at the Brookhaven National Laboratory, funded by the US Department of Energy, for assistance in data collection. We thank Dr. Ouathek Ouerfelli (MSKCC, New York) for the synthesis of iridium hexamine and E. Ennifar (IBMC, Strasbourg) for the discussion of the refinement strategy. D.J.P. was supported by funds from the National Institutes of Health grant GM66354.
Footnotes
Accession codes. Coordinates of the X-ray structures of the dG riboswitch have been deposited in the RCSB Protein Data Bank. The accession codes are as follows: bound to dG, 3SKI; bound to guanosine, 3SKZ; bound to GMP, 3SLQ; bound to dGMP, 3SLM; [Ir(NH3)6]3+-soaked, 3SKL; Cs+-soaked, 3SKW; Mn2+-soaked, 3SKT; and [Co(NH3)6]3+-soaked, 3SKR.
Author contributions A.P. and O.P. crystallized the riboswitch. O.P. determined the structures. O.P. and A.S. refined the structures and performed binding experiments. A.S. and D.J.P. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing financial interests The authors declare no competing financial interests.
Additional information Supplementary information is available online at http://www.nature.com/naturechemicalbiology/.
Reprints and permissions information are available at npg.nature.com/reprintsandpermissions.
References
- 1.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem. 2009;78:305–34. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serganov A, Patel DJ. Ribozymes, riboswitches and beyond: regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–33. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 5.Roth A, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 6.Corbino KA, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epshtein V, Mironov AS, Nudler E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. U S A. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 9.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. U S A. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serganov A. Determination of riboswitch structures: light at the end of the tunnel? RNA Biol. 2010;7:98–103. doi: 10.4161/rna.7.1.10756. [DOI] [PubMed] [Google Scholar]
- 11.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 12.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 13.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci U S A. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 15.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noeske J, et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci U S A. 2005;102:1372–7. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol. 2009;385:938–48. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gardner PP, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–40. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:339–50. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de la Pena M, Dufour D, Gallego J. Three-way RNA junctions with remote tertiary contacts: a recurrent and highly versatile fold. RNA. 2009;15:1949–64. doi: 10.1261/rna.1889509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–68. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J Am Chem Soc. 2006;128:14214–5. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–68. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol. 2006;359:754–68. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Jain N, Zhao L, Liu JD, Xia T. Heterogeneity and dynamics of the ligand recognition mode in purine-sensing riboswitches. Biochemistry. 2010;49:3703–14. doi: 10.1021/bi1000036. [DOI] [PubMed] [Google Scholar]
- 28.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15:177–82. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 29.Lu C, et al. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol. 2008;15:1076–1083. doi: 10.1038/nsmb.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 31.Edwards AL, Reyes FE, Heroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16:2144–55. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wacker A, et al. Structure and dynamics of the deoxyguanosine-sensing riboswitch studied by NMR-spectroscopy. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brenner MD, Scanlan MS, Nahas MK, Ha T, Silverman SK. Multivector fluorescence analysis of the xpt guanine riboswitch aptamer domain and the conformational role of guanine. Biochemistry. 2010;49:1596–605. doi: 10.1021/bi9019912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ottink OM, et al. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–12. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noeske J, Schwalbe H, Wohnert J. Metal-ion binding and metal-ion induced folding of the adenine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2007;35:5262–73. doi: 10.1093/nar/gkm565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 39.Noeske J, et al. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–83. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buck J, Noeske J, Wohnert J, Schwalbe H. Dissecting the influence of Mg2+ on 3D architecture and ligand-binding of the guanine-sensing riboswitch aptamer domain. Nucleic Acids Res. 2010;38:4143–53. doi: 10.1093/nar/gkq138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serganov A, Huang L, Patel DJ. Structural insights into amino acid binding and gene control by a lysine riboswitch. Nature. 2008;455:1263–1267. doi: 10.1038/nature07326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature. 2009;458:233–7. doi: 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delfosse V, et al. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010;38:2057–68. doi: 10.1093/nar/gkp1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixon N, et al. Reengineering orthogonally selective riboswitches. Proc Natl Acad Sci U S A. 107:2830–5. doi: 10.1073/pnas.0911209107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim JN, et al. Design and antimicrobial action of purine analogues that bind Guanine riboswitches. ACS Chem Biol. 2009;4:915–27. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulhbacher J, et al. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider TR, Sheldrick GM. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 2002;58:1772–9. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 48.Fortelle E.d.l., Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multi-wavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 49.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.