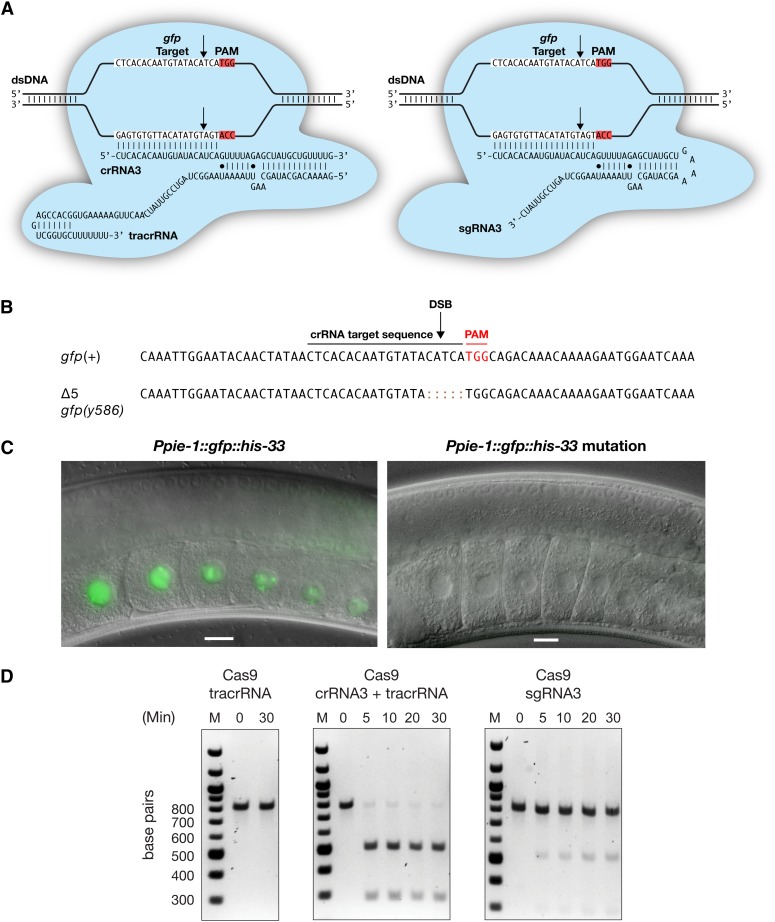
Figure 7.
Heritable, RNA-targeted gene disruption in the C. elegans germline using CRISPR/Cas9. (A) Schematic of Cas9 interacting with its dsDNA target, gfp, and either the dual crRNA:tracrRNA guide RNA (left) or an sgRNA (right), both having the same target DNA. Red highlights the TGG nucleotide protospacer-adjacent motif (PAM), and the arrow designates the DSB site, which occurs in genomic DNA complementary to the crRNA sequence. (B) DNA sequence of the gfp deletion resulting from NHEJ-mediated repair of the Cas9-induced DSB made from the Cas9 complex containing the dual crRNA:tracrRNA guide RNA. (C) Hermaphrodite gonads expressing the wild-type (left) or Cas9-mediated Δ5 mutant (right) version of the Ppie-1::gfp::his-33 transgene, which expresses GFP::histone 2B exclusively in the germline. Bars, 10 µm. (D) In vitro DNA cleavage assays comparing the effectiveness of Cas9-crRNA:tracrRNA and Cas9-sgRNA complexes. Double-stranded DNA cleavage was tested in time-course reactions, using the molar ratio of Cas9:guide RNA:target DNA as 0.5:1:0.5 µM. Reactions were conducted at 37°. The in vitro assays show that the dual RNA guides are more effective at promoting DNA cleavage than the sgRNAs, a finding that recapitulates our results in vivo demonstrating that the dual RNA guides are more effective at promoting mutations than the single RNA guides. M, 100-bp markers.