Abstract
The target of rapamycin (TOR) is a highly conserved protein kinase that regulates cell growth and metabolism. Here we performed a genome-wide screen to identify negative regulators of TOR complex 1 (TORC1) in Schizosaccharomyces pombe by isolating mutants that phenocopy Δtsc2, in which TORC1 signaling is known to be up-regulated. We discovered that Δnpr2 displayed similar phenotypes to Δtsc2 in terms of amino acid uptake defects and mislocalization of the Cat1 permease. However, Δnpr2 and Δtsc2 clearly showed different phenotypes in terms of rapamycin supersensitivity and Isp5 transcription upon various treatments. Furthermore, we showed that Tor2 controls amino acid homeostasis at the transcriptional and post-transcriptional levels. Our data reveal that both Npr2 and Tsc2 negatively regulate TORC1 signaling, and Npr2, but not Tsc2, may be involved in the feedback loop of a nutrient-sensing pathway.
Keywords: TORC1, Tor2, Npr2, Tsc2, nutrient response
THE target of rapamycin (TOR) plays vitally important roles in regulating cell growth and metabolism. TOR interacts with several proteins to form two structurally and functionally distinct complexes named TOR complex 1 (TORC1) and 2 (TORC2) (Laplante and Sabatini 2009, 2012). In response to environmental cues, TORC1 controls cell growth and differentiation by coordinating diverse cellular processes including transcription, translation, and autophagy. Research on TORC1 has generated a model of the complex TOR signaling network (Huang and Manning 2009; Orlova and Crino 2010; Laplante and Sabatini 2012). In the regulation of TORC1 signaling, four major signals have been identified, namely growth factors (insulin, IGF, etc.), energy status (AMP/ATP ratio), oxygen levels, and nutrients (amino acids) (Laplante and Sabatini 2009, 2012). In mammals, the tuberous sclerosis complex 1 and 2 (TSC1-TSC2) serves as a key point of signal integration. The growth factors stimulate TORC1 signaling via PI3K–Akt/PKB-mediated phosphoinhibition of TSC2 (Inoki et al. 2002; Manning et al. 2002). The energy starvation inhibits TORC1 signaling via AMPK-dependent phosphoactivation of TSC2 (Inoki et al. 2003). Then, TSC2 negatively regulates TORC1 activity by converting GTP-bound Rheb (Ras homolog enriched in brain) into its inactive GDP-bound state (Inoki et al. 2003; Tee et al. 2003). The amino acids, in particular the branched-chain amino acid leucine, positively regulate TORC1. The TORC1 signaling remains sensitive to amino acid deprivation in TSC2−/− cells (Nobukuni et al. 2005), indicating that the activation of TORC1 by amino acids is independent of TSC2. Recently, it was demonstrated that TORC1 responds to amino acid availability via mechanisms involving Rag GTPases (Sancak et al. 2008). In the presence of amino acids, the Rag GTPases interact with TORC1, thereby promoting the translocation of TORC1 to the lysosomal membranes and facilitating Rheb’s activation of TORC1 (Sancak et al. 2010).
In budding yeast Saccharomyces cerevisiae, TOR1 and TOR2 genes were originally identified as the targets of rapamycin, and mutations in TOR1 or TOR2 genes confer resistance to rapamycin (Heitman et al. 1991; Kunz et al. 1993). Studies using budding yeast have extensively enhanced our knowledge about TOR signaling; however, in this model there are no Tsc homologs. In contrast, in fission yeast Schizosaccharomyces pombe, Tsc1 and Tsc2 form a complex that acts as a GTPase activating protein (GAP) for the small GTPase Rhb1 (Rheb homolog) (van Slegtenhorst et al. 2004; Nakase et al. 2006), and Rhb1 physically associates with Tor2 in a GTP-dependent manner (Urano et al. 2005; Uritani et al. 2006). Recently, Nakashima et al. (2010, 2012) demonstrate that, similar to mammalian mTORC1–S6K–S6, the TORC1 (Tor2)–Psk1–Rps6 constitutes a nutrient-dependent signaling pathway. Valbuena et al. (2012) demonstrate that the Rag GTPases Gtr1 and Gtr2 activate TORC1 in response to amino acids. These studies provide evidence that fission yeast is a valuable model to study the conserved Tsc–Rheb–TOR signaling network.
In the present study, we performed a genome-wide screen in S. pombe to identify the genes that inactivate Tor2 by isolating deletion mutants that phenocopy tsc2 deletion. The Δnpr2 cells displayed similar phenotypes to Δtsc2 cells in terms of the growth defect on normal Edinburgh minimal medium (EMM) plates supplemented with leucine, canavanine resistance, and Cat1 mislocalization. However, Δnpr2 and Δtsc2 cells also showed several distinct phenotypes. Notably, upon rapamycin treatment, Isp5 transcriptional activity in Δnpr2 cells showed a marked increase that reached a level nearly 40- to 50-fold greater than that observed in Δtsc2 or wild-type (wt) cells, whereas upon nitrogen depletion the increase in Δnpr2 cells was significantly smaller than that observed in Δtsc2 or wild-type cells. Taken together, these results indicate that Tsc2 and Npr2 function differently as negative regulators of TORC1 signaling.
Materials and Methods
Strains, media, and genetic and molecular biology methods
The commercially available S. pombe nonessential gene haploid deletion strains (Kim et al. 2010) were purchased from Bioneer Corporation (http://pombe.bioneer.com/). The other strains used in this study are listed in Supporting Information, Table S1. The complete medium YPD (yeast extract–peptone–dextrose) and the minimal medium EMM have been described previously (Toda et al. 1996). YE plates (0.5% yeast extract, 3% glucose, 2% agar) are supplemented with 225 mg/liter adenine, histidine, leucine, uracil, and lysine to produce YES (yeast extract with supplements) plates. Gene disruptions are abbreviated by the gene preceded by Δ (for example, Δnpr2). Proteins are denoted by roman letters and only the first letter is capitalized (for example, Npr2).
Construction of Isp5 promoter reporter plasmids
Renilla luciferase was chosen as a reporter system, because it is minimally influenced by the level of ATP in living cells (Zhou et al. 2012). A 1038-bp DNA fragment in the 5′ flanking region of the isp5+ gene was amplified by PCR primers (sense primer 3940, 5′-AAC TGC AGG GGA TTT CAA GTC GGC CGC-3′; antisense primer 3941, 5′-CCG CTC GAG TTT AAT TTT TTG TTT GAT GG-3′). The amplified products containing 1038-bp Isp5 promoter were subcloned into the PstI/XhoI-digested pKB5878 (Zhou et al. 2012), a phRG(R2.2)-basic multicopy vector (Promega) that contains Renilla luciferase reporter gene. The resulting plasmid was registered as pKB8527 and used as Isp5 promoter reporter vector.
Real-time monitoring assays of Isp5 transcriptional activity
The Isp5 promoter activity was measured as described by Zhou et al. (2012) with minor modifications. Briefly, cells transformed with the reporter plasmid pKB8527 were cultured at 27° in EMM to mid-log phase and the optical density was adjusted to 0.3 at 660 nm. After incubation for 4 hr at 27°, 1 ml of cells was washed twice and resuspended in fresh EMM with or without NH4Cl, respectively. A final 5 mM coelenterazine (Promega, no. S200A) was added into the cells. Rapamycin (Sigma-Aldrich) was added to a final concentration of 0.2 μg/ml from a stock solution of 0.2 mg/ml in methanol, and Torin1 (Tocris Bioscience) was added to a final concentration of 200 nM from a stock solution of 1 mM in DMSO. Emission of light was detected and reported as relative light units (RLUs) using a luminometer (AB-2350; ATTO, Tokyo).
Construction of chromosome-borne Cat1–GFP under the control of its own promoter
The cat1+ gene was amplified by PCR with the genomic DNA of wild-type cells as a template. The sense primer was (3886) 5′-CGG GAT CCA TGT CCC ATA GCG ATT TTA ATA TGG-3′, and the antisense primer was (3887) 5′-CGG GAT CCG CGG CCG CCA CAG AAA ACC GAA CTG ATT TTC-3′. The amplified product containing the cat1+ gene was digested with BamHI, and the resulting fragment was subcloned into the BamHI site of BlueScriptSK (+) (Stratagene). Then, a BamHI/NotI fragment containing cat1+ was ligated to the BglII/NotI site of the C terminus of the GFP carrying the S65T mutation (Heim et al. 1995). The expression of pREP1–Cat1–GFP complemented the canavanine-resistant phenotype of the Δcat1 cells (data not shown). To obtain the chromosome-borne C-terminally tagged Cat1–GFP under the control of its own promoter, the DNA fragment containing the Cat1–GFP and ura4+ marker was integrated into the chromosome at the cat1+ gene locus of KP456 by homologous recombination after digesting the plasmid with a restriction enzyme.
Construction of GFP–Atg8 under pREP41 promoter
The atg8+ gene was amplified by PCR with the genomic DNA of wild-type cells as a template. The sense primer was (3956) 5′-CGG GAT CCA TG CGT TCT CAA TTC-3′, and the antisense primer was (3957) 5′-CGG GAT CCG CGG CCG CCT AAA AAG GAA ACA C-3′. The amplified product containing atg8+ was digested with BamHI/NotI, and ligated to the N terminus of the GFP carrying the S65T mutation under pREP41 promoter (Heim et al. 1995). For ectopic expression of proteins, we used the thiamine repressible nmt1 promoter (Maundrell 1993). Expression was induced by growing the cells in EMM without thiamine.
Gene deletion
A one-step gene disruption by homologous recombination was performed (Rothstein 1983). The cat1::ura4+ disruption was constructed as follows: The BamHI fragment containing cat1+ was subcloned into the BamHI site of pGEM7Zf (Promega). Then, a SmaI fragment containing ura4+ was inserted into the HincII site of the previous construct. The fragment containing the disrupted cat1+ gene was transformed into KP456. Stable integrants were selected on media lacking uracil, and disruption of the gene was checked by genomic Southern hybridization (data not shown).
The npr2::natMX6 disruption was constructed by the marker switching method (Hentges et al. 2005). The natMX6 was amplified by PCR with pFA6a-natMX6 as a template (Hentges et al. 2005). The sense primer was (MX4/6cassUP) 5′-GAC ATG GAG GCC CAG AAT AC-3′, and the antisense primer was (MX4/6cassDwn) 5′-TGG ATG GCG GCG TTA GTA TC-3′. The amplified product containing natMX6 cassette was transformed into npr2::KanMX4 cells and plated onto YES plates. The transformants were incubated at 27° for 1 day and replicated onto YES containing 100 μg/ml nourseothricin. The colonies were picked up, and tested for drug resistance. The cells that only showed nourseothricin resistance were registered as KP6366 (h− npr2::natMX6).
Miscellaneous methods
The fluorescence microscopy images were recorded and processed as described previously (Ma et al. 2009). Real-time reverse transcription–PCR was performed as described previously (Ryuko et al. 2012; Zhou et al. 2012). Database searches were performed using Pombe community database Pombase (http://www.pombase.org). Cell extract preparation and immunoblot analysis were performed as described previously (Sio et al. 2005). Anti-phospho-Akt substrate (PAS) polyclonal antibodies (no. 9611) were purchased from Cell Signaling Technology. Anti-α-tubulin (B5-1-2) antibodies were purchased from Sigma.
Results
Cell growth-based genomic library screen for negative regulators of TORC1
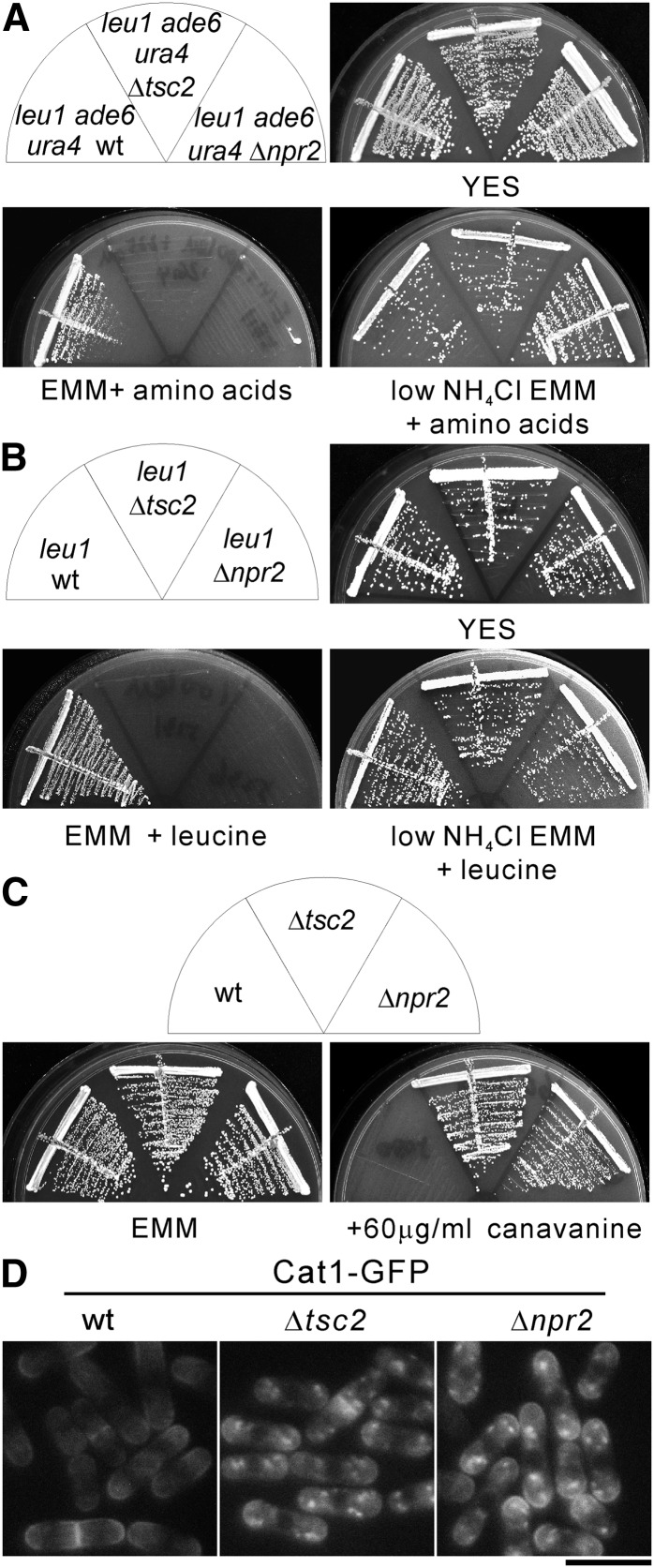
In fission yeast, several lines of evidence have demonstrated that Tsc2 functions as GAP for the small GTPase Rhb1 (van Slegtenhorst et al. 2004, 2005; Urano et al. 2005; Nakase et al. 2006), and Rhb1 positively regulates Tor2 activity (Urano et al. 2005; Uritani et al. 2006). Based on these reports we reasoned that isolation of the mutant strains that phenocopy Δtsc2 cells will identify novel negative regulators of Tor2. It is demonstrated that Δtsc2 cells have defects in amino acid uptake, thus resulting in slow growth when combined with auxotrophic mutations or resistance to the toxic arginine analog canavanine (Matsumoto et al. 2002; van Slegtenhorst et al. 2004). Prior to the comprehensive screening, we compared the growth of wild-type with Δtsc2 cells auxotrophic for leucine, adenine, and uracil (Bioneer) (Kim et al. 2010). As shown in Figure 1A, the Δtsc2 cells grew equally as well as wild-type cells on YES plates. However, Δtsc2 cells failed to grow on normal EMM (5 g/liter NH4Cl) media with amino acid supplements. It is demonstrated that poor nitrogen source rescued the growth defect by increasing amino acid permease expression (Weisman et al. 2005). This prompted us to investigate whether low NH4Cl (0.5 g/liter) EMM media can rescue the growth defect of Δtsc2 cells. On low NH4Cl EMM plates with amino acid supplements, the Δtsc2 cells grew as well as wild-type cells (Figure 1A), suggesting that the growth defect of Δtsc2 cells is caused by the defective amino acid uptake on normal EMM plates. Similarly, Δtsc2 cells auxotrophic for leucine (leu1 Δtsc2) failed to grow on normal EMM plates supplemented with leucine, but grew as well as wild-type cells auxotrophic for leucine (leu1 wt) on low NH4Cl EMM plates supplemented with leucine (Figure 1B). These results indicate that the growth defect of Δtsc2 cells is mainly due to leucine auxotrophy. Consistently, it is demonstrated that leu1-32 Δtsc2 failed to grow on PM media containing 40–200 μg/ml leucine (Matsumoto et al. 2002; Nakase et al. 2006). As reported previously (van Slegtenhorst et al. 2004; Nakase et al. 2006) and as shown in Figure 1C, the prototrophic Δtsc2 cells displayed canavanine resistance.
Figure 1.
The Δnpr2 and Δtsc2 cells display similar phenotypes. (A) The wild-type (KP93006), Δtsc2 (KP91087), and Δnpr2 (KP90390) cells auxotrophic for leucine, adenine, and uracil were streaked onto YES, EMM (5 g/liter NH4Cl), or low NH4Cl EMM (0.5 g/liter NH4Cl) supplemented with 100 mg/liter leucine, 225 mg/liter adenine, and 225 mg/liter uracil. The plates were incubated at 27° for 3 days (YES), 4 days (low NH4Cl EMM + amino acids), or 5 days (EMM + amino acids), respectively. (B) The leucine auxotrophic cells of wild-type (HM123), Δtsc2 (KP5131), and Δnpr2 (KP5236) were streaked onto YES, EMM (5 g/liter NH4Cl), or low NH4Cl EMM (0.5 g/liter NH4Cl) supplemented with 100 mg/liter leucine. The plates were incubated as described in Figure 1A. (C) The prototrophic cells of wild-type (KP5080), Δtsc2 (KP5128), and Δnpr2 (KP5237) were streaked onto EMM or EMM containing 60 μg/ml canavanine. The plates were incubated at 27° for 4 days (EMM) or 5 days (+60 μg/ml canavanine). (D) Subcellular localization of Cat1 in wild-type, Δtsc2, and Δnpr2 cells. The wild-type (KP5859), Δtsc2 (KP5826), and Δnpr2 (KP5822) cells expressing chromosome-borne Cat1–GFP under the control of its own promoter were grown to early log phase in EMM media at 27°. The fluorescence of the Cat1–GFP was examined. Bar, 10 μm.
Based on the phenotypes of Δtsc2 under various genetic backgrounds, we performed genome-wide screens as follows. In the preliminary screen, 3004 deletion strains auxotrophic for leucine, adenine, and uracil were streaked on YES, normal, or low NH4Cl EMM plates with amino acid supplements. The mutants that show low NH4Cl rescued growth defect were selected for the second screen. In the second screen, the candidates auxotrophic for leucine, adenine, and uracil were mated with prototrophic wild-type cells to get leucine auxotrophic or prototrophic strains. The leucine auxotrophic strains were tested for growth on normal or low NH4Cl EMM plates supplemented with leucine. The prototrophic strains were tested for growth on canavanine plates. Using the genome-wide screen, we isolated 13 deletion strains that phenocopy Δtsc2 cells. Here we will report the characterization of one of them, Npr2 (SPAC23H3.03c) encoding a protein of 409 amino acids that is highly similar to human NPRL2 (30.3% identity and 58.2% similarity) (Figure S1). The other genes will be described elsewhere.
The Δnpr2 and Δtsc2 displayed similar but distinct phenotypes
The Δnpr2 cells showed similar phenotypes to Δtsc2 cells. First, the Δnpr2 cells auxotrophic for leucine, adenine, and uracil failed to grow on normal EMM plates supplemented with appropriate amino acids (Figure 1A). Second, leucine auxotrophic Δnpr2 cells failed to grow on EMM plates supplemented with leucine, and low NH4Cl rescued the growth defect (Figure 1B). Third, prototrophic Δnpr2 cells displayed canavanine resistance (Figure 1C).
In fission yeast, the Tsc–Rhb1 signaling pathway controls basic amino acid uptake via the Cat1 permease, and Cat1 mislocalized into the intracellular organelles in Δtsc2 cells (Matsumoto et al. 2002). We constructed chromosome-borne Cat1–GFP under the control of its own promoter. We found that in wild-type cells, Cat1–GFP is mainly detected at the cell surface and is also enriched in the medial region and cell ends (Figure 1D). In Δnpr2 and Δtsc2 cells, Cat1–GFP is localized to intracellular small punctuated structures and, to a lesser extent, to plasma membrane (Figure 1D). These results indicate that Δnpr2 cells show very similar phenotypes to Δtsc2 cells.
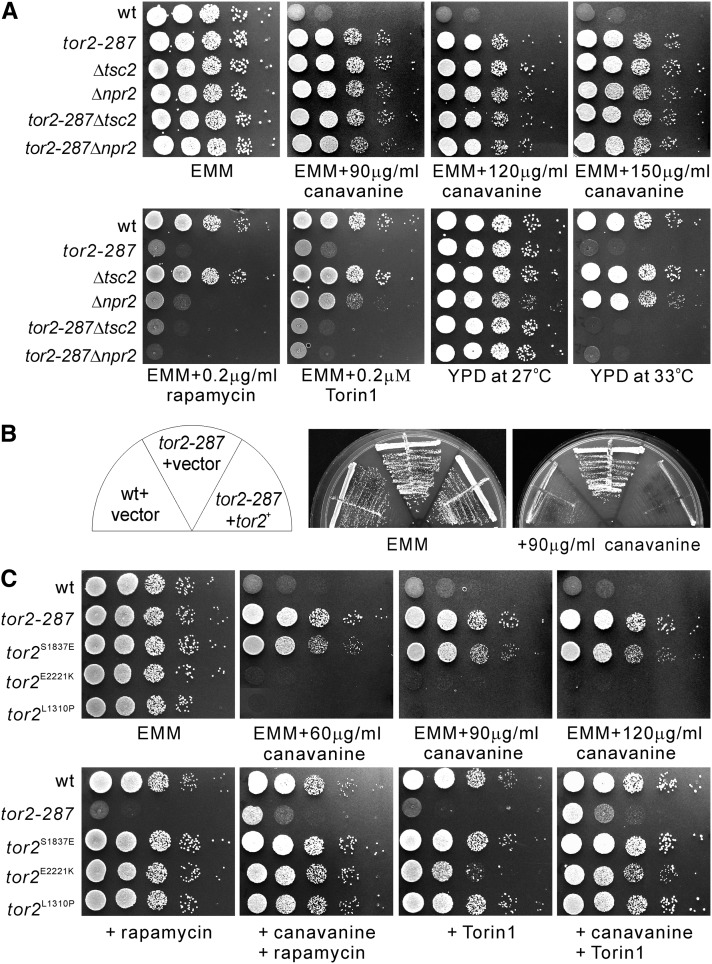
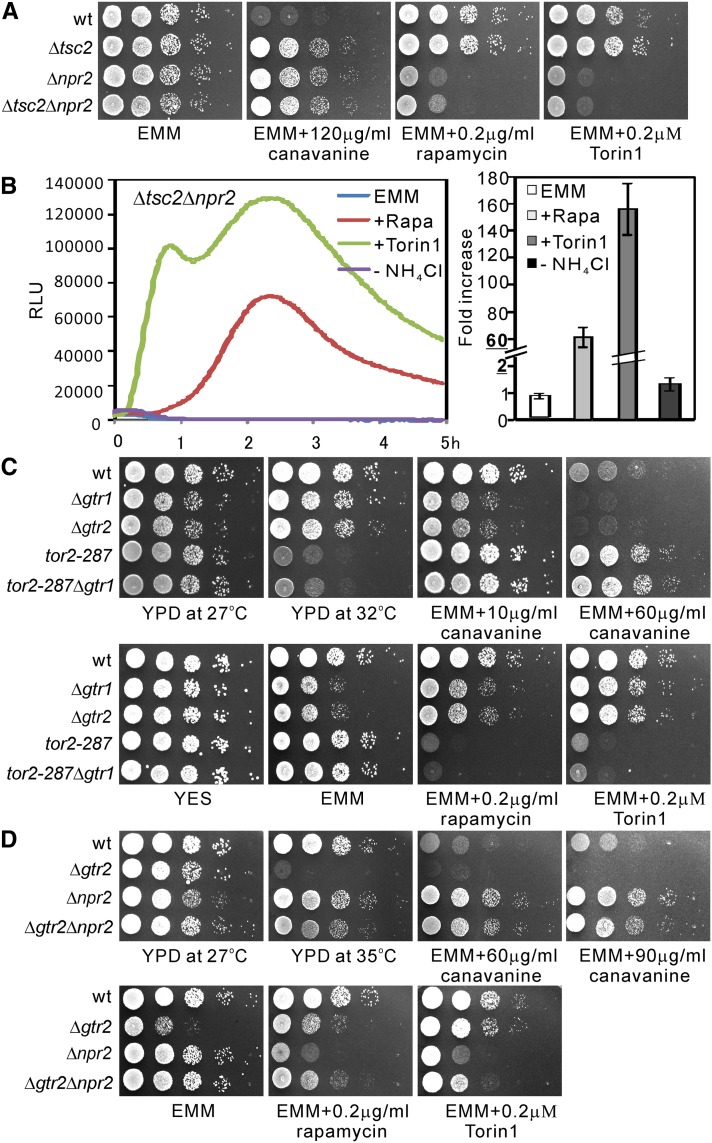
To examine the genetic interaction between Tor2, Tsc2, and Npr2, we constructed tor2-287Δtsc2 and tor2-287Δnpr2 double mutant cells and evaluated their temperature and rapamycin sensitivities as tor2-287 is sensitive to temperature and rapamycin (Hayashi et al. 2007). Results showed that both tor2-287Δtsc2 and tor2-287Δnpr2 double mutant cells displayed temperature- and rapamycin-sensitive phenotypes similar to tor2-287 mutant cells (Figure 2A, bottom). Since the tor2-287 mutation resulted in a rapamycin-sensitive phenotype, we expected that Δnpr2 and Δtsc2 cells would show mild resistance to rapamycin. Unexpectedly, Δnpr2 cells, but not Δtsc2 cells, showed supersensitivity to rapamycin. Similarly, Δnpr2 and tor2-287 cells, but not Δtsc2 cells, showed sensitivity to Torin1 (Figure 2A), which is a highly potent and selective ATP-competitive mTOR inhibitor (Thoreen et al. 2009).
Figure 2.
Tor2 dysfunction causes pleiotropic phenotypes. (A) Genetic interaction between Tor2, Tsc2, and Npr2. The tor2-287 mutation caused pleiotropic phenotypes. The indicated single and double knockout cells were spotted onto the plates as indicated and then incubated for 3 days at 33° or 4–5 days at 27°. The cells were spotted in serial 10-fold dilutions starting with OD660 = 0.3 of log-phase cells (5 μl). (B) The tor2+ gene rescued canavanine resistance of the tor2-287 mutants. The tor2-287 cells transformed with the control vector or the vector containing the tor2+ gene were streaked onto the plates as indicated and then incubated for 4–5 days at 27°. (C) The phenotypes of tor2 point mutation mutants. The indicated cells were spotted onto the indicated plates and incubated for 4–5 days at 27°.
Mutation or inhibition of Tor2 activity leads to canavanine resistance
We next examined the canavanine sensitivity of the tor2-287 single mutant and tor2-287Δnpr2 and tor2-287Δtsc2 double mutants. The results showed that the tor2-287 mutant cells displayed canavanine resistance, and tor2-287Δnpr2 and tor2-287Δtsc2 cells displayed similar canavanine resistance to the single mutant cells (Figure 2A, top). We confirmed that the expression of the tor2+ gene under its endogenous promoter from a multicopy plasmid suppressed canavanine resistance of tor2-287 mutant cells (Figure 2B), indicating that canavanine resistance is due to the tor2 mutation. Consistently, Murai et al. (2009) demonstrated that another Tor2 allele, the tor2-ts6 mutant, was resistant to canavanine.
Next, we examined several tor2 point mutants in which the wild-type tor2+ gene was replaced with a mutation on its chromosomal locus. The tor2L1310P and tor2E2221K mutants have activating mutations (Urano et al. 2007), and the tor2S1837E mutation has an inactivating mutation in the residue crucial for the binding between the FRB domain in TOR and the FKBP12–rapamycin complex (Nakashima et al. 2010). The inactivating tor2S1837E mutant, but not the activating mutant, displayed canavanine resistance (Figure 2C). Consistently, both rapamycin and Torin1 enhanced the canavanine resistance of wild-type cells (Figure 2C, wt).
Tor2 activity regulates transcription, intracellular trafficking, and protein level of Cat1
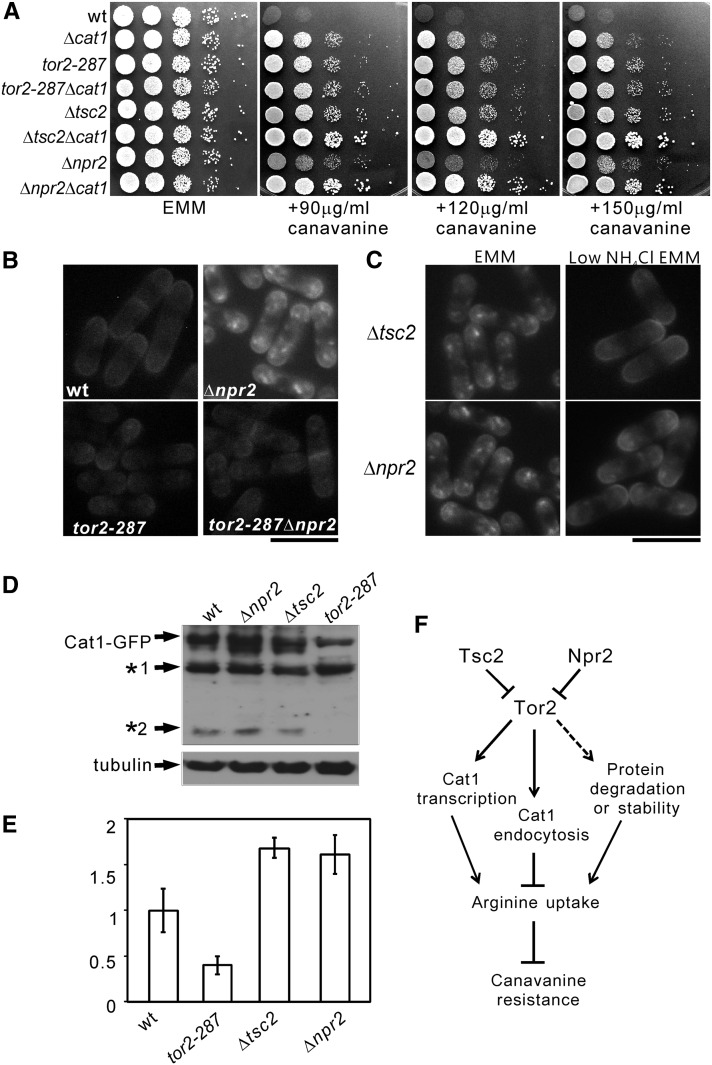
To examine whether canavanine resistance is due to the dysfunction of Cat1, a permease that transports arginine, we constructed a series of double mutants with Δcat1. As shown in Figure 3A, tor2-287Δcat1 cells did not show synergism in resistance to canavanine compared with the parental single mutant cells, indicating that the tor2-287 mutant displays canavanine resistance via Cat1. Notably, Δtsc2Δcat1 and Δnpr2Δcat1 cells showed synergism in resistance to canavanine compared with the parental single knockout cells (Figure 3A), indicating Δtsc2 and Δnpr2 displayed canavanine resistance not only via Cat1. To investigate whether the canavanine resistance in tor2-287 cells is due to Cat1 mislocalization, we looked at the localization of Cat1 in tor2-287 cells. The results showed Cat1 is prominently localized to plasma membrance (Figure 3B, tor2-287). In tor2-287Δnpr2 cells, Cat1 showed similar localization as that observed in tor2-287 cells (Figure 3B, tor2-287Δnpr2), suggesting that the tor2 mutation rescued Cat1 mislocalization in npr2 deletion background. Furthermore, our results show that low NH4Cl rescued the mislocalization of Cat1 in Δnpr2 and Δtsc2 cells (Figure 3C). We also noted that the fluorescence of Cat1–GFP in tor2-287 cells was weaker compared to that observed in wild-type cells (Figure 3B). This prompted us to check the protein level of Cat1–GFP. The results revealed that the protein level of Cat1–GFP in tor2-287 cells was lower than that in wild-type cells (Figure 3D). The two lower bands detected by immunoblot using anti-GFP antibody may represent protein degradation products of Cat1–GFP (Figure 3D, *1 and *2). Next, we examined the mRNA level of Cat1 using real time RT–PCR. In tor2-287 cells, Cat1 mRNA level was significantly decreased compared with that in wild-type cells (Figure 3E), indicating that Tor2 positively regulates Cat1 transcription. Consistently, the Cat1 mRNA level in Δtsc2 and Δnpr2 cells was slightly increased compared with that in wild-type cells (Figure 3E), suggesting a higher Tor2 activity in these cells even in normal EMM media. Based on these results, we hypothesize that Tor2 regulates Cat1 function at multiple steps, which may include transcription, membrane trafficking, and protein stability or degradation, and that Npr2 functions as a negative regulator of Tor2 (Figure 3F).
Figure 3.
Tor2 signaling regulates Cat1 function. (A) The Δnpr2Δcat1 and Δtsc2Δcat1 cells, but not tor2-287Δcat1 cells, were more resistant to canavanine compared with the parental single knockout cells. The indicated cells were spotted onto the plates as indicated and incubated for 4–5 days at 27°. (B) Subcellular localization of C-terminally tagged Cat1 in various cells. The wild-type (KP5859), tor2-287 (KP5955), Δnpr2 (KP5822), and tor2-287Δnpr2 (KP6385) cells expressing chromosome-borne Cat1–GFP under the control of its own promoter were grown to early log phase in EMM media at 27°. The fluorescence was examined as described in Figure 1D. Bar, 10 μm. (C) Low NH4Cl rescued Cat1 mislocalization in Δnpr2 and Δtsc2 cells. The cells expressing chromosome-borne Cat1–GFP under the control of its own promoter were grown to early log phase in EMM media at 27° and divided into two aliquots. One aliquot was continued to be incubated in EMM, and the other aliquot was washed twice, and shifted to low NH4Cl for 1 hr. The fluorescence was examined as described in Figure 1D. Bar, 10 μm. (D) The protein level of Cat1–GFP in various cells. The indicated cells were grown to early log phase in EMM media at 27° and then the cell extracts were subjected to electrophoresis using 8% polyacrylamide gel and were immunoblotted using anti-GFP antibodies to detect Cat1–GFP or using anti-α-tubulin antibodies to detect endogenous tubulin (loading control). *1 and *2 indicate possible degraded products of Cat1–GFP. (E) Tor2 activity affects Cat1 mRNA level. The indicated cells were grown overnight in EMM media to early log phase and then the cells were harvested. The RNA extract and real-time RT–PCR were performed as described in Materials and Methods. The data were analyzed by the comparative CT method and were obtained from three independent experiments. (F) Working model illustrating the possible roles of Npr2, Tsc2, and Tor2 in canavanine resistance. Npr2 and Tsc2 function as negative regulators of Tor2. Tor2 affects Cat1 expression, membrane trafficking, and protein stability or degradation.
The Δnpr2 and Δtsc2 cells showed different phenotypes
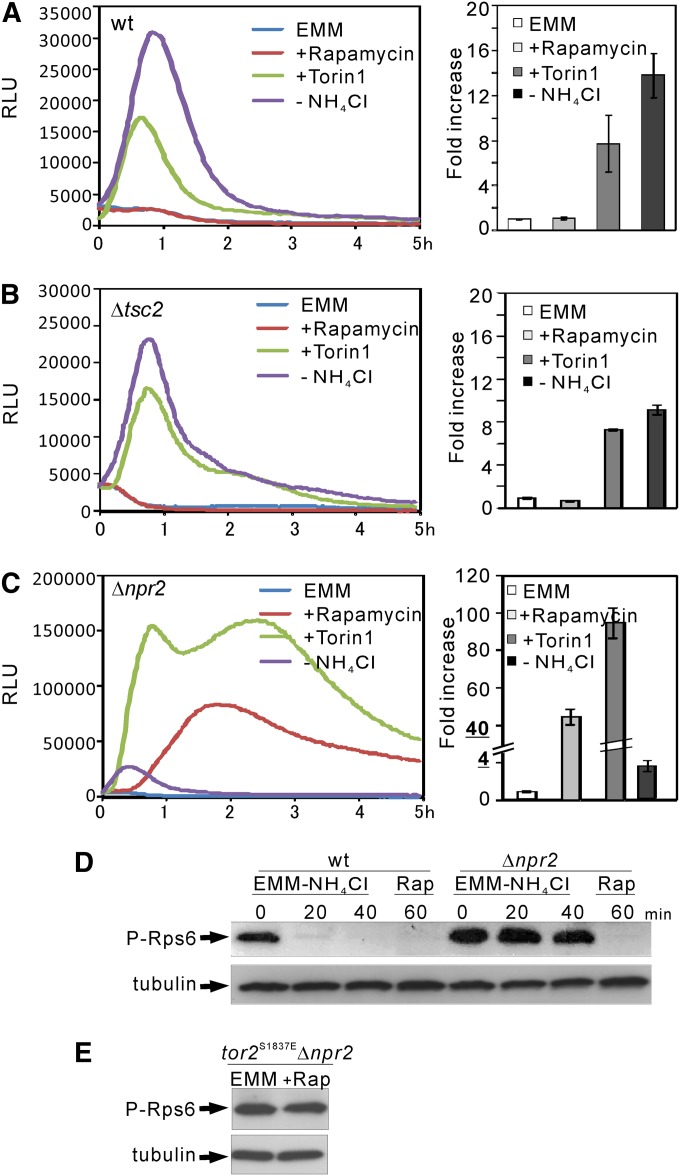
In fission yeast, the expression of amino acid permease Isp5 is induced by the tor2-ts6 mutation (Matsuo et al. 2007) and by the shift from EMM to proline media (Weisman et al. 2005). Deletion of the tor1+ gene reduced the level of Isp5 expression (Weisman et al. 2005). We previously monitored transcriptional activity in living fission yeast cells using luciferase reporter genes (Deng et al. 2006; Zhou et al. 2010, 2012). Here we fused the native promoter of the isp5+ gene with the Renilla luciferase reporter gene and estimated the activity of Tor2 using the Isp5–Renilla luciferase reporter system. Since Tor2 activation suppresses the transcription of Isp5 (Matsuo et al. 2007), and the shift from EMM to proline media induced Isp5 expression (Weisman et al. 2005), we expected that Tor2 inhibition by nitrogen depletion or rapamycin treatment should increase Isp5 transcriptional activity. The wild-type cells expressing the reporter vector were cultured and assayed as described in Materials and Methods. We found that in wild-type cells, Isp5 transcriptional activity was significantly induced by nitrogen depletion and Torin1 (Figure 4A, left). Nitrogen depletion and Torin1 treatment induced a 12- to 16- and a 6- to 10-fold increase in the ratio of accumulated RLU value up to 5 hr, respectively (Figure 4A, right). Notably, Isp5 transcriptional activity was not induced by rapamycin treatment (Figure 4A). Consistently, it was demonstrated that rapamycin treatment only led to subtle effects on gene expression, unless a poor nitrogen source was used (Rallis et al. 2013). In tor2-287 cells, the reporter activity was significantly higher even in EMM media (data not shown), indicating that decreased Tor2 activity induced the transcription of Isp5.
Figure 4.
The Δnpr2 and Δtsc2 cells show different phenotypes. (A) The transcriptional activity of the Isp5 promoter in wild-type cells. The wild-type cells harboring the reporter vector pKB8527 were grown to exponential phase and assayed as described in Materials and Methods. The luminescence was followed for 5 hr. The data shown are representative of multiple experiments. (Left) The luminescence, given as relative light units (RLUs), is plotted vs. time. (Right) The data represent the ratio of accumulated RLU value of each sample (nitrogen depletion, rapamycin treatment, or Torin1 treatment) to the basal (EMM). Standard deviations are from three independent experiments, and each sample was analyzed in triplicate. (B and C) The transcriptional activity of the Isp5 promoter in Δtsc2 and Δnpr2 cells. The Δtsc2 or Δnpr2 cells harboring the reporter vector pKB8527 were cultured and assayed as described in A. The data shown are representative of multiple experiments. (D) Nitrogen depletion-induced dephosphorylation of Rps6 is inhibited in Δnpr2 cells. The prototrophic cells of wild-type (KP5080) and Δnpr2 (KP5237) cells were grown at 27° to exponential phase in EMM (0 min). Then the cells were washed three times and transferred to liquid EMM-N for 20 and 40 min. Rapamycin was added to a final concentration of 0.2 μg/ml and incubated for 60 min in EMM media. The proteins were extracted as described in Materials and Methods. The cell lysates were subjected to SDS–PAGE and immunoblotted using anti-PAS polyclonal antibodies. Endogenous tubulin was immunoblotted using anti-α-tubulin antibodies as a loading control. (E) The effect of rapamycin was abolished in tor2S1837EΔnpr2 cells. Rapamycin treatment and immunoblot analysis was performed as described in D.
Next we monitored the reporter activity in Δtsc2 and Δnpr2 cells. In Δtsc2 cells, the level and pattern of reporter activity were almost the same as those observed in wild-type cells except that the nitrogen depletion-induced activity was slightly lower than that observed in wild-type cells (Figure 4B). Surprisingly, rapamycin treatment induced a 40- to 50-fold increase in Isp5 transcription in Δnpr2 cells vs. a 1.1- to 1.4-fold increase in wild-type cells, while nitrogen depletion induced a 3.5- to 4.5-fold increase in Δnpr2 cells vs. a 12- to 16-fold increase in wild-type cells (Figure 4, C vs. A). In Δfkh1Δnpr2 cells, Isp5 transcription activation induced by rapamycin was abolished, whereas that induced by Torin1 remained (Figure S2). This result is consistent with the finding that rapamycin inhibits Tor2 activity in an Fkh1-dependent manner, whereas Torin1 directly inhibits Tor2 activity. We also noted that in response to Torin1, the transcriptional activity in Δnpr2 cells showed an initial activation followed by the second activation at 2–3 hr after the treatment. This profile differed significantly compared to that observed with Δtsc2 cells. These results suggest that several factors are involved in the Tor-dependent regulation of the Isp5 transcription profile, and they differentially affect the transcription profile.
Recent studies demonstrate that TORC1–Psk1–Rps6 constitutes a nutrient-dependent signaling pathway, and that the phosphorylation of Rps6 is a readout of Tor2 activity (Nakashima et al. 2010, 2012). We evaluated Rps6 phosphorylation in Δnpr2 cells to test the role of Npr2 in regulating Tor2 activity. Rps6 phosphorylation is extremely high even after the shift to nitrogen-depletion media for 40 min, indicating that in Δnpr2 cells, dephosphorylation of Rps6 upon nitrogen depletion is markedly inhibited. To address whether the elevated level of Rps6 phosphorylation in Δnpr2 is due to the high activity of Tor2, we investigated the effect of rapamycin treatment on Rps6 phosphorylation. The results showed that Rps6 phosphorylation was completely abolished after rapamycin treatment for 60 min (Figure 4D). We also confirmed that in tor2S1837EΔnpr2 cells, rapamycin failed to decrease the elevated level of Rps6 phosphorylation (Figure 4E). Altogether, these results indicate that the elevated level of Rps6 phosphorylation in Δnpr2 is due to the high activity of Tor2.
Npr2 negatively regulates Tor2 activity downstream of Rag GTPase
The results above strongly suggest that similar to Tsc2, Npr2 also negatively regulates Tor2 activity. To confirm the relationship between Npr2 and Tsc2, we analyzed the phenotypes of Δtsc2Δnpr2 double knockout cells. We found Δtsc2Δnpr2 cells did not exhibit synergism in resistance to canavanine, compared with the parental single knockout cells (Figure 5A). In terms of Torin1 and rapamycin sensitivities, Δtsc2Δnpr2 cells showed the same phenotypes as Δnpr2 cells (Figure 5A). In Δtsc2Δnpr2 cells, rapamaycin-induced reporter activity is the same as that observed in Δnpr2 cells (Figure 5B). In fission yeast, the Rag GTPases Gtr1 and Gtr2 activate Tor2 in response to amino acids (Valbuena et al. 2012). This prompted us to confirm the epistasis relationship between Tor2 and Gtr. The results showed Δgtr1 and Δgtr2 cells displayed the same phenotypes in terms of slow growth on EMM plates and mild sensitivity to canavanine (Figure 5C). The tor2-287Δgtr2 cells displayed the same phenotypes as tor2-287 cells in terms of canavanine resistance, temperature-, rapamycin-, and Torin sensitivities (Figure 5C). These results indicate that Gtr2 functions upstream of Tor2. We next constructed Δnpr2Δgtr2 cells and investigated the relationship between Npr2 and Gtr2. The Δgtr2Δnpr2 cells displayed similar phenotypes to Δnpr2 cells (Figure 5D), indicating that Npr2 may function downstream of Gtr2. Based on the S. pombe predicted protein interaction database (www.bahlerlab.info/PInt), Npr2 may interact with TORC1 subunits (Tor2, Toc1, and Tco89) and Gtr1–Gtr2–Vam6 complex with a score >0.5. Together with our genetic epistasis analyses data, we propose that Npr2 functions downstream of Gtr1–Gtr2 and upstream of Tor2.
Figure 5.
Genetic interaction between Npr2, Tsc2, and Gtr2. (A) The Δtsc2Δnpr2 double knockout cells display similar phenotypes to Δnpr2 cells. The indicated cells were spotted onto the indicated plates and incubated for 4–5 days at 27°. (B) The Isp5 transcriptional activity was markedly increased in Δtsc2Δnpr2 double knockout cells in response to rapamycin and Torin1 treatment. The Δtsc2Δnpr2 cells harboring the reporter vector pKB8527 were grown and assayed as described in Figure 4A. (C) The tor2-287Δgtr2 double knockout cells displayed similar phenotypes to tor2-287 cells. The indicated cells were spotted onto the indicated plates and incubated for 3 days at 32° or 4–5 days at 27°. (D) The Δgtr2Δnpr2 double knockout cells displayed similar phenotypes to Δnpr2 cells. The indicated cells were spotted onto the indicated plates and incubated for 3 days at 35° or 4–5 days at 27°.
The role of Npr2 in the regulation of autophagy
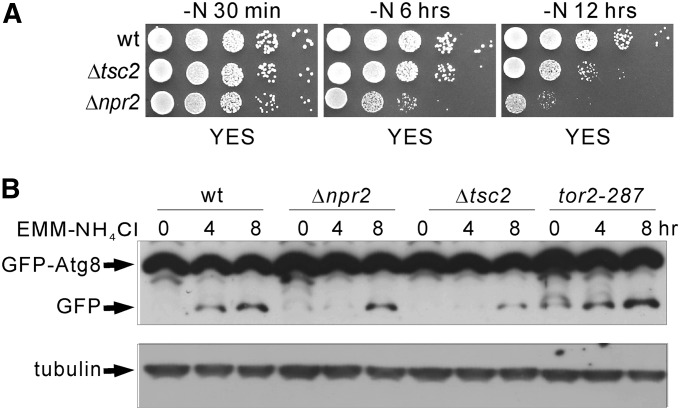
In budding yeast, NPR2 deletion exhibited impaired autophagy induction (Wu and Tu 2011). In fission yeast, it is demonstrated that autophagy contributes to the maintenance of cell viability upon nitrogen depletion (Kohda et al. 2007). These reports prompted us to compare the growth of wild-type, Δtsc2, and Δnpr2 cells harboring the leu1-32 mutation upon nitrogen deprivation on YES plates. As shown in Figure 6A, upon nitrogen deprivation for 30 min, the Δtsc2 and Δnpr2 grew as well as wild-type cells. Upon nitrogen deprivation for 6 hr, the growth of Δnpr2 cells was significantly slower compared with that of wild-type and Δtsc2 cells. Upon nitrogen deprivation for 12 hr, the growth of both Δnpr2 and Δtsc2 cells was markedly slower compared with that of wild type. Notably, the Δnpr2 cells showed more a severe growth defect compared with Δtsc2 cells. These results suggest that Tor2 signaling plays a role in regulating autophagy. However, the cell viability experiment upon nitrogen deprivation does not appear to solely reflect autophagy. Then, we examined the cleavage of GFP–Atg8p, which is used to monitor autophagy in fission yeast (Mukaiyama et al. 2009, 2010), to test whether autophagy induction is affected in Δnpr2 cells. As shown in Figure 6B, in wild-type cells, the cleavage from GFP–Atg8p to free GFP was detected upon nitrogen depletion for 4 hr. However, in both Δnpr2 and Δtsc2 cells, the cleavage was markedly inhibited. These results indicate that Tor2 signaling plays a role in regulating autophagy.
Figure 6.
The role of Npr2 in the regulation of autophagy. (A) Viability of wild-type, Δtsc2, and Δnpr2 cells harboring the leu1-32 mutation upon nitrogen deprivation. The indicated cells grown to logarithmic phase at 27° were collected, washed twice, and transferred to liquid EMM-N media. Aliquots of each culture were taken at the indicated time points, spotted onto YES plates in sequential 10-fold dilutions, and incubated at 27° for 3 days. (B) Cleavage of GFP–Atg8p. The multicopy plasmid encoding pREP41–GFP–Atg8p was introduced into the indicated cells. The transfomants were cultured in EMM to induce GFP–Atg8p expression. Then the cells were collected, washed three times, and transferred to liquid EMM-N media. Aliquots of each culture were taken at the indicated time points. The proteins were extracted as described in Materials and Methods. The cell lysates were subjected to SDS–PAGE and immunoblotted using anti-GFP antibodies. Endogenous tubulin was immunoblotted using anti-α-tubulin antibodies as a loading control.
Discussion
Fission yeast Npr2 and budding yeast Npr2p have similar but distinct functions
In budding yeast, Npr2p was first isolated as a nitrogen permeases regulatory protein (Rousselet et al. 1995), and recently Npr2p was demonstrated to be a subunit of Npr2–Npr3 (Neklesa and Davis 2009), Iml1p–Npr2p–Npr3p (Wu and Tu 2011), and SEA (Sea1p/Imh1p–Sea2p–Sea3p–Sea4p–Npr2p–Npr3p) complexes (Dokudovskaya et al. 2011). The Npr2–Npr3 complex was identified in a genome-wide screen as negative regulators of TORC1 complex (Neklesa and Davis 2009). The Iml1p–Npr2p–Npr3p complex is required for nonnitrogen-starvation-induced autophagy (Wu and Tu 2011). The SEA (Seh1-associated) complex is a conserved coatomer-related complex that associates dynamically with the vacuoles (Dokudovskaya et al. 2011). In fission yeast, as shown in Table S2 Npr2 binds with five of the six components of SEA complex based on the predicted protein interaction database (www.bahlerlab.info/PInt).
Our present study is the first identification and functional analysis of Npr2 in fission yeast. Similar to budding yeast, fission yeast Npr2 also functions as a negative regulator of Tor2 based on several lines of genetic and biochemical evidence. First, Δnpr2 cells failed to grow on leucine-supplemented EMM plates and showed canavanine resistance. Second, Δnpr2 cells showed an increase in intracellular localization of Cat1. Third, in Δnpr2 cells, Rps6 phosphorylation was extremely high even after the shift to nitrogen-depletion media for 40 min. Fourth, in Δnpr2 cells, the cleavage of GFP–Atg8 was markedly inhibited. These four phenotypes may be a result of increased Tor2 activity in Δnpr2 cells.
Despite these similarities, fission yeast Npr2 (SpNpr2) has two important differences from budding yeast Npr2p (ScNpr2p). First, ScNpr2p, but not SpNpr2, contains two characteristic PEST sequences, which are preferentially found in rapidly degraded regulatory proteins (Rousselet et al. 1995). Second, in budding yeast, deletion of ScNpr2p did not display rapamycin sensitivity (Wu and Tu 2011), whereas in fission yeast deletion of SpNpr2 resulted in rapamycin sensitivity.
Tsc2 and Npr2 play different roles in the regulation of Tor2 activity
We have shown that both Tsc2 and Npr2 function upstream of Tor2. One of the key questions that can be addressed in fission yeast, but not in budding yeast, is to list the differences between Δtsc2 and Δnpr2 cells. First, Δnpr2 cells displayed rapamycin sensitivity, whereas Δtsc2 cells did not. Second, rapamycin treatment induced a marked increase in Isp5 transcription in Δnpr2 cells, whereas it failed to induce an increase in Δtsc2 cells. Third, nitrogen depletion induced less increase in Isp5 transcription in Δnpr2 cells compared with Δtsc2 cells. These differences may be ascribed to differences in the level of Tor2 activation.
It is expected that an inhibitor would have less effect in the presence of high activity of its target, that is to say, Δtsc2 and Δnpr2 cells should display less sensitivity to rapamycin due to increased Tor2 activity. Unexpectedly, rapamycin treatment induced an extremely marked increase in Isp5 transcriptional activity in Δnpr2 cells, while no significant increase was observed in Δtsc2 or wild-type cells. Moreover, in Δtsc2 and wild-type cells Torin1 induced a marked increase in Isp5 transcriptional activity. The following mechanisms may be related to these effects of rapamycin and Torin1. First, as an allosteric inhibitor, rapamycin inhibits TOR activity by forming the FKBP12–rapamycin complex that binds directly to the FRB domain of TOR (Huang et al. 2003). We hypothesize that rapamycin may inhibit TOR only in its activated state or only when TOR reaches a certain state of activation, thus rapamycin may have a more prominent effect in the cells that have high TOR activity. In mammalian and fission yeast cells, the effects of rapamycin on cell growth and proliferation are less severe than its effects in budding yeast (Weisman et al. 1997; Neshat et al. 2001; Takeuchi et al. 2005). These differences may be ascribed to differences in TOR activity. Second, as an ATP-competitive inhibitor, Torin1 inhibits both TORC1 and TORC2 in mammalian cells. It also inhibits many other kinases, although Torin1 was at least 200-fold selective for mTOR over other kinases (Thoreen et al. 2009). In mammalian cells, Torin1, but not rapamycin, mimics many phenotypes caused by TOR inhibition in budding yeast (Thoreen et al. 2009). Here, we report for the first time the effects of Torin1 on the growth and the transcriptional activity in fission yeast. Both Torin1 and rapamycin had similar effect on the growth inhibition of tor2-287 mutant cells (Figure 2A), whereas Torin1, but not rapamycin, markedly induced Isp5 transcriptional activity (Figure 4A). These results suggest that the effects of Torin1 may also be explained by the inhibition of TORC1 and/or TORC2.
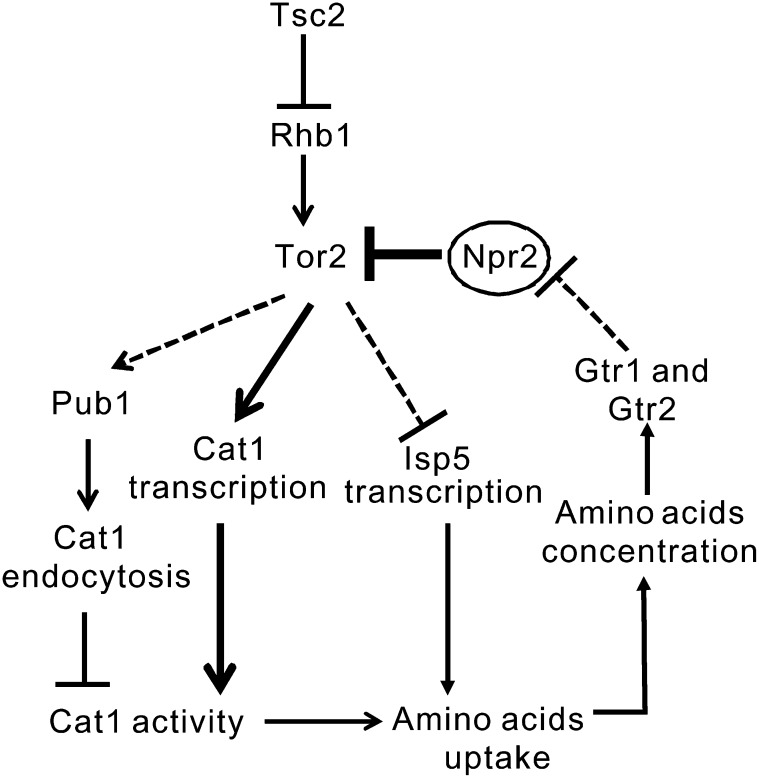
Our data demonstrate that Tsc2 and Npr2 function as negative regulators of Tor2 signaling in a parallel way, based on the following evidence: First, Δtsc2Δnpr2 cells showed the same phenotypes as Δnpr2 cells in terms of rapamycin and Torin1 sensitivities, and the pronounced increase in Isp5 transcriptional activity response to the two TORC1 inhibitors. Second, Δgtr2Δnpr2 cells showed similar phenotypes to Δnpr2 cells, indicating that Npr2 may function downstream of Gtr2. Consistently, S. pombe predicted protein interactions show Npr2 possibly interacts with TORC1 subunits and the Gtr1–Gtr2–Vam6 complex. In budding yeast, it is well established that the E3 ligase Rsp5 is required for the endocytosis of permeases (MacGurn et al. 2011). In fission yeast, the mislocalization of Cat1 in Δtsc2 cells was suppressed by the loss of the E3 ubiquitin ligase Rsp5 homolog, Pub1 (Aspuria and Tamanoi 2008). Based on our present study and previous work, we summarize our working model in Figure 7. We hypothesize that in fission yeast, Tor2 signaling regulates amino acid homeostasis by feedback mechanisms. First, Tor2 differently regulates the activity of multiple permeases at the transcriptional level. We found that Tor2 promotes Cat1 transcription and inhibits Isp5 transcription. This is consistent with the results in budding yeast that TOR promotes the activity of high-specificity amino acid permeases such as Tat2 and Hip1, and inhibits the synthesis and stability of the general amino acid permease Gap1 (Neufeld 2007). Second, Tor2 regulates Cat1 activity at the transcriptional and post-transcriptional levels. We hypothesize that the reduction in Tor2 activity decreased both Cat1 endocytosis and Cat1 transcription; however, the effects on Cat1 transcription are stronger than those on Cat1 endocytosis. This may explain why the tor2-287 mutant showed resistance to canavanine. Finally, increased Tor2 activity decreases the influx and level of amino acids, which in turn inhibits Tor2 activity, thus creating an autoinhibitory feedback loop that limits constitutive activation of Tor2 signaling. In this feedback loop, Npr2, but not Tsc2, appears to play key roles in the nutrient-sensing pathway that regulates amino acid homeostasis. Further studies are needed to elucidate the exact mechanisms through which Npr2 negatively regulates Tor2 signaling.
Figure 7.
Working model for the role of Npr2 in regulating Tor2 signaling. In S. pombe, when Tor2 activity is increased in the presence of high intracellular amino acid, Pub1 is activated to prompt the endocytosis of permease Cat1, thus resulting in a decreased Cat1 activity. On the other hand, increased Tor2 activity inhibits the transcription of the permease Isp5. Both of these two changes result in decreased uptake of amino acids, thereby decreasing intracellular amino acid concentration. Gtr1 and Gtr2 sense the low amino acid levels and down-regulate Tor2 activity. In this feedback loop, Npr2 functions between Gtr1–Gtr2 and Tor2. In the absence of Npr2, Gtr1–Gtr2 fail to down-regulate Tor2 activity, thus resulting in a constitutively active Tor2 signaling. However, in the absence of Tsc2, the negative feedback loop is intact, thus resulting in partially active Tor2 signaling due to a relative high Rhb1 activity. The solid lines indicate the processes that are proven by other studies and the present study. The dashed lines indicate hypotheses that are not yet proven.
In mammalian cells, the tuberous sclerosis complex 1 and 2 (TSC1–TSC2) inhibits mTORC1. Treatment of human patients with TSC with rapamycin or rapamycin analogs results in a decrease in the size of kidney and central nervous system tumors by ∼30–40%, with regrowth to the original size when the treatment is discontinued (Henske and McCormack 2012). Our data in fission yeast point to the possibility that strategies focused on Npr2-dependent regulation of mTOR activity could contribute to more complete and/or more durable therapeutic responses for patients with TSC and lymphangioleiomyomatosis, and for other tumors with mTORC1 activation, including many human malignancies.
Supplementary Material
Acknowledgments
We thank Takayoshi Kuno for helpful advice and discussions. We thank Mitsuhiro Yanagida, Fuyuhiko Tamanoi, Sergio Moreno, Tomohiro Matsumoto, Antony M. Carr, and National BioResource Project for providing yeast strains and plasmids. We thank Akio Nakashima at Kobe University, and Nicole A. Neuman and Damir Khabibullin at Brigham and Women’s Hospital and Harvard Medical School for helpful discussions and technical assistance. Financial support for this study was provided by Grant-in-Aid for Young Scientists (B) to Y.M. (no. 21790241) from the Japan Society for the Promotion of Science (http://www.jsps.go.jp/english/e-grants/grants.html).
Footnotes
Communicating editor: J. Heitman
Literature Cited
- Aspuria P. J., Tamanoi F., 2008. The Tsc/Rheb signaling pathway controls basic amino acid uptake via the Cat1 permease in fission yeast. Mol. Genet. Genomics 279: 441–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Sugiura R., Takeuchi M., Suzuki M., Ebina H., et al. , 2006. Real-time monitoring of calcineurin activity in living cells: evidence for two distinct Ca2+-dependent pathways in fission yeast. Mol. Biol. Cell 17: 4790–4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokudovskaya S., Waharte F., Schlessinger A., Pieper U., Devos D. P., et al. , 2011. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell Proteomics 10: M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., et al. , 2007. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells 12: 1357–1370. [DOI] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y., 1995. Improved green fluorescence. Nature 373: 663–664. [DOI] [PubMed] [Google Scholar]
- Heitman J., Movva N. R., Hall M. N., 1991. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253: 905–909. [DOI] [PubMed] [Google Scholar]
- Henske E. P., McCormack F. X., 2012. Lymphangioleiomyomatosis: a wolf in sheep’s clothing. J. Clin. Invest. 122: 3807–3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentges P., Van Driessche B., Tafforeau L., Vandenhaute J., Carr A. M., 2005. Three novel antibiotic marker cassettes for gene disruption and marker switching in Schizosaccharomyces pombe. Yeast 22: 1013–1019. [DOI] [PubMed] [Google Scholar]
- Huang J., Manning B. D., 2009. A complex interplay between Akt, TSC2 and the two mTOR complexes. Biochem. Soc. Trans. 37: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Bjornsti M. A., Houghton P. J., 2003. Rapamycins: mechanism of action and cellular resistance. Cancer Biol. Ther. 2: 222–232. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., Guan K. L., 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4: 648–657. [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., Guan K. L., 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17: 1829–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. U., Hayles J., Kim D., Wood V., Park H. O., et al. , 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat. Biotechnol. 28: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohda T. A., Tanaka K., Konomi M., Sato M., Osumi M., et al. , 2007. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells 12: 155–170. [DOI] [PubMed] [Google Scholar]
- Kunz J., Henriquez R., Schneider U., Deuter-Reinhard M., Movva N. R., et al. , 1993. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 73: 585–596. [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M., 2009. mTOR signaling at a glance. J. Cell Sci. 122: 3589–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M., 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Takeuchi M., Sugiura R., Sio S. O., Kuno T., 2009. Deletion mutants of AP-1 adaptin subunits display distinct phenotypes in fission yeast. Genes Cells 14: 1015–1028. [DOI] [PubMed] [Google Scholar]
- MacGurn J. A., Hsu P. C., Smolka M. B., Emr S. D., 2011. TORC1 regulates endocytosis via Npr1-mediated phosphoinhibition of a ubiquitin ligase adaptor. Cell 147: 1104–1117. [DOI] [PubMed] [Google Scholar]
- Manning B. D., Tee A. R., Logsdon M. N., Blenis J., Cantley L. C., 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10: 151–162. [DOI] [PubMed] [Google Scholar]
- Matsumoto S., Bandyopadhyay A., Kwiatkowski D. J., Maitra U., Matsumoto T., 2002. Role of the Tsc1-Tsc2 complex in signaling and transport across the cell membrane in the fission yeast Schizosaccharomyces pombe. Genetics 161: 1053–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M., 2007. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 27: 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell K., 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123: 127–130. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Kajiwara S., Hosomi A., Giga-Hama Y., Tanaka N., et al. , 2009. Autophagy-deficient Schizosaccharomyces pombe mutants undergo partial sporulation during nitrogen starvation. Microbiology 155: 3816–3826. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H., Nakase M., Nakamura T., Kakinuma Y., Takegawa K., 2010. Autophagy in the fission yeast Schizosaccharomyces pombe. FEBS Lett. 584: 1327–1334. [DOI] [PubMed] [Google Scholar]
- Murai T., Nakase Y., Fukuda K., Chikashige Y., Tsutsumi C., et al. , 2009. Distinctive responses to nitrogen starvation in the dominant active mutants of the fission yeast Rheb GTPase. Genetics 183: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y., Fukuda K., Chikashige Y., Tsutsumi C., Morita D., et al. , 2006. A defect in protein farnesylation suppresses a loss of Schizosaccharomyces pombe tsc2+, a homolog of the human gene predisposing to tuberous sclerosis complex. Genetics 173: 569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Sato T., Tamanoi F., 2010. Fission yeast TORC1 regulates phosphorylation of ribosomal S6 proteins in response to nutrients and its activity is inhibited by rapamycin. J. Cell Sci. 123: 777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A., Otsubo Y., Yamashita A., Sato T., Yamamoto M., et al. , 2012. Psk1, an AGC kinase family member in fission yeast, is directly phosphorylated and controlled by TORC1 and functions as S6 kinase. J. Cell Sci. 125: 5840–5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T. K., Davis R. W., 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5: e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neshat M. S., Mellinghoff I. K., Tran C., Stiles B., Thomas G., et al. , 2001. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc. Natl. Acad. Sci. USA 98: 10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T. P., 2007. TOR regulation: sorting out the answers. Cell Metab. 5: 3–5. [DOI] [PubMed] [Google Scholar]
- Nobukuni T., Joaquin M., Roccio M., Dann S. G., Kim S. Y., et al. , 2005. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc. Natl. Acad. Sci. USA 102: 14238–14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlova K. A., Crino P. B., 2010. The tuberous sclerosis complex. Ann. N. Y. Acad. Sci. 1184: 87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallis C., Codlin S., Bahler J., 2013. TORC1 signaling inhibition by rapamycin and caffeine affect lifespan, global gene expression, and cell proliferation of fission yeast. Aging Cell 12: 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J., 1983. One-step gene disruption in yeast. Methods Enzymol. 101: 202–211. [DOI] [PubMed] [Google Scholar]
- Rousselet G., Simon M., Ripoche P., Buhler J. M., 1995. A second nitrogen permease regulator in Saccharomyces cerevisiae. FEBS Lett. 359: 215–219. [DOI] [PubMed] [Google Scholar]
- Ryuko S., Ma Y., Ma N., Sakaue M., Kuno T., 2012. Genome-wide screen reveals novel mechanisms for regulating cobalt uptake and detoxification in fission yeast. Mol. Genet. Genomics 287: 651–662. [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., et al. , 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., et al. , 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sio S. O., Suehiro T., Sugiura R., Takeuchi M., Mukai H., et al. , 2005. The role of the regulatory subunit of fission yeast calcineurin for in vivo activity and its relevance to FK506 sensitivity. J. Biol. Chem. 280: 12231–12238. [DOI] [PubMed] [Google Scholar]
- Takeuchi H., Kondo Y., Fujiwara K., Kanzawa T., Aoki H., et al. , 2005. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 65: 3336–3346. [DOI] [PubMed] [Google Scholar]
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C., Blenis J., 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13: 1259–1268. [DOI] [PubMed] [Google Scholar]
- Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., et al. , 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284: 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Dhut S., Superti-Furga G., Gotoh Y., Nishida E., et al. , 1996. The fission yeast pmk1+ gene encodes a novel mitogen-activated protein kinase homolog which regulates cell integrity and functions coordinately with the protein kinase C pathway. Mol. Cell. Biol. 16: 6752–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano J., Comiso M. J., Guo L., Aspuria P. J., Deniskin R., et al. , 2005. Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol. Microbiol. 58: 1074–1086. [DOI] [PubMed] [Google Scholar]
- Urano J., Sato T., Matsuo T., Otsubo Y., Yamamoto M., et al. , 2007. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc. Natl. Acad. Sci. USA 104: 3514–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., et al. , 2006. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells 11: 1367–1379. [DOI] [PubMed] [Google Scholar]
- Valbuena N., Guan K. L., Moreno S., 2012. The Vam6 and Gtr1-Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J. Cell Sci. 125: 1920–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Slegtenhorst M., Carr E., Stoyanova R., Kruger W. D., Henske E. P., 2004. Tsc1+ and tsc2+ regulate arginine uptake and metabolism in Schizosaccharomyces pombe. J. Biol. Chem. 279: 12706–12713. [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., Mustafa A., Henske E. P., 2005. Pas1, a G1 cyclin, regulates amino acid uptake and rescues a delay in G1 arrest in Tsc1 and Tsc2 mutants in Schizosaccharomyces pombe. Hum. Mol. Genet. 14: 2851–2858. [DOI] [PubMed] [Google Scholar]
- Weisman R., Choder M., Koltin Y., 1997. Rapamycin specifically interferes with the developmental response of fission yeast to starvation. J. Bacteriol. 179: 6325–6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Nahari T., Kupiec M., 2005. Regulation of leucine uptake by tor1+ in Schizosaccharomyces pombe is sensitive to rapamycin. Genetics 169: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Tu B. P., 2011. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol. Biol. Cell 22: 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Ma Y., Sugiura R., Kobayashi D., Suzuki M., et al. , 2010. MAP kinase kinase kinase (MAPKKK)-dependent and -independent activation of Sty1 stress MAPK in fission yeast. J. Biol. Chem. 285: 32818–32823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Ma Y., Kato T., Kuno T., 2012. A measurable activation of the bZIP transcription factor Atf1 in a fission yeast strain devoid of stress-activated and cell integrity mitogen-activated protein kinase (MAPK) activities. J. Biol. Chem. 287: 23434–23439. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.