Abstract
That some plants benefit from being eaten is counterintuitive, yet there is now considerable evidence demonstrating enhanced fitness following herbivory (i.e., plants can overcompensate). Although there is evidence that genetic variation for compensation exists, little is known about the genetic mechanisms leading to enhanced growth and reproduction following herbivory. We took advantage of the compensatory variation in recombinant inbred lines of Arabidopsis thaliana, combined with microarray and QTL analyses to assess the molecular basis of overcompensation. We found three QTL explaining 11.4, 10.1, and 26.7% of the variation in fitness compensation, respectively, and 109 differentially expressed genes between clipped and unclipped plants of the overcompensating ecotype Columbia. From the QTL/microarray screen we uncovered one gene that plays a significant role in overcompensation: glucose-6-phosphate-1-dehydrogenase (G6PDH1). Knockout studies of Transfer-DNA (T-DNA) insertion lines and complementation studies of G6PDH1 verify its role in compensation. G6PDH1 is a key enzyme in the oxidative pentose-phosphate pathway that plays a central role in plant metabolism. We propose that plants capable of overcompensating reprogram their transcriptional activity by up-regulating defensive genes and genes involved in energy metabolism and by increasing DNA content (via endoreduplication) with the increase in DNA content feeding back on pathways involved in defense and metabolism through increased gene expression.
Keywords: overcompensation, QTL mapping, microarray, knockout mutants, complementation, fitness
THE loss of plant tissue to herbivores is an important selective agent shaping plant phenotypes. Most studies of plant adaptation, to date, have focused on the evolution of structural or chemical defensive traits that reduce or prevent tissue damage by herbivores (Berenbaum et al. 1986; Mauricio et al. 1997; Agrawal 1998). However, herbivores may also select for traits that allow plants to maintain fitness in the face of tissue loss (Stowe et al. 2000). Plant genotypes that can compensate for tissues lost with little or no decrement in fitness relative to those that are undamaged represent such an example and are termed “tolerant” (see Stowe et al. 2000 for a review). Interest in tolerance was motivated by empirical studies demonstrating that herbivore damage, under certain circumstances, can increase, rather than decrease, plant reproductive success (a specialized case termed “overcompensation,” i.e., increased flower, fruit, and seed production following herbivory). Specifically, studies by Paige and Whitham (1987) showed that when mule deer and elk removed 95% or more of the aboveground biomass of the monocarpic biennial scarlet gilia, Ipomopsis aggregata, the product of lifetime seed production, seed germination, and seedling survival averaged 3.0 times that of uneaten controls (see also Paige 1992, 1994, 1999). Since the initial study of Paige and Whitham (1987) evidence for increased flower, fruit, and seed production following herbivory has also been found for numerous plant species including Ipomopsis arizonica (Maschinski and Whitham 1989), Gentianella campestris, G. amarella (Nilsson et al. 1996; Lennartsson et al. 1997), Arabidopsis thaliana (Mauricio et al. 1997; Weinig et al. 2003), and Erysimum strictum (Rautio et al. 2005) to name but a few.
There is also evidence that genetic variation for tolerance/overcompensation exists. For example, studies comparing historically grazed and ungrazed populations of the plant G. campestris indicate that repeatedly grazed populations overcompensate, while ungrazed populations remain completely intolerant (Lennartsson et al. 1997). Furthermore, numerous genetic lineages within a plant species exhibit repeatable patterns of overcompensation, whereas others express only patterns of equal or undercompensation (Mauricio et al. 1997; Tiffin and Rausher 1999; Juenger and Bergelson 2000; Weinig et al. 2003). Although these observations provide evidence that genetic variation for compensation exists, little is known about the genetic mechanisms leading to enhanced growth and reproduction in plant species exhibiting growth compensation.
A recent study by Scholes and Paige (2011) showed that Arabidopsis sometimes responds to the removal of apical dominance at the whole-genome level through endoreduplication, the replication of the genome without mitosis, leading to endopolyploidy, an increase in cellular chromosome number. Different ecotypes of A. thaliana show different degrees of endoreduplication in response to loss of apical dominance, and the degree of endopolyploidy achieved is positively correlated with measures of fitness (i.e., the higher the DNA content the higher the fitness in the context of removal of apical dominance). Endoreduplication may have genetic effects that could lead to rapid regrowth and enhanced fitness by increasing gene expression through additional gene copies (Barow 2006). Although we know a great deal about the genetic basis of endoreduplication per se (Vlieghe et al. 2005; Imai et al. 2006; Yoshizumi et al. 2006) and have evidence that it plays a role in fitness compensation (Scholes and Paige 2011) we still do not know the underpinning genes or gene pathways affecting fitness compensation following endoreduplication in Arabidopsis (or any other plant species exhibiting growth compensation).
As a first step, we have taken advantage of the known compensatory variation in the annual plant A. thaliana, combined with commercially available microarrays and QTL analyses to begin to assess the molecular basis of overcompensation (increased fitness) following apical damage. In addition, we use a gene knockout and complementation approach to assess the phenotypic effects of one promising candidate gene uncovered from the microarray/QTL screen. Specifically, we (1) characterize fitness variation, following the removal of apical dominance, of recombinant inbred lines (RILs) generated from a cross between Landsberg erecta × Columbia; (2) determine seasonal variation in the compensatory response; (3) identify QTL responsible for the variation in compensation; (4) quantify differential gene expression underlying clipped and unclipped individuals of the Columbia ecotype using a commercially available microarray platform; (5) combine QTL and microarray data to narrow the genes responsible for the compensatory response; (6) evaluate the compensatory response of Transfer DNA (T-DNA) knockout lines of a promising candidate gene, glucose-6-phosphate-1-dehydrogenase (G6PDH1, At5g35790.1); (7) perform quantitative RT-PCR (qRT-PCR) on G6PDH1 to verify differences in expression between overcompensating (Columbia) and undercompensating (Landsberg erecta) plants; and (8) construct and assess the fitness response of a transgenic line for the complementation of G6PDH1. Twenty-five years ago we published the first empirical data supporting the idea that herbivore damage can lead to enhanced fitness (Paige and Whitham 1987). Here we present the first evidence for the molecular basis of this response.
Materials and Methods
Fitness variation
A total of 96 RILs (Lister and Dean 1993) of A. thaliana developed from a cross between Columbia (an overcompensating genotype) and Landsberg erecta (an undercompensating genotype) were used to assess fitness variation following the removal of apical dominance (to simulate mammalian herbivory) (Scholes and Paige 2011). The 96 F2 lines were advanced through inbreeding and single-seed descent for more than eight generations (Lister and Dean 1993) and are available through The Arabidopsis Information Resource. The RILs and their parental lines (Columbia and Landsberg erecta) were grown for two seasons (Spring 2007 and Fall 2008) in a greenhouse on the campus of the University of Illinois, Champaign, under 12 hr of light (∼100 µE/m2/sec) and dark. Plants were grown individually in 3.5-inch pots using LI Sunshine mix. Temperatures within the greenhouse ranged from 22° to 26°. Seeds/seedlings were kept moist during germination, and plants were watered daily to maintain soil moisture without saturating the soil. Plants were not fertilized. Ten plants per line (960 plants) were grown from seed, and half (five per line) were randomly chosen and clipped at a 6-cm inflorescence height down to ∼1 cm to simulate mammalian herbivory; the remaining five served as undamaged controls. At the end of the flowering season the numbers of siliques per plant were recorded. Our previous studies have shown that siliques are a good measure of plant fitness in that there are no significant differences in seed weights or germination success between clipped and unclipped plants of either Columbia or Landsberg erecta. In addition, clipped plants of Columbia produced significantly greater numbers of seed, whereas clipped plants of Landsberg erecta produced significantly fewer seeds (Scholes and Paige 2011) in comparison to unclipped controls. The seeds from unclipped plants collected during the first season were used to generate the second-season plants.
Potential differences in silique production were assessed using an Analysis of Variance (Systat 13) comparing plants with apical meristem damage to undamaged controls for each recombinant inbred line. Comparisons were made both within and between years to assess fitness variation among RILs and within-line repeatability across the 2 years. Silique counts were square-root-transformed to approximate normality. RILs were classified as under- (silique production significantly lower than the undamaged control), equal (silique production not statistically different from the undamaged control), or overcompensators (silique production significantly higher than the undamaged control) based on an Analysis of Variance for each RIL and year.
QTL analysis
QTL were identified by importing phenotypic (differential fitness data) and genotypic data sets (Nottingham Arabidopsis Stock Centre, AtEnsembl, http://atensembl.arabidopsis.info) into QTL Cartographer version 2.5 (Wang et al. 2010). Fitness data were pooled from 2 years using the average response across years and subtracting clipped from unclipped plants for each RIL. There were 14 of 96 lines for which we had only 1 year’s data; these were used in our QTL mapping study as well using Least Square (LS) means of fitness to adjust for unbalanced measures. A total of 141 markers equally distributed on all chromosomes with an average interval of ∼4.5 cM were selected (Zeng 1994). The data were initially analyzed using composite interval mapping (CIM) (Jansen and Stam 1994; Zeng 1994) to find QTL. Cofactors for CIM were selected from the forward and backward regression option. Significant QTL (LOD score of 2.5 and above) (Zeng 1994) from CIM were used to find other significant QTL and interactions among QTL elsewhere in the genome using multiple interval mapping. The model that minimized Akaike’s information criterion with penalty = 1 (Jansen 1993) was selected and applied to estimate the additive effect and the proportion of the fitness variation explained by each QTL. Effect sizes were calculated in QTL Cartographer version 2.5 (Wang et al. 2010) as the average percentage residual variance attributed to a QTL after removing the effects of covariates and all other QTL.
Although QTL can help in identifying regions of the genome responsible for compensation, it is difficult to identify specific candidate genes, as a single QTL likely contains hundreds of genes [a single QTL ranges from 10 to 20 cM in size with ∼1 cM of Arabidopsis covering 210 kb of the genome (Peters et al. 2001)] of which some may and some may not be responsible for observed patterns of fitness compensation. Considering the number of QTL obtained, we combined QTL mapping with microarray expression data to help in identifying potential candidate genes. Wayne and McIntyre (2002), for example, successfully combined data from QTL and microarrays to identify genes responsible for ovariole number in Drosophila melanogaster.
Microarray analysis
To identify potential candidate genes located within a QTL region, we carried out a microarray analysis on the Columbia ecotype (one that exhibits patterns of overcompensation) comparing clipped and unclipped individuals (data online at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc+GSE44781). Plants were grown in a greenhouse on the campus of the University of Illinois, Champaign, under 12 hr of light (∼100 µE/m2/sec) and dark. Forty plants were grown individually in 3.5-inch pots using LI Sunshine mix. Temperatures within the greenhouse ranged from 22° to 26°. Seeds/seedlings were kept moist during germination, and plants were watered daily to maintain soil moisture without saturating the soil. Plants were not fertilized. Axillary tissue was collected 6 days after clipping from both clipped (meristematic regrowth tissue) and unclipped plants (axillary meristematic tissue that naturally arises prior to clipping—similar in position, size, and location at the base of the unclipped plants’ primary inflorescence); three Affymetrix oligonucleotide arrays with eight pooled clipped plants/chip and two Affymetrix oligonucleotide arrays with eight pooled unclipped plants/chip were compared in this experiment. We used the Arabidopsis Affymetrix GeneChip containing >22,500 probe sets representing ∼24,000 gene sequences. This array is based on information from the International Arabidopsis Sequencing Project completed in December 2000 and is constructed by light-directed synthesis of oligonucleotides directly onto a glass “chip” approximately the size of glass coverslip. Each gene is represented on the array by a set of 20 oligonucleotide probes representing 25-mer sequences from some portion of the gene. Gene expression in a target sample is assessed by hybridization.
Total RNA was extracted with standard TRIzol (Life Technologies, Carlsbad, California) protocols from clipped (n = 3 chips) and unclipped (n = 2 chips) plants. The quality of the RNA was checked at 260 and 280 nm for determination of sample purity and concentration using a nano-drop. Messenger RNA was reverse-transcribed and labeled with the MessageAmp kit (Ambion, Austin, TX) and biotin-labeled dCTP and dGTP (ENZO Diagnostics, Farmingdale, NY). Affymetrix Arabidopsis GeneChip Arrays (Version 2.0) were hybridized at the University of Illinois Keck Center. Feature intensities on each chip were quantified with MAS 5.0 software. Following hybridization, the perfect match (PM) probes for all arrays were initially quantile-normalized with the Affy package in Bioconductor (Irizarry et al. 2003) to remove nonbiological variation among arrays. Only the PM data were used for the remainder of the analysis, and mismatch (MM) probes were ignored because they tend to increase random noise in the data. Data were analyzed using a t-test for each gene comparing clipped and unclipped plants. We controlled for multiple testing with a false discovery rate of P < 0.01.
Genes with significant expression upon clipping based on the microarray analysis were then analyzed for gene ontology. Biological process and molecular function information for each gene was obtained via AmiGo Slimmer Tool (v.1.8) analysis of the Gene Ontology (http://www.geneontology.org) database using the Plant GO Slim term set.
qRT-PCR was also performed on Columbia and Landsberg erecta plants to verify differences in expression in G6PDH1 between overcompensating and undercompensating ecotypes, respectively. Total RNA was isolated with TRIzol (Invitrogen, Carlsbad, CA) from rosette/cauline leaf material from clipped and unclipped Columbia and Landsberg erecta plants. Rosette/axillary meristematic tissues were collected at five time points [1 day before the inflorescence reached 6 cm (the height threshold for clipping), 1 day after, 5 days after, 15 days after, and at 50% flowering) in both clipped and unclipped plants to capture the time course of variation in gene expression patterns. Rosette leaves were collected for 1 day before and 1 day after treatments, and axillary meristems (regrowth tissue arising from the base of the plant off of the remaining portion of the primary inflorescence) were collected for the remaining time points. Of course, axillary meristems became more mature as time passed in terms of size and stage of development. The rationale for collecting axillary meristems at later points in time deals with the fact that plants translocate their nutrients to the developing tissues (axillary meristems, cauline leaves, siliques). The rationale for choosing samples before clipping was to check the actual change in gene expression following clipping and to assess any inherent differences between the genotypes in gene expression. The first-strand cDNA was synthesized using reverse transcriptase (SuperScript III, Invitrogen). The reverse transcription was carried out as recommended by the manufacturer. Three biological replicates and three technical replicates (i.e., three reads from each sample) were used for each ecotype and time interval. qRT-PCR was performed on each ecotype (Columbia and Landsberg erecta) and a reference gene from microarray data (ubiquitin) using SYBR green (a fluorescent dye). The G6PDH1 expression data were analyzed using the approach of Pfaffl (2001), where the ratio of a target gene (G6PDH1 in Columbia or Landsberg erecta) is expressed in a sample vs. a reference gene (ubiquitin), followed by an analysis of variance and linear contrasts comparing Columbia to Landsberg erecta. Expression data were square-root-transformed to approximate normality.
T-DNA knockout evaluation
A gene knockout experiment gives firsthand information on the role of a candidate gene in the response of interest (in this case, the degree of compensation) and a direct way of measuring the function of the gene product in situ. In Arabidopsis, T-DNA knockout plants are available for nearly all genes identified to date. A T-DNA inserted within the gene (in the 5′ UTR, ORF, or 3′ UTR) silences the gene, and plants harboring a T-DNA on both chromosomes are devoid of any gene product (or most of the gene product depending upon the position of the insert) for the gene of interest. Therefore, we assessed the role of the candidate gene (see results below) uncovered in our combined QTL mapping and microarray experiment above, using a T-DNA knockout approach followed by a clipping experiment and a fitness analysis. Mutant knockouts included G6PDH_1 (Sail_1252), G6PDH_2 (Salk_019323), and G6PDH_3 (Gabi_86405A). T-DNA inserts were confirmed by designing primers for the genomic region and T-DNA insertion using the T-DNA Primer Design Tool (Salk Institute Genomic Analysis Laboratory, http://signal.salk.edu/tdnaprimers.2.html). The primers LP–TGCCATTCATTTTTAAGCTGG, RP- AGATGCAAGGTAATGTGCACC and LB–ATATTGACCATCATACTCATTGC were used to genotype plants. The PCR reactions produced diagnostic banding patterns for homozygous, heterozygous, and wild-type individuals. Three homozygous knockout lines with differing T-DNA insertions were compared for fitness differences among clipped and unclipped plants. Columbia (an overcompensating plant that shares identical genetic background to the knockout plants except for the knocked-out gene) served as a control on the effects of the gene knockout. Plants were grown individually under 12 hr of light and dark (∼70 µE/m2/sec) in 3.5-inch pots with LI Sunshine mix in an environmental chamber on the campus of the University of Illinois. The temperature within the growth chamber was set at 22◦ C. Seeds/seedlings were kept moist during germination and plants were watered daily to maintain soil moisture without saturating the soil. Plants were not fertilized. A total of 40 plants per line were grown from seed, and half (20 per line) were randomly chosen and clipped to ∼1 cm above ground level (removing apical dominance) at 6 cm of inflorescence growth to simulate mammalian herbivory; the remaining 20 served as undamaged controls. Fitness comparisons were made in terms of the numbers of siliques produced. The data were square-root-transformed and analyzed using an analysis of variance (SAS v. 9.2; SAS, Cary, NC) followed by linear contrasts comparing clipped to unclipped plants within each treatment group so that we could assess whether knockout treatments altered the compensatory outcome from that of overcompensation observed in the Columbia wild type.
Complementation of G6PDH1
To further assess the effects of G6PDH1 on fitness compensation, we used a gene complementation approach wherein we replaced G6PDH1 in a T-DNA knockout line (G6PDH_3; Gabi_86405A). To amplify 6032 bp of G6PDH1 using Kod polymerase, we used the following forward and reverse primers: For1—CACCCGTGTCGACCTCCACTATTGCCTCAAGTTGATGTTGAGTTCCG and Rev1 —CCAATCTTCATCTTCGTCTTCATGGTACCTAACG. The region included ∼2.0 kb of the upstream promoter, exons, and introns and ∼1.1 kb of the downstream region. The PCR product was subcloned into a pENTR/d-TOPO vector as per the manufacturer’s recommendation and later cloned to pMDC 123 (Curtis and Grossniklaus 2003) using the gateway LR reaction (Left and Right recombination sites). As both entry clone and binary vector had the same selection marker (kanamycin), the entry clone was linearized using the restriction enzyme MluI. This enzyme linearizes without affecting the gene or the gateway-site-specific recombination sites. The gateway-site-specific recombination yielded the binary vector (pMHS 207), which was transformed to Agrobacterium tumefaciens strain GV3101:pMP90 by the freeze-thaw method (Holsters et al. 1978). Plant transformation was done using Clough and Bent’s (1998) floral dip protocol. Primary transformants were selected by spraying glufosinate at 250 mM concentration. Transgenic seed was carried through to the T2 generation for subsequent fitness analyses. Given that the transgenic line was grown in a separate experiment, under conditions identical to those used in comparing Columbia, Landsberg erecta, and three T-DNA knockout mutants to one another, a t-test was used to compare clipped to unclipped transgenic plants to see if restoring gene function led to a pattern of overcompensation as one would predict if G6PDH1 played a significant role in overcompensation.
Results
Fitness variation among RILs
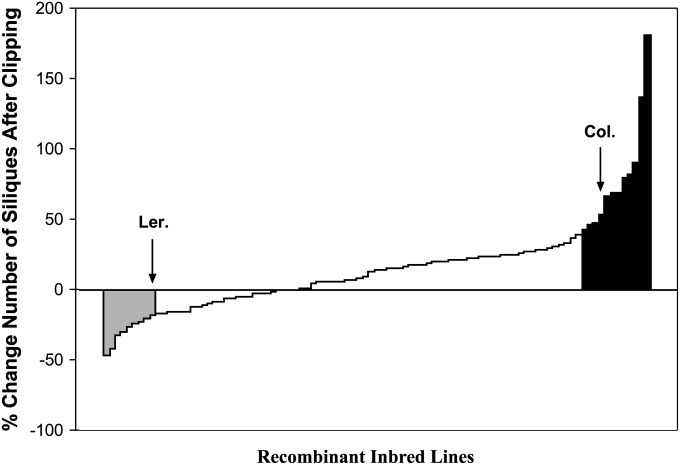
The RILs used from a cross between Columbia and Landsberg erecta showed variation in compensatory responses ranging from undercompensation to overcompensation within both seasons/years (Spring 2007: treatment × line interaction, F = 1.65, d.f. = 92, 654, P < 0.0001; Fall 2008: treatment × line interaction, F = 3.26, d.f. = 83, 640, P < 0.0001) (Figure 1). Although there was a significant treatment × line × year effect (F = 1.28, d.f. = 86, 1294, P = 0.045), the majority (68%) of lines had similar compensatory responses across years (48 lines equally compensated in both years, 4 overcompensated, and 2 undercompensated). Of the remaining lines for which we had 2 years of data, 19 shifted from overcompensation to equal compensation, 5 shifted from equal compensation to undercompensation, and only 1 shifted from overcompensation to undercompensation (P < 0.05 for those that changed category). There were 14 lines for which we had only one year’s data, of which 13 equally compensated and 1 overcompensated.
Figure 1.
Percentage change in number of siliques per line between clipped and unclipped plants for Columbia (right arrow) and Landsberg erecta (left arrow) parental lines and 93 recombinant inbred lines from a cross between Columbia and Landsberg erecta over 2 years; 14 lines had only 1 year of data. Solid area indicates overcompensating lines; shaded area indicates undercompensating lines.
QTL/microarray analyses
A total of three main QTL for compensation was found on chromosomes 1, 4, and 5 explaining 11.4, 10.1, and 26.7% of the variation in compensation, respectively (Table 1). The three QTL showed additive effects varying between 21.7 and 63.6 siliques and no evidence for epistatic interactions. All alleles increasing fitness were contributed by the Columbia ecotype (Table 1), although the compensatory response distribution suggests contributions from Landsberg erecta (i.e., evidence for transgressive segregation; see Figure 1).
Table 1. Estimates of QTL positions, effects, and interactions.
| Chromosome | Marker | Position (cM) | LOD | Additive effecta | b |
|---|---|---|---|---|---|
| 1 | 10 | 49 | 2.76 | 21.7 | 11.4 |
| 4 | 22 | 101 | 3.05 | 21.7 | 10.1 |
| 5 | 23 | 69 | 3.41 | 63.6 | 26.7 |
Significant QTL determined at LOD > 2.5.
All alleles increasing compensatory response originate from the Columbia ecotype.
: partial R2 provides an estimate of the amount of phenotypic variance explained by each QTL.
From the microarray analysis a total of 109 genes were found to be differentially expressed between clipped and unclipped plants of Columbia (see Supporting Information, File S1 and File S2 for gene list). A total of 30, 19, 17, 16, and 27 differentially expressed genes were located on chromosomes 1–5, respectively, between clipped and unclipped plants. Based on the gene ontology analysis, these genes can be generally classified into stress response genes, metabolic genes, and growth/reproductive genes (Table 2). When mapped with the QTL data, only a single gene colocalizes within one of the QTL markers (QTL 3 located on chromosome 5 at 69 cM; Table 1), a G6PDH1 (EC 1.1.1.49).
Table 2. Gene ontology analysis of 109 overexpressed genes in Columbia wild type after clipping.
| Biological process | No. of genes | Selected genes |
||||||
|---|---|---|---|---|---|---|---|---|
| Response to stress | 19 | ATP1a,b,c | CGL1d | FNR1a | GOLS2e | PDE345e | TCH4b,d | WR3c |
| Reproduction | 9 | AGL8a,f | GRH1g | GSH1e | MPK6h,i | RP1j | ||
| Carbohydrate metabolic process | 9 | CGL1d | CINV1b,g | G6PD1a,g | GALAKa,d,h | GOLS2e | IAR4e | |
| Transport | 9 | ATP1a,b,c | GDI2k | WR3c | ||||
| Response to biotic stimulus | 6 | CYP38e | FNR1a | GSH1e | MPK6h,i | WIN1d | ||
| Generation of precursor metabolites and energy | 5 | FNR1a | IAR4e | ORF291c | PDE345e | |||
| Flower development | 4 | AGL8a,f | GRH1g | GSH1e | MPK6h,i | |||
| Secondary metabolic process | 2 | GSH1e | MPK6h,i | |||||
| Photosynthesis | 1 | FNR1a | ||||||
| Cell differentiation | 1 | AGL8a,f | ||||||
| Growth | 1 | GRH1g | ||||||
Shown are a subset of biological processes and a selection of important genes. Superscripts indicate molecular function. See File S1 and File S2 for full gene ontology analysis.
Nucleotide/DNA/RNA binding.
Hydrolase activity.
Transporter activity.
Transferase activity.
Catalytic activity.
Sequence-specific DNA-binding transcription factor activity.
Protein binding.
Kinase activity.
Signal transducer activity.
Structural molecule activity.
Enzyme regulator activity.
T-DNA knockout fitness analyses and gene expression patterns
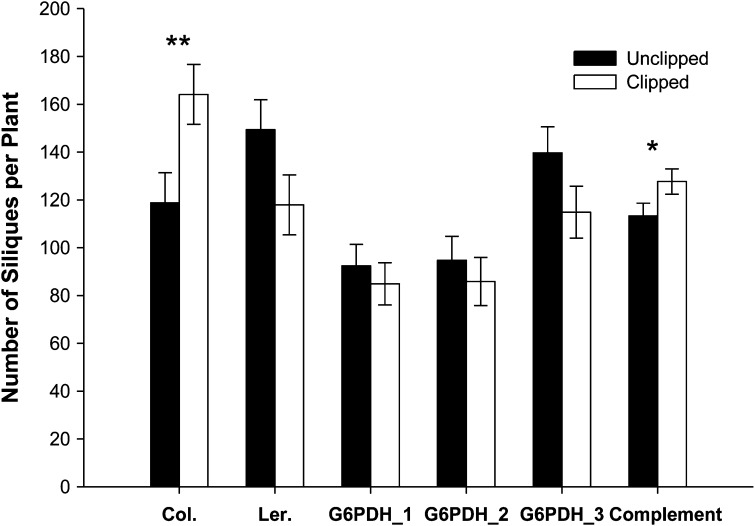
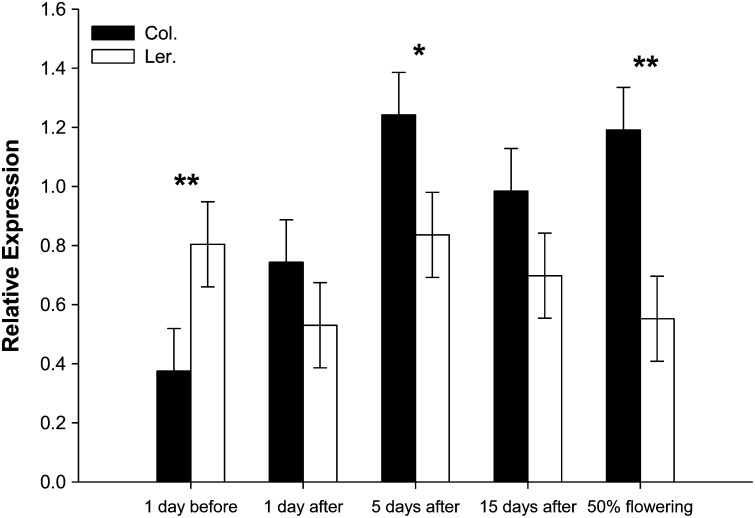
T-DNA knockout experiments verified the role of G6PDH1 in the compensatory response (Figure 2). Overall, results show a marginally significant clipping treatment × line interaction for silique production (F = 2.21, d.f. = 4, 114, P = 0.073). Results indicate that Landsberg erecta equally compensated with a nonsignificant trend toward undercompensation (P = 0.094; see Figure 2) while all three knockouts of G6PDH1 showed patterns of equal compensation [P = 0.471, P = 0.419, and P = 0.265 for knockouts 1 (Sail 1252), 2 (Salk 019323), and 3 (Gabi_86405A); see Figure 2], respectively, with a trend toward undercompensation, whereas Columbia overcompensated following clipping (P = 0.019). G6PDH1 expression data through time comparing Landsberg erecta, an undercompensating ecotype, and Columbia, an overcompensating ecotype, showed higher levels of expression (1.4- to 2.2-fold) in Columbia following the removal of apical dominance at all time points following clipping (overall expression differences between ecotypes: Columbia 0.907 ± 0.064 and Landsberg erecta 0.684 ± 0.064, F = 5.99, d.f. = 1, 20, P = 0.024; ecotype × time, F = 3.83, d.f. = 4, 20, P = 0.018) (Figure 3).
Figure 2.
Silique production for clipped and unclipped T-DNA knockout lines of G6PDH1_1–3 of Columbia, the two ecotypes, Landsberg erecta (Ler.), and Columbia (Col.) and a transgenic line complemented with G6PDH1 (G6PDH1_3; Gabi_86405A). Note that the gene complementation was conducted in a separate experiment and thus analyzed separately. Shown are means ± 1 SE. Asterisks indicate significance at **P < 0.05 and *P = 0.064.
Figure 3.
G6PDH1 gene expression through time before and after the removal (clipping) of the plants’ apical meristem, simulating mammalian herbivory, for Columbia wild type (Col.) and Landsberg erecta (Ler.). The G6PDH1 expression data were analyzed using the approach of Pfaffl (2001), where the ratio of a target gene (G6PDH1 in Columbia or Landsberg erecta) is expressed in a sample vs. a reference gene (ubiquitin). Shown are means ± 1 SE. Asterisks indicate significance at **P < 0.05 and *P < 0.06.
Complementation of G6PDH1
The results of our complementation studies support the role of G6PDH1 in the compensatory response. This transgenic line showed patterns more similar to the overcompensating Columbia line than either Landsberg erecta or the three knockout lines (see Figure 2). Clipped plants of the transgenic line complemented with G6PDH1 tended (P = 0.064) to produce more siliques/plant than unclipped plants (127.7 ± 5.0 fruits per plant) whereas unclipped plants produced 113.3 ± 5.3 fruits per plant (t = 1.975, d.f. = 1, 18, P = 0.064).
Discussion
Although there is evidence that genetic variation for fitness compensation exists, little is known about the genetic underpinnings leading to enhanced growth and reproduction in species exhibiting growth compensation following herbivory. Using a QTL analysis, we uncovered three QTL on chromosomes 1, 4, and 5, explaining 11.4, 10.1, and 26.7% of the variation in compensation, respectively. These three QTL showed additive effects increasing silique production by 21.7–63.6 siliques upon clipping. The increase in fitness is attributable to alleles contributed by the Columbia ecotype (Table 1) as opposed to alleles from Landsberg erecta, although, as noted above, the compensatory response distribution suggests contributions from Landsberg erecta (i.e., evidence for transgressive segregation) (Figure 1).
Furthermore, combining QTL and microarray analyses we have uncovered one gene that appears to play a significant role in the phenomenon of overcompensation in Arabidopsis, G6PDH1 (At5g35790.1). For a relatively important gene, residing in the middle of the QTL with the greatest effect size, the LOD score was relatively low (3.41, Table 1). This might be explained by not having markers very close to the estimated position of the QTL, having some missing data from the QTL analysis, or that the QTL might be involved in an important epistatic interaction within the area that may suppress the expected QTL effect (Darrah et al. 2006). Nonetheless, T-DNA knockouts, expression assays, and complementation studies confirm the importance of G6PDH1 in fitness compensation (see below).
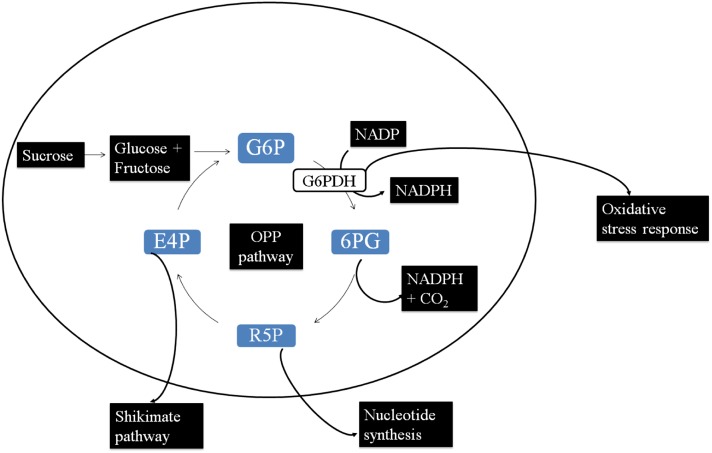
G6PDH1 is the key regulatory enzyme in the oxidative pentose phosphate pathway (OPPP) that plays a central role in plant metabolism by converting glucose to ribose-5-phosphate. The OPPP is a primary source of the reductant NADPH for biosynthetic processes such as the assimilation of nitrogen into amino acids, fatty-acid synthesis, and resistance to oxidative damage. Intermediates, such as ribose-5-phosphate, are also withdrawn from the OPP pathway for phenylpropanoid production via the shikimate pathway (Figure 4) (Kruger and Von Schaewen 2003; Scharte et al. 2009).
Figure 4.
Schematic representation of the cytosolic oxidative pentose-phosphate pathway, adapted from Hauschild and Von Schaewen (2003). G6P (glucose-6-phosphate) is oxidized by G6PDH to yield 6-phospho-gluconate (6PG) and in the process reduces NADP to NADPH and functions in reducing oxidative damage, eventually leading to the production of ribulose-5-phosphate (R5P) and erythrose-4-phosphate (E4P), which are used for nucleotide synthesis (essential in the synthesis of aromatic amino acids) and defensive chemistry (such as glucosinolates) via the shikimate pathway, respectively.
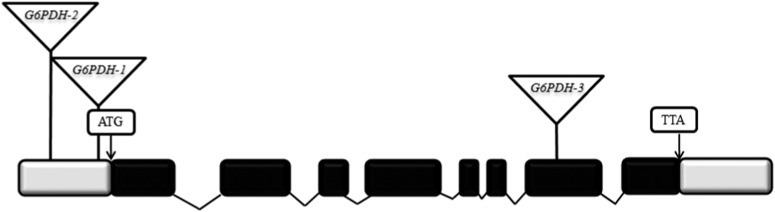
Knockout studies of three T-DNA insertion lines of G6PDH1 (sharing the same genetic background as Columbia) showed patterns of equal compensation, with a trend toward undercompensation, rather than overcompensation, as observed in the Columbia wild type (Figure 2). Two of the three T-DNA knockout mutants (G6PDH1-1, Sail 1252, and G6PDH1-2, Salk 019323) (Figure 2) showed overall lower levels of fitness (i.e., both clipped and unclipped plants), while the third, G6PDH1-3, Gabi_86405A, showed higher overall levels of fitness that were within the range of the wild types, suggesting potential positional effects of T-DNA insertion resulting in partial knockdown of G6PDH1-3. Specifically, the insertion of G6PDH1-3 is positioned within the ORF, rather than the UTRs of G6PDH1-1 and -2 (see Figure 5 for sites of insertion). Nonetheless, G6PDH1-3 plants also equally compensated with a trend toward undercompensation as in the other two knockout mutants.
Figure 5.
Schematic representation of G6PDH1 showing the position of each of the three T-DNA insertions. Exons (solid), introns (shaded), and start and stop codons are shown. The T-DNA inserts are represented by inverted triangles. See text for a discussion of potential positional effects.
In addition, G6PDH1 expression data through time comparing Landsberg erecta, an undercompensating ecotype, and Columbia, an overcompensating ecotype, showed higher levels of expression (1.4- to 2.2-fold) in Columbia following the removal of apical dominance; these data are consistent with our knockout experiments wherein lowering or knocking out G6PDH1 resulted in equal-to-undercompensation instead of overcompensation (Figure 3). There is considerable sequence variation in G6PDH1, with three nonsynonymous substitutions, each causing a change in an amino acid, between Columbia and Landsberg erecta that may explain the differential patterns of expression in G6PDH1 following apical damage and regrowth and perhaps the differences in compensation (Max Planck Institute for Developmental Biology, POLYMORPH Project, http://polymorph-clark20.weigelworld.org/cgi-bin/retrieve_cds_snp.cgi). In addition, our transgenic line complemented with G6PDH1 restored the compensatory response from equal compensation, with a trend toward undercompensation, in the knockout line (G6PDH1_3; Gabi_86405A) to overcompensation (at P = 0.064). We suspect that positional effects of the transgene or unmeasured environmental influences may have constrained the magnitude of the compensatory response typically observed in Columbia. Collectively, these results indicate the importance of G6PDH1 in regulating the compensatory response following the removal of apical dominance.
We propose that plants with the capability of overcompensating (increasing both biomass and fitness when compared to undamaged controls) reprogram their transcriptional activity in at least three important ways: through a suite of defensive mechanisms, through an increase in expression of genes involved in energy metabolism, and through an increase in DNA content (via endoreduplication; see Scholes and Paige 2011), with the increase in DNA content feeding back on pathways involved in defense and metabolism through increased gene expression. Initially, following apical damage, the G6PDH1 gene elicits a suite of defensive reactions that are likely associated with cellular damage from herbivory. These may include reactive oxygen species to ward off infection and induced chemical defenses, such as glucosinolates, via the shikimate pathway (Scharte et al. 2009). When analyzing genes that were significantly differentially expressed (from our microarray data), several of the genes affected were found to be enzymes (e.g., a suite of invertase genes, G6PDH1, and galactinol synthase) involved in carbohydrate metabolism, and these genes were significantly up-regulated and likely play a significant role in overcoming tissue loss. Of particular note, cytosolic invertase 1 is adjacent to a QTL located on chromosome 1. In addition, up-regulation of G6PDH1 ultimately leads to the biosynthesis of nucleic acids (see Figure 4), consistent with the significant increase in DNA content (through endoreduplication) observed in overcompensating ecotypes of A. thaliana when compared to undercompensating ecotypes (Scholes and Paige 2011). Interestingly, removal of apical dominance reduces the level of auxin leading to axillary bud break and stem regeneration, and low levels of auxin trigger an exit from mitotic cycles into the endocycle (Ishida et al. 2010). Thus, there is a direct link between endoreduplication and the removal of apical dominance.
Weinig et al. (2003) previously mapped QTL for resistance and tolerance (compensation) to apical meristem damage by rabbits under natural conditions of the field over two seasons in RILs from a Columbia × Landsberg erecta cross (Lister and Dean 1993) of Arabidopsis. Although QTL for resistance were found within each seasonal cohort, no QTL for tolerance were detected. This is in contrast to our study here, wherein we uncovered three QTL. We surmise that the differences in our findings can be attributed to the differences in natural herbivory under field conditions vs. artificial herbivory in a greenhouse. Natural herbivory resulted in wide variance in regrowth and fitness within any given line whereas our clipping experiments in the greenhouse resulted in far less variance, resulting in higher repeatability in fitness compensation, making it easier to uncover QTL. Whereas Weinig et al. (2003) interpreted this to mean that there were many genes of small effect involved in tolerance (compensation), our results indicate fewer genes of larger effect; i.e., in our study we uncovered a single gene of major effect, G6PDH1 (when knocked-out plants equally compensated with a trend toward undercompensation following apical damage, contributing significantly to the phenomenon of overcompensation). In both studies, there were also significant environmental effects (genotype × environment interactions) detected, with 25 of 79 lines for which we had two seasons of data responding differently in fitness compensation from one season to the next (all shifting to lower fitness levels). The remaining 54 lines all maintained the same level of fitness compensation.
Results here support the utility of using a combinatorial approach of QTL mapping and microarray data in uncovering potential candidate genes. As Wayne and McIntyre (2002, pp. 14,903) pointed out, “The use of microarray technology … allows an efficient, objective, quantitative evaluation of genes in the QTL and has the potential to reduce the overall effort needed in identifying genes causally associated with quantitative traits of interest.” Using these combined approaches, we uncovered a single differentially expressed gene colocated within one of three QTL regions in the recombinant inbred Lister–Dean lines created from a cross between Landsberg erecta and Columbia. Knockout and complementation studies of this candidate strongly suggest an important role of this gene and the pathway in which it resides in the compensatory response of Arabidopsis. Of course we want to be clear that this is likely not the only important gene involved in the compensatory response, given the additional QTL and the pathway in which G6PDH1 resides.
Gaining an understanding of the genetic basis of overcompensation (increased seed yield after damage), in particular, following apical damage should be of great interest to agriculturists who, through recent advances in genetic technology and selective breeding, might incorporate these traits into crop plants such as oilseed rape (Brassica napus), a close relative of Arabidopsis. The isoform of G6PDH1 could also be engineered in crops such as sugarcane or rice where ratoon cropping is conducted (ratoon cropping resembles simulated herbivory where the apical meristem is removed leading to increased plant yields through regrowth). Thus, our findings should be of great value in that the results of this study set the stage for genetically engineering or selecting plants that not only tolerate apical damage, but also actually increase seed yield from such damage. Furthermore, from an evolutionary perspective, the genetic basis of overcompensation uncovered here in the model system A. thaliana may be readily applied to natural systems, improving our understanding of plant regrowth following herbivory and the complexities of plant–animal interactions. With the results of this study, we are beginning to gain significant insights as to the underpinning genetic basis that contributes to the phenomenon of overcompensation.
Supplementary Material
Acknowledgments
We thank Sindhu Krishnankutty and Lauren Clayton for help in collecting fitness data; Osman Radwan, Steve Clough, and Bernarda Calla for help with RNA extractions and cDNA synthesis; and Jenny Drnevich for analyzing microarray data. Research was funded by National Science Foundation grants DEB-0522409, DEB-1010868, and DEB-1146085 to K.N.P.
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Agrawal A. A., 1998. Induced responses to herbivory and increased plant performance. Science 279: 1201–1202. [DOI] [PubMed] [Google Scholar]
- Barow M., 2006. Endopolyploidy in seed plants. Bioessays 28: 271–281. [DOI] [PubMed] [Google Scholar]
- Berenbaum M. R., Zangerl A. R., Nitao J. K., 1986. Constraints on chemical coevolution: wild parsnips and the parsnip webworm. Evolution 40: 1215–1228. [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F., 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U., 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrah C., Taylor B. L., Edwards K. D., Brown P. E., Hall A., et al. , 2006. Analysis of phase of LUCIFERASE expression reveals novel circadian quantitative trait loci in Arabidopsis. Plant Physiol. 140: 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauschild R., von Schaewen A., 2003. Differential regulation of glucose-6- phosphate dehydrogenase isoenzyme activities in potato. Plant Physiol. 133: 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M., de Waele D., Depicker A., Messens E., van Montagu M., et al. , 1978. Transfection and transformation of Agrobacterium tumefaciens. Molec. gen. Genet. 163: 181–187. [DOI] [PubMed] [Google Scholar]
- Imai K. K., Ohashi Y., Tsuge T., Yoshizumi T., Matsui M., et al. , 2006. The A-type cyclin CYCA2;3 is a key regulator of ploidy levels in Arabidopsis endoreduplication. Plant Cell 18: 382–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A., Hobbs B., Collin F., Beazer-Barclay Y. D., Antonellis K. J., et al. , 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Ishida T., Adachi S., Yoshimura M., Shimizu K., Umeda M., et al. , 2010. Auxin modulates the transition from the mitotic cycle to the endocycle in Arabidopsis. Development 137: 63–71. [DOI] [PubMed] [Google Scholar]
- Jansen R. C., 1993. Interval mapping of multiple quantitative trait loci. Genetics 135: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. C., Stam P., 1994. High resolution of quantitative traits into multiple loci via interval mapping. Genetics 136: 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juenger T., Bergelson J., 2000. Factors limiting rosette recruitment in scarlet gilia, Ipomopsis aggregata: seed and disturbance limitation. Oecologia 123: 358–363. [DOI] [PubMed] [Google Scholar]
- Kruger J. N., von Schaewen A., 2003. The oxidative pentose phosphate pathway: Structure and organization. Curr. Opin. Plant Biol. 6: 236–246. [DOI] [PubMed] [Google Scholar]
- Lennartsson T., Tuomi J., Nilsson P., 1997. Evidence for an evolutionary history of overcompensation in the grassland biennial Gentianella campestris (Gentianaceae). Am. Nat. 149: 1147–1155. [DOI] [PubMed] [Google Scholar]
- Lister C., Dean C., 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745–750. [DOI] [PubMed] [Google Scholar]
- Maschinski J., Whitham T. G., 1989. The continuum of plant responses to herbivory: the influence of plant association, nutrient availability, and timing. Am. Nat. 134: 1–19. [Google Scholar]
- Mauricio R., Rausher M. D., Burdick D. S., 1997. Variation in the defense strategies of plants: are resistance and tolerance mutually exclusive? Ecology 78: 1301–1311. [Google Scholar]
- Nilsson P., Tuomi J., Strom M. A., 1996. Bud dormancy as a bet hedging strategy. Am. Nat. 147: 269–281. [Google Scholar]
- Paige K. N., 1992. Overcompensation in response to mammalian herbivory: from mutualistic to antagonistic interactions. Ecology 73: 2076–2085. [Google Scholar]
- Paige K. N., 1994. Herbivory and Ipomopsis aggregata: differences in response, differences in experimental protocol: a reply to Bergelson and Crawley. Am. Nat. 143: 739–749. [Google Scholar]
- Paige K. N., 1999. Regrowth following ungulate herbivory in Ipomopsis aggregata: geographic evidence for overcompensation. Oecologia 118: 316–323. [DOI] [PubMed] [Google Scholar]
- Paige K. N., Whitham T. G., 1987. Flexible life history traits: shifts by scarlet gilia in response to pollinator abundance. Ecology 68: 1691–1695. [DOI] [PubMed] [Google Scholar]
- Peters J. L., Constandt H., Neyt P., Cnops G., Zethof J., et al. , 2001. A physical amplified fragment-length polymorphism map of Arabidopsis. Plant Physiol. 127: 1579–1589. [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W., 2001. A new mathematical model for relative quantification in real- time RT-PCR. Nucleic Acids Res. 29: 2002–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautio P., Huhta A.-P., Piippo S., Tuomi J., Juenger T., et al. , 2005. Overcompensation and adaptive plasticity of apical dominance in Erysimum strictum (Brassicaceae) in response to simulated browsing and resource availability. Oikos 111: 179–191. [Google Scholar]
- Scharte J., Schön H., Tjaden Z., Weis E., von Schaewen A., 2009. Isoenzyme replacement of glucose-6-phosphate dehydrogenase in the cytosol improves stress tolerance in plants. Proc. Natl. Acad. Sci. USA 106: 8061–8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes D. R., Paige K. N., 2011. Chromosomal plasticity: mitigating the impacts of herbivory. Ecology 92: 1691–1698. [DOI] [PubMed] [Google Scholar]
- Stowe K. A., Marquis R. J., Hochwender C. G., Simms E. L., 2000. The evolutionary ecology of tolerance to consumer damage. Annu. Rev. Ecol. Syst. 31: 565–595. [Google Scholar]
- Tiffin P., Rausher M. D., 1999. Genetic constraints and selection acting on tolerance to herbivory in the common morning glory Ipomoea purpurea. Am. Nat. 154: 700–716. [DOI] [PubMed] [Google Scholar]
- Vlieghe K., Boudolf V., Beemster G. T. S., Maes S., Magyar Z., et al. , 2005. The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr. Biol. 15: 59–63. [DOI] [PubMed] [Google Scholar]
- Wang S., Basten C. J., Zeng Z.-B., 2010. Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, North Carolina. [Google Scholar]
- Wayne M. L., McIntyre L. M., 2002. Combining mapping and arraying: an approach to candidate gene identification. Proc. Natl. Acad. Sci. USA 99: 14903–14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinig C., Stinchcombe J. R., Schmitt J., 2003. QTL architecture of resistance and tolerance traits in Arabidopsis thaliana in natural environments. Mol. Ecol. 12: 1153–1163. [DOI] [PubMed] [Google Scholar]
- Yoshizumi T., Tsumoto Y., Takiguchi T., Nagata N., Yamamoto Y. Y., et al. , 2006. Increased level of polyploidy1, a conserved repressor of CYCLINA2 transcription, controls endoreduplication in Arabidopsis. Plant Cell Online 18: 2452–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z.-B., 1994. Precision mapping of quantitative trait loci. Genetics 136: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.