CCL18 is an endogenous agonist of the human CCR8 receptor.
Abstract
The CC chemokine ligand 18 (CCL18) is one of the most highly expressed chemokines in human chronic inflammatory diseases. An appreciation of the role of CCL18 in these diseases has been hampered by the lack of an identified chemokine receptor. We report that the human chemokine receptor CCR8 is a CCL18 receptor. CCL18 induced chemotaxis and calcium flux of human CCR8-transfected cells. CCL18 bound with high affinity to CCR8 and induced its internalization. Human CCL1, the known endogenous CCR8 ligand, and CCL18 competed for binding to CCR8-transfected cells. Further, CCL1 and CCL18 induced heterologous cross-desensitization of CCR8-transfected cells and human Th2 cells. CCL18 induced chemotaxis and calcium flux of human activated highly polarized Th2 cells through CCR8. Wild-type but not Ccr8-deficient activated mouse Th2 cells migrated in response to CCL18. CCL18 and CCR8 were coexpressed in esophageal biopsy tissue from individuals with active eosinophilic esophagitis (EoE) and were present at markedly higher levels compared with esophageal tissue isolated from EoE patients whose disease was in remission or in normal controls. Identifying CCR8 as a chemokine receptor for CCL18 will help clarify the biological role of this highly expressed chemokine in human disease.
Chemokines are chemotactic cytokines that guide the directed migration of leukocytes in the steady-state and in inflammation through a subfamily of seven-transmembrane spanning G protein–coupled receptors. The CC chemokine ligand (CCL) 18 was identified in the late 1990s by several groups as a gene highly expressed in the lung and induced in alternatively activated macrophages (AAMs) and thus initially given the names PARC (pulmonary and activation-regulated chemokine), AMAC-1 (alternative macrophage activation-associated CC chemokine-1), DC-CK1 (dendritic cell chemokine 1), and macrophage inflammatory protein (MIP) 4 (Adema et al., 1997; Hieshima et al., 1997; Kodelja et al., 1998). CCL18 is highly expressed in many human chronic inflammatory diseases, which include pulmonary fibrosis and certain cancers, and a wide range of allergic diseases (Pivarcsi et al., 2004; Schutyser et al., 2005; de Nadaï et al., 2006; Chen et al., 2011; Lucendo et al., 2011). In some diseases, the level of circulating CCL18 has been demonstrated to be a biomarker for disease activity and outcome (Prasse et al., 2009). In patients with systemic sclerosis, for instance, levels of CCL18 have been associated with the complication of interstitial lung disease, and in patients with idiopathic pulmonary fibrosis and certain cancers, levels of CCL18 have been correlated with poor outcomes (Prasse et al., 2007, 2009; Chen et al., 2011). Understanding the role of CCL18 in these diseases has been hampered by the lack of an identified receptor and by the lack of a known murine orthologue.
CCL18 is secreted primarily by cells of the myeloid lineage and has been identified in alveolar macrophages, tolerogenic dendritic cells, AAMs, and keratinocytes (Hieshima et al., 1997; Kodelja et al., 1998; Pivarcsi et al., 2004; Bellinghausen et al., 2012). In macrophages, CCL18 is induced by the Th2 cytokines IL-4 and IL-13, which program macrophages to differentiate into AAMs (Kodelja et al., 1998). AAMs contribute to the healing phase of acute inflammatory reactions and to tissue remodeling and fibrosis in chronic inflammatory diseases. Concordant with the spectrum of AAM activity, the abundance of CCL18 has been found to correlate with disease severity in fibrotic diseases, such as pulmonary fibrosis and scleroderma, and diseases of dysregulated macrophage biology (Schutyser et al., 2005; Prasse et al., 2007, 2009). In allergic diseases, increased CCL18 is also frequently associated with eosinophil infiltration in affected tissues (Pivarcsi et al., 2004; Schutyser et al., 2005; de Nadaï et al., 2006; Lucendo et al., 2011).
CCL18 activity has been detected on peripheral blood lymphocytes (Adema et al., 1997; Hieshima et al., 1997). However, studies using cells transfected with CCR1-7, CXCR1-4, and CX3CR1 did not reveal a specific CCL18 signaling receptor but did find that CCL18 could antagonize CCR3 (Hieshima et al., 1997; Nibbs et al., 2000). Investigation into atopic dermatitis and allergic diseases enabled the first identification of a specific T cell subset that responded to CCL18, and thus provided clues to its possible chemokine receptor (Günther et al., 2005; de Nadaï et al., 2006). In atopic dermatitis, CCL18 is the most abundantly expressed chemokine in lesional skin (Pivarcsi et al., 2004; Günther et al., 2005). CCL18 bound to blood skin-tropic cutaneous leukocyte antigen expressing (CLA+) memory T cells from individuals with atopic dermatitis and induced migration of CD4+ T cell clones derived from atopic dermatitis lesional skin in vitro and into human skin transplanted in SCID mice in vivo (Günther et al., 2005). The signature homing receptors of skin-tropic T cells are CLA in combination with the chemokine receptors CCR4, CCR10, and CCR8 (Schaerli et al., 2004; Islam et al., 2011). More CCR8-expressing cells are found in inflamed skin of individuals with active atopic dermatitis compared with skin of healthy individuals (Gombert et al., 2005). In studies of human asthma, CCL18 was found to induce the migration of TCR-activated in vitro differentiated human Th2 cells (de Nadaï et al., 2006). CCL18 induced migration of human peripheral blood Th2 cells and regulatory T cells ex vivo (Bellinghausen et al., 2012; Chenivesse et al., 2012). Th2 but not Th1 cells are programmed to selectively express the chemokine receptors CCR4 and CCR8, a receptor profile shared with regulatory T cells (D’Ambrosio et al., 1998; Wei et al., 2010). Skin-homing CLA+ T cells, Th2 cells, and regulatory T cells thus all express CCR4 and CCR8. However, TCR activation is a prerequisite for functional CCR8 but not CCR4 expression on in vitro differentiated Th2 cells (D’Ambrosio et al., 1998). Here, we report that CCL18 is an endogenous agonist of the human CCR8 receptor.
RESULTS AND DISCUSSION
CCL18 induced migration of CCR8-transfected cells
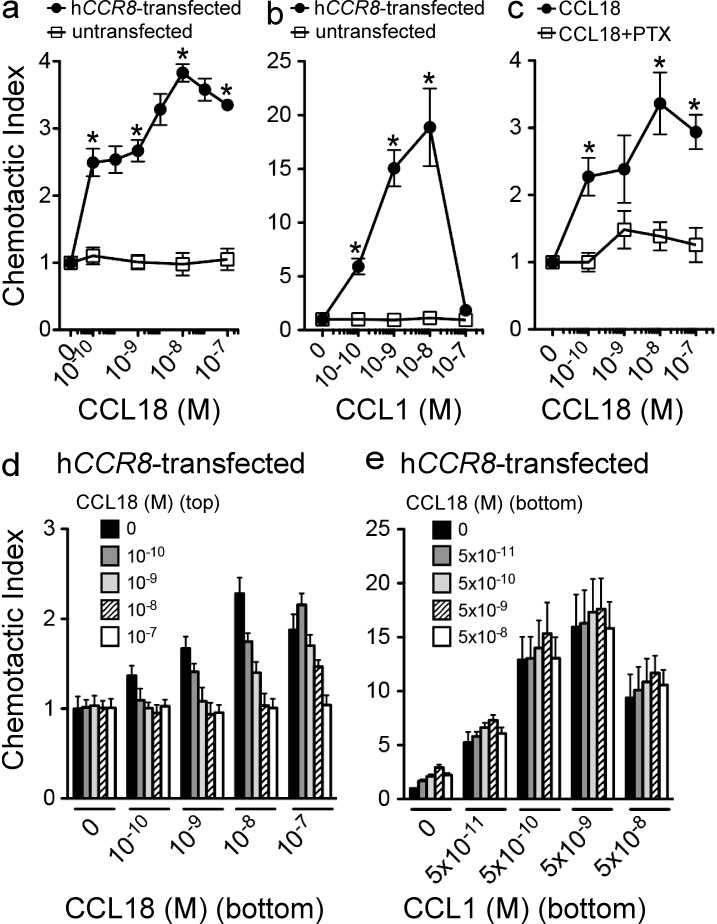
We assayed the ability of 4DE4 mouse pre–B cell lines transfected with human CCR8 to chemotax to CCL18 and observed peak migration at 10 nM (Fig. 1 a). Untransfected 4DE4 cells exhibited migration to CXCL12 (not depicted), but not to CCL18 (Fig. 1 a) or the known human CCR8 chemokine agonist CCL1 (also called I-309; Fig. 1 b). CCL1 also induced a peak migration of CCR8 transfectants at 10 nM but with a more robust response (Fig. 1 b). CCL18-induced chemotaxis in CCR8-transfected cells was inhibited by pertussis toxin (PTX), indicating that CCL18 induced the specific coupling of CCR8 to Gαi (Fig. 1 c). Checkerboard-type chemotaxis analysis revealed that CCL18 stimulated chemotaxis rather than chemokinesis (Fig. 1 d). CCL18 did not antagonize chemotaxis of CCR8-transfected cells to CCL1 when both chemokines were present in the bottom chamber in transwell chemotaxis assays (Fig. 1 e).
Figure 1.
CCR8 is sufficient for CCL18-induced migration. (a and b) Dose–response chemotaxis of hCCR8-transfected and untransfected 4DE4 cells to CCL18 (a) and hCCL1 (b). (c) Chemotaxis of PTX treated hCCR8-transfected cells to CCL18 (*, P < 0.05 by unpaired two-tailed t test in a–c). (d) Chemotaxis of hCCR8-transfected 4DE4 cells in a checkerboard-type transwell chemotaxis assay with varying concentrations of CCL18 in the bottom and top chamber. (e) Chemotaxis of hCCR8-transfected 4DE4 cells to varying concentrations of CCL1 mixed with varying concentrations of CCL18 in the bottom chamber (P = NS for CCL18 by two-way ANOVA). Data in all panels are representative of at least three independent experiments (mean ± SEM).
CCL18 induced calcium flux in CCR8-transfected cells
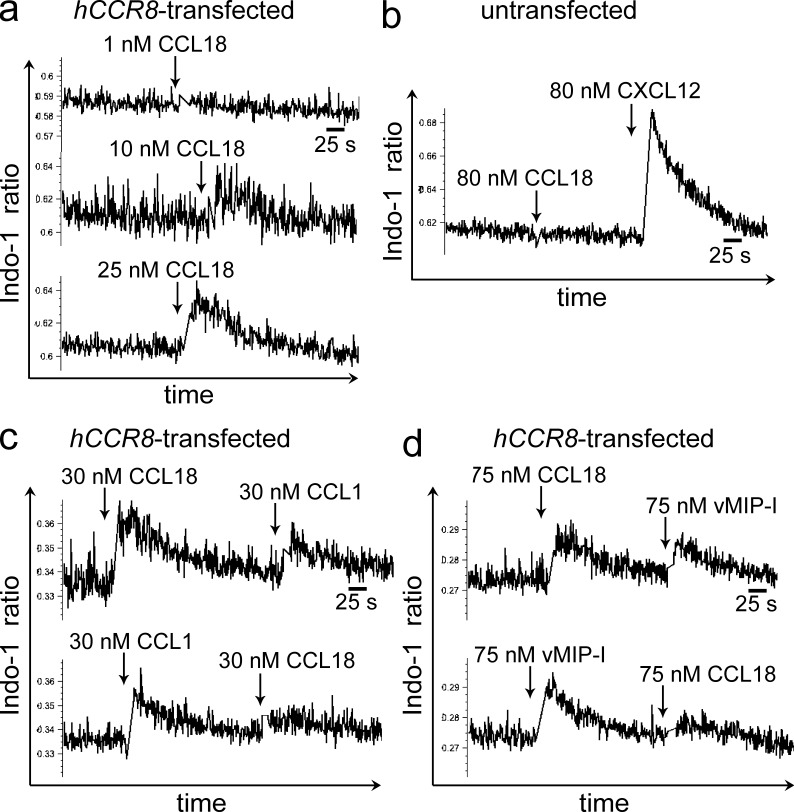
CCR8 transfectants showed a rapid and robust dose-dependent increase in intracellular calcium upon stimulation with CCL18 (Fig. 2 a). In control experiments, CCL18 did not induce a calcium flux in untransfected 4DE4 cells, although they responded to CXCL12 with a robust calcium flux (Fig. 2 b). Heterologous desensitization of ligand-induced calcium flux is routinely used to assess activity at a shared receptor. Stimulation of CCR8 transfectants with CCL18 inhibited, albeit incompletely, subsequent signaling to CCL1, reflecting CCL18-induced partial desensitization of the CCR8 receptor (Fig. 2 c). Stimulation of CCR8 transfectants with CCL1 completely inhibited subsequent signaling to CCL18 (Fig. 2 c). We then assayed the ability of CCL18 to desensitize another well characterized agonist of the human CCR8 receptor, HHV-8 (human herpes virus 8)–encoded chemokine viral MIP (vMIP) 1 (Dairaghi et al., 1999). vMIP-1–induced calcium signaling was partially desensitized by CCL18, and vMIP-I completely desensitized CCL18-induced signaling (Fig. 2 d). We also assayed the L1.2 murine pre–B cell line stably transfected with human CCR8 for chemotaxis and calcium flux to CCL18 but found that they responded weakly to CCL18 despite responding to CCL1. We do not have a clear explanation for the discrepancy in the magnitude of CCL18 responsiveness between L1.2 and 4DE4 CCR8 transfectants but consequently pursued all our functional studies on 4DE4 CCR8 transfectants.
Figure 2.
CCL18 induces calcium flux in CCR8-transfected cells. (a) Dose–response calcium flux of hCCR8-transfected cells to CCL18. (b) Calcium flux of untransfected 4DE4 cells to CCL18 and CXCL12. (c and d) Heterologous cross-desensitization of hCCR8-transfected cells to CCL18 and CCL1 (c), and to CCL18 and vMIP-I (d). a–d are representative of three to eight independent experiments.
CCL18 promoted internalization of CCR8
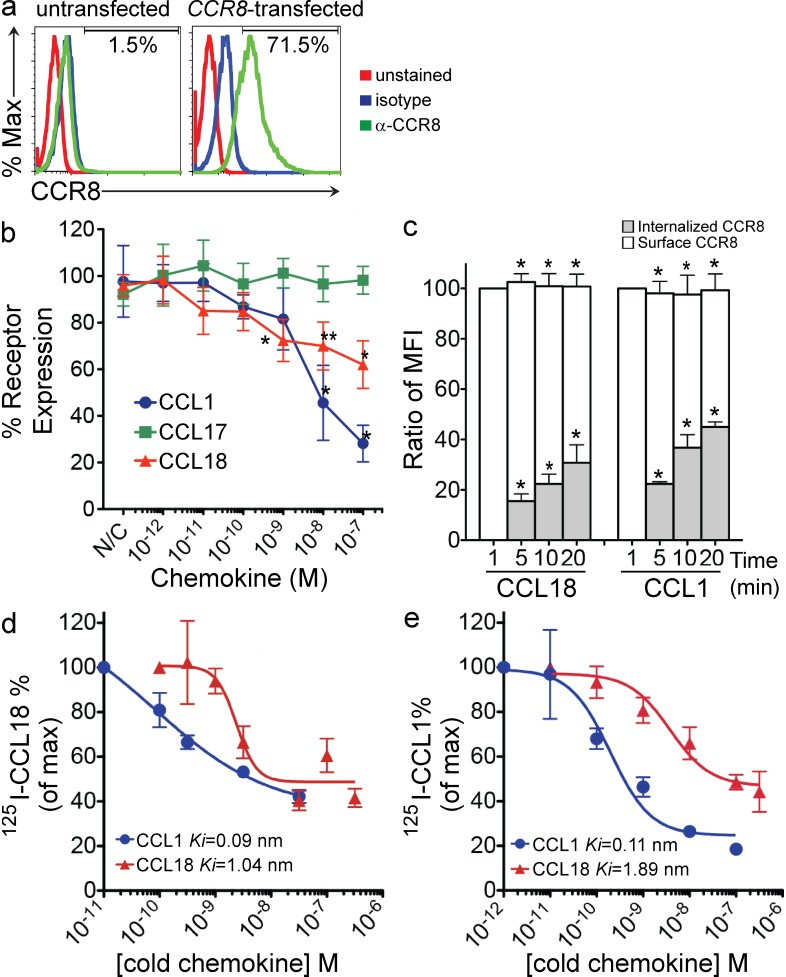
Chemokine receptors typically undergo internalization upon chemokine agonist binding. Robust surface expression of CCR8 on CCR8-transfected cells was detected at baseline but not on untransfected 4DE4 cells (Fig. 3 a). Surface CCR8 expression on CCR8-transfected cells was reduced after treatment with increasing concentrations of CCL18 or CCL1 but not CCL17 (a CCR4 but not CCR8 agonist) for 20 min at 37°C (Fig. 3 b). CCL18 and CCL1 binding to CCR8-transfected cells at 4°C did not affect mAb binding to CCR8. To demonstrate that CCL18 induced CCR8 internalization, we followed the kinetics of surface, total, and internalized (total − surface) CCR8 levels in CCR8-transfected cells after treatment with CCL18 or CCL1 (10−8 M). After treatment with CCL18 or CCL1, surface CCR8 levels decreased contemporaneously with increased internalized CCR8 levels, whereas total CCR8 levels were not affected by either treatment (Fig. 3 c).
Figure 3.
CCL18 induces internalization of CCR8 and inhibits 125I-CCL18 and 125I-hCCL1 binding to CCR8. (a) Representative FACS plots of CCR8 surface expression on hCCR8-transfected and untransfected cells. (b) Dose-dependent internalization of surface CCR8 in hCCR8-transfected cells stimulated with CCL18 and hCCL1 but not CCL17 (*, P < 0.05; **, P = 0.06, for CCL18 vs. CCL17 and CCL1 vs. CCL17 by unpaired two-tailed t test). (c) Kinetics of internalized versus surface CCR8 expression in hCCR8-transfected cells stimulated with CCL18 (10−8 M) or hCCL1 (10−8 M) for 1, 5, 10, or 20 min at 37°C (*, P < 0.05 for internalized or surface CCR8 treated with CCL18 or CCL1 at 37°C compared with 4°C control by unpaired two-tailed Student’s t test). Data are expressed as mean ± SEM, representative of at least five experiments in a–c. (d) Inhibition of 0.2 nM 125I-CCL18 binding to hCCR8-transfected cells with increasing concentrations of unlabeled CCL18 and hCCL1. Representative of two (cold hCCL1) and five (cold CCL18) experiments (P = 0.01 for CCL18 versus CCL3 and P ≤ 0.0001 for CCL1 versus CCL3 by two-way ANOVA). (e) Inhibition of 0.1 nM 125I-hCCL1 binding to hCCR8-transfected cells with unlabeled CCL18 and hCCL1. Representatives of three (cold hCCL1) and five (cold CCL18) experiments are shown; data are shown as mean ± SEM in d and e.
CCL18 binds to CCR8
Competitive ligand binding experiments with 125I-labeled CCL18 and 125I-labeled hCCL1 enabled us to determine specific high affinity binding of CCL18 to CCR8. In direct competition binding experiments, CCL18 competed for 125I-CCL18 binding to CCR8-transfected cells with a Ki of 1.04 nM, whereas human CCL1 competed for 125I-CCL18 binding with a Ki of 0.09 nM (Fig. 3 d). In heterologous cross-competition binding experiments, CCL18 competed for 125I-CCL1 binding to CCR8 with a Ki of 1.89 nM, and CCL1 competed for 125I-CCL1 binding with a Ki of 0.114 nM, which is consistent with published CCL1 Ki values (Fig. 3 e). hCCL3, the human chemokine with the greatest homology to CCL18, did not compete for either 125I-CCL18 or 125I-hCCL1 binding to CCR8 (not depicted). The Ki determined for CCL18 is comparable to the published Kd of 1.9 nM measured on lymphocytes in saturation binding experiments and also to the Ki values determined for vMIP-I (Hieshima et al., 1997; Dairaghi et al., 1999).
CCL18 induced migration and calcium flux of highly polarized Th2 cells
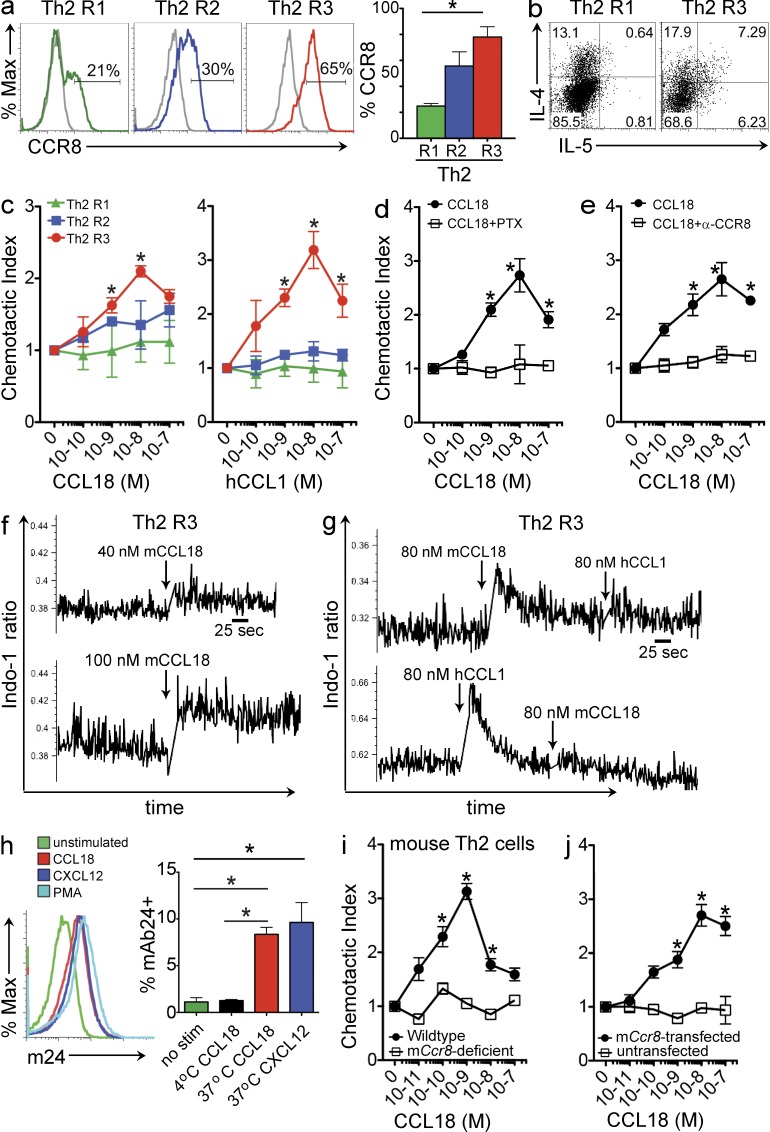
CCR8 induction in murine Th2 cells at levels high enough to detect agonist function occurs transiently after TCR activation of Th2 cells generated by multiple rounds of polarization (D’Ambrosio et al., 1998; Islam et al., 2011). Human Th2 cells also exhibited increased CCR8 expression with successive rounds of Th2 polarization (Fig. 4 a) with a concomitant increase in IL-4 and IL-5 expression (Fig. 4 b). Highly polarized human Th2 cells generated by three rounds of polarization with subsequent TCR activation (Th2 R3) migrated in response to CCL18 and CCL1, whereas less polarized Th2 cells that expressed lower levels of CCR8 did not (Fig. 4 c). Peak migration for both CCL18 and CCL1 was at 10 nM, similar to what was observed on CCR8-transfected cells. CCL18-induced chemotaxis of Th2 R3 cells was inhibited by PTX (Fig. 4 d). A blocking mAb to human CCR8 specifically inhibited migration of Th2 R3 cells to CCL18 (Fig. 4 e). Th2 R3 cells also showed robust calcium flux to CCL18 in a dose-dependent manner (Fig. 4 f). Stimulation of highly polarized Th2 cells with CCL18 inhibited subsequent signaling to CCL1, and CCL18-induced calcium signaling in Th2 cells was desensitized by CCL1 signaling (Fig. 4 g).
Figure 4.
CCL18 induces calcium flux and CCR8-dependent migration of activated highly polarized Th2 cells. (a) Representative histograms of CCR8 expression on human Th2 cells that have undergone one (Th2 R1), two (Th2 R2), or three (Th2 R3) rounds of Th2 polarization. (Left) CCR8 mAb staining and isotype control are depicted with colored and gray lines, respectively. (Right) Quantitation of CCR8 surface expression on human Th2 cells. (*, P < 0.05 for CCR8 on Th2 R1 vs. Th2 R3 by unpaired two-tailed Student’s t test). (b) Representative cytokine profiles of Th2 cells. (c) Dose–response chemotaxis of activated human Th2 cells to CCL18 (left) or hCCL1 (right; *, P < 0.05 for Th2 R1 vs. Th2 R3 by unpaired two-tailed Student’s t test). (d) Comparison of chemotaxis of 100 ng/ml PTX–treated activated human Th2 R3 cells and untreated controls. (e) Activated human Th2 R3 cells were treated with 1 µg/ml CCR8 mAb and compared with untreated activated Th2 R3 cells in chemotaxis assay to CCL18. a–e are representative of at least three independent experiments (mean ± SEM). (f) Dose-dependent calcium flux response of human Th2 R3 cells to CCL18 stimulation. (g) Heterologous cross-desensitization of the calcium flux response of human Th2 R3 cells to CCL18 and hCCL1 stimulation. f and g are representative of three to five separate experiments. (h) Representative histograms of surface expression of the LFA-1 activation epitope m24 on activated human Th2 R3 cells treated with CCL18, CXCL12, PMA, or not treated (left). Quantitation of m24 expression on activated Th2 R3 cells after CCL18 or CXCL12 stimulation (right). (i) Chemotaxis of activated wild-type and Ccr8-deficient mouse Th2 cells to CCL18. (j) Chemotaxis of mCcr8-transfected and untransfected Baf/3 cells to CCL18 stimulation. h–j are representative of at least three independent experiments (mean ± SEM; *, P < 0.05 by unpaired two-tailed Student’s t test for d, e, and h–j).
CCL18 induced LFA-1 activation
Chemokine signaling can induce “inside-out” activation of integrins. In T cells, chemokine activation of LFA-1 results in a conformational change that can be detected by mAb24, which recognizes the appearance of a new LFA-1 epitope m24 (Schürpf and Springer, 2011). CCL18 induced expression of the m24 epitope, detected by mAb24, which was comparable to that induced by CXCL12 or PMA on activated highly polarized Th2 cells at 37°C. This increase in mAb24 binding was not detected at 4°C or in the absence of stimulation (Fig. 4 h).
CCL18 activates mouse CCR8
Activated highly differentiated wild-type mouse Th2 cells migrated to CCL18 in a dose-dependent bell-shaped migration curve with peak migration at 1 nM (Fig. 4 i). Migration was abrogated in Ccr8-deficient Th2 cells in which migration to the positive control CXCL12 remained intact and comparable to wild-type Th2 cells (not depicted). Moreover, mCcr8-transfected but not untransfected Baf/3 murine B cells migrated in response to CCL18 with peak migration at 10 nM (Fig. 4 j). The peak migration induced by CCL18 was comparable to the peak migration induced in these cells by mCCL1 and the recently identified second chemokine ligand for murine CCR8, mouse CCL8 (mCCL8 or mouse monocyte chemoattractant protein [MCP] 2; Islam et al., 2011).
Human CCL18 and murine CCL8 are functional analogues
Although mCCL8 was originally given the orthologous designation to human CCL8 (hCCL8), we recently established they are not functional orthologues. hCCL8 is not a CCR8 ligand but is a CCR2 ligand, whereas mCCL8 is a CCR8 ligand but is not a CCR2 ligand (Islam et al., 2011). Synteny mapping of mCCL8 and hCCL8, which are located in the mouse and human MCP chemokine clusters, respectively, has also revealed they are not orthologues (Zlotnik and Yoshie, 2012). In fact, mCCL8 lacks a human orthologue, and both hCCL8 and CCL18 lack rodent orthologues (Homologene, MGI).
This has led us to hypothesize that mCCL8 and CCL18 may be functional analogues. Consistent with this hypothesis, CCL18 and mCCL8 have been reported to be highly induced in AAMs (Kodelja et al., 1998; Schutyser et al., 2005; Thomas et al., 2012; Egawa et al., 2013). Additionally, mCCL8 was the most abundant chemokine induced in the lung in a bleomycin model of pulmonary fibrosis by microarray analysis (Liu et al., 2011), and CCL18 is the most highly expressed chemokine in idiopathic pulmonary fibrosis (Prasse et al., 2009).
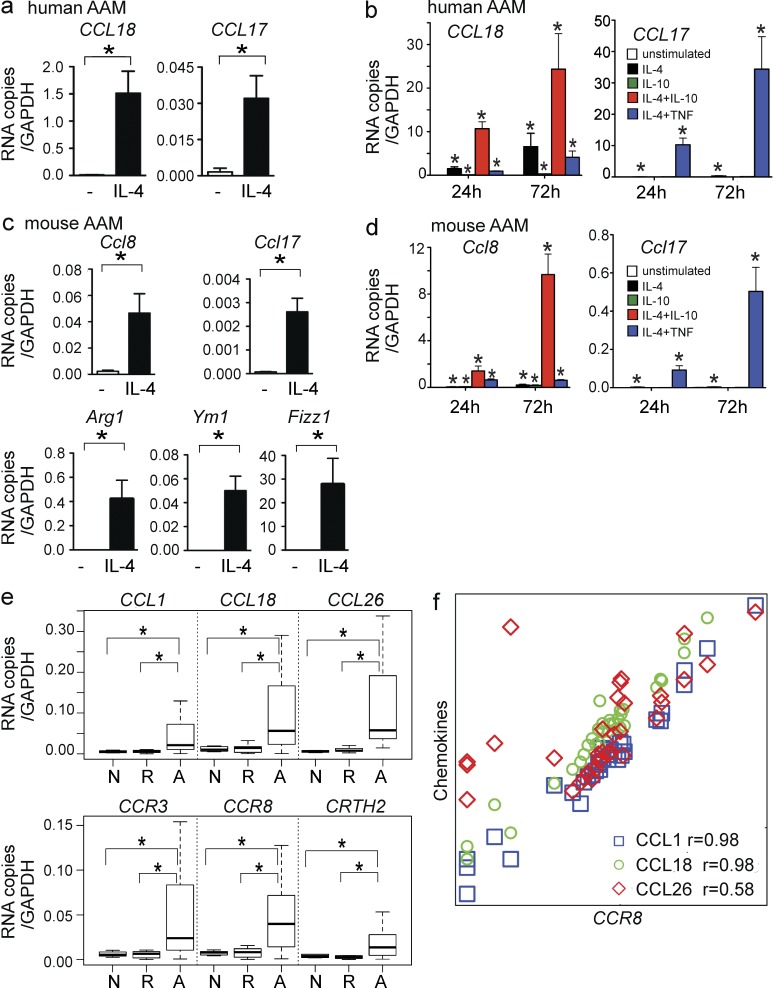
We thus determined if cytokines known to regulate CCL18 induction might also regulate mCCL8 induction in AAMs in an analogous fashion. As expected, human CCL18 and another human Th2-associated chemokine CCL17 were both induced in human AAMs in response to IL-4, but CCL18 was induced to much greater levels (Fig. 5 a). Consistent with published studies (Kodelja et al., 1998), we found that IL-10 weakly induced CCL18 in macrophages. However, as noted by others, a combination of IL-4 and IL-10 synergistically induced CCL18 (Pechkovsky et al., 2010), which is in distinct contrast to IL-4 and TNF (Fig. 5 b). In contrast, IL-10 treatment did not induce CCL17 and instead inhibited IL-4–induced CCL17 expression in AAMs, whereas TNF and IL-4 synergistically induced CCL17 (Fig. 5 b).
Figure 5.
CCL18 expression in AAM and in human EoE. (a) Induction of CCL18 and CCL17 RNA in human AAM by IL-4 at 24 h. (b) Comparison of 24- and 72-h CCL18 and CCL17 induction in human AAM after IL-4, IL-10, and TNF treatment. (c) 24-h induction of Ccl8 and Ccl17 in mouse AAM by IL-4. (d) Comparison of 24 and 72 h Ccl8 and Ccl17 induction in mouse AAM after treatment with IL-4, IL-10, and TNF. a–d are representative of at least three independent experiments (mean ± SEM; *, P < 0.05 by unpaired two-tailed Student’s t test for a–d). (e) Expression of CCL18 and CCR8 and other Th2-associated chemokines (top) and receptors (bottom) in esophageal biopsy tissue recovered from individuals with active EoE (A), individuals with EoE in remission (R), and normal controls (N). Data are presented in box-and-whiskers plots with bars indicating fifth and 95th percentiles (*, P < 0.05 by Mann-Whitney test). (f) Relationship of CCR8 mRNA expression to the expression of CCL1 (blue square), CCL18 (green circle), and CCL26 (red diamond) in the human esophageal samples from all groups studied (r represents Spearman correlation).
In mouse AAMs, we found that Ccl8 and Ccl17 were also induced by IL-4. AAM differentiation was confirmed by the specific induction of the mouse AAM markers Arg1, Ym1, and Fizz1 (Fig. 5 c). In a striking parallel to CCL18 induction in human AAMs, IL-4 and IL-10 both independently induced Ccl8, and importantly, IL-4 and IL-10 in combination, but not IL-4 and TNF, synergistically induced Ccl8 expression (Fig. 5 d). In contrast, regulation of Ccl17 induction in mouse AAMs mirrored that of human CCL17 in human AAMs and was distinct from that of CCL18 and Ccl8. Thus, IL-10, an inhibitory cytokine, paradoxically amplified IL-4 induction of both Ccl8 and CCL18. These data provide evidence that CCL18 and Ccl8 are regulated similarly in AAMs, which lends further support to the hypothesis that mCCL8 and CCL18 are functional analogues.
CCL18 and CCR8 are induced in human eosinophilic inflammation
Previously, we found that CCR8 and mCCL8 were essential for the induction of chronic eosinophilic skin inflammation in vivo (Islam et al., 2011). To investigate whether CCL18 and its receptor CCR8 are involved in chronic human eosinophilic inflammation, we examined esophageal biopsy tissue of patients with eosinophilic esophagitis (EoE; Table S1) for expression of Th2-associated chemokines (CCL18, CCL1, and CCL26) and receptors (CCR8, CCR3, and CRTH2). CCL18 and CCR8 mRNA were up-regulated in subjects with active EoE compared with subjects whose disease was in remission and to normal controls (Fig. 5 e). CCL1, CCL26, CCR3, and CRTH2 mRNA were also up-regulated in active EoE. Moreover, CCL18 and CCL1 mRNA levels in esophageal tissue correlated strongly with CCR8 mRNA levels (Fig. 5 f). In contrast, the eosinophil chemoattractant CCL26, which is known to be abundantly expressed in EoE (Blanchard et al., 2007) and is a CCR3 ligand, was not as highly correlated with CCR8 (Fig. 5 f). Thus, CCL18 and its receptor CCR8 are coexpressed in human diseased tissue during active eosinophilic inflammation, consistent with our findings in the mouse.
Phylogeny of CCL18
CCL18 has orthologues only in primates (Homologene). Sequence analysis revealed that CCL18 shares the most amino acid identity (65%) with hCCL3 (MIP-1α), which lacks activity on CCR8. It is hypothesized that the CCL18 gene, which is located within the MIP chemokine gene cluster, arose through duplication and fusion of two CCL3-like genes with deletion and selective use of exons after the speciation of primates and rodents (Schutyser et al., 2005). Consistent with this hypothesis, vMIP-1, the potent virally encoded CCR8 agonist also believed to have arisen from the MIP gene cluster, is more homologous to CCL18 (46% identity) than it is to the other CCR8 agonist hCCL1 (25% identity). Alignment of the mature amino acid sequences of the CCR8 agonists CCL18, CCL1, and vMIP-1 revealed that only CCL18 and vMIP-I share a continuous stretch of 20-aa identity. The human and mouse MIP and MCP chemokines are closely linked gene clusters on human chromosome 17 and mouse chromosome 11. Inflammatory cluster chemokines often tend to evolve rapidly, and tandem gene duplication and diversification are believed to have occurred within the MCP and MIP clusters independently after the speciation of rodents and primates (Zlotnik and Yoshie, 2012). This may explain why these two chemokine clusters are not orthologous between humans and mice and why mouse CCL8 and human CCL18 lack identified orthologues by phylogenetic analysis and synteny mapping. Instead, functional analogues need to be identified based on similar biological function, such as similar regulation and receptor usage, as we have demonstrated for human CCL18 and mouse CCL8.
Our study shows that CCR8 has two endogenous chemokine ligands. Ligands for the same chemokine receptor often have distinct yet cooperative and complementary functions in vivo (Zlotnik and Yoshie, 2012). In the CCR8 system, CCL1 appears to be the more potent agonist. However, CCL18 appears to be more highly expressed in inflammation. CCL18 is detected at two log higher levels than CCL1 in lesional atopic skin, which is analogous to the two log higher expression of mCCL8 compared with mCCL1 in inflamed atopic mouse skin (Pivarcsi et al., 2004; Gombert et al., 2005; Islam et al., 2011). While CCL18 is expressed in dermal dendritic cells and keratinocytes, CCL1 is expressed by epidermal Langerhans cells and skin endothelial cells (Gombert et al., 2005). CCL1 may therefore promote entry through the cutaneous vasculature while CCL18 may promote retention of CCR8-expressing cells in inflamed tissue. Thus, we predict that CCL1 and CCL18 likely have nonredundant roles in vivo in part dictated by their pattern of expression and their activity on CCR8-expressing cells.
In certain human cancers (e.g., breast and ovarian), tumor-associated macrophages, an AAM-like population which undergoes additional differentiation by T reg cell–derived IL-10, abundantly produce CCL18 (Bonecchi et al., 2011). It is notable that the induction of both CCL18 and mCCL8 is markedly augmented by IL-10. In human breast cancer, CCL18 expression in blood and cancer stroma was associated with reduced patient survival (Chen et al., 2011). CCL18 promoted tumor cell invasiveness and metastasis, which was shown to be mediated by PITPNM3 (phosphatidylinositol transfer protein 3; Chen et al., 2011). PITPNM3 is a six-transmembrane receptor distinct from seven-transmembrane chemokine receptors (Bonecchi et al., 2011). We did not detect PITPNM3 RNA or protein expression in our CCR8-transfected 4DE4 B cell like cell lines or in Th2 R3 cells, although we did detect PITPNM3 RNA and protein in the MDA-MB-231 epithelial breast cancer cell line (unpublished data). Therefore, the CCL18-induced CCR8 activation that we described in transfected cells and primary T cells was independent of PITPNM3.
Identifying CCR8 as a receptor for CCL18 offers new insights into the role of this important human chemokine identified in many chronic inflammatory diseases. In a recent transcription profiling study, CCR8 was found to be the chemokine receptor most highly expressed in induced T reg cells (Wei et al., 2010). Recruitment of CCR8-expressing T reg cells into an IL-10–rich tumor milieu may possibly explain the association of CCL18 with increased metastasis and poor prognosis. The ability of CCL18 to recruit CCR8-expressing Th2 cells may explain the association of this chemokine with many allergic diseases. Our data support this hypothesis by showing the coexpression of CCL18 and CCR8 in human diseased esophageal tissue in individuals with active EoE. This may also be of particular importance in atopic dermatitis where CCL18 is abundantly expressed and likely facilitates the recruitment of CCR8-expressing skin-homing Th2 cells, as we have seen for mCCL8 (Islam et al., 2011). Further, the attraction of Th2 cells by CCL18-secreting AAM may provide a positive-feedback loop for the maintenance of the AAM phenotype in chronic inflammation. These are a few examples in which understanding the receptor for CCL18 will allow a greater appreciation of its role in human disease. Identifying CCR8 as the CCL18 receptor and mCCL8 as its rodent functional analogue will also enable the study of the CCR8 receptor–ligand system in murine disease models to better understand CCL18-mediated human disease pathology.
MATERIALS AND METHODS
Human studies.
Peripheral blood was obtained from healthy volunteers, and fresh buffy coats were obtained from the Massachusetts General Hospital blood bank. For endoscopy studies, patients were referred for upper gastrointestinal symptoms suggestive of reflux or EoE (Table S1). Normal controls had a normal appearing esophagus on endoscopy and had no histological findings of inflammation on esophageal biopsy. In accordance with the consensus guidelines, active EoE was defined by the presence of >15 eosinophils per 40× high power field (hpf) in at least one esophageal biopsy (of a total six obtained from the proximal, middle, and distal esophagus) despite a minimum 6-wk course of high dose proton pump inhibitors (Liacouras et al., 2011). EoE patients in remission were on either elimination diet or on topical steroid treatment, and esophageal biopsy samples contained fewer than five eosinophils/hpf in all six biopsies. All human subject protocols were approved by the Partners Institutional Review Board. Informed consent was obtained from all study participants.
Mice.
C57BL/6 mice were purchased from the National Cancer Institute (Bethesda, MD). CCR8−/− mice (from S. Lira, Mount Sinai School of Medicine, New York) in the C57BL/6 background were housed under specific pathogen-free conditions. All protocols were approved by the Massachusetts General Hospital Subcommittee on Research Animal Care.
Human and mouse CCR8 receptor–transfected cells.
Limiting dilution clones were generated from a CCL18-responsive CCR8-transfected 4DE4 cell line (provided by J. Pease, Imperial College, London, UK) grown in low concentrations of G418 (0.1–0.4 mg/ml; Tiffany et al., 1997). We found that these clones responded better to CCL18 when grown in low G418 concentrations (and, in fact, seemed to lose activity at G418 concentrations ≥1 mg/ml) and were therefore maintained in 0.1 mg/ml G418. 4DE4 cells were provided by P. Murphy (National Institutes of Health, Bethesda, MD). hCCR8-L1.2 transfected cell line was provided by M. Locati and A. Mantovani (University of Milan, Milan, Italy). Cells were treated with 5 mM butyric acid overnight in some calcium flux, binding, and chemotaxis assays to increase surface expression of CCR8. Mouse Ccr8-transfected Baf/3 cells were generated as previously described (Islam et al., 2011).
Generation of human monocyte-derived DCs.
DCs were generated from PBMCs isolated from fresh human buffy coats by Histopaque-1077 density gradient (Sigma-Aldrich). PBMCs were resuspended in C10 media (RPMI complete medium with 10% fetal calf serum) and incubated for 2–3 h at 37°C and washed gently with PBS. The remaining adherent cells were cultured in 10 ml C10 media supplemented with 50 ng/ml of GM-CSF (BioLegend) and 50 ng/ml IL-4 (PeproTech) at 37°C. Additional GM-CSF and IL-4 was added on days 2, 4, and 5. On day 6, DCs were rested for 24 h after change of media, and then incubated with C10 media containing 100 ng/ml of LPS (Sigma-Aldrich) for 24–48 h.
Generation of Th2 cells and induction of highly polarized Th2 cells.
CD4+ T cells were isolated from Histopaque-1077 density gradient PBMCs by two negative selection columns: (1) CD4+ T cell Biotin Antibody Cocktail (Miltenyi Biotec), followed by incubation with anti-biotin microbeads (Miltenyi Biotec) to isolate CD4+ T cells; and (2) CD45RO PE (BD) staining, followed by incubation with anti-PE microbeads to isolate naive (CD45RO−) CD4+ T cells. DCs and T cells were cultured in C10 media with 50 ng/ml IL-4 and 10 µg/ml anti–IFN-γ (BioLegend) for 5 d at 37°C. At day 5, new C10 supplemented with 250 ng/ml IL-2 (Roche) was added. For repolarization of Th2 cells at day 7 to generate Th2 R2 cells, Th2 R1 cells were repolarized with irradiated human PBMCs, 1 µg/ml anti-CD3 (BioLegend), 1 µg/ml anti-CD28 (BioLegend), 50 ng/ml IL-4, and 10 µg/ml anti–IFN-γ (BioLegend). A similar procedure was used to generate Th2 R3 cells from Th2 R2 cells. Prior to assays, Th2 cells were activated with 2 µg/ml plate-bound mAb to CD3 and 1 µg/ml soluble mAb to CD28 for 24 h. Cells were then washed, replated, and rested for 4–6 h in low–IL-2 medium for migration or IL-2–free medium for calcium flux. Highly polarized and activated mouse Th2 cells were generated for chemotaxis assays in a similar manner as previously described (Islam et al., 2011).
Generation and activation of human and mouse macrophages.
Human monocytes were isolated from peripheral blood by negative selection with the monocyte isolation kit II (Miltenyi Biotec) and differentiated into macrophages by culturing in the presence of 50 ng/ml m-CSF (PeproTech) in C10 media for 7 d in 24- or 12-well plates. Human AAMs were obtained by culturing these macrophages in fresh C10 supplemented with 25 ng/ml IL-4 (PeproTech), 10 ng/ml IL-10 (Miltenyi Biotec), and 20 ng/ml TNF (Miltenyi Biotec) for an additional 24–72 h. Bone marrow cells isolated from femurs and tibias were cultured in non–tissue culture–treated 100-mm bacterial culture Petri dishes (Thermo Fisher Scientific) in C10 media supplemented with 10 ng/ml murine M-CSF (PeproTech) and fed with fresh media on days 4 and 8. Cells were harvested at day 8 and replated in 24- or 12-well plates in M-CSF–containing media and stimulated with 20 ng/ml IL-4, 10 ng/ml IL-10, and 20 ng/ml TNF and harvested after 24 or 72 h.
Quantitation of integrin LFA-1 activation in highly polarized Th2 cells.
Prior to assays, human Th2 cells were activated with 2 µg/ml plate-bound mAb to CD3 and 1 µg/ml soluble mAb to CD28 for 24 h. Cells were then washed, replated, and rested for 4–5 h in low–IL-2 medium before assays. For LFA-1 activation assays, cells were washed and stained in 1% human serum/RPMI with 10 µg/ml mAb24 (Hycult Biotech) in the presence of 200 nM CCL18, 200 nM CXCL12, or 50 ng/ml PMA or in the absence of stimulation for 30 min at 37°C, washed, and then stained with 1 µg/ml secondary goat anti–mouse PE for 20 min, washed, fixed, and acquired on a FACSCalibur. As a control, CD3/CD28-activated Th2 cells were stained with 10 µg/ml mAb24 in the presence of 200 nM CCL18 at 4°C for 30 min, followed by secondary antibody staining. As an additional control, Th2 cells that were not activated with CD3/CD28 were stimulated with 200 nM CCL18 and stained with 10 µg/ml mAb24 antibody at 37°C for 30 min followed by secondary antibody staining. CCL18 did not induce m24 epitope expression on these resting Th2 cells.
Flow cytometry and intracytoplasmic cytokine staining.
For CCR8 staining, cells were stained with the 433H mAb to human CCR8, which was purified from the hybridoma (American Type Culture Collection), followed by PE-conjugated goat anti–mouse IgG(H+L) (SouthernBiotech; Islam et al., 2011). Intracytoplasmic staining was performed using a standard protocol (Islam et al., 2011). Briefly, cells were stained with PE-conjugated mAb to IL-4 and allophycocyanin-conjugated mAb to IL-5 (BD), or appropriate fluorochrome-conjugated IgG1κ isotype-control (BD). Cells were acquired on a FACSCalibur and analyzed with FlowJo (9.5.2).
Chemotaxis assays.
CCR8-transfected cells, untransfected cells, and human Th2 cells (2.5 × 104-105 per well) in RPMI containing 0.5% BSA were placed on the top of a 96-well ChemoTx chemotaxis apparatus with 5-µm pores (NeuroProbe). CCL18, human CCL1, and CXCL12 (R&D Systems) were added to the bottom well. For “checkerboard-type” chemotaxis assays, CCL18 was added to the top well in varying concentrations. The apparatus was incubated at 37°C and 5% CO2 for 3–4 h, and the cells migrating at each concentration of chemoattractant were counted using an inverted microscope at the end of the experiment or after fixation with 10% buffered formalin. The number of cells migrating at each concentration of chemokine was normalized to the number of cells migrating in the presence of medium alone (chemokinesis) to calculate the chemotactic index for each leukocyte type. PTX (Sigma-Aldrich) or the 433H neutralizing mAb to human CCR8 was used in some assays (Islam et al., 2011).
Calcium flux assays.
CCL18, human CCL1 and CXCL12, or vMIP-I (R&D Systems) were used in calcium flux assays. Calcium flux assays were performed on a UV laser–equipped 13-color LSR II flow cytometer (BD) as previously described (Islam et al., 2011). Indo-1 AM (Molecular Probes) fluorescence was analyzed with the UV A detector at 530/30 and UV B detector at 440/40 for free and bound probe with Indo-1 (blue) and Indo-1 (violet), respectively. Data were analyzed with standardized settings using the calcium flux kinetic software analysis platform of FlowJo (8.8).
Receptor binding assays.
Binding assays were performed with 100,000 cells in a total volume of 100 µl binding buffer in 0.65 µm 96-well Durapore glass fiber filter plates (EMD Millipore) that were first wetted with binding buffer before assays. Binding buffer consisted of 50 mM Hepes, 1 mM CaCl2, 5 mM MgCl2, 0.5% BSA, and 125 mM NaCl, adjusted to a pH of 7.22 for competition binding experiments with 125I-hCCL1, and adjusted to a pH of 7.4 with the addition of 0.05% sodium azide for competition binding experiments with 125I-hCCL18. Cells were grown to log phase to a concentration of 1.2–1.7 million/ml at the time of the assay. Binding assays were performed for 1.5 h with continuous shaking at 140 rpm, and were performed at room temperature and at 37°c for experiments with 125I-hCCL1 and 125I-CCL18, respectively. At the end of the assay, plates were washed four times with ice-cold binding buffer. Radioactivity was counted after addition of scintillation fluid in a TopCount NXT counter (Perkin-Elmer). 125I-hCCL1 and 125I-CCL18 were purchased from Perkin-Elmer. Because CCL18 had been reported to self-aggregate at high concentrations (Hieshima et al., 1997), we adapted published binding protocols used for other chemokines also reported to self-aggregate, such as CCL5 and CCL3, to perform our direct competition binding experiments with 125I-CCL18. Binding data were analyzed with Prism 5 (GraphPad Software).
Internalization assays.
Flow cytometry using the 433H CCR8 mAb was used to evaluate CCR8 receptor internalization. 5 × 106 cells CCR8-transfected 4DE4 cells were resuspended in 100 µl of ice-cold culture medium in duplicate, followed by addition of varying concentrations of CCL18, hCCL1, or hCCL17 (R&D Systems). One sample was incubated for 20 min at 37°C, whereas the other was kept at 4°C before washing, staining, and receptor expression analysis by flow cytometry. Receptor expression was calculated using the following equation: 100 × [(mean fluorescence of chemokine treated cells at 37°C − mean fluorescence of negative control cells)/(mean fluorescence of chemokine treated cells at 4°C − mean fluorescence of negative control cells)], where negative control cells were buffer-treated cells. To determine the kinetics of surface and total CCR8 expression, 5 × 106 cells were resuspended in 100 µl of ice-cold culture medium in duplicate, followed by the addition of 10−8 M CCL18, 10−8 M CCL1, or media at 37 or 4°C). One set of samples was directly stained for CCR8 (surface CCR8), whereas the other was fixed and permeabilized and stained for CCR8 (total CCR8). The difference between total and surface CCR8 was defined as internalized CCR8. Change in expression of CCR8 is shown as the ratio of mean fluorescence (mean fluorescence of CCR8 + ligand/mean fluorescence of negative control). As controls, no significant internalization was seen when CCR8-transfected cells were stimulated with CCL18 and hCCL1 at 4°C or in cells incubated with media at 37°C.
RNA extraction and RT-PCR analysis for esophageal specimens and macrophages.
Total RNA was extracted from cryopreserved esophageal biopsy samples using mirVana Isolation Kit (P/N: 1560; Ambion) according to the manufacturer’s protocol. Power SYBR green qRT-PCR assay (Applied Biosystems) was used for mRNA quantification using StepOnePlus Real-Time PCR System (Applied Biosystems). The following primers were used: CCL26, 5′-CCAAGACCTGCTGCTTCCAA-3′ and 5′-GAATTCATAGCTTCGCACCCA-3′; CRTH2, 5′-AAAAGGCTCGGGAAGGTTAAATG-3′ and 5′-ACCGGGGAACCAAGAGAGAG-3′; CCR8, 5′-GGTCATCCTGGTCCTTGTGG-3′ and 5′-CAGGGCCAGGTTCAAGAGG-3′; CCR3, 5′-GCAAGCATCTGGACCTGGTC-3′ and 5′-GGTTCATGCAGCAGTGGGA-3′; CCL1, 5′-TGCAGATCATCACCACAGCC-3′ and 5′-GTCCACATCTTCCGGCCA-3′; CCL18, 5′-TGGCAGATTCCACAAAAGTTCA-3′ and 5′-GGATGACACCTGGCTTGGG-3′; and GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. Using the comparative Ct method, relative gene expression was calculated as 2−ΔΔCt where ΔCt = Ct (gene of interest) − Ct (normalizer = GAPDH) and the ΔΔCt = Ct (sample) − Ct (calibrator = mean ΔCt of normals). Harvested mouse and human macrophages were harvested and stored in RLT buffer and purified RNA was isolated using the RNeasy kit (QIAGEN). Real-time PCR was performed as above with the mouse and human primers described below. Mouse primers: Arg1, 5′-CAGAAGAATGGAAGAGTCAG-3′ and 5′-CAGATATGCAGGGAGTCACC-3′; Ym1, 5′-TCACAGGTCTGGCAATTCTTCTG-3′ and 5′-TTTGTCCTTAGGAGGGCTTCCTCG-3′; Fizz1, 5′-GGTCCCAGTGCATATGGATGAGACCATAGA-3′ and 5′-CACCTCTTCACTCGAGGGACAGTTGGCAGC-3′; Ccl8, 5′-TCTACGCAGTGCTTCTTTGCC-3′ and 5′-AAGGGGGATCTTCAGCTTTAGTA-3′; Ccl17, 5′-CCCATGAAGACCTTCACCTC-3′ and 5′-CATCCCTGGAACACTCCACT-3′; and Gapdh, 5′-GGCAAATTCAACGGCACAGT-3′ and 5′-AGATGGTGATGGGCTTCCC-3′. Human primers: CCL18, 5′-GGTGTCATCCTCCTAACCAAGAGA-3′ and 5′-GCTGATGTATTTCTGGACCCACTT-3′; and CCL17, 5′-CCATCGTTTTTGTAACTGTGCAG-3′ and 5′-TGCATTCTTCACTCTCTTGTTGTTG-3′.
Statistical analysis.
Statistical analysis was performed using Student’s unpaired two-tailed t test for means, except where indicated in Fig. 1 e (two-way ANOVA to assess the effect of multiple concentrations of CCL18 on CCL1-induced chemotaxis), Fig. 3 c (two-way ANOVA to compare the binding displacement of 125I-CCL18 by multiple concentrations of cold CCL18 and cold CCL1 to that induced by the non-CCR8 ligand CCL3), and Fig. 5 e (Mann-Whitney U test for nonparametric comparison of means among normal controls, patients with inactive EoE, and patients with active EoE). Spearman’s correlation coefficients were used for nonparametric testing of statistical dependence between pairs of predefined EoE-associated variables. P < 0.05 was considered significant.
Online supplemental material.
Table S1 shows a summary of study subjects who underwent endoscopic biopsy after being referred for suspected reflux or EoE. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20122842/DC1.
Supplementary Material
Acknowledgments
The authors thank P. Murphy, J. Pease, M. Locati, and A. Mantovani for cell lines and S. Lira for CCR8-deficient mice. We would also like to thank A. Katz and Q. Yuan for providing clinical samples, and M. Deruaz and G. Campanella for assistance with binding assays.
This work is funded by National Institutes of Health grant R37AI040618 and U19AI095261 to A.D. Luster.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AAM
- alternatively activated macrophage
- CCL
- CC chemokine ligand
- EoE
- eosinophilic esophagitis
- MCP
- monocyte chemoattractant protein
- MIP
- macrophage inflammatory protein
- PTX
- pertussis toxin
- vMIP
- viral MIP
References
- Adema G.J., Hartgers F., Verstraten R., de Vries E., Marland G., Menon S., Foster J., Xu Y., Nooyen P., McClanahan T., et al. 1997. A dendritic-cell-derived C-C chemokine that preferentially attracts naive T cells. Nature. 387:713–717 10.1038/42716 [DOI] [PubMed] [Google Scholar]
- Bellinghausen I., Reuter S., Martin H., Maxeiner J., Luxemburger U., Türeci O., Grabbe S., Taube C., Saloga J. 2012. Enhanced production of CCL18 by tolerogenic dendritic cells is associated with inhibition of allergic airway reactivity. J. Allergy Clin. Immunol. 130:1384–1393 10.1016/j.jaci.2012.08.039 [DOI] [PubMed] [Google Scholar]
- Blanchard C., Mingler M.K., Vicario M., Abonia J.P., Wu Y.Y., Lu T.X., Collins M.H., Putnam P.E., Wells S.I., Rothenberg M.E. 2007. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J. Allergy Clin. Immunol. 120:1292–1300 10.1016/j.jaci.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Bonecchi R., Locati M., Mantovani A. 2011. Chemokines and cancer: a fatal attraction. Cancer Cell. 19:434–435 10.1016/j.ccr.2011.03.017 [DOI] [PubMed] [Google Scholar]
- Chen J., Yao Y., Gong C., Yu F., Su S., Chen J., Liu B., Deng H., Wang F., Lin L., et al. 2011. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 19:541–555 10.1016/j.ccr.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenivesse C., Chang Y., Azzaoui I., Ait Yahia S., Morales O., Plé C., Foussat A., Tonnel A.B., Delhem N., Yssel H., et al. 2012. Pulmonary CCL18 recruits human regulatory T cells. J. Immunol. 189:128–137 10.4049/jimmunol.1003616 [DOI] [PubMed] [Google Scholar]
- D’Ambrosio D., Iellem A., Bonecchi R., Mazzeo D., Sozzani S., Mantovani A., Sinigaglia F. 1998. Selective up-regulation of chemokine receptors CCR4 and CCR8 upon activation of polarized human type 2 Th cells. J. Immunol. 161:5111–5115 [PubMed] [Google Scholar]
- Dairaghi D.J., Fan R.A., McMaster B.E., Hanley M.R., Schall T.J. 1999. HHV8-encoded vMIP-I selectively engages chemokine receptor CCR8. Agonist and antagonist profiles of viral chemokines. J. Biol. Chem. 274:21569–21574 10.1074/jbc.274.31.21569 [DOI] [PubMed] [Google Scholar]
- de Nadaï P., Charbonnier A.S., Chenivesse C., Sénéchal S., Fournier C., Gilet J., Vorng H., Chang Y., Gosset P., Wallaert B., et al. 2006. Involvement of CCL18 in allergic asthma. J. Immunol. 176:6286–6293 [DOI] [PubMed] [Google Scholar]
- Egawa M., Mukai K., Yoshikawa S., Iki M., Mukaida N., Kawano Y., Minegishi Y., Karasuyama H. 2013. Inflammatory monocytes recruited to allergic skin acquire an anti-inflammatory M2 phenotype via basophil-derived interleukin-4. Immunity. 38:570–580 10.1016/j.immuni.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Gombert M., Dieu-Nosjean M.C., Winterberg F., Bünemann E., Kubitza R.C., Da Cunha L., Haahtela A., Lehtimäki S., Müller A., Rieker J., et al. 2005. CCL1-CCR8 interactions: an axis mediating the recruitment of T cells and Langerhans-type dendritic cells to sites of atopic skin inflammation. J. Immunol. 174:5082–5091 [DOI] [PubMed] [Google Scholar]
- Günther C., Bello-Fernandez C., Kopp T., Kund J., Carballido-Perrig N., Hinteregger S., Fassl S., Schwärzler C., Lametschwandtner G., Stingl G., et al. 2005. CCL18 is expressed in atopic dermatitis and mediates skin homing of human memory T cells. J. Immunol. 174:1723–1728 [DOI] [PubMed] [Google Scholar]
- Hieshima K., Imai T., Baba M., Shoudai K., Ishizuka K., Nakagawa T., Tsuruta J., Takeya M., Sakaki Y., Takatsuki K., et al. 1997. A novel human CC chemokine PARC that is most homologous to macrophage-inflammatory protein-1 alpha/LD78 alpha and chemotactic for T lymphocytes, but not for monocytes. J. Immunol. 159:1140–1149 [PubMed] [Google Scholar]
- Islam S.A., Chang D.S., Colvin R.A., Byrne M.H., McCully M.L., Moser B., Lira S.A., Charo I.F., Luster A.D. 2011. Mouse CCL8, a CCR8 agonist, promotes atopic dermatitis by recruiting IL-5+ T(H)2 cells. Nat. Immunol. 12:167–177 10.1038/ni.1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodelja V., Müller C., Politz O., Hakij N., Orfanos C.E., Goerdt S. 1998. Alternative macrophage activation-associated CC-chemokine-1, a novel structural homologue of macrophage inflammatory protein-1 alpha with a Th2-associated expression pattern. J. Immunol. 160:1411–1418 [PubMed] [Google Scholar]
- Liacouras C.A., Furuta G.T., Hirano I., Atkins D., Attwood S.E., Bonis P.A., Burks A.W., Chehade M., Collins M.H., Dellon E.S., et al. 2011. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J. Allergy Clin. Immunol. 128:3–20: e6, quiz :21–22 10.1016/j.jaci.2011.02.040 [DOI] [PubMed] [Google Scholar]
- Liu T., Baek H.A., Yu H., Lee H.J., Park B.H., Ullenbruch M., Liu J., Nakashima T., Choi Y.Y., Wu G.D., et al. 2011. FIZZ2/RELM-β induction and role in pulmonary fibrosis. J. Immunol. 187:450–461 10.4049/jimmunol.1000964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucendo A.J., Arias A., De Rezende L.C., Yagüe-Compadre J.L., Mota-Huertas T., González-Castillo S., Cuesta R.A., Tenias J.M., Bellón T. 2011. Subepithelial collagen deposition, profibrogenic cytokine gene expression, and changes after prolonged fluticasone propionate treatment in adult eosinophilic esophagitis: a prospective study. J. Allergy Clin. Immunol. 128:1037–1046 10.1016/j.jaci.2011.08.007 [DOI] [PubMed] [Google Scholar]
- Nibbs R.J., Salcedo T.W., Campbell J.D., Yao X.T., Li Y., Nardelli B., Olsen H.S., Morris T.S., Proudfoot A.E., Patel V.P., Graham G.J. 2000. C-C chemokine receptor 3 antagonism by the beta-chemokine macrophage inflammatory protein 4, a property strongly enhanced by an amino-terminal alanine-methionine swap. J. Immunol. 164:1488–1497 [DOI] [PubMed] [Google Scholar]
- Pechkovsky D.V., Prasse A., Kollert F., Engel K.M., Dentler J., Luttmann W., Friedrich K., Müller-Quernheim J., Zissel G. 2010. Alternatively activated alveolar macrophages in pulmonary fibrosis-mediator production and intracellular signal transduction. Clin. Immunol. 137:89–101 10.1016/j.clim.2010.06.017 [DOI] [PubMed] [Google Scholar]
- Pivarcsi A., Gombert M., Dieu-Nosjean M.C., Lauerma A., Kubitza R., Meller S., Rieker J., Muller A., Da Cunha L., Haahtela A., et al. 2004. CC chemokine ligand 18, an atopic dermatitis-associated and dendritic cell-derived chemokine, is regulated by staphylococcal products and allergen exposure. J. Immunol. 173:5810–5817 [DOI] [PubMed] [Google Scholar]
- Prasse A., Pechkovsky D.V., Toews G.B., Schäfer M., Eggeling S., Ludwig C., Germann M., Kollert F., Zissel G., Müller-Quernheim J. 2007. CCL18 as an indicator of pulmonary fibrotic activity in idiopathic interstitial pneumonias and systemic sclerosis. Arthritis Rheum. 56:1685–1693 10.1002/art.22559 [DOI] [PubMed] [Google Scholar]
- Prasse A., Probst C., Bargagli E., Zissel G., Toews G.B., Flaherty K.R., Olschewski M., Rottoli P., Müller-Quernheim J. 2009. Serum CC-chemokine ligand 18 concentration predicts outcome in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 179:717–723 10.1164/rccm.200808-1201OC [DOI] [PubMed] [Google Scholar]
- Schaerli P., Ebert L., Willimann K., Blaser A., Roos R.S., Loetscher P., Moser B. 2004. A skin-selective homing mechanism for human immune surveillance T cells. J. Exp. Med. 199:1265–1275 10.1084/jem.20032177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürpf T., Springer T.A. 2011. Regulation of integrin affinity on cell surfaces. EMBO J. 30:4712–4727 10.1038/emboj.2011.333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutyser E., Richmond A., Van Damme J. 2005. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J. Leukoc. Biol. 78:14–26 10.1189/jlb.1204712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G.D., Rückerl D., Maskrey B.H., Whitfield P.D., Blaxter M.L., Allen J.E. 2012. The biology of nematode- and IL4Rα-dependent murine macrophage polarization in vivo as defined by RNA-Seq and targeted lipidomics. Blood. 120:e93–e104 10.1182/blood-2012-07-442640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffany H.L., Lautens L.L., Gao J.L., Pease J., Locati M., Combadiere C., Modi W., Bonner T.I., Murphy P.M. 1997. Identification of CCR8: a human monocyte and thymus receptor for the CC chemokine I-309. J. Exp. Med. 186:165–170 10.1084/jem.186.1.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Vahedi G., Sun H.W., Watford W.T., Takatori H., Ramos H.L., Takahashi H., Liang J., Gutierrez-Cruz G., Zang C., et al. 2010. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 32:840–851 10.1016/j.immuni.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik A., Yoshie O. 2012. The chemokine superfamily revisited. Immunity. 36:705–716 10.1016/j.immuni.2012.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.