Targeted deletion of CD103+CD11b+ LP DCs results in reduced LP Th17 cells at steady state, but has no impact on Citrobacter infection or the composition of the intestinal microbiota.
Abstract
Dendritic cells (DCs) in the intestinal lamina propria (LP) are composed of two CD103+ subsets that differ in CD11b expression. We report here that Langerin is expressed by human LP DCs and that transgenic human langerin drives expression in CD103+CD11b+ LP DCs in mice. This subset was ablated in huLangerin-DTA mice, resulting in reduced LP Th17 cells without affecting Th1 or T reg cells. Notably, cognate DC–T cell interactions were not required for Th17 development, as this response was intact in huLangerin-Cre I-Aβfl/fl mice. In contrast, responses to intestinal infection or flagellin administration were unaffected by the absence of CD103+CD11b+ DCs. huLangerin-DTA x BatF3−/− mice lacked both CD103+ LP DC subsets, resulting in defective gut homing and fewer LP T reg cells. Despite these defects in LP DCs and resident T cells, we did not observe alterations of intestinal microbial communities. Thus, CD103+ LP DC subsets control T cell homeostasis through both nonredundant and overlapping mechanisms.
DCs form a dense network at barrier surfaces such as skin, lung, and the intestinal lamina propria (LP). LP DCs acquire antigens from both commensal microbes and invading pathogens. They are thought to direct and regulate local innate immune responses, as well as determine the balance between tolerogenic and inflammatory adaptive responses (Iwasaki, 2007; Coombes and Powrie, 2008).
LP APCs can be phenotypically divided into two major developmentally distinct populations. The first, CD103−CD11b+ CX3CR1hi cells, derive from Ly6Chi monocyte precursors and share a common transcriptome with tissue-resident macrophages (Bogunovic et al., 2009; Varol et al., 2009; Miller et al., 2012). These cells produce IL-10, which is thought to be required for FoxP3+ regulatory T cell (T reg cell) maintenance in the LP (Denning et al., 2007; Hadis et al., 2011). However, they do not express CCR7 in the steady state and their ability to migrate to mesenteric lymph nodes (MLNs) remains controversial (Schulz et al., 2009; Diehl et al., 2013). The second population, CD103+ DCs, develops from a dedicated Flt3L-dependent conventional DC precursor and has a transcriptome similar to other DC lineages (Bogunovic et al., 2009; Varol et al., 2009; Miller et al., 2012). These cells express CCR7 and migrate to MLNs under steady-state and inflammatory conditions (Schulz et al., 2009). They have been shown to transport Salmonella enterica into the mesenteric LN (MLN) and produce retinoic acid (RA), inducing differentiation of CCR9+ gut-homing T reg cells both in vitro and in vivo (Coombes et al., 2007; Sun et al., 2007; Jaensson et al., 2008; Bogunovic et al., 2009; Semmrich et al., 2011).
Importantly, CD103+ DCs can be subdivided into two ontogenetically distinct subsets based on the expression of CD11b (Bogunovic et al., 2009). CD103+CD11b− DCs depend on the transcription factors BatF3, IRF8, and Id2 (Ginhoux et al., 2009; Edelson et al., 2010). Despite the absence of CD103+CD11b− DCs in BatF3−/− mice, alterations in bulk T cell homeostasis or intestinal inflammation are not observed (Edelson et al., 2010). Development of the second CD103+ DC subset, CD103+CD11b+ DC, requires Notch2 signaling (Lewis et al., 2011). These DCs are able to induce differentiation of Th17 cells in vitro, and the frequency of Th17 cells is reduced in the LP of CD11c-Cre Notch2fl/fl mice (Denning et al., 2011; Fujimoto et al., 2011; Lewis et al., 2011). In addition to this adaptive function, CD103+CD11b+ DCs are thought to exert innate immune functions through their ability to detect flagellin via Toll-like receptor 5 (TLR5; Uematsu et al., 2008; Fujimoto et al., 2011). Flagellin administration induces IL-22 from innate lymphoid cells in the LP and is thought to enhance innate resistance to intestinal pathogens (Van Maele et al., 2010; Kinnebrew et al., 2010). Elaboration of IL-22 depends on TLR5 and DC-derived IL-23. Reduced IL-22 production in Flt3−/− mice and the expression of TLR5 by CD103+CD11b+ DCs has suggested that this DC subset is required for IL-22 production (Kinnebrew et al., 2012). Additionally, IL-23–dependent IL-22 is required for innate resistance to Citrobacter rodentium, a mouse model for enteropathogenic Escherichia coli infection (Zheng et al., 2008).
Mouse models that allow for in vivo deletion of DC subsets are valuable tools to study DC function (Chow et al., 2011). However, multiple DC subsets are often affected, preventing the attribution of particular functions to an individual subset. Flt3−/− mice have greatly reduced numbers of CD103+CD11b+ DCs in the LP, but ∼40% of CD103+CD11b− DCs, as well as a statistically significant number of CD103−CD11b+ cells, are also absent (Bogunovic et al., 2009). Likewise, CD11c-Cre Notch2fl/fl mice lack CD103+CD11b+ DC, but have a concomitant increase in CD103+CD11b− LP DC, along with a loss of splenic CD11b+ Esamhi DCs (Lewis et al., 2011).
To investigate the function of DC subsets in the skin, we previously generated mice that ablate epidermal Langerhans cells (LCs) based on transgenic expression of human Langerin (huLangerin-DTA mice; Kaplan et al., 2005). In this study, we report that, in addition to LCs, CD103+CD11b+ LP DCs selectively express human Langerin (huLangerin) and are absent in these mice. Because all other DCs in the LP and MLN are intact, we use huLangerin-DTA mice, as well as Batf3−/− mice that lack CD103+CD11b− DC, to dissect the in vivo roles of specific CD103+ DC subsets in establishing innate and adaptive immune responses.
RESULTS
CD103+CD11b+ LP DCs express human Langerin
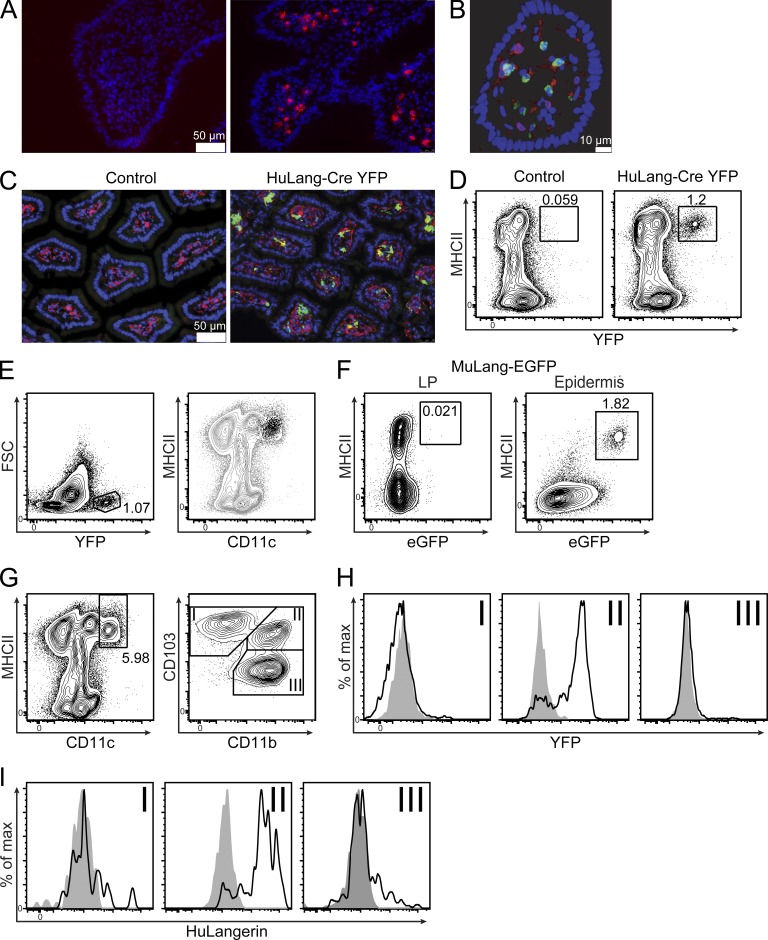
LP DCs in C57BL/6 mice do not express Langerin under steady-state conditions (Chang and Kweon, 2010). In contrast, Langerin expressing DCs have been reported in human LP (Kaser et al., 2004; Chikwava and Jaffe, 2004). To confirm Langerin expression in human LP, we examined fresh samples of human intestine by immunofluorescence. We observed numerous cells expressing Langerin that coexpressed MHC-II, identifying them as LP APCs (Fig. 1, A and B).
Figure 1.
LP DCs express human Langerin. (A) Human ileal biopsies were stained with DAPI (blue) and isotype control (left) or anti-huLangerin antibody (right, red). (B) Human colonic villus stained with DAPI (blue) and antibodies against HLA-DR (red) and Langerin (green). (C) Ileum from wild-type littermates (control) or huLangerin-Cre x Rosa26-Stopfl-YFP (HuLang-Cre YFP) mice stained with anti-YFP (green), anti-MHCII (red), and DAPI (blue). (D) Flow cytometric analysis of LP cells isolated from mice of the indicated genotypes. (E) Small intestinal LP suspensions from HuLang-Cre YFP mice were gated on live single cells before gating for total YFP+ cells (left). YFP+ cells (black dots) were then compared for expression of MHCII and CD11c with total live gated cells (gray contour, right). (F) Cell suspensions from the indicated tissues of murine Langerin-EGFP mice were analyzed for MHCII and GFP expression. (G) Live single cells from HuLang-Cre YFP mice gated on CD11chi and MHCII+ cells and subgated for CD103 and CD11b. (H) YFP expression in the indicated subsets from HuLang-Cre YFP (black line) or littermate control (shaded gray) mice. (I) huLangerin expression in the indicated DC subsets isolated from transgene+ (black line) or littermate control (shaded gray) mice. All data are representative of at least 2 independent experiments.
In mice, endogenous Langerin is expressed primarily by LCs and CD103+ dermal DCs in the skin. In contrast, human LCs express Langerin, but DCs in the dermis do not (Haniffa et al., 2012). We previously reported that the human promoter of the gene langerin drives expression in murine LCs, but not CD103+ dermal DCs (Bursch et al., 2007). Because human Langerin transgenic mice appear to recapitulate the human pattern of Langerin expression in the skin, we considered the possibility that human Langerin might be expressed by LP DCs in these transgenic mice.
We previously generated huLangerin-Cre x Rosa26-Stopfl-YFP mice (henceforth referred to as huLang-Cre YFP) that faithfully report the expression of transgenic human Langerin (Kaplan et al., 2007). Immunofluorescent microscopy of the small intestine of huLang-Cre YFP mice revealed numerous cells coexpressing MHC-II and huLangerin in the LP that were not present in control littermates (Fig. 1 C). huLangerin-expressing cells could also be detected in the colon, but at a reduced frequency (unpublished data). YFP+ MHC-II+ cells could be easily detected by flow cytometry of collagenase-digested small intestinal LP (Fig. 1 D). Importantly, all YFP+ cells were CD11chi and MHC-II+, identifying them as LP APCs (Fig. 1 E). In contrast, reporter mice for murine Langerin (muLangerin-eGFP; Kissenpfennig et al., 2005) confirmed that murine langerin was not expressed in MHCII+ cells of the LP, although it was easily detected in epidermal LCs (Fig. 1 F). Thus, the human BAC containing the gene for langerin that we have previously used to generate several lines of transgenic mice recapitulates the human pattern of Langerin expression and drives expression in intestinal LP DCs.
To determine which DC subsets express huLangerin, single-cell suspensions were generated from the small intestinal LP of huLang-Cre YFP and control mice. To identify LP DCs, we gated on CD11chi MHC-II+ cells, further dividing them into three subsets (I–III) based on their expression of CD103 and CD11b (Fig. 1 G) (Bogunovic et al., 2009). We did not observe YFP expression in CD103+CD11b− DCs, CD103−CD11b+ macrophages, or control littermates (Fig. 1 H). In contrast, we observed robust YFP expression in CD103+CD11b+ DCs isolated from huLang-Cre YFP mice. Because YFP expression in these Cre reporter mice could potentially result from huLangerin transgene expression in a DC precursor population, we stained DCs from transgenic mice (Bobr et al., 2010) with a huLangerin-specific antibody. As expected, we observed huLangerin protein expression in CD103+CD11b+ DC, but not in other DC/macrophage subsets or transgene-negative controls (Fig. 1 I). Thus, the gene for human langerin drives YFP and Langerin expression selectively in CD103+CD11b+ LP DCs of BAC transgenic mice. Because we observed few huLangerin+ DCs in the colon, which agrees with previous data showing that CD103+CD11b+ DCs are predominantly present in the small bowel (Denning et al., 2011), we focused on the small intestinal LP for the remainder of our studies.
huLangerin-DTA mice lack CD103+CD11b+ LP DCs
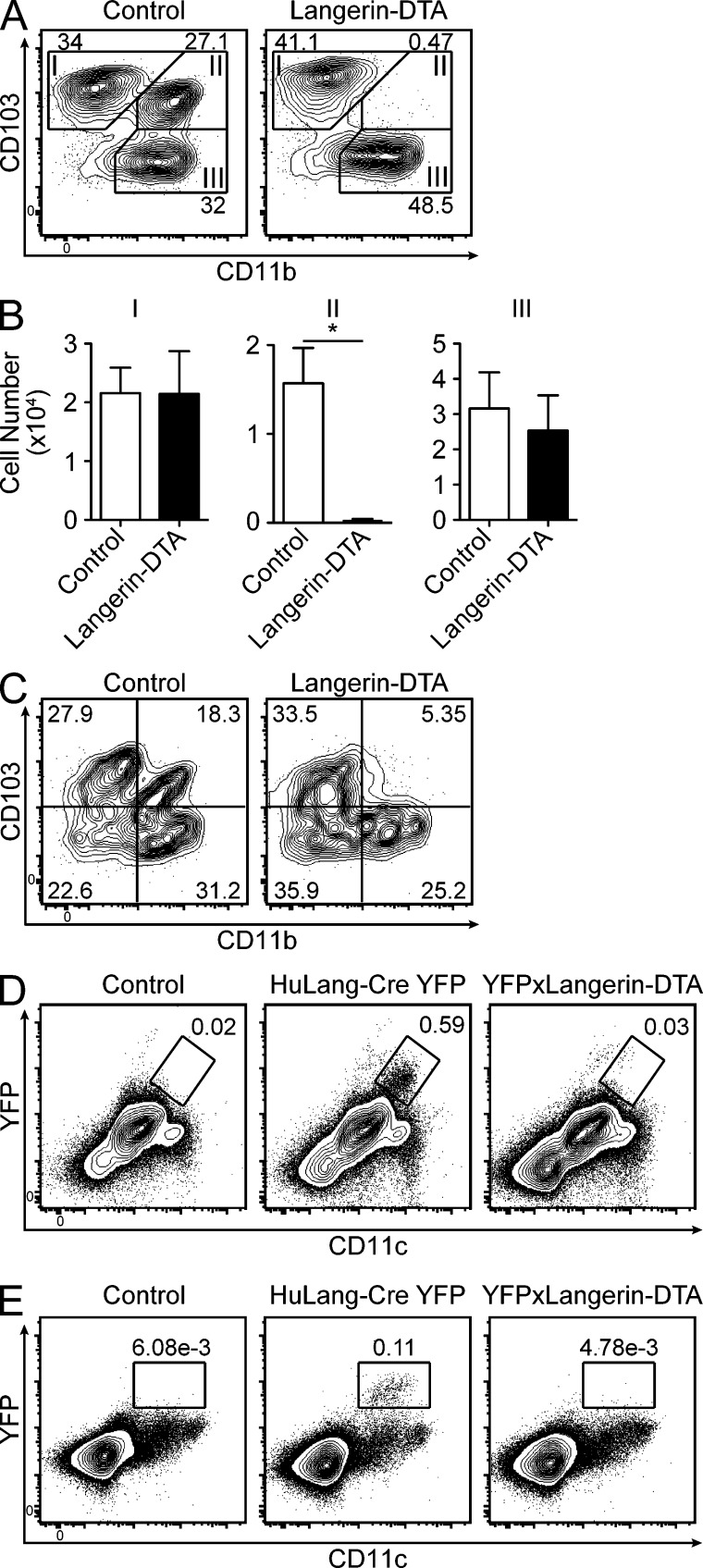
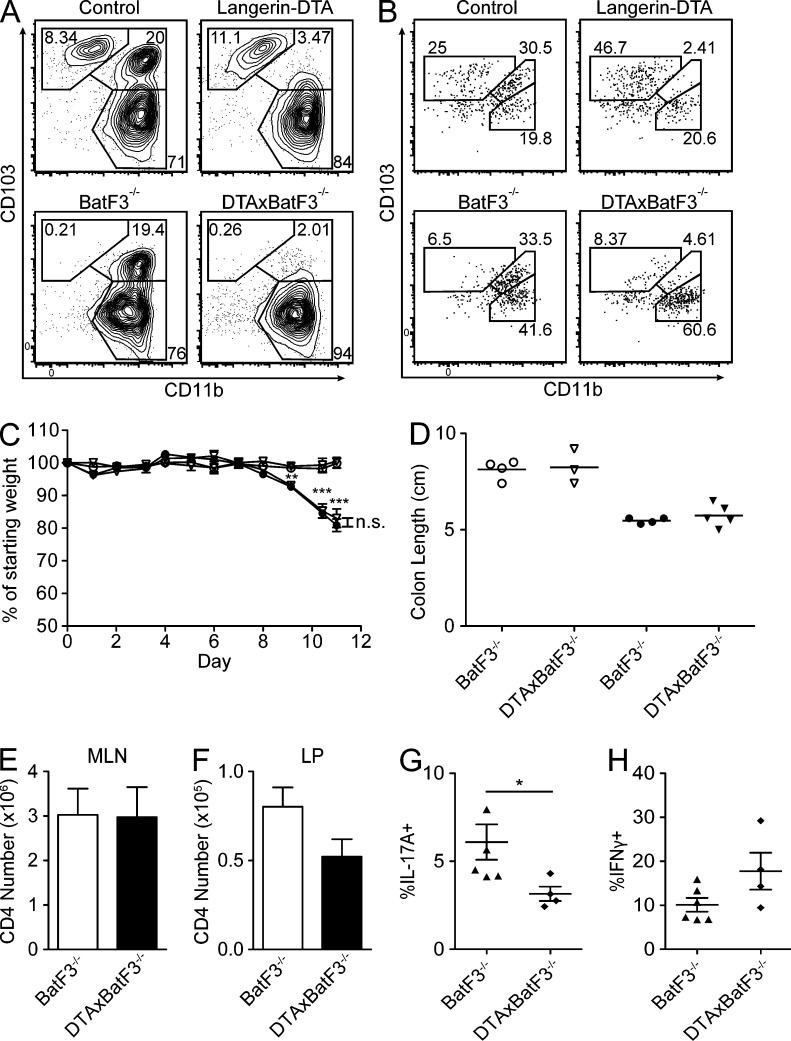
huLangerin-DTA BAC transgenic mice were engineered to express an attenuated form of diphtheria toxin driven by the same human Langerin promoter used by huLangerin-Cre mice. In the skin, these mice have a selective ablation of epidermal LCs (Kaplan et al., 2005). To determine whether LP DCs are also ablated in these mice, we compared DC populations in the small intestinal LP of huLangerin-DTA and littermate control mice. Using the same gating strategy shown in Fig. 1 G, we noted a near-complete absence of CD11chi MHC-II+ CD103+CD11b+ DCs (Fig. 2, A and B, subset II). Importantly, the numbers of CD103+CD11b− DCs (subset I) and CD103−CD11b+ macrophages (subset III) in the LP were unaffected. We noted a similar absence of CD103+CD11b+ DCs in the draining MLNs of huLangerin-DTA mice (Fig. 2 C).
Figure 2.
CD103+CD11b+ LP DCs are ablated in huLangerin-DTA mice. (A) Flow cytometry of CD11chi MHCII+ gated LP cells isolated from littermate control or huLangerin-DTA mice analyzed for CD103 and CD11b expression. (B) The number of cells for each subset in A is shown (n = 6–8 mice per genotype). (C) MLN cells analyzed as in A. (D–E) Flow cytometry of live single cells from LP (D) or MLN (E) isolated from littermates of the indicated genotypes. All data are representative of at least three experiments. Statistics were calculated using an unpaired Student’s t test (*, P ≤ 0.05).
CD103 expression by DCs can be regulated by environmental stimuli (Dresch et al., 2012). Therefore, to confirm ablation of CD103+CD11b+ DCs in huLangerin-DTA mice, we bred huLangerin-DTA mice with YFP reporter mice. As expected, a population of CD11c+ cells expressing YFP could be detected in the LP of huLang-Cre YFP but not control mice. These cells were absent from the LP of huLang-Cre YFP x huLangerin-DTA mice (Fig. 2 D). The same was observed in the MLNs (Fig. 2 E). These findings in independently derived transgenic lines confirm that huLangerin drives transgene expression in CD103+CD11b+ DCs in the LP and MLN. This DC subset is selectively ablated in huLangerin-DTA transgenic mice, providing a novel and powerful tool to study the function of CD103+CD11b+ DCs in vivo.
Functional characterization of huLangerin-DTA mice
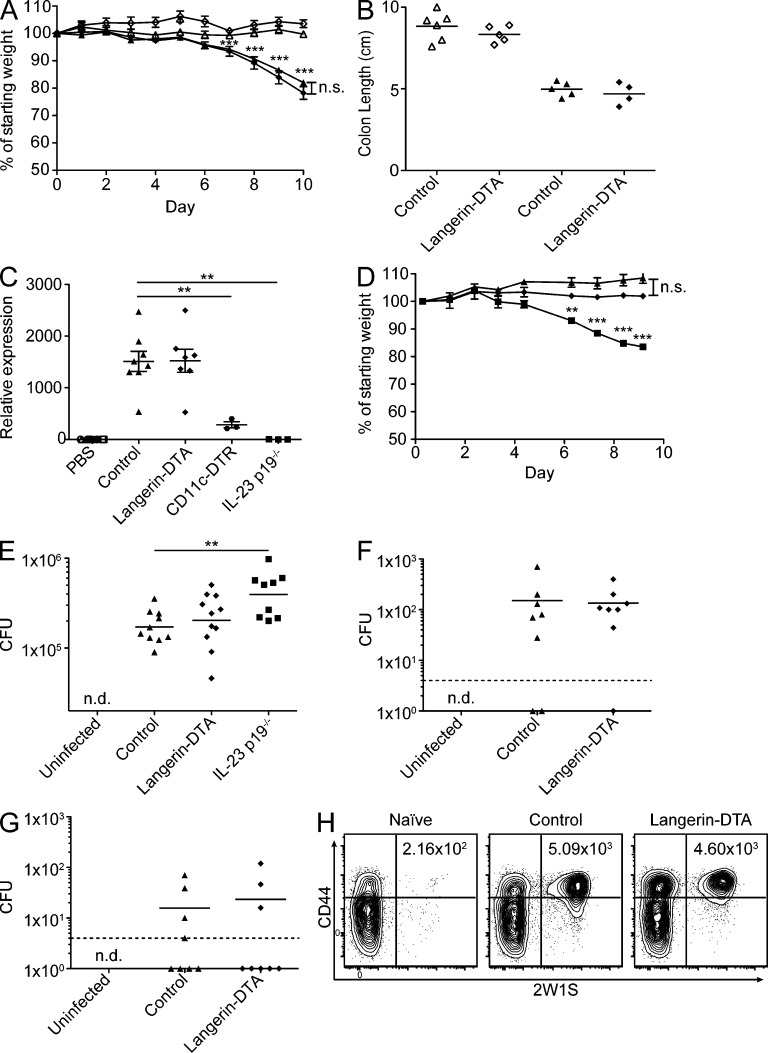
huLangerin-DTA mice have been well characterized (Kaplan et al., 2005; Igyarto et al., 2009). They are long-lived and do not develop spontaneous autoimmunity. Because the ablation of CD103+CD11b+ LP DCs in these mice was previously unappreciated, we tested their response to an intestinal inflammatory challenge. Dextran-sodium sulfate (DSS) generates an acute hemorrhagic enterocolitis and is a standard assay of the intestinal inflammatory response (Wirtz et al., 2007). We administered DSS in the drinking water of huLangerin-DTA and littermates for 10 d and monitored their weight loss. As expected, treated groups lost a significant amount of weight. However, huLangerin-DTA mice lost weight at the same rate as littermate controls when treated with either 2% (unpublished data) or 5% DSS (Fig. 3 A). Treated control and huLangerin-DTA animals also exhibited an identical degree of colonic shortening, a measure of colitis severity (Fig. 3 B). Thus, CD103+CD11b+ LP DCs are not required for the development of, or protection from, DSS enterocolitis.
Figure 3.
CD103+CD11b+ DCs are not required for IL-22 production or resistance to Citrobacter or Salmonella. (A) Groups of 4–6 littermate control (triangles) and huLangerin-DTA (diamonds) mice were given water (open symbols) or 5% DSS (filled symbols) ad libitum for the indicated number of days and monitored for weight loss. Data are presented as the mean ± SEM. (B) Colon length upon sacrifice at day 10. Representative data from 1 of 2 independent experiments is shown. (C) Animals were treated intravenously with PBS (open symbols) or 1 µg flagellin (closed symbols) and euthanized 2 h later for LP mRNA extraction and qPCR analysis. CD11c-DTR mice were treated with 1 µg diphtheria toxin 18 h before flagellin injection. Data are normalized to HPRT and displayed as fold change over PBS-treated mice. (D) Control (triangles) or huLangerin-DTA (diamonds) littermates and IL-23 p19−/− mice (squares) were infected with 2 × 109 C. rodentium and monitored for weight loss. Data are presented as the mean ± SEM with at least 3 mice per group pooled from 2 independent experiments. One animal succumbed due to unrelated causes and was excluded from the analysis. (E) Mice were pretreated with 20 mg streptomycin sulfate 24 h before infection with 5 × 107 Salmonella strain SL1344 by gavage. MLNs were harvested 48 h later and analyzed for CFU. (F and G) Mice were orogastrically infected with 108 AroA− Salmonella-2W1S. Indicated organs were harvested 10 d later for CFU. Dashed lines represent the limit of detection. (H) Spleens and MLN were pooled from uninfected (Naive) and Salmonella-2W1S infected mice after 10 d and magnetically enriched for 2W1S-specific cells. Flow plots represent the binding of 2W1S-I-Aβ tetramer by CD4 T cells. Numbers represent the total number of tetramer-specific cells. Flow plots in H are representative of three experiments. All symbols represent individual mice pooled from two (C, F, and G) or four (E) independent experiments. Lines in E–G represent the geometric mean. Statistics were calculated using unpaired Student’s t tests (nd, not detected; **, P ≤ 0.01; ***, P ≤ 0.001).
CD103+CD11b+ DCs express TLR5 (Uematsu et al., 2008; Fujimoto et al., 2011) and are reportedly required for IL-23–dependent amplification of IL-22 production by gut innate lymphoid cells in response to intravenously administered flagellin (Kinnebrew et al., 2012). Because these studies were performed in mice with defects in multiple gut DC lineages (e.g., CD11c-DTR and Flt3−/− mice), we tested the response to intravenous flagellin in huLangerin-DTA mice. As expected, we observed a dramatic increase in LP IL-22 mRNA expression in the small intestine of control mice 2 h after treatment with flagellin. This increase was absent in IL-23 p19−/− mice and significantly reduced in CD11c-DTR mice treated with diphtheria toxin. IL-22 production was unaffected in huLangerin-DTA mice (Fig. 3 C), indicating that CD103+CD11b+ DCs are not required for innate immune responses to i.v. flagellin.
To determine whether CD103+CD11b+ DCs are required for IL-22–IL-23–dependent immune responses during infection, we orogastrically inoculated mice with 2 × 109 C. rodentium. As previously reported (Basu et al., 2012; Mangan et al., 2006), IL-23 p19−/− mice lost a significant amount of weight beginning at day 4 after infection (Fig. 3 D) and succumbed in 8–12 d (unpublished data). However, huLangerin-DTA mice and their wild-type littermates did not lose weight or succumb to C. rodentium infection (Fig. 3 D). We considered that, though they may be an important source of IL-23, CD103+CD11b+ DCs might not be required for protection from Citrobacter because of the lack of TLR5 ligation by this aflagellate pathogen (Khan et al., 2008). We therefore infected mice with S. enterica serovar Typhimurium, using an antibiotic pretreatment model of acute S. enterica–induced gastroenteritis (Barthel et al., 2003). We observed that CFU of S. enterica in the MLN was significantly increased in IL-23 p19−/− mice 48 h after infection, but was unaffected in huLangerin-DTA mice (Fig. 3 E). Thus, CD103+CD11b+ DCs do not appear to be required for resistance to IL-22–IL-23–dependent infections.
To investigate whether adaptive immunity to S. enterica requires CD103+CD11b+ DCs, we orally infected mice with S. enterica that expresses a previously characterized 2W1S CD4 T cell epitope, allowing us to track a pathogen-specific adaptive immune response during infection (Moon et al., 2007; Nelson et al., 2013). 10 d after inoculation, near the peak of the response, huLangerin-DTA mice bore a bacterial burden equivalent to that of littermate controls in both the MLN (Fig. 3 F) and spleen (Fig. 3 G). This was also true at later time points (unpublished data). As expected, Samonella-2W1S infection led to an ∼20-fold increase in the number of 2W1S-specific endogenous CD4 T cells. Notably, the lack of CD103+CD11b+ LP DCs did not impair the development of a pathogen-specific CD4 T cell response (Fig. 3 H). Thus, CD103+CD11b+ LP DCs are not required for adaptive immune responses to a flagellated gut pathogen.
CD103+CD11b+ DCs are required for Th17 homeostasis
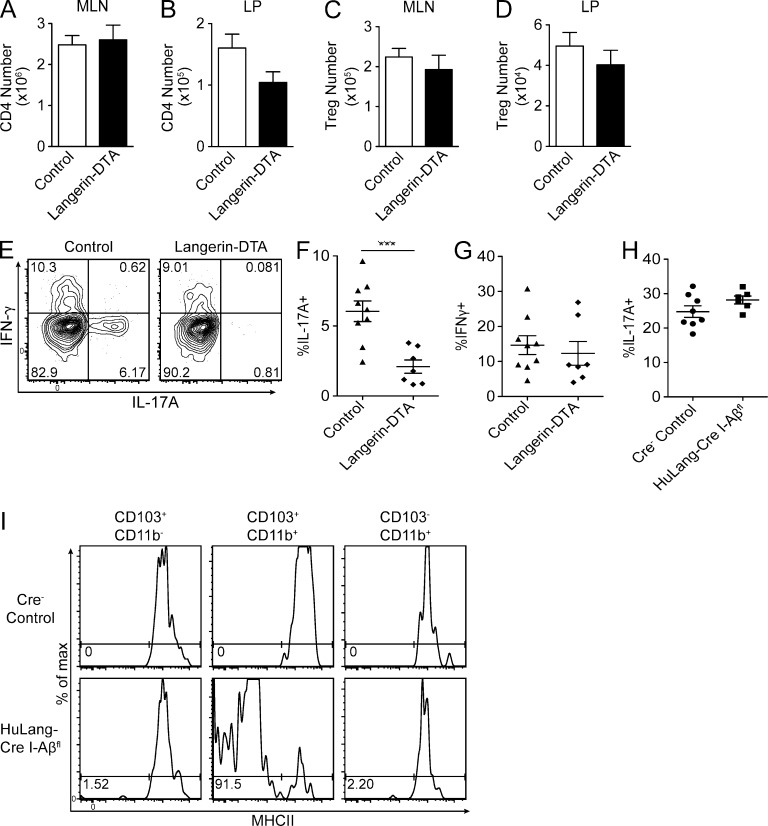
To determine whether the absence of CD103+CD11b+ DCs affects steady-state helper T cell development in vivo, we compared the numbers of CD4 T cell subsets in huLangerin-DTA and control mice. We found that CD4 T cell numbers in the MLN were not altered by the absence of CD103+CD11b+ DCs (Fig. 4 A). In the small intestinal LP, however, we noted a nonsignificant trend toward fewer CD4 T cells in huLangerin-DTA mice (Fig. 4 B). The number of Foxp3+ T reg cells in the MLN and LP was unaffected (Fig. 4, C and D).
Figure 4.
LP Th17 cells require CD103+CD11b+ DCs through an MHC II independent mechanism. (A–D) CD4 cells gated as CD3ε+ TCRγδ− CD8α− CD4+ were isolated and enumerated from indicated tissues by flow cytometry. T reg cells were detected by intracellular staining for FoxP3 directly ex vivo. Data are presented as mean ± SEM and pooled from five independent experiments (n = 8–10 mice per group). (E) LP suspensions were restimulated ex vivo for 3 h with PMA/ionomycin and stained for intracellular cytokines. Representative flow plots gated on LP CD4 T cells as in A–D. (F–H) Frequency of restimulated CD4+ LP T cells expressing indicated cytokines by flow cytometry. (I) Surface expression of MHCII on small intestinal LP DC subsets gated as CD3ε− B220− CD64− CD11c+ cells and subgated for CD103 and CD11b, as in Fig. 1 G. Flow plots represent two independent experiments. Symbols represent individual mice and are pooled from 5 (E–G) or 3 (H) independent experiments. Statistics were calculated by unpaired Student’s t tests, with Welch’s correction for unequal variance in B and D. ***, P ≤ 0.001.
To examine the requirement of CD103+CD11b+ LP DCs for other T helper subsets, we compared the expression of IL-17A and IFN-γ in PMA/ionomycin-stimulated cells isolated from the LP of huLangerin-DTA and littermate control mice. We observed that the frequency of CD4 T cells expressing IL-17 was greatly reduced in huLangerin-DTA mice, whereas the expression of IFN-γ was unaffected (Fig. 4, E–G). As previously noted, CD4 T cells expressing IL-17 in the MLN were rare (Atarashi et al., 2008). The observed differences were not caused by differential carriage of segmented filamentous bacteria because littermates were used as controls. In addition, we confirmed equivalent carriage of segmented filamentous bacteria by quantitative PCR (unpublished data). Finally, we examined the LP of huLangerin-Cre x I-Aβfl/fl mice. Notably, these mice had a normal frequency of Th17 cells in the LP compared with controls (Fig. 4 H), despite the fact that >90% of CD103+CD11b+ DCs lacked expression of MHC-II (Fig. 4 I). Thus, Th17 development in the LP depends on the presence of CD103+CD11b+ DCs but does not require their cognate interaction with CD4 T cells.
CD103+ DCs are required for LP T reg cells
Batf3−/− mice lack CD103+CD11b− DCs in the LP, as well as other peripheral tissues. Despite this deficiency, no abnormalities in T cell or T reg cell homeostasis, or steady-state intestinal inflammation, were noted (Edelson et al., 2010). We generated huLangerin-DTA x BatF3−/− animals (henceforth referred to as DTAxBatF3−/−). As expected, in the resulting DTAxBatF3−/− mice, both subsets of CD103+ DCs were constitutively deleted in the LP and MLN (Fig. 5, A and B). DTAxBatF3−/− animals were long lived, did not develop autoimmunity (unpublished data), and did not show altered susceptibility to DSS (Fig. 5, C and D). We observed that the additional absence of CD103+CD11b+ DCs did not alter the number CD4 T cells in the MLN (Fig. 5 E). However, as we observed with huLangerin-DTA mice, there was a nonsignificant trend toward fewer total CD4 T cells in the LP of DTAxBatF3−/− animals (Fig. 5 F).
Figure 5.
CD103+CD11b+ DCs are non-redundant for Th17 development, whereas CD103+CD11b− DCs are dispensable. (A and B) Flow cytometry of CD11chi MHCII+ cells from LP (A) or MLN (B) of the indicated genotypes. Data represent 3 experiments (n = 3–5 per group). (C) Groups of 3–5 BatF3−/− (circles) and DTAxBatF3−/− (triangles) littermate mice were given water (open symbols) or 5% DSS (filled symbols) ad libitum for the indicated number of days and monitored for weight loss. Data are presented as the mean ± SEM (D) Colon length upon sacrifice at day 10. Symbols represent individual mice and lines represent the mean. C and D represent one of two experiments. (E and F) Cells were isolated from indicated tissues and CD4 T cell numbers enumerated as previously described. (n = 10–12 mice per group). (G and H) Frequency of restimulated CD4 LP T cells producing the indicated cytokines. Data are presented as mean ± SEM and symbols represent individual mice. Animals in E–H are pooled from three independent experiments. Statistics were calculated by unpaired Student’s t tests, using Welch’s correction in C. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
We next examined the presence of Th17 and Th1 cells in the small intestinal LP. We compared DTAxBatF3−/− mice with littermate BatF3−/− controls to ensure that any observed changes were not caused by alterations in the gut microbiota. We observed reduced Th17 cells in DTAxBatF3−/− mice compared with Batf3−/− mice. However, Th1 cells in the LP were not affected (Fig. 5, G and H). These data are quite similar to those obtained with huLangerin-DTA mice not bred to Batf3−/− animals (Fig. 4 F). Because the additional deletion of CD103+CD11b− DCs did not alter the presence of Th17 or Th1 cells in the LP, these data demonstrate that CD103+CD11b+ DCs are specifically required for steady-state Th17 development.
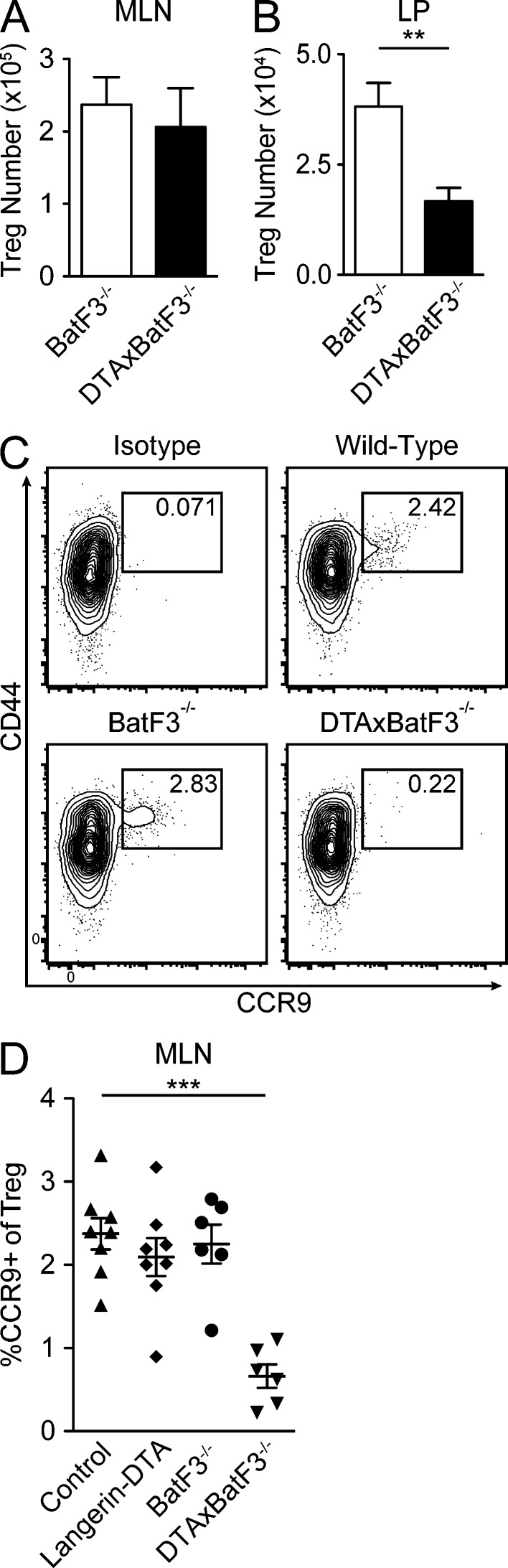
As T reg cell numbers are unaffected in huLangerin-DTA mice (Fig. 4, C and D) and BatF3−/− mice (Edelson et al., 2010), we next examined T reg cell development in DTAxBatF3−/− mice. The number of FoxP3+ T reg cells in the MLN was unaffected in DTAxBatF3−/− mice lacking both CD103+ DC subsets (Fig. 6 A). However, there was a significant reduction in the number of FoxP3+ T reg cells in the LP of DTAxBatF3−/− mice compared with BatF3−/− littermates (Fig. 6 B). We next investigated expression of gut-homing chemokine receptors on FoxP3+ T reg cells in the MLN. The frequency of T reg cells expressing the RA-inducible gut-homing receptor CCR9 was greatly diminished in DTAxBatF3−/− mice compared with both wild-type mice and mice lacking only one CD103+ DC subset (Fig. 6, C and D). We therefore conclude that CD103+CD11b+ and CD103+CD11b− DCs are individually redundant but are together required to imprint gut-homing receptors on T reg cells in the MLN, thereby maintaining normal numbers of T reg cells in the LP.
Figure 6.
CD103+ DCs are required for LP T reg cells. (A and B) Cell suspensions from the indicated tissues were stained ex vivo for intracellular FoxP3 and enumerated by flow cytometry (n = 10–12 mice per group). (C) Representative flow plots of MLNs gated on CD3ε+ CD8α− CD4+ FoxP3+ cells and stained with isotype control (top left, wild-type mouse) or anti-CCR9 antibody (indicated genotypes). (D) Frequency of CCR9+ MLN T reg cells in mice of indicated genotype. All data are presented as mean ± SEM and symbols represent individual mice. Animals are pooled from three different experiments. Statistics were calculated by unpaired Student’s t tests, using Welch’s correction in B. **, P ≤ 0.01; ***, P ≤ 0.001.
CD103+CD11b+ DCs do not shape the microbiome
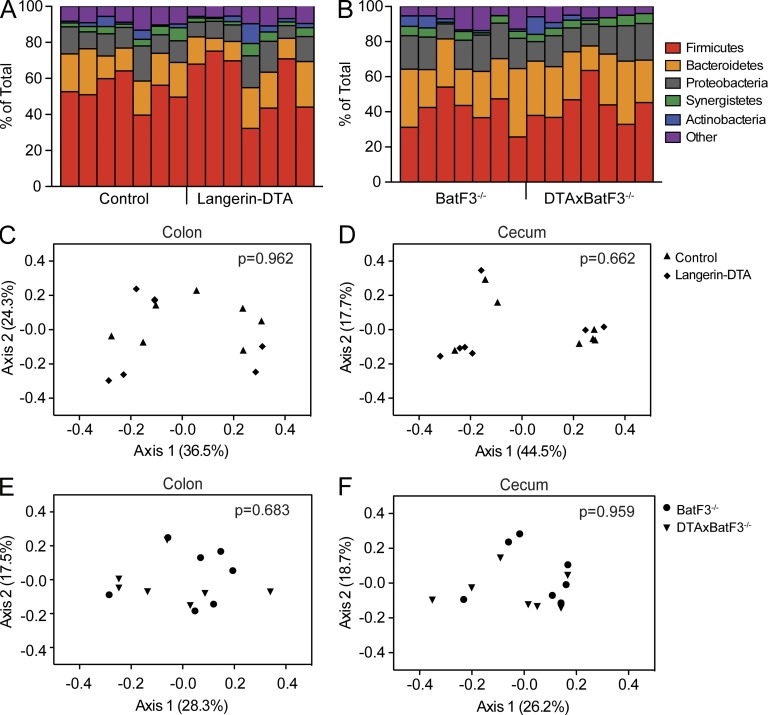
Numerous studies have demonstrated that the presence of specific commensal microorganisms in the intestine direct host immune development (Chung et al., 2012; Honda and Littman, 2012). In contrast, the capacity of the host immune system to shape the intestinal microbiome has been less robustly examined. We have now characterized two genetic mouse models in which intestinal DC subsets have been ablated, leading to either reduced numbers of Th17 cells (huLangerin-DTA mice) or reduced numbers of both Th17 and T reg cells (DTAxBatF3−/− mice) in the LP. To test whether these immune alterations affect the intestinal microbiome, we obtained fresh fecal pellets and scraped cecal contents from age- and sex-matched huLangerin-DTA and wild-type littermate controls. We also sampled DTAxBatF3−/− mice and BatF3−/− littermate controls. We then performed high-throughput sequencing on 16S bacterial rDNA amplified from these samples. A total of 1.11 × 108 paired-end sequence reads (2 × 100 nt) were generated, with 2.17 × 105 sequence reads per sample meeting our quality criteria. From these reads, 19,592 operational taxonomic units (OTUs) were identified among all samples, with a mean of 2,338 ± 261 OTUs present in individual samples. Sequence analyses identified a total of 28 phyla, which were dominated by Firmicutes (49 ± 7%), Bacteroidetes (21 ± 8%), and Proteobacteria (13 ± 5%; Fig. 7, A and B).
Figure 7.
Loss of CD103+CD11b+ LP DCs does not alter commensal microbial communities. (A and B) Phylogenetic classification of 16S rDNA sequence reads obtained from colonic fecal samples of wild-type/huLangerin-DTA littermates (A) or BatF3−/−/DTAxBatF3−/− littermates (B). Bars represent bacterial phyla present in individual mice (7 mice per genotype). (C–F) Principal component plots comparing overall bacterial composition in colonic (C and E) or cecal (D and F) stool samples. Symbols in C–F represent individual mice. P-values represent unweighted UniFrac comparisons of huLangerin-DTA or DTAxBatF3−/− animals to their respective wild-type or BatF3−/− littermates at each anatomical site.
Nonmetric multidimensional scaling (NMDS) on the Bray-Curtis distance matrix was used to visualize microbial community structure along axes of maximal variance (Bray and Curtis, 1957). Unweighted UniFrac analysis revealed that the loss of CD103+CD11b+ DCs and LP Th17 cells in huLangerin-DTA mice did not significantly affect the microbial composition in colonic (Fig. 7 C) or cecal (Fig. 7 D) contents (P = 0.962 and P = 0.662, respectively). There were also no significant differences in Shannon and Simpson diversity indices between groups (unpublished data). Of note, we did observe clustering of different litters, most evident in the cecum, demonstrating the importance of parental transmission in shaping steady-state gut microbial ecology. Similarly, the reduction of T reg cells and Th17 cells in the LP attendant with deletion of all CD103+ DCs in DTAxBatF3−/− animals also did not significantly alter composition of the colonic (Fig. 7 E) or cecal (Fig. 7 F) microbiota (P = 0.683 and P = 0.959, respectively). Thus, the absence of CD103+CD11b+ LP DCs, as well as reduced numbers of Th17 and T reg cells in the LP, does not significantly affect the composition of the steady-state enteric microflora.
DISCUSSION
Here, we report that huLangerin-DTA mice that were developed to target LCs in the skin also have a robust and selective ablation of CD103+CD11b+ LP DCs. Unlike mice with genetic ablation of factors required for DC subset development, transgenic expression of diphtheria toxin results in a highly specific ablation of individual DC subsets without alterations in the numbers of other DC subsets. The absence of CD103+CD11b+ DCs in the LP and mesenteric LN in huLangerin-DTA mice allowed us to examine the functional requirements of this subset in vivo.
LP Th17 cells contribute to autoimmune disease in humans and in animal models (Korn et al., 2009; Honda and Littman, 2012). Given this pathogenic potential, it is important to understand the mechanisms promoting LP Th17 differentiation. In agreement with other models that target CD103+CD11b+ DCs (Lewis et al., 2011; Persson et al., 2013; Schlitzer et al., 2013), we observed that the frequency of Th17 cells in the LP was greatly reduced in the absence of CD103+CD11b+ DCs. Other LP-resident T cell subsets were unaffected. Specific commensal species and bacterial products are sufficient to generate Th17 cells in the LP (Atarashi et al., 2008; Ivanov et al., 2009). Thus, the reduction in Th17 cells could potentially result from a decrease in the overall number of DCs presenting commensal antigens. Notably, Th17 development was not affected in Batf3−/− mice that lack CD103+CD11b− DCs, and DTAxBatF3−/− mice had a reduction of Th17 cells similar to that observed in huLangerin-DTA mice. Thus, CD103+CD11b+ DCs have a nonredundant role in LP Th17 development that is independent of CD103+CD11b− DCs.
The cytokines required for Th17 differentiation in the LP are controversial, but include IL-1β, IL-6, and TGFβ (Atarashi et al., 2008; Ivanov et al., 2008; Hu et al., 2011; Shaw et al., 2012). Consistent with a reduction of Th17 cells in huLangerin-DTA mice, CD103+CD11b+ DCs produce ∼5–10 fold more these cytokines than CD103+CD11b− DCs (unpublished data; Denning et al., 2011; Fujimoto et al., 2011). Remarkably, Th17 differentiation was intact in huLangerin-Cre I-Aβfl/fl mice. Thus, direct antigen presentation by CD103+CD11b+ DCs is not required for Th17 development, arguing that this DC subset is rather required to establish the appropriate cytokine milieu for Th17 differentiation. Though reported to occur during certain infections (Perona-Wright et al., 2010), DCs driving T helper differentiation under steady-state conditions through noncognate interactions in vivo is a novel observation. The identity of the cell type that presents antigen remains unclear. Interestingly, Th17 cells can be generated by commensals even in transgenic mice that presumably lack T cell antigen receptors capable of recognizing antigen derived from these microbes, suggesting that cognate DC–T cell interactions may not be required (Lochner et al., 2011).
Earlier reports have suggested that CD103+CD11b+ DCs are required for flagellin-induced expression of IL-22 and to transport bacterial antigens from the LP to the MLN during infection with S. typhimurium (Bogunovic et al., 2009; Kinnebrew et al., 2012). In contrast to these studies, huLangerin-DTA mice showed no defects in flagellin-induced IL-22 production or in protection from the IL-22/IL-23–dependent pathogen C. rodentium. huLangerin-DTA mice also mounted appropriate innate and adaptive immune responses to S. typhimurium. These differences may be due to different DC deletion strategies used in these studies and to potential functional overlap between known DC and/or macrophage subsets. Alternatively, these discrepancies may point to a functional importance for Flt3-dependent CD103− DCs in the LP that have been recently identified (Cerovic et al., 2013).
CD103+ DCs generate T reg cells in vitro through a RA-dependent mechanism (Coombes et al., 2007; Sun et al., 2007; Jaensson et al., 2008). The particular CD103+ DC subset responsible for T reg cell development in vivo has not been identified because both CD103+ LP DC subsets, as well as CX3CR1+ macrophages, have the capacity to metabolize RA and to induce T reg cells in vitro (Guilliams et al., 2010; Denning et al., 2011). In agreement with others, we show that, in vivo, neither CD103+ DC subset is individually required for T reg cell homeostasis (Edelson et al., 2010; Lewis et al., 2011). The reduction of T reg cells in DTAxBatF3−/− mice reconciles the in vitro and in vivo data by demonstrating that, though mutually redundant, CD103+ DC subsets are jointly required to imprint CCR9+ T reg cells in the MLN and thereby maintain normal T reg cell numbers in the LP. Whether CD103+ DCs are required for antigen presentation or are an obligate source of RA cannot be distinguished. It is surprising that, despite a reduction in LP T reg cells, we did not observe enhanced steady-state or DSS-induced inflammatory pathology in DTAxBatF3−/− mice. This may result from the concomitant reduction in Th17 cells or the microbial flora found in our animal facility.
The significance of intestinal commensal microorganisms in establishing and maintaining normal gut immune maturation is very well appreciated (Honda and Littman, 2012). In contrast, the role played by the intestinal immune system in determining the constituents of the commensal microbial community has been less rigorously explored. Altered intestinal microbial communities that result from genetic defects have been reported for Nalp6−/− and Tbet−/− Rag2−/− mice (Elinav et al., 2011; Garrett et al., 2007). Altered commensalism in these mice occurs in the setting of spontaneous inflammatory bowel disease. Because huLangerin-DTA and DTAxBatF3−/− mice do not develop inflammation, they can be used to test whether DCs and LP-resident T cells regulate steady-state intestinal commensal communities. Remarkably, despite the absence of CD103+ LP DCs and defects in LP T reg cells and Th17 cells, we did not observe any alteration in the intestinal microbiome of these mice. This is not the result of a technical limitation, as we were able to detect differences resulting from vertical transmission of maternal flora (Ubeda et al., 2012). Our data thus support a model in which the adaptive immune system, with the exception of IgA (Fagarasan et al., 2002), has a limited role in shaping the intestinal microbiome.
In this study, we report novel in vivo tools and provide strong evidence that distinct intestinal DC subsets make essential contributions to the maturation of the intestinal adaptive immune system. Future work with these tools will provide additional insight into the mechanisms through which specific LP DC subsets interact with both commensal and pathogenic microbes to shape immune responses, key considerations for the development of future therapeutics.
MATERIALS AND METHODS
Mice.
All strains were backcrossed for at least eight generations onto the C57BL/6 genetic background. huLangerin-Cre Rosa26-Stopfl-YFP (Kaplan et al., 2007), muLangerin-EGFP (Kissenpfennig et al., 2005), huLangerin-DTA (Kaplan et al., 2005), and IL-23 p19−/− (Ghilardi et al., 2004) mice have all been previously described. BatF3−/− mice (Hildner et al., 2008) were a gift from K. Murphy (Washington University, St. Louis, MO). Experiments were performed with 6–12-wk-old age- and sex-matched, cohoused littermates. Mice were housed in specific pathogen–free microisolator cages and fed irradiated food and acidified water. The University of Minnesota Institutional Animal Care and Use Committee approved all animal protocols.
Immunohistochemistry.
Tissues were washed in PBS and fixed in 10 volumes of 1% phosphate-buffered paraformaldehyde containing 75 mM l-lysine (Sigma-Aldrich) and 10 mM NaIO4 (Sigma-Aldrich). Tissues were then washed and dehydrated in 15% sucrose to preserve tissue architecture. Fixed, dehydrated samples were embedded in OCT compound (Tissue-Tek) before freezing, cutting, and mounting. Sections were then blocked and stained with fluorochrome-conjugated and biotinylated antibodies and DAPI. Fluorochrome-conjugated streptavidin secondary was used to detect biotinylated antibodies. Stained tissues were imaged using a DM5500B epifluorescent microscope (Leica).
LP cell isolation.
Small intestines were dissected and cleaned in situ of mesenteric fat and connective tissue. Peyer’s patches were removed with scissors and the entire small intestine was cut into 1 cm pieces for digestion. These pieces were first washed in HBSS before incubation at 37°C for 20 min in 3% RPMI containing 1 mM DTT, and 5 mM EDTA. The supernatant, containing the IEL fraction, was discarded. The remaining LP fraction was then washed twice in RPMI with 2 mM EDTA before digestion with 0.1 mg/ml Liberase TL (Roche) and 0.05 mg/ml DNase (Sigma-Aldrich) for 30 min at 37°C. LP suspensions were passed through a 70-µm filter, washed, and resuspended in 5 ml Histopaque-1077 (Sigma-Aldrich) to enrich for lymphocytes. Samples were overlaid with 2 ml RPMI before room-temperature centrifugation at 2,000 g for 30 min with no break. The interface was collected and cells were washed before staining and analysis by flow cytometry.
Flow cytometry.
Single-cell suspensions from lymph nodes or small intestine LP were stained with fluorochrome-conjugated antibodies to CD3ε, CD4, CD8α, CD44, CCR9, CD11c, CD64, B220, I-A/I-E (MHCII), CD103, and CD11b purchased from BioLegend. Fixable viability dye and antibodies against FoxP3, IFN-γ, and IL-17A were purchased from eBioscience, anti-TCRγδ was obtained from BD, anti-huLangerin was purchased from Dendritics, and anti-GFP was obtained from Rockland. For evaluating cytokine expression, cells were stimulated for 3 h with PMA (50 ng/ml) and ionomycin (1.5 µM; Sigma-Aldrich) in complete DMEM media in the presence of brefeldin A for the final 2.5 h. Intracellular staining for FoxP3, GFP, or cytokines was performed with Cytofix/Cytoperm kit (BD) in accordance with the manufacturer’s instructions. Detection of 2W1S-specific CD4 T cells was performed as previously described (Moon et al., 2007) with an I-Aβ-2W1S tetramer, a gift from M. Jenkins. Stained samples were analyzed on an LSRFortessa flow cytometer (BD), and data were processed using FlowJo software (Tree Star). Cell numbers were calculated from flow cytometry frequencies using hemocytometer counts of Trypan blue–excluding cells.
Flagellin injection and LP mRNA extraction.
1 µg ultrapure flagellin (InvivoGen) in sterile PBS was injected intravenously through the tail vein. 2 h later, a 1.5-cm segment of proximal small intestine 2 cm distal to the pylorus was dissected and opened longitudinally with scissors. Peyer’s patches were removed from this segment before incubation at 37°C for 15 min in HBSS containing 25 mM Hepes, 1 mM DTT, and 10 mM EDTA to remove the epithelium. Segments were then pulse-vortexed eight times and washed in sterile PBS before stabilization in RNAlater (QIAGEN) for mRNA extraction and analysis. To deplete LP DCs, CD11c-DTR mice were injected i.p. with 1 µg diphtheria toxin 18 h before flagellin injection.
DSS colitis.
Animals were treated with 2 or 5% wt/vol DSS (MP Biomedicals) in the drinking water and monitored daily for weight loss, as previously described (Wirtz et al., 2007).
Infections.
C. rodentium strain DBS100 was purchased from the American Type Culture Collection. Bacteria were subcloned and cultured for 12 h overnight in Luria-Bertani media, harvested by centrifugation, and suspended in sterile PBS. Bacterial concentration was estimated by measuring optical density at 600 nm (OD 600) and confirmed by plating serial dilutions. Mice were starved for 12 h before orogastric inoculation with 2 × 109 bacteria in 200 µl PBS by gavage.
For S. enterica infection, attenuated AroA− S. enterica serovar Typhimurium expressing the 2W1S epitope was a gift from M. Jenkins (University of Minnesota, Minneapolis, MN) and K. Smith (University of Washington, Seattle, WA). Either this strain or the naturally streptomycin resistant wild-type S. enterica serovar Typhimurium strain SL1344 were subcloned and cultured as above in the presence of 0.1 mg/ml streptomycin. Bacteria were harvested and animals infected as above with 5 × 107–108 S. enterica. To induce S. enterica-dependent enterocolitis, animals were treated as previously described (Barthel et al., 2003). In brief, mice were pretreated by gavage with 20 mg streptomycin sulfate in 100 µl sterile PBS. 24 hours later, mice were gavaged with 100 µl 5% NaHCO3, pH 9.0, before orogastric inoculation with 5 × 107 S. typhimurium strain SL1344. Live bacterial loads were determined at indicated time points after infection by homogenizing tissues in 0.1% Triton X-100 buffer and plating serial dilutions on MacConkey agar.
Fecal sample collection and bacterial DNA extraction.
Immediately after euthanization, fresh fecal pellets were obtained from the distal colon. Ceca were then opened with sterile scissors and cecal contents were scraped out with forceps. All samples were immediately frozen and stored in sterile microfuge tubes at −80°C. DNA was extracted from samples using the PowerSoil DNA extraction kit (MoBio) in accordance with the manufacturer’s instructions. Approximately 0.3 g of each sample was used for extraction.
16S rDNA amplification and pyrosequencing.
DNA extracts were normalized to a concentration of 2.5 ng µl−1 by dilution in nuclease-free water and used as template for a PCR assay targeting the hypervariable V6 region of the 16S rDNA using the 967F/1046R primer set (Sogin et al., 2006). Each sample was amplified with a reverse primer containing a unique 6-bp nucleotide sequence on the 5′ terminus to allow samples to be pooled for sequencing and separated later (Sogin et al., 2006). Library construction and paired-end sequencing (2 × 100 nt) of amplicons was performed by the BioMedical Genomics Center at the University of Minnesota (Saint Paul, MN) using the Illumina HiSeq platform.
16S sequence analysis and statistics.
Raw sequence reads were pair-end aligned, separated by sample, and trimmed for quality as described previously using mothur software v. 1.27.0 (Schloss et al., 2009). Sequences of abundance <2 over the entire dataset were excluded from analysis. Sequences were aligned to the RDP taxonomic database (Cole et al., 2009), and analysis of OTUs was performed at a sequence cutoff of 0.03. The number of sequence reads associated with each group was subsampled to that of the smallest group (containing 217,483 sequences) for comparisons of β diversity and relative taxonomic abundance. Alpha diversity indices (Shannon, nonparametric Shannon, and Simpson indices) were calculated using mothur. The Bray-Curtis measure of dissimilarity was used to construct distance matrices among sites (Bray and Curtis, 1957). Unweighted UniFrac analyses (Lozupone and Knight, 2005) and analysis of molecular variance (AMOVA; Excoffier et al., 1992) were also calculated using mothur. Diversity indices and relative taxon abundance were compared via two-way analysis of variance (ANOVA) followed by Tukey’s post-hoc test at α = 0.05 using SPSS Statistics software v. 19.0 (IBM).
Quantitative real-time PCR.
RNA was extracted from RNAlater-stabilized tissues using an RNeasy mini kit (QIAGEN) according to the manufacturer’s instructions. RNA was reverse-transcribed using a high-capacity cDNA RT kit and analyzed via qPCR with TaqMan Gene Expression Assays and an ABI 7900HT (Applied Biosystems) as previously described (Haley et al., 2012). Data were normalized to Hprt expression, and relative expression values calculated using the comparative Ct method. Data are presented as 2−ΔΔCt.
Acknowledgments
We thank M. Jenkins and K. Smith for providing Salmonella strains and M. Jenkins for 2W1S tetramer. R. Nelson, B. Chicoine, B. Jarrett, and Z. Zelickson provided technical assistance. We also thank the University of Minnesota Research Animal Resources staff for expert animal care and P. Champoux and T. Martin of the Flow Cytometry Core Facility at the Center for Immunology for assistance with flow cytometry experiments. The University of Minnesota Supercomputing Institute provided resources for microbiome sequence analysis.
This work was supported by grants from the National Institutes of Health (NIH; AR056632 and AR060744; D.H. Kaplan), a Dermatology Foundation research award (B.Z. Igyártó), and an American Skin Association research award (B.Z. Igyártó). D.H. Kaplan is also supported by the Dr. Al Zelickson Family endowed professorship. N.E. Welty was supported by the Warren and Henrietta Warwick fellowship, the University of Minnesota NIH MSTP grant T32 GM008244, and an NIH F30 fellowship (DK097856).
N. Ghilardi is a full-time employee of Genentech, a member of the Roche group. The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- DSS
- dextran sodium sulfate
- huLangerin
- human Langerin
- LC
- Langerhans cell
- LP
- lamina propria
- MLN
- mesenteric LN
- RA
- retinoic acid
- TLR5
- Toll-like receptor 5
- T reg cell
- regulatory T cell
References
- Atarashi K., Nishimura J., Shima T., Umesaki Y., Yamamoto M., Onoue M., Yagita H., Ishii N., Evans R., Honda K., Takeda K. 2008. ATP drives lamina propria T(H)17 cell differentiation. Nature. 455:808–812 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- Barthel M., Hapfelmeier S., Quintanilla-Martínez L., Kremer M., Rohde M., Hogardt M., Pfeffer K., Rüssmann H., Hardt W.-D. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839–2858 10.1128/IAI.71.5.2839-2858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R., O’Quinn D.B., Silberger D.J., Schoeb T.R., Fouser L., Ouyang W., Hatton R.D., Weaver C.T. 2012. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 37:1061–1075 10.1016/j.immuni.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobr A., Olvera-Gomez I., Igyarto B.Z., Haley K.M., Hogquist K.A., Kaplan D.H. 2010. Acute ablation of Langerhans cells enhances skin immune responses. J. Immunol. 185:4724–4728 10.4049/jimmunol.1001802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray J.R., Curtis J.T. 1957. An ordination of the upland forest communities of southern wisconsin. Ecol. Monogr. 27:325–349 10.2307/1942268 [DOI] [Google Scholar]
- Bursch L.S., Wang L., Igyarto B., Kissenpfennig A., Malissen B., Kaplan D.H., Hogquist K.A. 2007. Identification of a novel population of Langerin+ dendritic cells. J. Exp. Med. 204:3147–3156 10.1084/jem.20071966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerovic V., Houston S.A., Scott C.L., Aumeunier A., Yrlid U., Mowat A.M., Milling S.W.F. 2013. Intestinal CD103(-) dendritic cells migrate in lymph and prime effector T cells. Mucosal Immunol. 6:104–113 10.1038/mi.2012.53 [DOI] [PubMed] [Google Scholar]
- Chang S.-Y., Kweon M.-N. 2010. Langerin-expressing dendritic cells in gut-associated lymphoid tissues. Immunol. Rev. 234:233–246 10.1111/j.0105-2896.2009.00878.x [DOI] [PubMed] [Google Scholar]
- Chikwava K., Jaffe R. 2004. Langerin (CD207) staining in normal pediatric tissues, reactive lymph nodes, and childhood histiocytic disorders. Pediatr. Dev. Pathol. 7:607–614 10.1007/s10024-004-3027-z [DOI] [PubMed] [Google Scholar]
- Chow A., Brown B.D., Merad M. 2011. Studying the mononuclear phagocyte system in the molecular age. Nat. Rev. Immunol. 11:788–798 10.1038/nri3087 [DOI] [PubMed] [Google Scholar]
- Chung H., Pamp S.J., Hill J.A., Surana N.K., Edelman S.M., Troy E.B., Reading N.C., Villablanca E.J., Wang S., Mora J.R., et al. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 149:1578–1593 10.1016/j.cell.2012.04.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam-Syed-Mohideen A.S., McGarrell D.M., Marsh T., Garrity G.M., Tiedje J.M. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Powrie F. 2008. Dendritic cells in intestinal immune regulation. Nat. Rev. Immunol. 8:435–446 10.1038/nri2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R.R., Arancibia-Cárcamo C.V., Hall J., Sun C.-M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid–dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning T.L., Wang Y.-C., Patel S.R., Williams I.R., Pulendran B. 2007. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8:1086–1094 10.1038/ni1511 [DOI] [PubMed] [Google Scholar]
- Denning T.L., Norris B.A., Medina-Contreras O., Manicassamy S., Geem D., Madan R., Karp C.L., Pulendran B. 2011. Functional specializations of intestinal dendritic cell and macrophage subsets that control Th17 and regulatory T cell responses are dependent on the T cell/APC ratio, source of mouse strain, and regional localization. J. Immunol. 187:733–747 10.4049/jimmunol.1002701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl G.E., Longman R.S., Zhang J.-X., Breart B., Galan C., Cuesta A., Schwab S.R., Littman D.R. 2013. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature. 494:116–120 10.1038/nature11809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresch C., Leverrier Y., Marvel J., Shortman K. 2012. Development of antigen cross-presentation capacity in dendritic cells. Trends Immunol. 33:381–388 10.1016/j.it.2012.04.009 [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207:823–836 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., Flavell R.A. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 131:479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasan S., Muramatsu M., Suzuki K., Nagaoka H., Hiai H., Honjo T. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 298:1424–1427 10.1126/science.1077336 [DOI] [PubMed] [Google Scholar]
- Fujimoto K., Karuppuchamy T., Takemura N., Shimohigoshi M., Machida T., Haseda Y., Aoshi T., Ishii K.J., Akira S., Uematsu S. 2011. A new subset of CD103+CD8alpha+ dendritic cells in the small intestine expresses TLR3, TLR7, and TLR9 and induces Th1 response and CTL activity. J. Immunol. 186:6287–6295 10.4049/jimmunol.1004036 [DOI] [PubMed] [Google Scholar]
- Garrett W.S., Lord G.M., Punit S., Lugo-Villarino G., Mazmanian S.K., Ito S., Glickman J.N., Glimcher L.H. 2007. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 131:33–45 10.1016/j.cell.2007.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghilardi N., Kljavin N., Chen Q., Lucas S., Gurney A.L., De Sauvage F.J. 2004. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J. Immunol. 172:2827–2833 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Liu K., Helft J., Bogunovic M., Greter M., Hashimoto D., Price J., Yin N., Bromberg J., Lira S.A., et al. 2009. The origin and development of nonlymphoid tissue CD103+ DCs. J. Exp. Med. 206:3115–3130 10.1084/jem.20091756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Crozat K., Henri S., Tamoutounour S., Grenot P., Devilard E., de Bovis B., Alexopoulou L., Dalod M., Malissen B. 2010. Skin-draining lymph nodes contain dermis-derived CD103(-) dendritic cells that constitutively produce retinoic acid and induce Foxp3(+) regulatory T cells. Blood. 115:1958–1968 10.1182/blood-2009-09-245274 [DOI] [PubMed] [Google Scholar]
- Hadis U., Wahl B., Schulz O., Hardtke-Wolenski M., Schippers A., Wagner N., Müller W., Sparwasser T., Förster R., Pabst O. 2011. Intestinal tolerance requires gut homing and expansion of FoxP3+ regulatory T cells in the lamina propria. Immunity. 34:237–246 10.1016/j.immuni.2011.01.016 [DOI] [PubMed] [Google Scholar]
- Haley K., Igyártó B.Z., Ortner D., Bobr A., Kashem S., Schenten D., Kaplan D.H. 2012. Langerhans cells require MyD88-dependent signals for Candida albicans response but not for contact hypersensitivity or migration. J. Immunol. 188:4334–4339 10.4049/jimmunol.1102759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniffa M., Shin A., Bigley V., McGovern N., Teo P., See P., Wasan P.S., Wang X.-N., Malinarich F., Malleret B., et al. 2012. Human tissues contain CD141hi cross-presenting dendritic cells with functional homology to mouse CD103+ nonlymphoid dendritic cells. Immunity. 37:60–73 10.1016/j.immuni.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Littman D.R. 2012. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30:759–795 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Troutman T.D., Edukulla R., Pasare C. 2011. Priming microenvironments dictate cytokine requirements for T helper 17 cell lineage commitment. Immunity. 35:1010–1022 10.1016/j.immuni.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igyarto B.Z., Jenison M.C., Dudda J.C., Roers A., Müller W., Koni P.A., Campbell D.J., Shlomchik M.J., Kaplan D.H. 2009. Langerhans cells suppress contact hypersensitivity responses via cognate CD4 interaction and Langerhans cell-derived IL-10. J. Immunol. 183:5085–5093 10.4049/jimmunol.0901884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Frutos Rde.L., Manel N., Yoshinaga K., Rifkin D.B., Sartor R.B., Finlay B.B., Littman D.R. 2008. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 4:337–349 10.1016/j.chom.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A. 2007. Mucosal dendritic cells. Annu. Rev. Immunol. 25:381–418 10.1146/annurev.immunol.25.022106.141634 [DOI] [PubMed] [Google Scholar]
- Jaensson E., Uronen-Hansson H., Pabst O., Eksteen B., Tian J., Coombes J.L., Berg P.-L., Davidsson T., Powrie F., Johansson-Lindbom B., Agace W.W. 2008. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 205:2139–2149 10.1084/jem.20080414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D.H., Jenison M.C., Saeland S., Shlomchik W.D., Shlomchik M.J. 2005. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 23:611–620 10.1016/j.immuni.2005.10.008 [DOI] [PubMed] [Google Scholar]
- Kaplan D.H., Li M.O., Jenison M.C., Shlomchik W.D., Flavell R.A., Shlomchik M.J. 2007. Autocrine/paracrine TGFβ1 is required for the development of epidermal Langerhans cells. J. Exp. Med. 204:2545–2552 10.1084/jem.20071401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser A., Ludwiczek O., Holzmann S., Moschen A.R., Weiss G., Enrich B., Graziadei I., Dunzendorfer S., Wiedermann C.J., Mürzl E., et al. 2004. Increased expression of CCL20 in human inflammatory bowel disease. J. Clin. Immunol. 24:74–85 10.1023/B:JOCI.0000018066.46279.6b [DOI] [PubMed] [Google Scholar]
- Khan M.A., Bouzari S., Ma C., Rosenberger C.M., Bergstrom K.S.B., Gibson D.L., Steiner T.S., Vallance B.A. 2008. Flagellin-dependent and -independent inflammatory responses following infection by enteropathogenic Escherichia coli and Citrobacter rodentium. Infect. Immun. 76:1410–1422 10.1128/IAI.01141-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew M.A., Ubeda C., Zenewicz L.A., Smith N., Flavell R.A., Pamer E.G. 2010. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J. Infect. Dis. 201:534–543 10.1086/650203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnebrew M.A., Buffie C.G., Diehl G.E., Zenewicz L.A., Leiner I., Hohl T.M., Flavell R.A., Littman D.R., Pamer E.G. 2012. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 36:276–287 10.1016/j.immuni.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C.H., Douillard P., Leserman L., Kaiserlian D., et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654 10.1016/j.immuni.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., Kuchroo V.K. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L., et al. 2011. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 35:780–791 10.1016/j.immuni.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochner M., Bérard M., Sawa S., Hauer S., Gaboriau-Routhiau V., Fernandez T.D., Snel J., Bousso P., Cerf-Bensussan N., Eberl G. 2011. Restricted microbiota and absence of cognate TCR antigen leads to an unbalanced generation of Th17 cells. J. Immunol. 186:1531–1537 10.4049/jimmunol.1001723 [DOI] [PubMed] [Google Scholar]
- Lozupone C., Knight R. 2005. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., Weaver C.T. 2006. Transforming growth factor-β induces development of the T(H)17 lineage. Nature. 441:231–234 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Miller J.C., Brown B.D., Shay T., Gautier E.L., Jojic V., Cohain A., Pandey G., Leboeuf M., Elpek K.G., Helft J., et al. ; Immunological Genome Consortium 2012. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 13:888–899 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J.J., Chu H.H., Pepper M., McSorley S.J., Jameson S.C., Kedl R.M., Jenkins M.K. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity. 27:203–213 10.1016/j.immuni.2007.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R.W., McLachlan J.B., Kurtz J.R., Jenkins M.K. 2013. CD4+ T cell persistence and function after infection are maintained by low-level peptide:MHC class II presentation. J. Immunol. 190:2828–2834 10.4049/jimmunol.1202183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona-Wright G., Mohrs K., Mohrs M. 2010. Sustained signaling by canonical helper T cell cytokines throughout the reactive lymph node. Nat. Immunol. 11:520–526 10.1038/ni.1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., Agace W.W. 2013. IRF4 transcription-factor-dependent CD103(+)CD11b(+) dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 38:958–969 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W.S., See P., Shin A., Wasan P.S., et al. 2013. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 38:970–983 10.1016/j.immuni.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz O., Jaensson E., Persson E.K., Liu X., Worbs T., Agace W.W., Pabst O. 2009. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206:3101–3114 10.1084/jem.20091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semmrich M., Plantinga M., Svensson-Frej M., Uronen-Hansson H., Gustafsson T., Mowat A.M., Yrlid U., Lambrecht B.N., Agace W.W. 2011. Directed antigen targeting in vivo identifies a role for CD103+ dendritic cells in both tolerogenic and immunogenic T-cell responses. Mucosal Immunol. 5:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw M.H., Kamada N., Kim Y.G., Núñez G. 2012. Microbiota-induced IL-1β, but not IL-6, is critical for the development of steady-state TH17 cells in the intestine. J. Exp. Med. 209:251–258 10.1084/jem.20111703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M.L., Morrison H.G., Huber J.A., Mark Welch D., Huse S.M., Neal P.R., Arrieta J.M., Herndl G.J. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc. Natl. Acad. Sci. USA. 103:12115–12120 10.1073/pnas.0605127103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.-M., Hall J.A., Blank R.B., Bouladoux N., Oukka M., Mora J.R., Belkaid Y. 2007. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J. Exp. Med. 204:1775–1785 10.1084/jem.20070602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C., Lipuma L., Gobourne A., Viale A., Leiner I., Equinda M., Khanin R., Pamer E.G. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209:1445–1456 10.1084/jem.20120504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uematsu S., Fujimoto K., Jang M.H., Yang B.-G., Jung Y.-J., Nishiyama M., Sato S., Tsujimura T., Yamamoto M., Yokota Y., et al. 2008. Regulation of humoral and cellular gut immunity by lamina propria dendritic cells expressing Toll-like receptor 5. Nat. Immunol. 9:769–776 10.1038/ni.1622 [DOI] [PubMed] [Google Scholar]
- Van Maele L., Carnoy C., Cayet D., Songhet P., Dumoutier L., Ferrero I., Janot L., Erard F., Bertout J., Leger H., et al. 2010. TLR5 signaling stimulates the innate production of IL-17 and IL-22 by CD3(neg)CD127+ immune cells in spleen and mucosa. J. Immunol. 185:1177–1185 10.4049/jimmunol.1000115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.-D., Shakhar G., Jung S. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Wirtz S., Neufert C., Weigmann B., Neurath M.F. 2007. Chemically induced mouse models of intestinal inflammation. Nat. Protoc. 2:541–546 10.1038/nprot.2007.41 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Valdez P.A., Danilenko D.M., Hu Y., Sa S.M., Gong Q., Abbas A.R., Modrusan Z., Ghilardi N., de Sauvage F.J., Ouyang W. 2008. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14:282–289 10.1038/nm1720 [DOI] [PubMed] [Google Scholar]