VEGF dampens the expression of microRNA-1, which drives inflammation in part via increasing the expression of Mpl.
Abstract
Asthma, the prototypic Th2-mediated inflammatory disorder of the lung, is an emergent disease worldwide. Vascular endothelial growth factor (VEGF) is a critical regulator of pulmonary Th2 inflammation, but the underlying mechanism and the roles of microRNAs (miRNAs) in this process have not been defined. Here we show that lung-specific overexpression of VEGF decreases miR-1 expression in the lung, most prominently in the endothelium, and a similar down-regulation occurs in lung endothelium in Th2 inflammation models. Intranasal delivery of miR-1 inhibited inflammatory responses to ovalbumin, house dust mite, and IL-13 overexpression. Blocking VEGF inhibited Th2-mediated lung inflammation, and this was restored by antagonizing miR-1. Using mRNA arrays, Argonaute pull-down assays, luciferase expression assays, and mutational analysis, we identified Mpl as a direct target of miR-1 and showed that VEGF controls the expression of endothelial Mpl during Th2 inflammation via the regulation of miR-1. In vivo knockdown of Mpl inhibited Th2 inflammation and indirectly inhibited the expression of P-selectin in lung endothelium. These experiments define a novel VEGF–miR-1–Mpl–P-selectin effector pathway in lung Th2 inflammation and herald the utility of miR-1 and Mpl as potential therapeutic targets for asthma.
Asthma, the prototypic type 2 inflammatory disorder of the lung, is an emergent disease worldwide (Asher et al., 2006; Kim et al., 2010). Several studies have demonstrated that vascular endothelial growth factor (VEGF) and endothelial signaling play critical roles in the lung Th2 inflammation (Hoshino et al., 2001a,b; Lee et al., 2004; Simcock et al., 2007; Tuder and Yun, 2008; Asosingh and Erzurum, 2009). Most prominent were studies demonstrating that transgenic expression of VEGF in the lung leads to eosinophil-rich inflammation, mucus metaplasia, and airway remodeling and that selective VEGF receptor 2 (VEGFR2) blockade abrogates aeroallergen-induced pulmonary inflammation (Lee et al., 2004). However, the mechanisms of VEGF contribution to this inflammatory cascade are poorly understood (Voelkel et al., 2006; Tuder and Yun, 2008).
MicroRNAs (miRNAs) regulate gene expression by base pairing with conserved sites within the 3′ untranslated region (UTR) of their target mRNAs (Bartel, 2009). Each miRNA can potentially bind to hundreds of mRNAs in the cell and thus regulate a multitude of functional and structural changes in a cell- and organ-specific manner (Giraldez et al., 2005; Chen and Rajewsky, 2006; Sokol et al., 2008). Although the mechanisms of translational regulation by miRNAs have not been fully defined, it is known that miRNAs function through formation of a ribonucleoprotein complex called the miRNA-induced silencing complex (mi-RISC). mi-RISC contains a guide miRNA bound to one of four mammalian Argonaute proteins and is one of the effector arms of the RNA interference pathway causing mRNA deadenylation and decay or inhibition of translation (Jackson and Standart, 2007). Of these, Argonaute 2 (Ago2) is the most abundantly expressed Argonaute in mammalian tissues (Wang et al., 2012) and is the only mammalian Argonaute with endonucleolytic activity (Liu et al., 2004). Recent studies have demonstrated that miRNAs play important roles in a wide variety of biological processes (Giraldez et al., 2005; Chen and Rajewsky, 2006; Sokol et al., 2008; Fish et al., 2008; Wang et al., 2008; Suárez et al., 2008; Bonauer et al., 2009; Nicoli et al., 2010). However, the roles of miRNA in Th2 inflammation and the inflammatory effects of VEGF have not been investigated.
We hypothesized that the effects of VEGF in Th2 inflammation are mediated by its ability to alter endothelial miRNA and their downstream targets. To test this hypothesis, we evaluated the miRNA and miRNA target alterations in VEGF transgenic mice and models of aeroallergen-induced Th2 inflammation. We found that lung-targeted VEGF down-regulated miR-1, most prominently in the endothelium, and that a similar down-regulation takes place in lung-targeted IL-13 transgenic and aeroallergen-sensitized and challenged mice. We next demonstrated that intranasal delivery of miR-1 decreased Th2- and IL-13–stimulated inflammation and that antagonizing miR-1 rescued Th2 immunity in the context of VEGFR2 blockade. We also found a direct target of miR-1, Mpl, and defined the critical role of Mpl in Th2 inflammation by silencing this gene in Th2-inflamed lungs. Lastly, we showed that Mpl, similar to its role in platelets, regulates the expression of P-selectin in the lung endothelium. Overall, these experiments define a novel VEGF-driven, miR-1–Mpl–P-selectin effector pathway in lung Th2 inflammation.
RESULTS
VEGF down-regulates lung endothelial miR-1 in Th2 inflammation
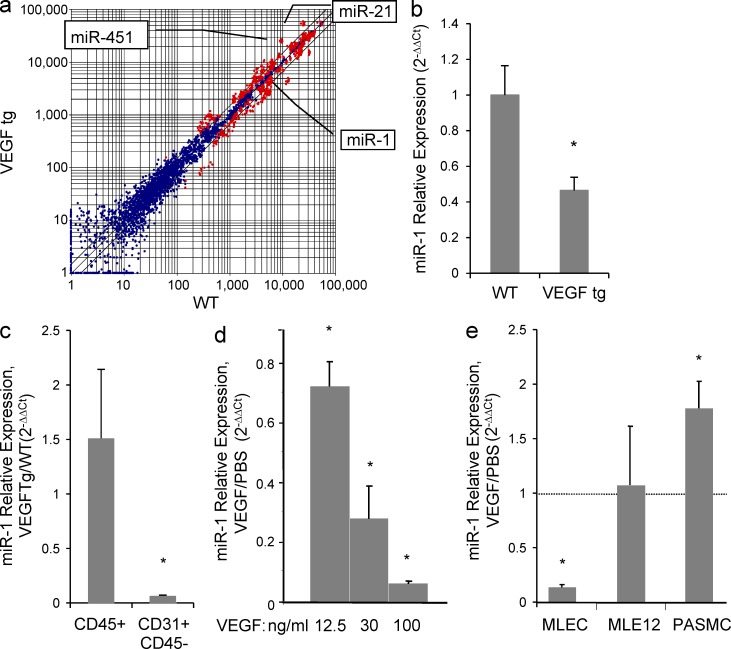
To define the relationships between VEGF and miRNA, we evaluated the expression of miRNAs in VEGF transgenic mice and models of Th2 inflammation. Microarray analysis and TaqMan stem loop quantitative RT-PCR (qRT-PCR) showed that in the lungs of CC10-rtTA-VEGF165 transgenic mice, the levels of miR-1 were decreased by ∼50–60% (Fig. 1, a and b). A comparison of miR-1 levels in isolated endothelial (CD31+, CD45−) and hematopoietic (CD45+) cells from these lungs showed that, in contrast to hematopoietic cells, the levels of miR-1 in endothelial cells were diminished by >90% (Fig. 1 c). In accord with these findings, recombinant VEGF decreased the levels of miR-1 in primary mouse lung endothelial cells (MLECs) cultured in vitro in a concentration-dependent manner (Fig. 1 d). This down-regulation was at least partially MLEC specific because VEGF did not decrease the levels of miR-1 in mouse pulmonary artery smooth muscle cells (PASMCs) or a lung epithelial cell line (MLE12; Fig. 1 e).
Figure 1.
Effect of VEGF on lung miRNAs. (a) VEGF was overexpressed in the lung epithelium of CC10-rtTA-VEGF transgenic mice. RNA from the lungs of these mice (VEGFtg) and their WT litters was analyzed by microarray. miRNAs that were changed significantly are indicated (n = 3 in each group; P < 0.01). Blue indicates P > 0.01, and red indicates P < 0.01. (b) Relative expression of miR-1 in these lungs was measured by TaqMan qRT-PCR (values normalized to WT; two experiments; n = 5 in each group; *, P < 0.04). (c) Relative expression of miR-1 was measured by TaqMan qRT-PCR in lung endothelial (CD31+, CD45−) or hematopoietic (CD45+) cells isolated from these lungs (values normalized to WT; three experiments; n = 3–4 per group; *, P < 0.05). (d) Recombinant VEGF was added to MLECs in culture, and relative expression of miR-1 was measured by TaqMan qRT-PCR (values normalized to PBS-treated group; two experiments; n = 4 in each group; *, P < 0.05). (e) Recombinant VEGF was added to MLECs, lung epithelial cells (MLE12), and PASMCs, and relative expression of miR-1 was measured as in a (values normalized to the corresponding PBS-treated group; three experiments; n = 4 in each group; *, P < 0.05). All error bars represent mean ± SEM.
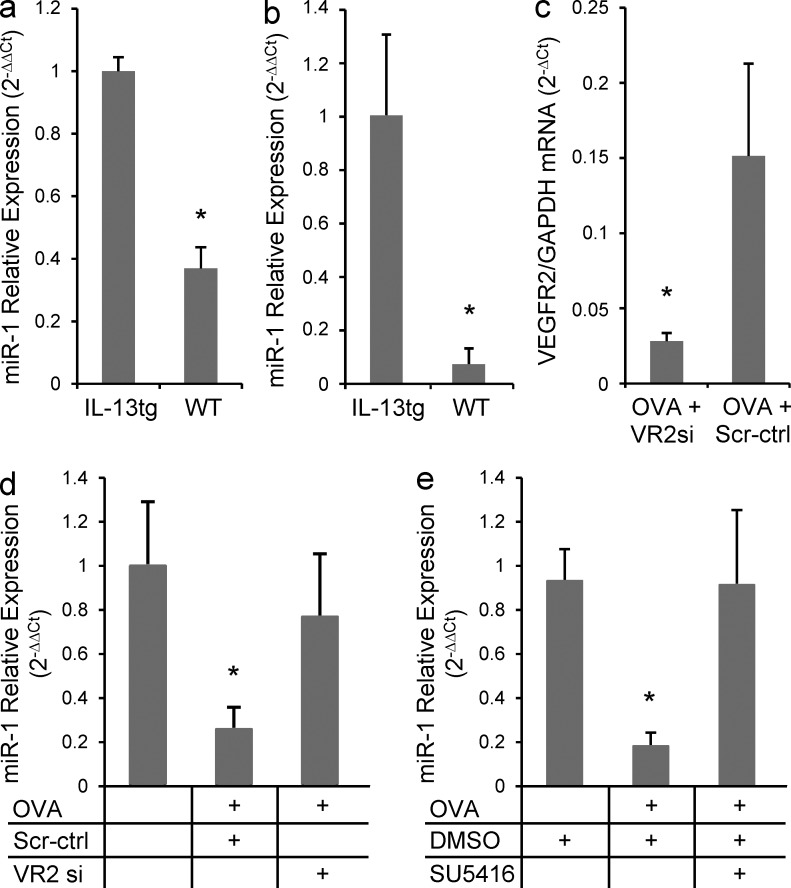
To test the Th2 relevance of this down-regulation, we next analyzed the expression of miR-1 in two well established models of lung Th2 inflammation. Similar to VEGF transgenic mice, overexpression of IL-13 in the lung epithelium inhibited expression of miR-1 (Fig. 2 a), and this inhibition was more prominent in endothelial cells (Fig. 2 b). To investigate the consistency of this down-regulation in aeroallergen-induced Th2 inflammation and show the role of VEGF, we measured the level of miR-1 in lung endothelium of OVA-sensitized and -challenged mice treated with intranasal or systemic VEGFR2 blockers. We first showed that intranasal delivery of VEGFR2 siRNA decreased expression of VEGFR2 in the lung endothelium by >70% (Fig. 2 c). As shown in Fig. 2 (d and e), OVA sensitization and challenge reduced the expression of endothelial miR-1 by >70% and VEGFR2 blockade in the lung (by intranasal delivery; Fig. 2 d) or systemically (i.p. delivery of SU5416; Fig. 2 e) abrogated this down-regulation. These experiments demonstrate that airway Th2 inflammation down-regulates miR-1 via a VEGF-dependent mechanism.
Figure 2.
The effect of lung Th2 inflammation of VEGFR2 siRNA on miR-1 expression. (a) IL-13 expression was induced in the lung epithelium of CC10-rtTA-Il-13 transgenic mice for 7 d, and miR-1 expression was measured by TaqMan qRT-PCR in the whole lung RNA from these mice and their WT litters (values normalized to WT; three experiments; n = 3 or more in each group; *, P < 0.0001). (b) Relative expression of miR-1 was measured in endothelial cells isolated from these lungs (two experiments; n = 3 or 4 mice in each group; *, P < 0.04). (c) C57BL/6 mice were sensitized with OVA and challenged for 3 d, 3 wk after the sensitization. Intranasal VEGFR2 siRNA (VR2si) or scrambled control (scr-ctrl) was delivered every day before the OVA challenge. Lung endothelial cells were isolated after the last challenge, and the level of VEGFR2 mRNA was measured by real-time qRT-PCR (two experiments; n = 4 or more in each group; *, P < 0.02). (d) Relative expression of miR-1 was measured by TaqMan qRT-PCR in the endothelial cells isolated from mice treated as described in c (values are normalized to PBS-challenged control group; two experiments; n = 4 or more in each group; VR2si group compared with both OVA and scr-ctrl groups: *, P < 0.05). (e) C57BL/6 mice were sensitized and challenged with OVA as described in c and received i.p. SU5416 or vehicle (DMSO) the day before the first challenge and every day afterward. Expression of miR-1 was measured by TaqMan qRT-PCR in lung endothelial cells (values normalized to vehicle control; two experiments; n = 5 in each group; SU5416 compared with both other groups: *, P < 0.05). Relative expression of miR-1 is measured in all experiments by comparing it with the internal control (sno202) and normalizing to a calibrator (WT or PBS treatment). Error bars represent mean ± SEM.
miR-1 controls Th2 inflammation in the lung
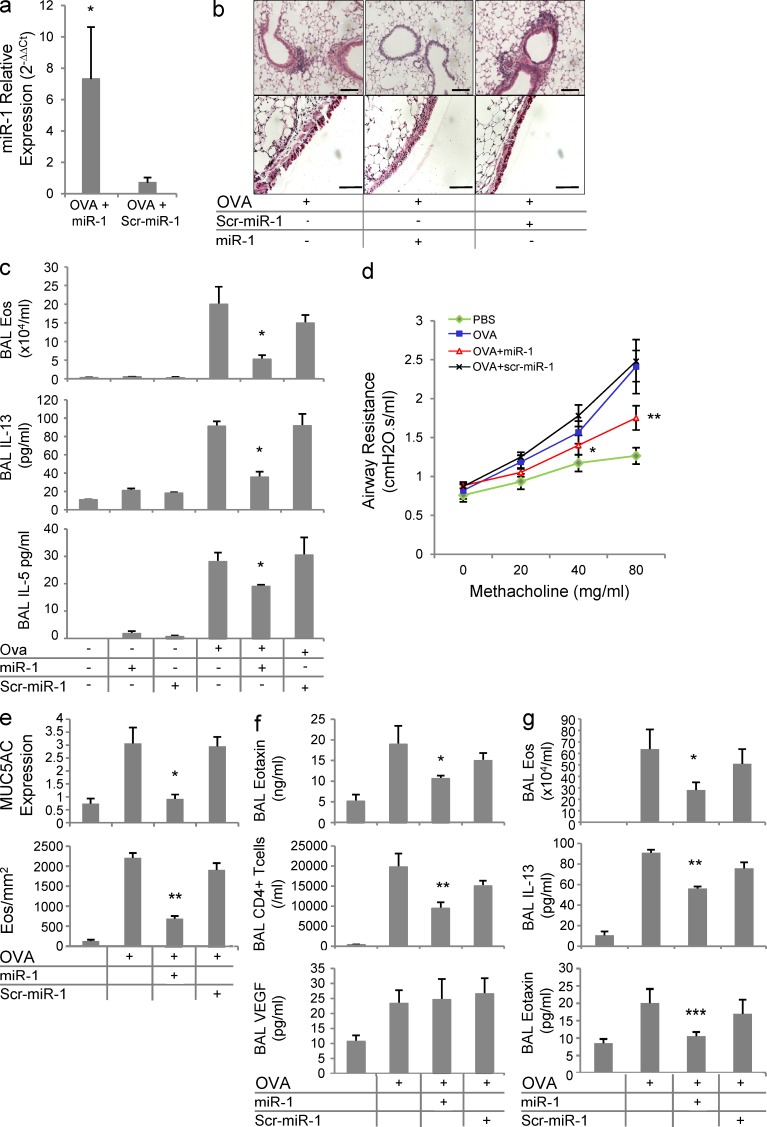
To further define the roles of miR-1 in Th2 inflammation, we evaluated the effects of miR-1 delivery in the OVA model. A double-stranded miR-1 mimic or its negative control (scr–miR-1) were delivered intranasally to mice at the time of OVA challenge. Using this method, miR-1 levels increased by >10-fold in the lung endothelium, confirming the efficiency of delivery (Fig. 3 a). In these experiments, mice that received the vehicle or scrambled miR-1 control manifested significant increases in tissue inflammation, eosinophilia, and type 2 cytokine accumulation in the bronchoalveolar lavage fluid (BAL) and airway hyperresponsiveness (AHR) in response to methacholine challenge (Fig. 3, a–d). However, miR-1 delivery at the time of OVA challenge ameliorated each of these Th2 hallmark responses (Fig. 3, a–d). These changes were accompanied by a significant decrease in mucus (MUC5) hypersecretion and lung tissue eosinophilia, further demonstrating that miR-1 inhibitory effects are not limited to BAL (Fig. 3 e). In agreement with the above experiments, the secretion of eotaxin and the accumulation of pulmonary CD4+ T cells were also significantly inhibited, whereas the level of VEGF in the BAL remained unchanged (Fig. 3 f), suggesting that the delivered miR-1 is not directly targeting VEGF. Levels of serum OVA-specific IgE also did not differ among different OVA-challenged groups (not depicted), suggesting that, as expected, miR-1 delivery at the time of the antigenic challenge does not affect T cell sensitization. In accord with these observations, miR-1 delivery also ameliorated ongoing Th2 inflammation. As shown in Fig. 3 g, the effectiveness of intranasal miR-1 was not limited to its prophylactic administration and intranasal delivery even 24 h after the first antigenic challenge inhibited the BAL eosinophilia, IL-13, and eotaxin secretion.
Figure 3.
The role of miR-1 in the lung Th2 inflammation. (a) C57BL/6 mice were sensitized and challenged with OVA as described in Fig. 2 c. Intranasal miR-1 or scrambled miR-1 (scr–miR-1) was delivered the day before the first challenge and every day afterward. The level of miR-1 was measured by TaqMan qRT-PCR in lung endothelial cells (normalized to the control group; n = 4 in each group; *, P < 0.05). (b) Lungs from these mice were stained with H&E (top) to show the level of peribronchial inflammation and PAS (bottom) to show the extent of mucus metaplasia (dark red) in the large airways. Bars, 50 µm. (c) BALs were collected from these mice, and cytospin preps were stained with H&E to determine the number of eosinophils (top). Concentrations of IL-13 (middle) and IL-5 (bottom panel) were measured by ELISA (n = 5 in each group; three experiments; miR-1 + OVA groups were compared with the other OVA-challenged groups by one way ANOVA: *, P < 0.01 for eosinophils and IL-13; and *, P < 0.05 for IL-5). (d) Airway responses were measured by forced oscillation technique. Mean airway resistance (R, cmH2O.s/ml) was measured after exposing ventilated mice to increasing concentrations of methacholine (values are derived from two experiments with 5 or more mice per group; total of 11–13 mice per group; miR-1 group was compared with OVA and OVA + scr–miR-1 groups: *, P = 0.05; **, P = 0.03). (e) MUC5AC (top) was measured in the BAL of these mice by slot blot hybridization and normalized to control (PBS challenged) group. Lung eosinophils (bottom) were quantified by staining the lung sections with Congo red and counting the eosinophils in the peribronchial areas (three experiments; n = 5 mice per group; miR-1 group compared with both OVA and OVA + scr–miR-1 groups: *, P < 0.002; **, P < 0.0002). (f) In the BAL of these mice, eotaxin (top) and VEGF (bottom) were measured by ELISA, and CD4+ cells (middle) were quantified by FACS analysis (three experiments; n = 5 mice per group; miR-1–treated group was compared with both OVA and OVA + scr–miR-1 groups: *, P < 0.05; **, P < 0.025). (g) C57BL/6 mice were sensitized and challenged with OVA as described in Fig. 2 c, and intranasal miR-1 or scr–miR-1 was delivered after each challenge. The number of eosinophils (top) and levels of IL-13 and eotaxin (middle and bottom) were measured in the BAL as described in c (three experiments; n = 4 or more per group; miR-1 group compared with OVA and OVA + scr–miR-1 groups: *, P = 0.029; **, P = 0.033; ***, P = 0.0127). Error bars represent mean ± SEM.
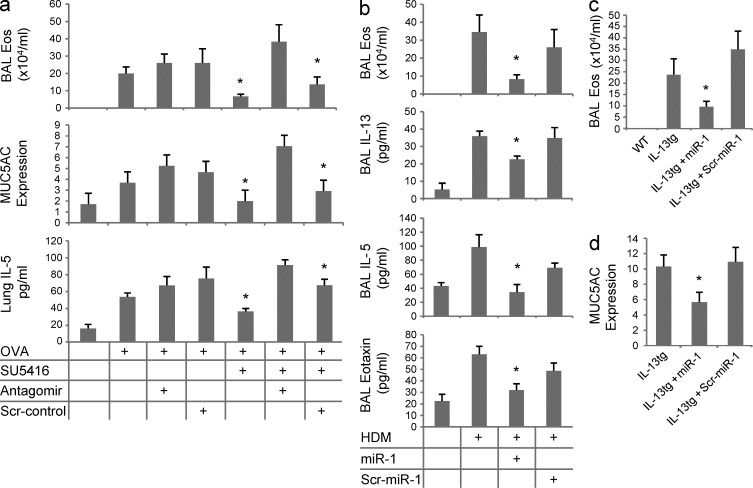
To further investigate the roles of miR-1 in Th2 inflammation, we also characterized the effects of miR-1 blockade. As can be seen in Fig. 4 a, the intranasal administration of an miR-1 antagomir reversed the inhibitory effects of VEGFR blockade and rescued OVA-induced eosinophilic inflammation, mucus metaplasia, and IL-5 secretion. Collectively, these observations demonstrate that miR-1 down-regulation plays a critical role in many facets of pulmonary Th2 inflammation.
Figure 4.
miR-1 is a critical component of Th2 inflammation. (a) C57BL/6 mice were sensitized and challenged with OVA as described in Fig. 2 c. Intranasal miR-1 antagomir or scrambled control (scr-control) and SU5416 or vehicle (DMSO) were delivered the day before the first challenge and every day afterward. BALs were collected, and eosinophils (*, P < 0.05), MUC5AC (*, P < 0.0001), and lung IL-5 (*, P < 0.002) were measured as described in Fig. 3 (two experiments; n = 5 in each group; OVA-challenged groups were compared using one-way ANOVA, and the p-values were corrected for multiple comparisons). (b) C57BL/6 mice were sensitized with HDM and challenged intranasally for 3 d in the third week after sensitization. Intranasal miR-1 or scr–miR-1 was delivered the day before the first challenge and every day afterward. BALs were collected, and eosinophils (*, P < 0.05), IL-13 (*, P < 0.05), IL-5 (*, P < 0.015), and eotaxin (*, P < 0.0015) were measured as described in Fig. 3 (graphs represent data from three experiments with n = 5 mice per group; miR-1 groups were compared with both scr–miR-1 and vehicle groups). (c and d) IL-13 expression was induced in the lung epithelium of IL-13 transgenic mice (as described in Fig. 2 a). Intranasal miR-1 was administered the day before starting doxycycline and every other day afterward. After 7 d, BALs were collected, and the number of eosinophils (*, P < 0.04) and MUC5AC relative expression (*, P < 0.01) were measured as described in Fig. 3 (relative expression of MUC5AC was calculated by normalizing MUC5AC levels in each transgenic group to WT litters; three experiments; n = 3 or more in each group; miR-1 group was compared with the other two transgenic groups). Error bars represent mean ± SEM.
miR-1 regulation in other models of type 2 inflammation
To determine whether the effects of miR-1 were limited to the OVA model or dependent on antigenic sensitization, we tested the effects of miR-1 in house dust mite (HDM) and lung IL-13 transgenic models, respectively. In keeping with our findings in the OVA model, intranasal delivery of miR-1 to HDM-sensitized and -challenged mice at the time of antigen challenge significantly decreased eosinophilic inflammation and type 2 cytokine and eotaxin elaboration (Fig. 4 b). As previously reported (Zhu et al., 1999), overexpression of IL-13 in the lung epithelium induces features of Th2 inflammation such as eosinophilic inflammation and mucus metaplasia without any prior antigenic sensitization or challenge. As shown in Fig. 4 (c and d), intranasal miR-1 inhibited both of these antigen-independent type 2 responses. These experiments clearly demonstrate that the regulatory effects of miR-1 are not OVA specific and can be seen in other aeroallergen-induced and antigen-independent inflammatory responses.
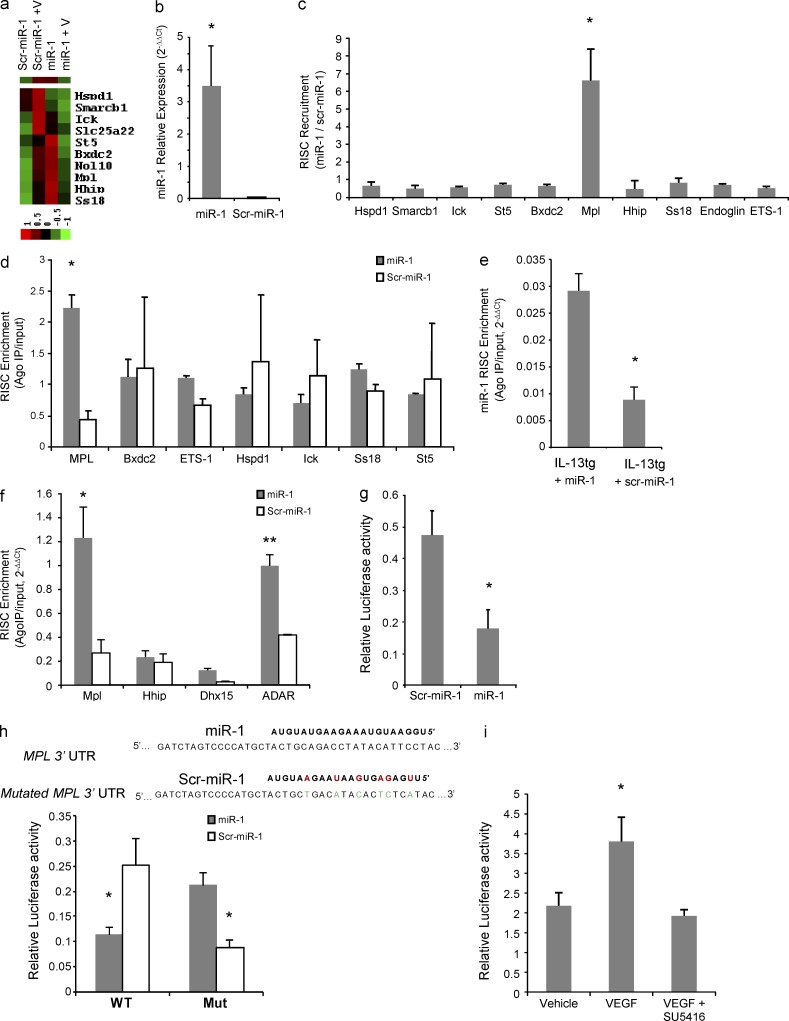
Identification of miR-1 targets by expression profiling and RISC analysis
Because of the incomplete base pairing between miRNAs and their targets, identification of genuine miRNA targets is often fraught with difficulties and most of the available prediction algorithms produce an inordinate number of potential targets for each miRNA. Appreciating these limitations, we chose a two-step strategy to identify a more limited and specific number of candidate genes with a higher possibility of genuine targeting. Because miRNAs often lower the levels of their target mRNAs (Lim et al., 2005; Ziegelbauer et al., 2009), we first used an array-based expression profiling of lung endothelial cells to identify potential targets of miR-1. We screened for genes that were up-regulated in response to VEGF and whose up-regulation was inhibited by miR-1 (i.e., in the miR-1–transfected cells). Using three available prediction algorithms (Targetscan, miRANDA, and Pictar), we then produced a comprehensive list of all possible miR-1 targets. A comparison between the genes that passed both of the aforementioned steps (i.e., were differentially regulated by VEGF in the presence of miR-1 and were predicted to be a target of miR-1 by a prediction algorithm) produced a list of 10 candidates (Fig. 5 a). To interrogate these potential miR-1 targets, we first asked whether miR-1 recruits the corresponding mRNAs to the RISC. MLECs were transfected with miR-1 or scr–miR-1, and the cell lysates were immune-precipitated with anti-Ago2. Recruitment of specific mRNAs to the RISC was assayed by real-time qRT-PCR on these immunoprecipitates (IPs). We first confirmed the efficiency of miR-1 transfection and its presence in the RISC by measuring the level of miR-1 in the Ago2-IP (Fig. 5 b). The concentrations of seven of the candidate genes (Bxdc2, Ick, Ss18, St5, Smarcb1, Mpl, and Hspd1) and two other putative miR-1 targets, Ets1 (v-ets erythroblastosis virus E26 oncogene homologue 1) and Endoglin, with known roles in angiogenesis, were compared in the Ago2-IPs from miR-1–transfected cells with those of controls. As shown in Fig. 5 c, among all of the genes analyzed, miR-1 only recruited Mpl mRNA to the RISC. We further confirmed the miR-1–dependent recruitment of Mpl mRNA in the more stringent enrichment assay (Fig. 5 d). In this assay, the specific recruitment of targets to mi-RISC is analyzed by measuring the enrichment of mRNAs in the IP compared with the total cytoplasmic (input) fraction. Once again, Mpl mRNA was uniquely enriched in the IP fraction, suggesting that Mpl is a direct target of miR-1.
Figure 5.
Identification and validation of an miR-1 target. (a) MLECs were transfected with miR-1 or scrambled control (scr–miR-1) and stimulated with VEGF or PBS. RNA was extracted from the cells and hybridized to an mRNA microarray. Heat map shows the 10 genes that were selected as potential targets of miR-1 based on their response to VEGF stimulation and the effect of miR-1 transfection on this response. The dataset was log transformed, normalized, and median centered. The bar shows the color scale used in the heat map (values are in log base). (b–d) MLECs were transfected with miR-1 or scr–miR-1. Cells were lysed, and the lysate was immune-precipitated with anti-Ago2. (b) The level of miR-1 that coimmunoprecipitated with Ago2 was measured by TaqMan qRT-PCR and normalized to the scrambled control (data from two experiments; n = 4 in each group; *, P < 0.0001). (c) The level of mRNAs that coimmunoprecipitated with Ago2 (Ago2-IP) were measured by real-time qRT-PCR and normalized to the values for the scrambled control. The ratios represent miR-1–dependent RISC recruitment and are expressed as 2−ΔΔCt (data from three experiments; n = 4 in each group; values were compared by one-way ANOVA and corrected for multiple comparisons: *, P < 0.0002). (d) Relative concentration of each candidate mRNA in the Ago-IP fraction was divided by its relative concentration in the total cellular RNA (input fraction), and presented as 2−ΔΔCt (RISC enrichment analysis). Values for miR-1 were compared with those for scr–miR-1 (two experiments; n = 4 each group; *, P < 0.0005). (e and f) IL-13 expression was induced in IL-13 transgenic mice, and intranasal miR-1 was delivered as described in Fig. 4 c. RISC enrichment for miR-1 (*, P = 0.011) and its putative target genes (* and **, P < 0.03) were analyzed in whole lung lysates after Ago2-IP as described in c (two experiments; n = 3 per group). (g) Full-length Mpl 3′ UTR was cloned downstream from the firefly luciferase gene. This vector was cotransfected with a control Renilla luciferase vector and miR-1 or scr–miR-1, and relative luciferase activity (firefly luciferase/Renilla luciferase expression) was measured with a luminometer (three experiments; n = 5 each group; *, P < 0.0001). (h) A 56-nt fragment of Mpl 3′ UTR containing the miR-1 binding site with (Mut) or without (WT) compensatory mutations was cloned downstream of the firefly luciferase gene, and relative luciferase activity was measured as described in g (two experiments; n = 4 each; *, P < 0.005). Red letters indicate mutations in scr–miR-1, and green letters indicate compensatory mutations in Mpl 3′ UTR. (i) MLECs were transfected with firefly luciferase–Mpl3′ UTR (reporter construct) and Renilla luciferase vectors and stimulated with 100 ng/ml VEGF with or without 1 µM SU5416 (three experiments; n = 3 or more in each group; VEGF group was compared with the other two groups: *, P < 0.05). Hspd1, heat shock 60-kD protein 1 (chaperonin); Smarcb1, SWI/SNF-related matrix-associated actin-dependent regulator of chromatin, subfamily b, member 1; Ick, intestinal cell (MAK-like) kinase; Slc25a22, solute carrier family 25 (mitochondrial carrier: glutamate member 22); St5, suppression of tumorigenicity 5; Bxdc2, Brix domain–containing protein 2; Nol10, nucleolar protein 10; Ss18, synovial sarcoma translocation, chromosome 18; Ets1:v-ets, erythroblastosis virus E26 oncogene homologue 1. All error bars represent mean ± SEM.
We next examined the validity of these observations in the whole lung and after intranasal miR-1 delivery. Lung epithelial IL-13 transgenic mice were treated with intranasal miR-1 or scr–miR-1, and enrichment of Mpl mRNA and three other putative targets of miR-1 (ADAR [adenosine deaminase RNA specific], Hhip [hedgehog-interacting protein], and Dhx15 [DEAH box polypeptide 15]) were analyzed as above. As shown in Fig. 5 (e and f), miR-1 is enriched in the RISC after intranasal delivery and recruits Mpl and ADAR mRNAs to this compartment. ADAR is a previously described target of miR-1 in fibroblasts and cardiac myocytes (Lim et al., 2005; Srivastava, 2006; Vinther et al., 2006), and enrichment of ADAR in the lung RISC further confirms that miR-1 reaches the mi-RISC upon intranasal delivery.
The VEGF–miR-1 axis regulates Mpl expression in the lung endothelial cells through the direct interaction of miR-1 with the Mpl 3′ UTR
To validate the aforementioned targeting in a direct study, we first cloned the full-length Mpl 3′ UTR downstream of a firefly luciferase gene and compared the levels of firefly luciferase expression (relative to the control Renilla luciferase) after cotransfection with miR-1 or its negative control. In these experiments, miR-1 reduced the level of firefly luciferase by >50% (Fig. 5 g). To further confirm the site of this interaction, we cloned a 50-nt piece of the Mpl 3′ UTR containing the predicted miR-1 target site (Fig. 5 h, top) into the firefly luciferase vector and showed that, similar to above, cotransfection of this vector with miR-1 inhibits the relative expression of firefly luciferase. We then introduced compensatory mutations in this isolated miR-1 site so that it matched the negative control (scr–miR-1). As expected, miR-1 lost its inhibitory effect on this mutated target site and instead scr–miR-1, which was complementary to this site, inhibited the luciferase expression (Fig. 5 h).
To investigate the specific effect of VEGF on Mpl expression, we transfected the MLECs with the Mpl reporter vector (firefly luciferase–Mpl 3′ UTR). Stimulation of these cells with VEGF increased and inhibition of VEGF signaling with its specific receptor inhibitor (SU5416) decreased the Mpl-UTR–regulated luciferase expression (Fig. 5 i).
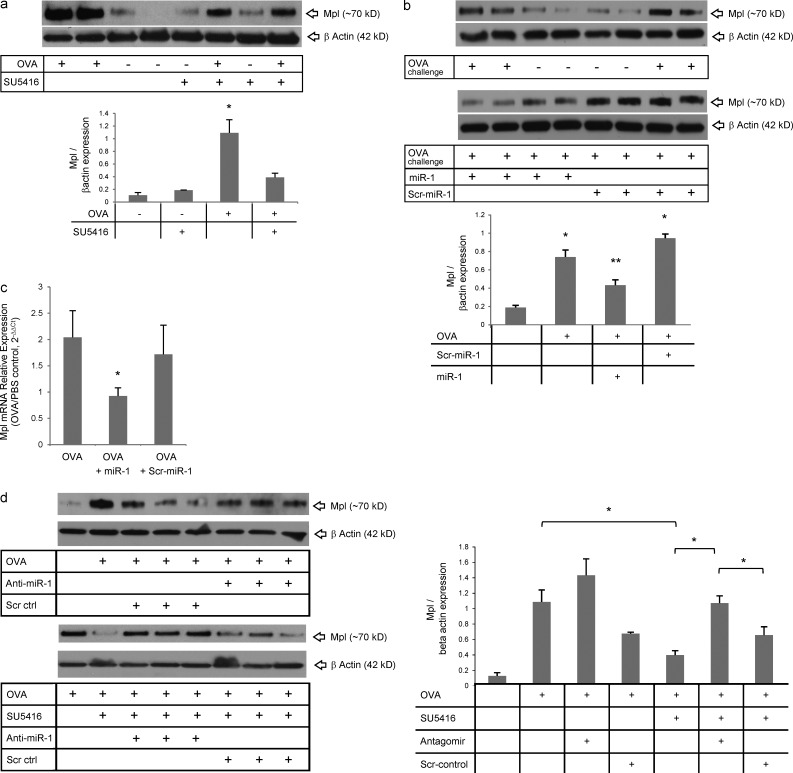
In Th2 inflammation, VEGF controls the expression of Mpl via down-regulation of miR-1
We first examined the effects of allergic airway inflammation on lung Mpl expression and the role of VEGF in this context. OVA sensitization and challenge increased the levels of lung Mpl to four to five times the levels in controls, and VEGF blockade, in this context, decreased this induction by 60% (Fig. 6 a). Intranasal delivery of miR-1 at the time of antigen challenge had a similar effect and decreased the levels of Mpl in the lung by 50% (Fig. 6 b). To test whether this down-regulation occurs in the endothelium, we measured the levels of Mpl mRNA in lung endothelial cells isolated from miR-1 (or control RNA)–treated mice. As shown in Fig. 6 c, endothelial Mpl mRNA was decreased by 50% after miR-1 intranasal delivery. To confirm the role of the VEGF–miR-1 axis in the up-regulation of Mpl in Th2 inflammation, we characterized the effects of miR-1 antagomir in this model. Similar to its effects on lung inflammation and Th2 cytokines, antagonizing miR-1 reversed the effect of VEGF blockade on Mpl expression in the OVA-sensitized and -challenged mice (Fig. 6 d). In combination, these findings demonstrate that Mpl up-regulation in Th2 inflammation is VEGF dependent and mediated by a VEGF-induced decrease in miR-1.
Figure 6.
VEGF controls the expression of Mpl in Th2 inflammation through regulation of miR-1. (a) C57BL/6 mice were sensitized to and challenged with OVA and received SU5416 (or vehicle) the day before the first challenge and every day afterward. Whole lung lysates from these mice were blotted with anti-Mpl or –β-actin antibodies. The top panel shows a representative Western blot, and the bottom panel shows the quantification based on densitometry (three experiments; n = 3 or more replicates in each; OVA-challenged groups were compared statistically: *, P < 0.05). (b) C57BL/6 mice were sensitized and challenged with OVA and received intranasal miR-1 as described in Fig. 3. Whole lung lysates were blotted as described in a. The top panels show representative gels, and the bottom panel shows quantification based on densitometry (two experiments; n = 4 in each; *, all groups were compared with PBS-challenged mice; **, miR-1–treated mice were compared with scr–miR-1 and vehicle [OVA] groups: both * and **, P < 0.03). (c) Expression of Mpl in the lung endothelial cells from these mice was measured by real-time qRT-PCR (values normalized to those in PBS-challenged mice and presented as 2−ΔΔCt; graphs represent two experiments; n = 4 or more in each group; *, P < 0.03). (d) C57BL/6 mice were sensitized and challenged with OVA and received SU5416 and miR-1 antagomir as described in Fig. 4 a. Whole lung lysates were blotted as described in a. The left panels show representative gels with three replicates in each group. The graph on the right shows quantification based on densitometry (two experiments; n = 4 or more in each group; OVA groups were compared by one-way ANOVA and multiple comparisons: *, P < 0.0001; three of the statistically significant comparisons are shown with horizontal bars). Error bars represent mean ± SEM.
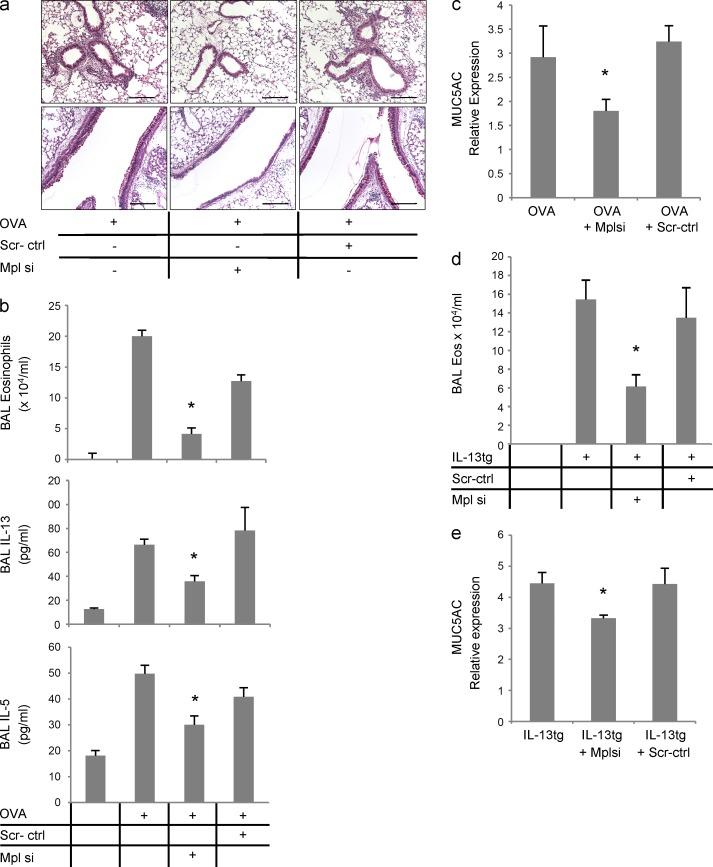
Mpl is required for lung Th2 inflammation
To directly examine the role of Mpl, we next tested the effects of intranasal siRNA against Mpl in the Th2 models. The efficiency of knockdown by this method was confirmed by showing that levels of lung Mpl protein were decreased by >80% after siRNA delivery (not depicted). Mpl knockdown led to a significant reduction in lung inflammation and mucus metaplasia (Fig. 7 a), BAL eosinophilia and Th2 cytokine secretion (Fig. 7 b), and mucin secretion (Fig. 7 c). Similarly, in IL-13 transgenic mice, Mpl knockdown decreased lung eosinophilia and mucin secretion (Fig. 7, d and e). These findings demonstrate that Mpl plays a critical role in the pathogenesis of Th2 responses in the lung.
Figure 7.
The role of Mpl in the lung Th2 inflammation. (a–c) C57BL/6 mice were sensitized and challenged with OVA as described in Fig. 2 c and received intranasal Mpl siRNA or scrambled control (scr-ctrl) the day before the first challenge and every day afterward. (a) Lung sections were stained with H&E (top) for peribronchial inflammation and PAS (bottom) for mucus metaplasia (dark red) in the large airways. Bars, 50 µm. (b) BALs were collected from these mice and analyzed for eosinophilia (H&E stain of cytospin preps, top), IL-13, and IL-5 (ELISA, middle and bottom; three experiments; n = 4 or more in each group; OVA-challenged Mplsi-treated mice were compared with the OVA + scrambled control and vehicle [OVA] groups: for all comparisons *, P < 0.05). (c) Levels of MUC5AC in the BALs of these mice were measured by slot blot hybridization and normalized to the values in the PBS-challenged control group (graphs represent two experiments; n = 5 in each group; *, P < 0.001). (d and e) IL-13 expression was induced in IL-13 transgenic mice as described in Fig. 2 a, and intranasal Mpl siRNA or scr-ctrl was delivered on the day before starting the induction and every other day afterward. After 7 d, BALs were collected, and eosinophilia and MUC5AC expression was determined as described in Fig. 3 (b and e, respectively; values were normalized to the levels in the WT group; graphs represent data from two experiments; n = 3 or more in each group; for both *, P < 0.04). Error bars represent mean ± SEM.
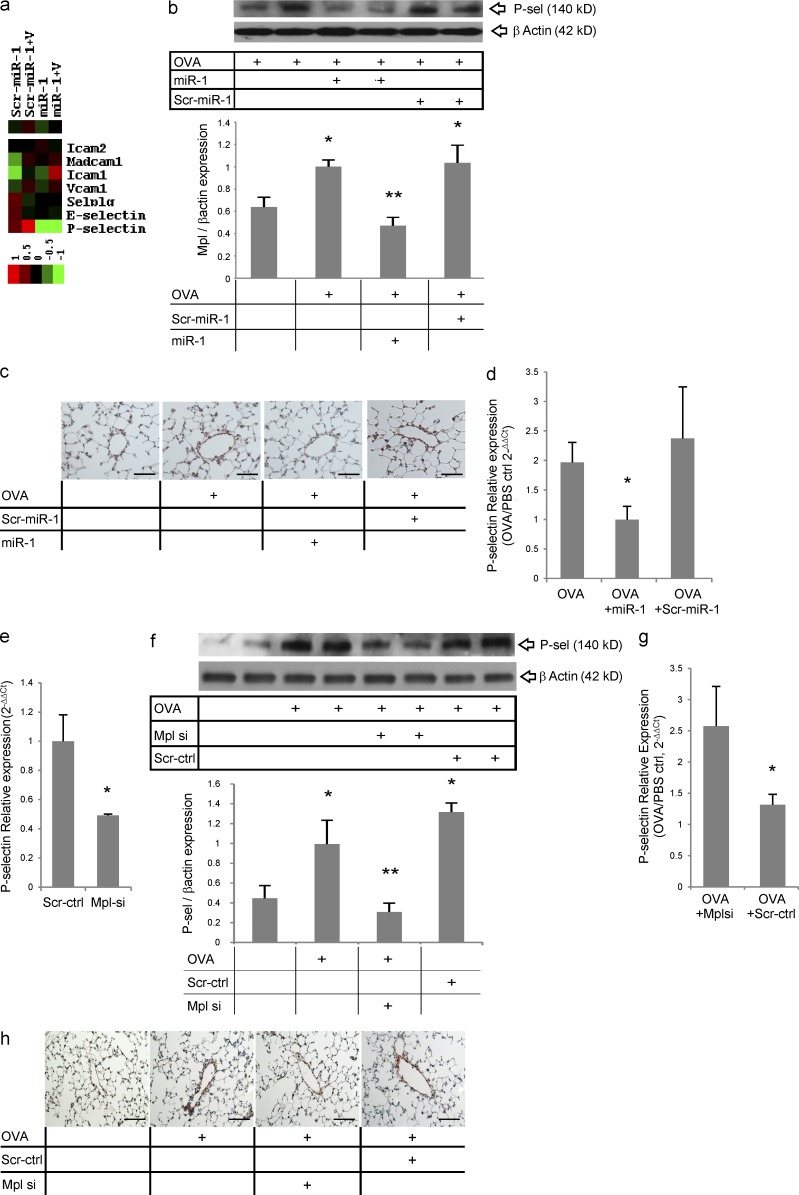
The miR-1–Mpl axis controls the expression of P-selectin in lung endothelium
At sites of inflammation, endothelial cells facilitate leukocyte entry into tissues via the expression of leukocyte adhesion molecules (Ley et al., 2007). Thus, we analyzed the effects of miR-1 and Mpl on the expression of endothelial adhesion molecules during Th2 inflammation. Evaluations of mRNA expression arrays of MLECs transfected with miR-1 (or its negative control) and stimulated with VEGF (Fig. 8 a) demonstrated that miR-1 specifically and strongly inhibited the expression of P-selectin in lung endothelium. Western blot and histological analysis of the OVA-sensitized and -challenged lungs showed a significant down-regulation of P-selectin after miR-1 delivery in the whole lung lysate (Fig. 8 b), which was quite prominent in lung venules (Fig. 8 c). Finally, measurement of the P-selectin mRNA level in lung endothelial cells isolated from these mice (Fig. 8 d) confirmed that miR-1 delivery decreases the endothelial expression of P-selectin in the OVA-sensitized and -challenged mice. However, the P-selectin 3′ UTR does not contain an miR-1–binding site, and P-selectin mRNA is not recruited to the RISC after miR-1 transfection (not depicted), suggesting that the inhibitory effects of miR-1 on P-selectin are indirect and mediated through its target, Mpl. In accord with this hypothesis, Mpl knockdown decreased the level of P-selectin mRNA in MLECs (Fig. 8 e). More importantly, intranasal delivery of Mpl siRNA caused a significant down-regulation of P-selectin expression in the OVA-sensitized and -challenged lungs (Fig. 8 f). P-selectin mRNA level in endothelial cells isolated from these lungs was significantly decreased (Fig. 8 g), and expression of P-selectin in the venules was diminished (Fig. 8 h), showing that this down-regulation occurs in the endothelium. In keeping with the known ability of Mpl to regulate platelet P-selectin (Tibbles et al., 2002) and the inhibitory effect of miR-1 on P-selectin expression, these experiments strongly suggest that the VEGF–miR-1 axis controls the expression of endothelial P-selectin through regulation of Mpl. Importantly, they also suggest that the VEGF–miR-1–Mpl axis mediates its effects on lung Th2 inflammation, at least in part, by regulating this critical adhesion molecule.
Figure 8.
The effect of the miR-1–Mpl axis on lung P-selectin. (a) MLECs were transfected with miR-1 and stimulated with VEGF, and extracted RNA was hybridized to an mRNA microarray as described in Fig. 5 a. Heat map shows the expression levels of various adhesion molecules under the four experimental conditions (the dataset was normalized and median centered; Icam1 and 2, intercellular adhesion molecule 1 and 2; Madcam1, mucosal vascular adressin cell adhesion molecule 1; Vcam1, vascular cell adhesion molecule 1; SELPLG, P-selectin glycoprotein ligand-1; the bar shows the color scale used in the heat map; values are in log base). (b) C57BL/6 mice were sensitized and challenged with OVA and received miR-1 or scr–miR-1 as described in Fig. 3. Whole lung lysates from these mice were blotted with anti–P-selectin and –β-actin antibodies. The top panel shows a representative blot of these lungs, and the graph shows quantification based on densitometry (data from two experiments; n ≥ 4 in each; *, all groups were compared with PBS-challenged mice; **, miR-1–treated mice were compared with the scr–miR-1 and vehicle [OVA] groups; both * and **, P < 0.04). (c) Representative images of lungs from mice treated as described in b and stained with anti–P-selectin antibody. (d) P-selectin expression was measured by real-time qRT-PCR in lung endothelial cells isolated from these mice (levels in OVA-challenged mice were normalized to PBS controls and presented as 2−ΔΔCt; n = 4 or more in each group; *, P < 0.03). (e) MLECs were transfected with Mpl siRNA or scrambled control (scr-ctrl). The levels of P-selectin/GAPDH mRNA were measured by real-time qRT-PCR, normalized to the scrambled control, and expressed as 2−ΔΔCt (two experiments; n = 3 or more; *, P < 0.05). (f) C57BL/6 mice were sensitized and challenged with OVA and treated with Mpl siRNA or scr-ctrl as described in Fig. 7 a. Whole lung lysates from these mice were blotted as described in b. The top panel shows a representative gel, and the bottom panel shows quantification based on densitometry (two experiments; n = 4 or more in each group; *, all groups were compared with PBS-challenged mice; **, miR-1–treated mice were compared with the scr-ctrl and vehicle [OVA] groups; both * and **, P < 0.01). (g) P-selectin expression was measured by real-time qRT-PCR in lung endothelial cells isolated from these mice (values from Mplsi-treated mice were normalized to those in scr-ctrl–treated group and expressed as 2−ΔΔCt; graphs represent data from two experiments; n = 5 in each group; *, P < 0.05). (h) Representative images of lungs from these mice stained with anti–P-selectin antibody. Bars, 50 µm. Error bars represent mean ± SEM.
The VEGF–miR-1–Mpl axis in human endothelium
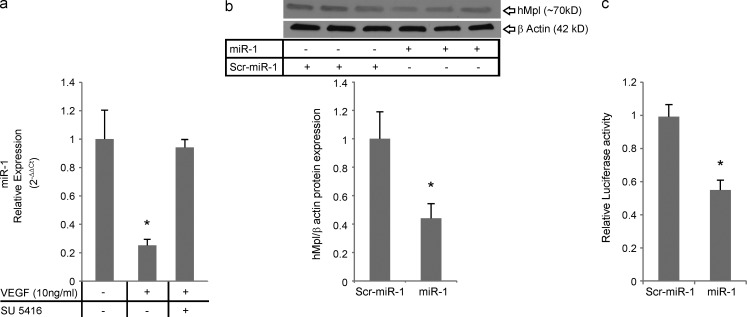
To investigate the relevance of the VEGF–miR-1–Mpl axis in human pathologies, we first determined whether VEGF regulated the expression of miR-1 in human primary endothelial cells (HUVECs). As shown in Fig. 9 a, VEGF down-regulated miR-1 in human endothelial cells and inhibition of VEGF signaling abrogated this down-regulation. We next asked whether miR-1 targets Mpl in human primary endothelial cells. As shown in Fig. 9 b, miR-1 decreased the expression of Mpl in HUVECs by >50%. Experiments were next undertaken to determine whether this inhibitory effect was mediated through the miR-1 interaction with Mpl 3′ UTR. Because human Mpl (hMpl) 3′ UTR is longer than that of the mouse and has two predicted miR-1–binding sites that differ from that in the mouse, human 3′ UTR was cloned and used in this study. In these experiments, we cotransfected the miR-1 mimic with a reporter plasmid containing firefly luciferase cloned upstream from the human full-length Mpl 3′ UTR. As shown in Fig. 9 c, miR-1 cotransfection inhibited the expression of firefly luciferase (represented as relative luciferase activity) by ∼50%. Thus, these observations demonstrate that the VEGF–miR-1–Mpl pathway is active in human cells.
Figure 9.
The VEGF–miR-1–Mpl axis in human endothelial cells. (a) HUVECs were stimulated with 10 ng/ml recombinant human VEGF and treated with SU5416 or vehicle (DMSO). miR-1 relative expression was measured by TaqMan qRT-PCR, normalized to vehicle controls, and expressed as 2−ΔΔCt (three experiments; n = 3 or more in each group; VEGF-treated groups were compared; *, P < 0.01). (b) HUVECs were transfected with miR-1 or scrambled control (scr–miR-1). Protein lysates from these cells were blotted with anti–hMpl antibody. The top panel shows a representative gel, and the bottom graph shows the densitometric analysis (values are normalized to the control; two experiments; three replicates in each; *, P = 0.03). (c) MLE12 cells were cotransfected with miR-1 (or scrambled miR-1) and a dual-luciferase reporter vector. This reporter vector expressed hMpl 3′ UTR downstream from the firefly luciferase mRNA and a separate Renilla luciferase mRNA as internal control. Values for firefly/Renilla luciferase luminescence are expressed as relative luciferase activity (two experiments; n = 3 or more each; *, P < 0.001). Error bars represent mean ± SEM.
DISCUSSION
The prevalence of asthma has increased considerably in the past three decades (Barnes, 2010; Kim et al., 2010). However, despite extensive research in this area, disease-modifying interventions for asthma are distinctly lacking (Holgate and Polosa, 2008; Barnes, 2010; Akdis, 2012). VEGF is a multifaceted, highly conserved cytokine with far-reaching effects on biological processes, including angiogenesis, carcinogenesis, and inflammation (Ferrara et al., 2003). We hypothesized that the inflammatory effects of VEGF are mediated, at least in part, through the regulation of an miRNA pathway. Specifically, we hypothesized that VEGF-induced alterations in endothelial cell miRNA play an important role in the pathogenesis of pulmonary Th2 inflammation. In the present study, we have defined a novel and critical effector pathway in Th2 inflammation in which VEGF inhibits the expression of endothelial miR-1, leading to an accumulation of its direct target, Mpl, and thereby increased expression of P-selectin in the lung endothelium. This pathway controls the accumulation of Th2 effector cells and eosinophils in the lung after antigenic challenge and regulates the major pathophysiological features of pulmonary Th2 responses. These experiments validated the functionality of this pathway by demonstrating that miR-1 is down-regulated and Mpl is up-regulated during Th2 inflammation, miR-1 supplementation inhibits the Th2 inflammation and Mpl expression and antagonizing miR-1 rescues both the Th2 inflammation and Mpl expression after VEGF blockade, and finally, Mpl silencing, similar to miR-1 delivery, abrogates lung Th2 responses and down-regulates the expression of endothelial P-selectin.
Recent human studies have demonstrated that VEGF and VEGFRs are highly expressed in the lungs, BAL, and sputum of asthmatic subjects and that their levels correlate with disease severity (Hoshino et al., 2001b; Redington et al., 2001; Simcock et al., 2007; Tuder and Yun, 2008; Asosingh and Erzurum, 2009). In experimental models, the overexpression of VEGF induced Th2 inflammation, and VEGF blockade inhibited aeroallergen-induced inflammation (Lee et al., 2004). However, despite the abundance of evidence supporting the role of VEGF in asthma, the mechanism of this VEGF effect has not been adequately studied, and the clinical use of VEGF blockers in this disorder has never been realized. Our study defines the VEGF–miR-1–Mpl axis in Th2 inflammation and suggests that this axis is also active in human asthma. By so doing, they introduce a novel VEGF-driven pathway that can be targeted to control Th2 and asthma-like responses.
Several studies have addressed the identities and effects of miRNAs involved in VEGF-induced angiogenesis (Fish et al., 2008; Suárez et al., 2008; Wang et al., 2008; Bonauer et al., 2009; Nicoli et al., 2010). However, the roles of miRNAs in VEGF-induced inflammation and the mechanistic details of this contribution have never been investigated. Our experiments address these deficiencies. The down-regulation of endothelial miR-1 by VEGF, IL-13, and allergen-induced inflammation and the ability of miR-1 antagonists to overcome the effects of VEGFR blockade on Th2 inflammation establish the existence of a robust miR-1 pathway downstream of VEGF in the lung. The direct targeting of the Mpl 3′ UTR by miR-1 and the demonstration that Mpl knockdown inhibits eosinophilic inflammation, mucus metaplasia, Th2 cytokine production, and endothelial P-selectin expression defines a molecular mechanism for this pathway.
Preliminary investigations have implicated miRNAs in Th2 inflammation. These studies defined an miRNA induced by TLR-4 in lung epithelium (Mattes et al., 2009), highlighted up-regulation of miR-21 by IL-13 (Lu et al., 2009), and demonstrated that Let7 targets IL-13 3′ UTR in vitro and possibly in vivo (Polikepahad et al., 2010; Kumar et al., 2011). However, as valuable as these findings have been, they have only addressed isolated steps in the Th2 cascade. By using two lung-targeted transgenics and two widely used aeroallergen inflammation models, we have been able to identify and validate an miRNA pathway downstream from VEGF and VEGFR2 and demonstrate that it plays a critical role in the elicitation of lung Th2 inflammatory responses. We have also highlighted the relationship between the miR-1 target, Mpl, and one of its downstream molecules, P-selectin.
There are two interesting endothelial-related findings in our studies. First, although miR-1 was administered via an intranasal route, it readily reached the pulmonary endothelium. This demonstrates that an intranasal or aerosol route can be used to deliver miRNA to the pulmonary endothelium. Second, our experiments demonstrate that in Th2 inflammation, the level of endothelial miR-1 is controlled by VEGF signaling and correlates closely with the extent of inflammation. We found the strongest down-regulatory effect of VEGF in the endothelium, showed that VEGF blockade inhibits the endothelial miR-1 down-regulation, and found a genuine, biologically valid target of miR-1 in the lung endothelial cells. These data strongly suggest that lung endothelium is a critical integration site for antigen-triggered pulmonary Th2 responses. This observation is in line with a recent study defining endothelial cells as central orchestrators of cytokine responses induced by influenza infection (Teijaro et al., 2011).
We used four different approaches to identify a target of miR-1. First, by combining in silico prediction and differential mRNA expression analysis, we narrowed the list of potential miR-1 targets to 10 genes that are induced by VEGF, inhibited in the presence of miR-1, and contain an miR-1–responsive element in their 3′ UTR (Fig. 5 a). This method has been previously used to screen and identify miRNA targets (Lim et al., 2005; Ziegelbauer et al., 2009). Second, we used Ago2 immunoprecipitation to identify mRNAs that are directly targeted by miR-1, i.e., are recruited to RISC by miR-1. We used two RISC recruitment assays to analyze the selective recruitment of a gene to the RISC in the presence of miR-1 compared with its scrambled control (Fig. 5 c) and more specifically show selective enrichment of this gene in the Ago2-IP fraction compared with the cytoplasmic fraction (Fig. 5 d). Both of these assays have been widely used to detect the direct targeting of genes by miRNAs (Liu et al., 2004; Karginov et al., 2007; Hendrickson et al., 2008; Fabian et al., 2009; Fasanaro et al., 2009; Challagundla et al., 2011; Thomson et al., 2011; Riley et al., 2012). Third, we showed direct base pairing between miR-1 and target gene 3′ UTR by luciferase assay (Fig. 5 e) and mutational analysis (Fig. 5 f). Lastly, we confirmed the biological validity of this targeting by defining the effects of VEGF, miR-1, and an miR-1 antagomir on target expression (Figs. 5 d and 6). These experiments demonstrated that Mpl is the direct target of VEGF–miR-1 axis in Th2 inflammation.
Mpl, the receptor for thrombopoietin, is known for its roles in megakaryopoiesis and hematopoietic stem cell differentiation (Chou and Mulloy, 2011). It is expressed on endothelial cells and progenitors (Cardier and Dempsey, 1998; Amano et al., 2005; Jin et al., 2006; Eguchi et al., 2008) and affects the accumulation of plasma cells in bone marrow and the differentiation of mast cells (Migliaccio et al., 2007; Winter et al., 2010). However, a direct proinflammatory role for Mpl has not been previously appreciated. Our study provides new insights into the biology of Mpl by demonstrating that Mpl plays an essential role in Th2 inflammation and endothelial P-selectin expression.
P-selectin is one of the main adhesion molecules expressed on the surface of the lung endothelium (Broide et al., 1998a,b), and it is known that Mpl induces expression of P-selectin in platelets (Tibbles et al., 2002). Human studies have also shown a close correlation between the levels of VEGF and cellular adhesion molecules such as ICAM-1, VCAM-1, and E-, L-, and P-selectins (Azimi-Nezhad et al., 2013) and that VEGF induces the expression of VCAM-1 and E-selectin and promotes the adhesion of leukocyte to the endothelium (Ismail et al., 2012). The present study adds to our knowledge in this area by demonstrating that in Th2 inflammation, endothelial P-selectin expression is a component of the VEGF–miR-1 pathway that is controlled through Mpl modulation. Specifically, they demonstrate that endothelial miR-1 is inhibited by VEGF, leading to increased Mpl and P-selectin expression at sites of Th2 inflammation. They also demonstrate that the administration of miR-1 decreases the levels of Mpl and P-selectin, eosinophilic inflammation, mucus metaplasia, Th2 cytokine elaboration, eotaxin production, and AHR. These observations suggest that the Th2 inhibitory effects of miR-1 or Mpl siRNA are mediated, at least in part, by the ability of these interventions to alter the adhesion molecule repertoire of the endothelium and, in turn, the entry of T cells and or eosinophils into the lung. Our demonstration that the accumulation of CD4+ T cells was decreased while levels of antigen-specific IgE were not altered by miR-1 further supports this concept. The inhibitory effect of miR-1 on the antigen-independent, eosinophil-rich inflammatory response in IL-13 transgenic mice further supports the concept that miR-1 is not altering T cell sensitization or Th2 skewing.
The idea that endothelial events are critical regulators of Th2 inflammation is in accord with a sizable literature that has demonstrated that P-selectin plays an important role in recruiting T cells and eosinophils to the lung (Broide et al., 1998a,b; Symon et al., 1994; Lukacs et al., 2002; Stephens and Chaplin, 2002; Larbi et al., 2003; Ulfman et al., 2003; Banerjee, 2011). Antigen-sensitized and -challenged P-selectin–null mice fail to develop airway hyperreactivity or airway eosinophilia, whereas the levels of IgE in their serum are comparable with their WT littermate controls (Lukacs et al., 2002). Asthmatic endothelial P-selectin can also directly activate eosinophil attachment to the endothelium (Johansson and Mosher, 2011). Eosinophils are required for and can significantly augment the severity of the allergen-induced inflammation by secreting Th2 cytokines (Rosenberg et al., 2007), activating and recruiting T cells (MacKenzie et al., 2001; Mattes et al., 2002; Jacobsen et al., 2008; Walsh et al., 2008), and modulating dendritic cell responses (Jacobsen et al., 2008). Additional studies will be required to further define the mechanisms that the VEGF–miR-1–Mpl–P-selectin pathway uses to regulate pulmonary Th2 responses.
It is often crucial to investigate whether the pathways discovered in animal models are actually operative in human cells and tissues. Because the Mpl miR-1–responsive element in human differs from that in mice, this investigation is of even higher importance. To address this issue, we undertook experiments using primary human endothelial cells. These experiments demonstrated that VEGF decreased the levels of miR-1 in human cells. They also demonstrated that, despite the sequence differences, miR-1 inhibits hMpl via interacting with its 3′ UTR. When viewed in combination, these experiments demonstrate that the VEGF–miR-1–Mpl axis is operative in humans.
In summary, this study highlights a novel endothelial miRNA–based pathway that plays an important role in the genesis of Th2- and IL-13–induced responses in the lung. Specifically, they demonstrate that VEGF contributes to Th2 inflammation by down-regulating miR-1 in the endothelium, thereby increasing the expression of Mpl and its downstream target, P-selectin. This in turn contributes to recruitment of activated T cells and eosinophils into the lung and subsequent eosinophilic inflammation, Th2 cytokine elaboration, mucus metaplasia, and AHR. Additional investigations of this pathway, its relationship to other VEGF- and miR-1–mediated responses, and its roles in health and disease are warranted.
MATERIALS AND METHODS
Microarray analysis.
miRNA profiling of the mouse lungs was performed through the microarray service of LC Sciences after harvesting the lungs and RNA extraction by mirVana RNA Isolation kit (Ambion). Lung RNA from lung epithelial VEGF transgenic mice and their WT litters were labeled with Cy5 and Cy3, respectively, and hybridized on a microarray that detected miRNA transcripts listed in Sanger miRBase Release 9.1. For the array analysis, the signal values were derived by background subtraction and normalization. For all the detectable and acceptable signals (i.e., signal intensity higher than 3× background, spot coefficient of variation <0.5, and signals from >50% of repeating probes on the chip), the median value of the repeating probes were calculated, different values were averaged, and the differential ratios were presented in log2 scale. The statistical significance was analyzed by Student’s t test, and the differentials reported as significant if P < 0.01. Array data were deposited in the GEO database (accession no. GSE50644).
miRNA and mRNA analysis by qRT-PCR.
miRNA analyses were performed by TaqMan real-time qRT-PCR assay (Applied Biosystems) on a 7500 Fast Real-Time PCR machine and according to the manufacturer’s instructions. Based on Applied Biosystems’ technical recommendation, sno202 was used as the internal control. In each set, the test sample was compared with a calibrator (WT control or PBS treatment) and expressed as relative expression or 2−ΔΔCt.
mRNA analyses were performed using the SYBER Green real-time RT-PCR kit and according to the manufacturer’s instructions (Applied Biosystems) on a 7500 Fast Real-Time PCR machine. When the concentrations of the extracted RNA were <5 ng/µl (e.g., after MACS [magnetic-activated cell sorting] for endothelial cells on a single lung), the mRNA or miRNA of interest and the corresponding normalization gene were preamplified as described previously (Tang et al., 2006).
Isolation of endothelial cells from the mouse lung by MACS.
Endothelial (CD31+, CD45− or CD105+, CD45−) and hematopoietic (CD45+) cells were isolated using Miltenyi Biotec magnetic columns or Dynabead Tosyl–activated Dynabeads and Dynamag15 magnet (Invitrogen) and corresponding antibodies from the same manufacturers and according to the instructions. In brief, lungs were harvested from mice, sliced to small pieces in PBS buffer on ice, and filtered through 70-µm and then 40-µm nylon meshes to make a single cell suspensate, and RBCs were lysed using Sigma-Aldrich RBC lysis buffer. Cells were then counted by hemocytometer before incubation with the antibody-coated magnetic beads. Magnetic labeling and separations were performed according to the instructions. Total lung cell suspensates were first depleted of CD45+ cells and then positively selected for CD31+ or CD105+ cells. The enrichment and purity of the endothelial cells were checked by FACS analysis of the selected cells and qRT-PCR for VCAM on the RNAs extracted from the whole lung cells, CD45+, CD45−/CD31+, and CD45−/CD31+ cells.
Allergen sensitization and challenge.
Sensitization and challenge with OVA was performed as described previously (Lee et al., 2009). In brief, C57BL/6 mice were sensitized to a mixture of OVA and Alum (Imject; Sigma-Aldrich) by i.p. injection, boosted with a second injection after 1 wk, and challenged with aerosolized OVA for 3 d, 3 wk after the initial sensitization. Mice were injected i.p. with 20 µg/g SU5416 (dissolved in DMSO; Sigma-Aldrich) or given intranasal miR-1 or Mpl siRNA (synthesized by Integrated DNA Technologies) the night before the first challenge and every night after the challenge afterward. In all of the experiments involving Su5416, the control mice were injected with DMSO i.p. HDM allergy was induced by i.p. injection of a mixture of HDM (Greer Labs; ∼1 µg DerP1 per mouse) and alum (Imject; Sigma-Aldrich; 3 mg per mouse) on day 0 (sensitization), followed by a boost dose of the same mixture i.p. on day 7 and antigenic challenge with intranasal HDM (∼2 µg DerP1) on days 17–19 after sensitization. miRNA delivery was similar to above.
Transgenic mice and intranasal delivery.
All animal experiments were performed under protocol #2009-07586 approved by the Yale Institutional and Animal Care and Use Committee. All of the mice used in this manuscript are on a C57BL/6 background, and the WT controls of the transgenic mice were their littermates. All of the transgenic animals have been crossed with WTs for more than six generations. The generation and phenotype of the CC10-rtTA-VEGF and CC10-rtTA–IL-13 transgenic mice in which expression of human VEGF165 and mouse IL-13, respectively, is under the control of an inducible promoter has been described previously (Zhu et al., 1999; Lee et al., 2004). Mice were grown to 6–8 wk, and expression of the transgene was induced by adding 1 g/liter doxycycline to their drinking water for 7–10 d. For intranasal delivery, mice were anesthetized by i.p. injection of 0.5–0.7 ml ketamine/xylazine. A total of 50 µl of double-stranded RNA (1 µg/µl) in siRNA hybridization buffer (Thermo Fisher Scientific) was administered by placing the tip of the pipetter near each nostril and injecting 10–20 µl each time as described previously (Zhang et al., 2004).
BAL and lung tissue analyses.
Cytokines in the BAL or lung lysates were measured using R&D Systems ELISA kits and according to the manufacturer’s instructions. Mucus hypersecretion was assessed in the BAL by slot blot analysis with antibody against MUC5AC as described previously (Zheng et al., 2008). For the general assessment of lung inflammation and mucus metaplasia, lung sections were stained with hematoxylin and eosin (H&E) and periodic acid-Schiff (PAS), respectively. Eosinophils in the lung tissues were counted in the bronchovascular areas after staining the lung sections with Congo Red. Four or more typical areas were delineated in each sample, the surface area was measured by ImageJ (National Institutes of Health), and the number of the eosinophils was expressed as the mean cells/mm2. CD4+ T cells in the BAL were detected by FACS analysis as described before (Niu et al., 2007). In brief, 1–5 × 105 cells were stained with CD4-APC and CD11b-PE (1:1,000 and 1:2,000; both from BD) in staining buffer (0.1% BSA and 0.05% NaN3 in PBS) for 20 min, washed three times, and fixed in 1% paraformaldehyde (in PBS) and kept at 4°C overnight before FACS analysis or analyzed directly after staining. Sorting was performed on a FACSCalibur (BD), and the data were analyzed using FlowJo (version 9; Tree Star). CD11b marker was used to exclude alveolar macrophages.
Measurement of AHR.
Airway responses were measured by the forced oscillation technique using FlexiVent (SCIREQ). Mice were anesthetized with 70 mg/kg pentobarbital and 1.8 g/kg urethane followed by 0.5 mg/kg pancuronium bromide. A tracheostomy was performed and a 19-gauge metal needle was inserted, and mice were ventilated with a tidal volume of 10 ml/kg at a frequency of 150 breaths/min and a positive end-expiratory pressure of 3 cm H2O. Each mouse was challenged with saline (0 mg/ml) followed by increasing concentrations of methacholine aerosol (10–80 mg/ml) generated with an in-line nebulizer and administered directly through the ventilator for 10 s. Mean airway resistance (R) measures were calculated at each methacholine concentration.
RISC analysis.
The recruitment of miRNAs and mRNAs to RISC was analyzed by Ago2 coimmunoprecipitation as described previously (Vasudevan and Steitz, 2007) with modifications. MLECs were grown in 10-cm plates and transfected with double-stranded miR-1 as described above. 48 h after transfection, cells were collected by scraping in cold PBS, spun down (110 g for 10 min at 4°C), and resuspended in 250 µl of cold lysis buffer (10 mM Hepes, pH 7.0, 150 mM KCl, 2 mM MgCl2, 2 mM DTT, 10% glycerol, 0.05% NP-40, and 1 mM PMSF). Part of this lysate was kept as “input fraction,” and the rest was immunoprecipitated with anti-Ago2 antibody (EMD Millipore) and protein G beads (GE Healthcare). The beads were directly added to TRIzol after washing three times (wash buffer is made by adding tRNA to the lysis buffer to the final concentration of 100 µg/ml) and the RNA extracted with mirVana kit after one round of chloroform extraction.
miRNA and siRNA.
Mature miR-1 mimic was made by hybridizing the following two RNA oligonucleotides: M-1-s, 5′-UGGAAUGUAAAGAAGUAUGUA-3′; M-1-as, 5′-CAUACUUCUUUACAUUCACUA-3′. The scrambled control was made by hybridizing the following: Delta-s, 5′-UUGAGAGUGAAUAAGAAUGUA-3′; Delta-as, 5′-CAUUCUUAUUCACUCUCCCUA-3′. This control contains sequence scrambled at only six positions. miR-1 antagomir and its scramble control were designed and synthesized by Regulus and contained the following sequences with phosphorothioate modification of the backbone: miR-1 antagomir, 5′-UACAUACUUCUUUACAUUCCA-3′; scrambled control, 5′-CCUUCCCUGAAGGUUCCUCCUU-3′. Mpl siRNA, VEGFR2 siRNA, and scrambled controls were purchased from Invitrogen.
Cell culture.
MLECs were isolated from mouse lungs as described previously (Zhang et al., 2006). P2-P6 cells were used for the experiments. MLE12 cells were purchased from ATCC, and PASMCs were a gift from P. Lee’s laboratory at Yale Pulmonary and Critical Care Department. MLECs were grown in DMEM/F12 (Invitrogen) supplemented with 20% FCS. MLE12s and PASMCs were grown in DMEM (Invitrogen) supplemented with 2% and 10% FCS, respectively.
Cells were starved in 0.1–0.5% FCS supplemented medium for 24 h before the addition of VEGF. Cultured cells were lysed directly by adding TRIzol (Invitrogen) to the wells, and RNA was purified extracted by the mirVana isolation kit after a chloroform extraction. Su5416 was added to the medium to the final concentration of 1 µM.
Transfections were performed using Lipofectamine 2000 (Invitrogen) and Opti-MEM reagents and according the protocol recommended by the manufacturer for DNA or siRNA transfection, respectively. The medium was changed the next day, starvation started on the second day, and VEGF was added to the medium on the third day after transfection.
Plasmids, cloning, and luciferase assays.
Mpl 3′ UTR was cloned into the multiple cloning site of PGL3-MCS (PO98) that was a gift from K. Riley in J. Steitz’s laboratory (Yale University School of Medicine, New Haven, CT). This plasmid was constructed by cloning a fragment containing BglII–EcoRI–EcoRV–NdeI–PstI–SpeI–XhoI digestion sites into the Xba1 site of the PGL3 vector from Promega. In brief, mouse Mpl 3′ UTR was amplified from mouse total genomic DNA (Promega) using the two primers F, 5′-GCAGCAGAGATCTAGGCAGTCCCCATG-3′; and R, 5′-TTCAGAGAATTCTTATTGGAGGTCTGCCAC-3′, which added a BglII and an EcoR1 site to the ends of the fragment, respectively. This PCR fragment was gel purified and cloned into PGEM-T-easy (Promega), cut with BglII and EcoRI, and cloned into the corresponding sites in PGL3-MCS. The miR-1–binding site from the Mpl 3′ UTR was constructed by hybridizing the two DNA oligonucleotides F, 5′-GATCTAGTCCCCATGCTACTGCAGACCTATACATTCCTACACACTACCTTATCCAG-3′; and R, 5′-AATTCTGGATAAGGTAGTGTGTAGGAATGTATAGGTCTGCAGTAGCATGGGGACTA-3′, digesting the resulting double-stranded DNA with BglII and EcoRI, and cloning the digested fragment into PGL3-MCS as described above. The fragment containing a mutated binding site corresponding to the scrambled control RNA was constructed as above by hybridizing the following two oligonucleotides: F, 5′-GATCTAGTCCCCATGCTACTGCTGACATACACTCTCATACACACTACCTTATCCAG-3′; and R, 5′-AATTCTGGATAAGGTAGTGTAGGAGAGTGTATGTCAGCAGTAGCATGGGGACTA-3′. P-CMV-Renilla luciferase was a gift from K. Riley in J. Steitz’s laboratory. Luciferase activity in the MLE12 cells transfected with the above vectors was measured by the Dual-Glo luciferase assay kit (Promega) according to the instructions.
HUVECs were purchased from the Keck facility at Yale University and maintained in M199, 20% FBS, and EGS (1:1,000; purchased from the Keck facility). hMpl 3′ UTR plasmid was purchased from Genecopoeia (miRNA 3′ UTR target clones). Luciferase assay on the cells transfected with this plasmid was performed using luc-pair luciferase assay kit (Genecopoeia) reagents according to the manufacturer’s instructions.
Western blot analysis.
Lungs were lysed in RIPA lysis buffer (Thermo Fisher Scientific) with added protease inhibitor (Roche), and the proteins were separated on a 4–20% gradient Tris polyacrylamide gel, transferred at 100 V for 1 h at 4°C. The blots were then blocked with 5% nonfat dry milk in TBST and incubated with 1:1,000 primary antibody (anti-TPOR/cMpl; EMD Millipore) overnight before adding the secondary antibody (anti–rabbit; Santa Cruz Biotechnology, Inc.) and Femto Supersignal reagent (Thermo Fisher Scientific) and exposing to film.
mRNA expression arrays.
Cellular RNA for the array was extracted using the mirVana kit according to the instructions. The isolated mRNA was sent to the Keck center at Yale for hybridization to Illumina arrays. The signal values were reported as mean of repeating probes. The heat maps for the mRNA expression array were generated using Cluster and Treeview software. The data were polished for the clustering by normalizing rows and columns and median centering the columns.
Gel densitometry.
Densitometries were performed on TIFF images of radiographical films exposed to Western or slot blots. We used National Institutes of Health ImageJ software analysis and assigned values to each lane by integrating the area under the curve.
Statistical analyses.
In all comparisons, the mean and standard error of the datasets were calculated and presented in the graphs using Excel 2007 (Microsoft). Unless otherwise specified, statistical significance was analyzed by Student’s t test. ANOVA tests were performed using Prism software (GraphPad Software). All error bars represent SEM.
Acknowledgments
We wish to thank Joan Steitz and Patty Lee for reviewing this manuscript and providing invaluable guidance and support. We thank Robert Homer for help and instructions on analyzing the histopathology slides. We thank Aimee Jackson from Regulus Therapeutics for providing the miR-1 antagomir and its negative control. We also would like to thank Shobha Vasudevan and Kasandra Riley for providing experimental advice and constructs as described in the manuscript.
This work was supported by National Institutes of Health grants K99/R00 HL098695-01 and 1 R01 HL 078744.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AHR
- airway hyperresponsiveness
- BAL
- bronchoalveolar lavage fluid
- HDM
- house dust mite
- hMpl
- human Mpl
- IP
- immunoprecipitate
- miRNA
- microRNA
- MLEC
- mouse lung endothelial cell
- PAS
- periodic acid-Schiff
- PASMC
- pulmonary artery smooth muscle cell
- qRT-PCR
- quantitative RT-PCR
- UTR
- untranslated region
- VEGF
- vascular endothelial growth factor
References
- Akdis C.A. 2012. Therapies for allergic inflammation: refining strategies to induce tolerance. Nat. Med. 18:736–749 10.1038/nm.2754 [DOI] [PubMed] [Google Scholar]
- Amano H., Hackett N.R., Rafii S., Crystal R.G. 2005. Thrombopoietin gene transfer-mediated enhancement of angiogenic responses to acute ischemia. Circ. Res. 97:337–345 10.1161/01.RES.0000179534.17668.f8 [DOI] [PubMed] [Google Scholar]
- Asher M.I., Montefort S., Björkstén B., Lai C.K., Strachan D.P., Weiland S.K., Williams H.; ISAAC Phase Three Study Group 2006. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet. 368:733–743 10.1016/S0140-6736(06)69283-0 [DOI] [PubMed] [Google Scholar]
- Asosingh K., Erzurum S.C. 2009. Angioplasticity in asthma. Biochem. Soc. Trans. 37:805–810 10.1042/BST0370805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi-Nezhad M., Stathopoulou M.G., Bonnefond A., Rancier M., Saleh A., Lamont J., Fitzgerald P., Ndiaye N.C., Visvikis-Siest S. 2013. Associations of vascular endothelial growth factor (VEGF) with adhesion and inflammation molecules in a healthy population. Cytokine. 61:602–607 10.1016/j.cyto.2012.10.024 [DOI] [PubMed] [Google Scholar]
- Banerjee E.R. 2011. Triple selectin knockout (ELP-/-) mice fail to develop OVA-induced acute asthma phenotype. J. Inflamm. (Lond.). 8:19 10.1186/1476-9255-8-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P.J. 2010. New therapies for asthma: is there any progress? Trends Pharmacol. Sci. 31:335–343 10.1016/j.tips.2010.04.009 [DOI] [PubMed] [Google Scholar]
- Bartel D.P. 2009. MicroRNAs: target recognition and regulatory functions. Cell. 136:215–233 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonauer A., Carmona G., Iwasaki M., Mione M., Koyanagi M., Fischer A., Burchfield J., Fox H., Doebele C., Ohtani K., et al. 2009. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 324:1710–1713 10.1126/science.1174381 [DOI] [PubMed] [Google Scholar]
- Broide D.H., Humber D., Sullivan S., Sriramarao P. 1998a. Inhibition of eosinophil rolling and recruitment in P-selectin- and intracellular adhesion molecule-1-deficient mice. Blood. 91:2847–2856 [PubMed] [Google Scholar]
- Broide D.H., Sullivan S., Gifford T., Sriramarao P. 1998b. Inhibition of pulmonary eosinophilia in P-selectin- and ICAM-1-deficient mice. Am. J. Respir. Cell Mol. Biol. 18:218–225 10.1165/ajrcmb.18.2.2829 [DOI] [PubMed] [Google Scholar]
- Cardier J.E., Dempsey J. 1998. Thrombopoietin and its receptor, c-mpl, are constitutively expressed by mouse liver endothelial cells: evidence of thrombopoietin as a growth factor for liver endothelial cells. Blood. 91:923–929 [PubMed] [Google Scholar]
- Challagundla K.B., Sun X.X., Zhang X., DeVine T., Zhang Q., Sears R.C., Dai M.S. 2011. Ribosomal protein L11 recruits miR-24/miRISC to repress c-Myc expression in response to ribosomal stress. Mol. Cell. Biol. 31:4007–4021 10.1128/MCB.05810-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. 2006. Natural selection on human microRNA binding sites inferred from SNP data. Nat. Genet. 38:1452–1456 10.1038/ng1910 [DOI] [PubMed] [Google Scholar]
- Chou F.S., Mulloy J.C. 2011. The thrombopoietin/MPL pathway in hematopoiesis and leukemogenesis. J. Cell. Biochem. 112:1491–1498 10.1002/jcb.23089 [DOI] [PubMed] [Google Scholar]
- Eguchi M., Masuda H., Kwon S., Shirakura K., Shizuno T., Ito R., Kobori M., Asahara T. 2008. Lesion-targeted thrombopoietin potentiates vasculogenesis by enhancing motility and enlivenment of transplanted endothelial progenitor cells via activation of Akt/mTOR/p70S6kinase signaling pathway. J. Mol. Cell. Cardiol. 45:661–669 10.1016/j.yjmcc.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 35:868–880 10.1016/j.molcel.2009.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasanaro P., Greco S., Lorenzi M., Pescatori M., Brioschi M., Kulshreshtha R., Banfi C., Stubbs A., Calin G.A., Ivan M., et al. 2009. An integrated approach for experimental target identification of hypoxia-induced miR-210. J. Biol. Chem. 284:35134–35143 10.1074/jbc.M109.052779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N., Gerber H.P., LeCouter J. 2003. The biology of VEGF and its receptors. Nat. Med. 9:669–676 10.1038/nm0603-669 [DOI] [PubMed] [Google Scholar]
- Fish J.E., Santoro M.M., Morton S.U., Yu S., Yeh R.F., Wythe J.D., Ivey K.N., Bruneau B.G., Stainier D.Y., Srivastava D. 2008. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 15:272–284 10.1016/j.devcel.2008.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F. 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 308:833–838 10.1126/science.1109020 [DOI] [PubMed] [Google Scholar]
- Hendrickson D.G., Hogan D.J., Herschlag D., Ferrell J.E., Brown P.O. 2008. Systematic identification of mRNAs recruited to argonaute 2 by specific microRNAs and corresponding changes in transcript abundance. PLoS ONE. 3:e2126 10.1371/journal.pone.0002126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holgate S.T., Polosa R. 2008. Treatment strategies for allergy and asthma. Nat. Rev. Immunol. 8:218–230 10.1038/nri2262 [DOI] [PubMed] [Google Scholar]
- Hoshino M., Nakamura Y., Hamid Q.A. 2001a. Gene expression of vascular endothelial growth factor and its receptors and angiogenesis in bronchial asthma. J. Allergy Clin. Immunol. 107:1034–1038 10.1067/mai.2001.115626 [DOI] [PubMed] [Google Scholar]
- Hoshino M., Takahashi M., Aoike N. 2001b. Expression of vascular endothelial growth factor, basic fibroblast growth factor, and angiogenin immunoreactivity in asthmatic airways and its relationship to angiogenesis. J. Allergy Clin. Immunol. 107:295–301 10.1067/mai.2001.111928 [DOI] [PubMed] [Google Scholar]
- Ismail H., Mofarrahi M., Echavarria R., Harel S., Verdin E., Lim H.W., Jin Z.G., Sun J., Zeng H., Hussain S.N. 2012. Angiopoietin-1 and vascular endothelial growth factor regulation of leukocyte adhesion to endothelial cells: role of nuclear receptor-77. Arterioscler. Thromb. Vasc. Biol. 32:1707–1716 10.1161/ATVBAHA.112.251546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Standart N. 2007. How do microRNAs regulate gene expression? Sci. STKE. 2007:re1 10.1126/stke.3672007re1 [DOI] [PubMed] [Google Scholar]
- Jacobsen E.A., Ochkur S.I., Pero R.S., Taranova A.G., Protheroe C.A., Colbert D.C., Lee N.A., Lee J.J. 2008. Allergic pulmonary inflammation in mice is dependent on eosinophil-induced recruitment of effector T cells. J. Exp. Med. 205:699–710 10.1084/jem.20071840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin D.K., Shido K., Kopp H.G., Petit I., Shmelkov S.V., Young L.M., Hooper A.T., Amano H., Avecilla S.T., Heissig B., et al. 2006. Cytokine-mediated deployment of SDF-1 induces revascularization through recruitment of CXCR4+ hemangiocytes. Nat. Med. 12:557–567 10.1038/nm1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M.W., Mosher D.F. 2011. Activation of beta1 integrins on blood eosinophils by P-selectin. Am. J. Respir. Cell Mol. Biol. 45:889–897 10.1165/rcmb.2010-0402OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karginov F.V., Conaco C., Xuan Z., Schmidt B.H., Parker J.S., Mandel G., Hannon G.J. 2007. A biochemical approach to identifying microRNA targets. Proc. Natl. Acad. Sci. USA. 104:19291–19296 10.1073/pnas.0709971104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.Y., DeKruyff R.H., Umetsu D.T. 2010. The many paths to asthma: phenotype shaped by innate and adaptive immunity. Nat. Immunol. 11:577–584 10.1038/ni.1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Ahmad T., Sharma A., Mabalirajan U., Kulshreshtha A., Agrawal A., Ghosh B. 2011. Let-7 microRNA-mediated regulation of IL-13 and allergic airway inflammation. J. Allergy Clin. Immunol. 128:1077–1085 10.1016/j.jaci.2011.04.034 [DOI] [PubMed] [Google Scholar]
- Larbi K.Y., Dangerfield J.P., Culley F.J., Marshall D., Haskard D.O., Jose P.J., Williams T.J., Nourshargh S. 2003. P-selectin mediates IL-13-induced eosinophil transmigration but not eotaxin generation in vivo: a comparative study with IL-4-elicited responses. J. Leukoc. Biol. 73:65–73 10.1189/jlb.0302126 [DOI] [PubMed] [Google Scholar]
- Lee C.G., Link H., Baluk P., Homer R.J., Chapoval S., Bhandari V., Kang M.J., Cohn L., Kim Y.K., McDonald D.M., Elias J.A. 2004. Vascular endothelial growth factor (VEGF) induces remodeling and enhances TH2-mediated sensitization and inflammation in the lung. Nat. Med. 10:1095–1103 10.1038/nm1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.G., Hartl D., Lee G.R., Koller B., Matsuura H., Da Silva C.A., Sohn M.H., Cohn L., Homer R.J., Kozhich A.A., et al. 2009. Role of breast regression protein 39 (BRP-39)/chitinase 3-like-1 in Th2 and IL-13–induced tissue responses and apoptosis. J. Exp. Med. 206:1149–1166 10.1084/jem.20081271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K., Laudanna C., Cybulsky M.I., Nourshargh S. 2007. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7:678–689 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 433:769–773 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 305:1437–1441 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- Lu T.X., Munitz A., Rothenberg M.E. 2009. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J. Immunol. 182:4994–5002 10.4049/jimmunol.0803560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukacs N.W., John A., Berlin A., Bullard D.C., Knibbs R., Stoolman L.M. 2002. E- and P-selectins are essential for the development of cockroach allergen-induced airway responses. J. Immunol. 169:2120–2125 [DOI] [PubMed] [Google Scholar]
- MacKenzie J.R., Mattes J., Dent L.A., Foster P.S. 2001. Eosinophils promote allergic disease of the lung by regulating CD4(+) Th2 lymphocyte function. J. Immunol. 167:3146–3155 [DOI] [PubMed] [Google Scholar]
- Mattes J., Yang M., Mahalingam S., Kuehr J., Webb D.C., Simson L., Hogan S.P., Koskinen A., McKenzie A.N., Dent L.A., et al. 2002. Intrinsic defect in T cell production of interleukin (IL)-13 in the absence of both IL-5 and eotaxin precludes the development of eosinophilia and airways hyperreactivity in experimental asthma. J. Exp. Med. 195:1433–1444 10.1084/jem.20020009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattes J., Collison A., Plank M., Phipps S., Foster P.S. 2009. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc. Natl. Acad. Sci. USA. 106:18704–18709 10.1073/pnas.0905063106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliaccio A.R., Rana R.A., Vannucchi A.M., Manzoli F.A. 2007. Role of thrombopoietin in mast cell differentiation. Ann. N. Y. Acad. Sci. 1106:152–174 10.1196/annals.1392.024 [DOI] [PubMed] [Google Scholar]
- Nicoli S., Standley C., Walker P., Hurlstone A., Fogarty K.E., Lawson N.D. 2010. MicroRNA-mediated integration of haemodynamics and Vegf signalling during angiogenesis. Nature. 464:1196–1200 10.1038/nature08889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N., Le Goff M.K., Li F., Rahman M., Homer R.J., Cohn L. 2007. A novel pathway that regulates inflammatory disease in the respiratory tract. J. Immunol. 178:3846–3855 [DOI] [PubMed] [Google Scholar]
- Polikepahad S., Knight J.M., Naghavi A.O., Oplt T., Creighton C.J., Shaw C., Benham A.L., Kim J., Soibam B., Harris R.A., et al. 2010. Proinflammatory role for let-7 microRNAS in experimental asthma. J. Biol. Chem. 285:30139–30149 10.1074/jbc.M110.145698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redington A.E., Roche W.R., Madden J., Frew A.J., Djukanovic R., Holgate S.T., Howarth P.H. 2001. Basic fibroblast growth factor in asthma: measurement in bronchoalveolar lavage fluid basally and following allergen challenge. J. Allergy Clin. Immunol. 107:384–387 10.1067/mai.2001.112268 [DOI] [PubMed] [Google Scholar]
- Riley K.J., Rabinowitz G.S., Yario T.A., Luna J.M., Darnell R.B., Steitz J.A. 2012. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 31:2207–2221 10.1038/emboj.2012.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg H.F., Phipps S., Foster P.S. 2007. Eosinophil trafficking in allergy and asthma. J. Allergy Clin. Immunol. 119:1303–1310 10.1016/j.jaci.2007.03.048 [DOI] [PubMed] [Google Scholar]
- Simcock D.E., Kanabar V., Clarke G.W., O’Connor B.J., Lee T.H., Hirst S.J. 2007. Proangiogenic activity in bronchoalveolar lavage fluid from patients with asthma. Am. J. Respir. Crit. Care Med. 176:146–153 10.1164/rccm.200701-042OC [DOI] [PubMed] [Google Scholar]
- Sokol N.S., Xu P., Jan Y.N., Ambros V. 2008. Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev. 22:1591–1596 10.1101/gad.1671708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava D. 2006. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 126:1037–1048 10.1016/j.cell.2006.09.003 [DOI] [PubMed] [Google Scholar]
- Stephens R., Chaplin D.D. 2002. IgE cross-linking or lipopolysaccharide treatment induces recruitment of Th2 cells to the lung in the absence of specific antigen. J. Immunol. 169:5468–5476 [DOI] [PubMed] [Google Scholar]
- Suárez Y., Fernández-Hernando C., Yu J., Gerber S.A., Harrison K.D., Pober J.S., Iruela-Arispe M.L., Merkenschlager M., Sessa W.C. 2008. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc. Natl. Acad. Sci. USA. 105:14082–14087 10.1073/pnas.0804597105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symon F.A., Walsh G.M., Watson S.R., Wardlaw A.J. 1994. Eosinophil adhesion to nasal polyp endothelium is P-selectin-dependent. J. Exp. Med. 180:371–376 10.1084/jem.180.1.371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Hajkova P., Barton S.C., Lao K., Surani M.A. 2006. MicroRNA expression profiling of single whole embryonic stem cells. Nucleic Acids Res. 34:e9 10.1093/nar/gnj009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Walsh K.B., Cahalan S., Fremgen D.M., Roberts E., Scott F., Martinborough E., Peach R., Oldstone M.B., Rosen H. 2011. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 146:980–991 10.1016/j.cell.2011.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson D.W., Bracken C.P., Goodall G.J. 2011. Experimental strategies for microRNA target identification. Nucleic Acids Res. 39:6845–6853 10.1093/nar/gkr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbles H., Vassilev A., Uckun F.M. 2002. A dual function anti-leukemic agent with anti-thrombotic activity. Leuk. Lymphoma. 43:1121–1127 [DOI] [PubMed] [Google Scholar]
- Tuder R.M., Yun J.H. 2008. Vascular endothelial growth factor of the lung: friend or foe. Curr. Opin. Pharmacol. 8:255–260 10.1016/j.coph.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulfman L.H., Joosten D.P., van Aalst C.W., Lammers J.W., van de Graaf E.A., Koenderman L., Zwaginga J.J. 2003. Platelets promote eosinophil adhesion of patients with asthma to endothelium under flow conditions. Am. J. Respir. Cell Mol. Biol. 28:512–519 10.1165/rcmb.4806 [DOI] [PubMed] [Google Scholar]
- Vasudevan S., Steitz J.A. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 128:1105–1118 10.1016/j.cell.2007.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinther J., Hedegaard M.M., Gardner P.P., Andersen J.S., Arctander P. 2006. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 34:e107 10.1093/nar/gkl590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel N.F., Vandivier R.W., Tuder R.M. 2006. Vascular endothelial growth factor in the lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 290:L209–L221 10.1152/ajplung.00185.2005 [DOI] [PubMed] [Google Scholar]
- Walsh E.R., Sahu N., Kearley J., Benjamin E., Kang B.H., Humbles A., August A. 2008. Strain-specific requirement for eosinophils in the recruitment of T cells to the lung during the development of allergic asthma. J. Exp. Med. 205:1285–1292 10.1084/jem.20071836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhang Z., O’Loughlin E., Lee T., Houel S., O’Carroll D., Tarakhovsky A., Ahn N.G., Yi R. 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev. 26:693–704 10.1101/gad.182758.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Aurora A.B., Johnson B.A., Qi X., McAnally J., Hill J.A., Richardson J.A., Bassel-Duby R., Olson E.N. 2008. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 15:261–271 10.1016/j.devcel.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter O., Moser K., Mohr E., Zotos D., Kaminski H., Szyska M., Roth K., Wong D.M., Dame C., Tarlinton D.M., et al. 2010. Megakaryocytes constitute a functional component of a plasma cell niche in the bone marrow. Blood. 116:1867–1875 10.1182/blood-2009-12-259457 [DOI] [PubMed] [Google Scholar]
- Zhang X., Shan P., Jiang D., Noble P.W., Abraham N.G., Kappas A., Lee P.J. 2004. Small interfering RNA targeting heme oxygenase-1 enhances ischemia-reperfusion-induced lung apoptosis. J. Biol. Chem. 279:10677–10684 10.1074/jbc.M312941200 [DOI] [PubMed] [Google Scholar]
- Zhang X., Shan P., Jiang G., Zhang S.S., Otterbein L.E., Fu X.Y., Lee P.J. 2006. Endothelial STAT3 is essential for the protective effects of HO-1 in oxidant-induced lung injury. FASEB J. 20:2156–2158 10.1096/fj.06-5668fje [DOI] [PubMed] [Google Scholar]
- Zheng T., Liu W., Oh S.Y., Zhu Z., Hu B., Homer R.J., Cohn L., Grusby M.J., Elias J.A. 2008. IL-13 receptor alpha2 selectively inhibits IL-13-induced responses in the murine lung. J. Immunol. 180:522–529 [DOI] [PubMed] [Google Scholar]
- Zhu Z., Homer R.J., Wang Z., Chen Q., Geba G.P., Wang J., Zhang Y., Elias J.A. 1999. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest. 103:779–788 10.1172/JCI5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer J.M., Sullivan C.S., Ganem D. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41:130–134 10.1038/ng.266 [DOI] [PMC free article] [PubMed] [Google Scholar]