Abstract
Genome-wide association studies (GWAS) and other genomic technologies have accelerated the discovery of genes and genomic regions contributing to common human ocular disorders with complex inheritance. Age-related macular degeneration (AMD), diabetic retinopathy (DR), glaucoma and myopia account for the majority of visual impairment worldwide. Over 19 genes and/or genomic regions have been associated with AMD. Current investigations are assessing the clinical utility of risk score panels and therapies targeting disease-specific pathways. DR is the leading cause of blindness in the United States and globally is a major cause of vision loss. Genomic investigations have identified molecular pathways associated with DR in animal models which could suggest novel therapeutic targets. Three types of glaucoma, primary-open-angle glaucoma (POAG), angle-closure glaucoma and exfoliation syndrome (XFS) glaucoma, are common age-related conditions. Five genomic regions have been associated with POAG, three with angle-closure glaucoma and one with XFS. Myopia causes substantial ocular morbidity throughout the world. Recent large GWAS have identified >20 associated loci for this condition. In this report, we present a comprehensive overview of the genes and genomic regions contributing to disease susceptibility for these common blinding ocular disorders and discuss the next steps toward translation to effective gene-based screening tests and novel therapies targeting the molecular events contributing to disease.
INTRODUCTION
Common age-related ocular disorders with complex inheritance are responsible for the majority of blindness worldwide. In the United States alone, >3 million individuals over the age of 40 (1) are visually impaired as a consequence of these conditions, and this number is expected to triple by 2020 (2). Effective disease surveillance and treatment will become increasingly important as the population ages. The identification of genetic risk factors contributing to common complex disease is the first step toward the development of gene-based screening tests and novel therapies targeted to the molecular events responsible for disease. Genome-wide association studies (GWAS) and other genomic analyses have successfully identified risk alleles for a large number of common complex human disorders (3), including diseases affecting vision. In this review, we summarize the recent advances in the genomics of the four disorders that are leading causes of visual impairment: age-related macular degeneration (AMD), diabetic retinopathy (DR), glaucoma and myopia (near-sightedness).
AGE-RELATED MACULAR DEGENERATION
AMD is a progressive neurodegenerative disease that diminishes the quality of life for millions of elderly individuals worldwide. In the United States, advanced AMD accounts for more than half of all blindness (4). Phenotypically, macular degeneration progresses from early stages (characterized by abnormalities in the retinal pigment epithelium and accumulation of extracellular deposits called ‘drusen’ in the macular region of the retina) to advanced or late stages with retinal neovascularization and scarring (especially in the macular region) and atrophy of the retinal pigment epithelium (geographic atrophy) (Fig. 1). AMD has both genetic and environmental contributions; smoking and age are the most consistently observed non-genetic risk factors (5). The genetic component of AMD is significant and has been well established (6–8), with great progress having been made in the past several years toward identifying contributing genetic loci. Significant loci include variants in CFH (9–11) and in several additional complement genes including complement 2 (C2), complement factor B (CFB) (12), complement 3 (C3) (13) and complement factor I (CFI) (14). Outside the complement genes, the major known genetic contributor to AMD risk resides in the ARMS2/HTRA region (15–18). More recently, variants near TIMP3, LIPC and CETP have been associated with AMD (19,20). Though several high-effect loci have been identified, these likely account for less than two-thirds of the genetic component of AMD (21). Significant gene–environment interactions have been identified for CFH and smoking (22) as well as CFH and antibodies to the bacterial pathogen Chlamydophila pneumonia (23). Both smoking and C. pneumonia would be expected to influence the inflammatory pathways that include CFH. The majority of GWAS have been carried out with advanced AMD cases; however, a recent GWAS investigated early-stage AMD cases and confirmed the involvement of CFH and ARMS2/HTRA in early-stage disease and also provided suggestive evidence for risk alleles specific to early AMD (24). Since preventative therapies would ideally be targeted to early-stage disease, further study of the genetic and environmental risk factors associated with disease onset is warranted. The AMDGene Consortium formed in 2010 (21), performed a genome-wide association study using 7650 advanced AMD cases and 51 844 controls, with follow-up in an additional 9531 cases and 8230 controls. Joint analysis detected 19 loci attaining genome-wide significance at P < 5 × 10–8. Significant loci included 12 previously identified variants in ARMS2-HTRA1, CFH, C2-CFB, C3, TIMP3, APOE, CETP, VEGFA, TNFRSF10A, LIPC, CFI, COL10A1, with odds ratios (ORs) ranging from 1.13 (COL10A1) to 2.71 (ARMS2-HTRA1). An additional seven variants in COL8A1-FILIP1L, IER3-DDR1, SLC16A8, TGFBR1, RAD51B, ADAMTS9 and B3GALTL met genome-wide significance for the first time, with ORs ranging from 1.11 (RAD51B) to 1.28 (COL8A10-FILIP1L) (21). Pathway analyses highlighted lipid metabolism, complement activation, angiogenesis and inflammation (Fig. 2).
Figure 1.
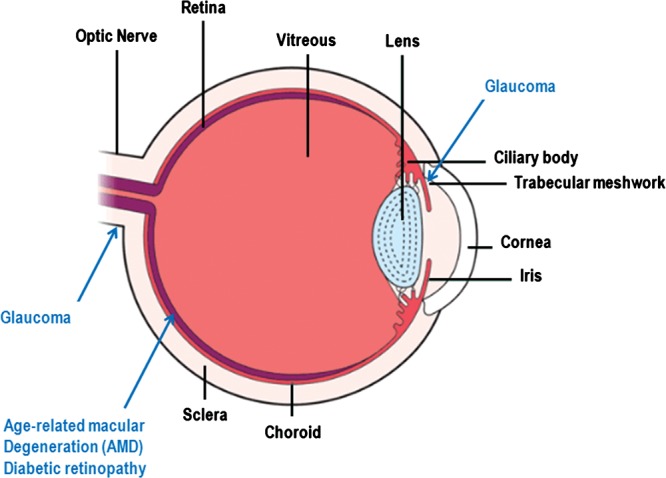
Schematic diagram of the eye. The basic structures of the eye are labeled in black font and the ocular tissues affected by the disorders discussed in this review are labeled in blue font. The function of the eye is to transduce a light signal to an electrical impulse. Light enters the eye through the cornea is focused by the lens and then stimulates the photoreceptors of the retina. The photoreceptors transduce the light signal into an electrical impulse that travels through the optic nerve to the brain. AMD affects the retina, and in particular the macular region of the retina responsible for high acuity vision. The retinal pigment epithelium (also a site of damage in AMD) is the outermost layer of the retina just beneath the photoreceptors. Diabetic retinopathy also affects the retina, frequently causing damage to the macular region and also other parts of the retina. Elevated IOP in glaucoma is caused by a reduction in the rate of removal of intraocular fluid by the trabecular meshwork. The visual compromise in glaucoma is caused by damage to the optic nerve and elevated IOP is a significant risk factor for optic nerve degeneration. Myopia, or near-sightedness, is created when the image focal point occurs in front of the retina (not shown on this figure). This can occur when the eye is too long for it's inherent focusing power.
Figure 2.
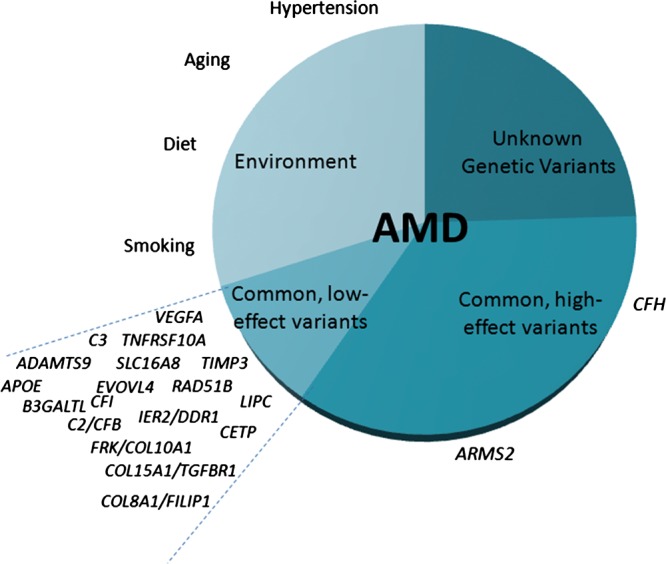
Genes and genomic regions associated with age-related macular degeneration (AMD). Genetic and environmental risk factors are presented. CFH (complement factor H) and ARMS2 (age-related maculopathy susceptibility 2) are major genetic risk factors for the disease.
The International Age related Macular Degeneration Genomics Consortium (IAMDGC) is currently assessing the ∼50 000 person dataset typed on a custom Illumina Exome array, which was designed to bridge the gap between association studies of common variants and sequencing studies of rare variants. Analysis of the ∼500 000 markers will provide improved coverage of known disease susceptibility loci through fine-mapping, enhance the power for discovery of disease loci in the largest single-variant analysis yet performed in AMD and facilitate assessment of rare coding variation and copy number variants. These data will also support further exploration of gene–environment interactions and gene association studies in early-stage as well as late-stage disease.
DIABETIC RETINOPATHY
DR is the leading cause of blindness in Americans between 20 and 74 years of age (25) and is rapidly becoming a common cause of visual impairment in developing countries (26). Diabetes causes injury to retinal blood vessels promoting a neovascularization response that causes further retinal damage especially due to retinal hemorrhage (Fig. 1). The frequency and severity of DR is heterogeneous (27,28). Known risk factors, most notably duration of diabetes and glycemic control, explain some, but not all, of the observed heterogeneity (28–30). Genetic variation may explain some of the remaining heterogeneity in DR development. Heritability has been estimated to be as high as 27% for DR and 52% for proliferative diabetic retinopathy (PDR), the more extreme phenotype (31–33).
Genomic investigations have confirmed and revealed pathways associated with DR. Retinal whole-genome microarray analyses in animal models of diabetes have detected gene expression changes indicating that pro-inflammatory, anti-vascular barrier and neurodegenerative pathways are involved in the disease (34). Candidate gene association studies have explored the contributions from these pathways to disease in humans however; the results have not been reliably reproduced (35–38). For example, strong evidence has been presented for an association between the T allele of rs1617640 in the erythropoietin promoter and PDR (39); however, a second, albeit smaller, study found the opposite allele of this same single nucleotide polymorphism (SNP) to be associated with DR risk (40). TCF7L2, a consistent risk locus for type 2 diabetes (T2D), has been studied in DR with both positive (41,42) and negative results (43).
GWAS for DR have also not produced any consistent risk loci. The first two published GWAS for DR, one in a Caucasian type 1 diabetes (T1D) population and the other in a Mexican-American T2D population, generated new candidate loci but these loci did not reach genome-wide significance (44,45). Replication of the loci from the T1D study was subsequently attempted without success (46). A third GWAS for DR reported variants that were associated with genome-wide significant P values in a Chinese T2D cohort but there was no independent replication attempted (47). The most recent GWAS of DR in Chinese participants with T2D also did not reveal any genome-wide significant loci (48).
There are several reasons why GWAS have yet to yield consistent findings. The genetic effects are likely to be modest and require large sample sizes to be identified. Data sets from diverse populations have not yet been combined to this end. Another explanation for the inability to replicate DR associations may lie in the heterogeneity among studies with regards to DR case and control definitions, participants' mean duration of diabetes and degree of glycemic control, and the underlying type of diabetes. Larger genomic studies with harmonized phenotyping, particularly examining the extreme and more heritable phenotype of PDR, will be important for uncovering true risk loci.
GLAUCOMA
Glaucoma is a neurodegenerative condition causing irreversible damage to the optic nerve. Most patients with vision loss due to optic nerve degeneration also have elevated intraocular pressure (IOP) caused by abnormal intraocular fluid dynamics (Fig. 1). The most common type of glaucoma, primary-open-angle glaucoma (POAG), has a significant heritability with a sibling risk between 5 and 10 times the population risk (49). Advances in genomic technologies coupled with the formation of consortia contributing appropriate numbers of cases and controls have facilitated genome-wide association studies identifying genes contributing to ocular quantitative traits related to glaucoma pathogenesis (IOP, cup/disc ratio (CDR) optic nerve size and central corneal thickness (CCT)), as well as genes associated with POAG, angle-closure glaucoma and glaucoma associated with exfoliation syndrome (XFS).
Quantitative traits related to glaucoma development are highly heritable and show substantial variation in human populations. IOP is a quantitative trait that is the only modifiable risk factor for glaucoma. Recent genome-wide analyses using normal populations have identified two genes significantly associated with IOP, GAS7 and TMCO1 (50). Similar analyses for optic nerve parameters associated with glaucoma risk have identified CDKN2BAS and SIX1SIX6 as genetic risk factors contributing to CDR (51), and ATOH7 as an important determinant of optic nerve size (52). Populations from around with world have been used to identify genetic factors contributing to CCT, one of the most heritable of the ocular quantitative traits (53–55). A recent study from the International Glaucoma Genetics Consortium identified 16 loci significantly associated with CCT (56) and showed that the collagen and extracellular matrix pathways are important regulators of CCT (Fig. 3).
Figure 3.
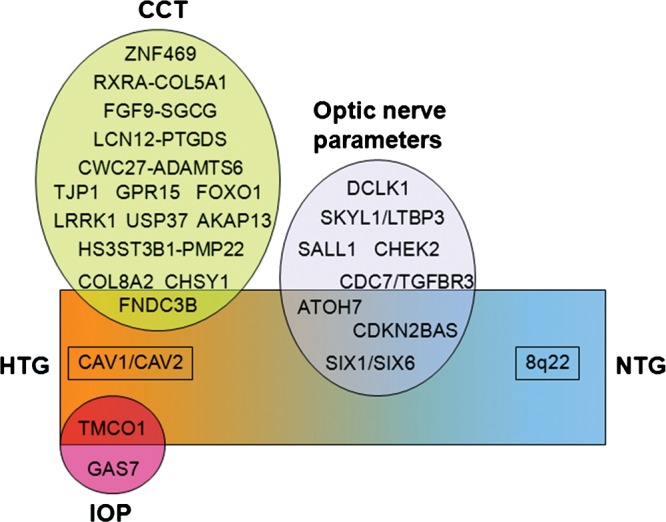
Genes and genomics regions associated with glaucoma and the related quantitative traits. Variants associated with ocular quantitative traits related to glaucoma and (ellipses) and POAG are presented (rectangle). POAG can be divided into high-tension (HTG) and normal-tension (NTG) subgroups. Of the two genes associated with elevated IOP, one (TMCO1) is also associated with HTG. One gene associated with thin CCT (FNDC3B) is also associated with HTG. Three genomic regions associated with optic nerve parameters (CDKN2BAS, ATOH7 and SIX1/SIX6) are associated with HTG, and one of these, CDKN2BAS, is associated with both HTG and NTG. The CAV1/CAV2 genomic region is associated with HTG but not with any quantitative trait. The 8q22 genomic region is associated with NTG only.
POAG is the most common type of glaucoma in the Western world. The disease results in a relentless progressive destruction of the optic nerve eventually causing permanent visual loss. Therapeutic strategies are currently limited to reducing optic nerve destruction by lowering IOP. Neuro-protective therapies are not currently available and a major goal of glaucoma genomic research is to identify potential therapeutic targets based on information about genes that influence susceptibility to optic nerve disease. Several GWAS for POAG have been completed. A study from Iceland using 1263 cases and over 34 877 controls identified variants in the CAV1/CAV2 intergenic region associated with POAG (57). This finding has been replicated in a study of Caucasian cases and controls from the USA (GLAUGEN) (58). Using 590 advanced glaucoma cases and 3956 controls, the CDKN2BAS (previously associated with CDR) and TMCO1 (also associated with IOP) genes were found to be associated with POAG in a study of Australian cases and controls (59). The CDKN2BAS and SIX1/SIX6 genes were also associated with POAG in 3500 cases and controls analyzed in a meta-analysis of the GLAUGEN and NEIGHBOR studies (60). POAG patients affected by the ‘normal-tension’ subtype of glaucoma (NTG) have increased susceptibility to optic nerve degeneration. The NEIGHBOR/GLAUGEN meta-analysis included a NTG subgroup analysis that showed significant association with CDKN2BAS as well as a highly conserved regulatory region 8q22 (60). These results suggest that CDKN2BAS, regulating expression of CDKN2B, an inhibitor of CDK4, and the 8q22 region primarily influence optic nerve degeneration for glaucoma and could be targets for neuro-protective therapies.
Angle-closure glaucoma is a major cause of blindness in Asia. The condition results when intraocular fluid cannot be removed by the trabecular meshwork (Fig. 1) because access is anatomically blocked by an abnormal configuration of the iris and other intraocular structures. Using 1854 angle-closure cases and 9608 controls from five different Asian populations a GWAS identified three associated loci: PLEKHA7, COL11A1 and an intergenic region between PCMTD1 and ST18 on chromosome 8q (61). All three of these loci were replicated in a second study using cases and controls from Nepal and Australia (62). PLEKHA7, encoding pleckstrin homology domain-containing protein 7, maintains a protein complex that regulates paracellular permeability of several ocular tissues including structures involved in angle-closure glaucoma (iris, trabecular meshwork). COL11A1 encodes one of the two alpha chains of type XI collagen and mutations in this gene are known to cause Stickler syndrome and other related disorders (63). Interestingly, myopia or near-sightedness is a feature of the Stickler syndromes, and the opposite refractive error (hyperopia) is more commonly associated with angle-closure glaucoma (64). COL11A1 is expressed in the scleral tissue which is implicated in both myopia and hyperopia (65,66). Little is currently known about the genes flanking the intergenic third locus on chromosome 8q.
XFS is an age-related disease characterized by the progressive accumulation of a fibrillar extracellular material in the trabecular meshwork where it causes a decrease in fluid removal and a corresponding increase inIOP. A GWAS using Icelandic cases and controls identified LOXL1 (lysyl oxidase like 1) as a major genetic risk factor for XFS (67), a finding that has been replicated in populations worldwide (68). LOXL1 is necessary for development and maintenance of elastin (69). Recent studies suggest that LOXL1 variants may influence disease development by reducing LOXL1 gene expression (70,71), causing a compromise of elastic structures in the eye. The frequency of LOXL1 risk alleles is high in both affected and unaffected individuals arguing that other factors, which could genetic or environmental, must also contribute to the disease (68).
MYOPIA
Myopia is the most common ocular disorder worldwide, with a significant ocular morbidity and impact on global public health. It also carries a huge economic burden, estimated to be $139 billion a year in the United States. The condition develops when the refractive power of the eye is not sufficient to place the focal point in the plane of the retina so that images come into focus in front of the retina (Fig. 1). While temporal and geographical changes in prevalence (affecting >80% young adults in urban East Asia) suggest important environmental influences (72), myopia is highly heritable. Before GWAS, numerous myopia loci were identified, but there were no known non-syndromic myopia genes. GWAS studies have involved either high-grade ‘pathological’ myopia case–control studies, or analysis of quantitative ‘healthy variation’ of refractive error using population-based cohorts.
The first high myopia GWAS, published from Japan in 2009, identified a locus on chromosome 11q24, albeit not at genome-wide significance (73). The following year two studies of over 4000 participants in the discovery phase each identified a single locus at genome-wide significance on chromosome 15, at 15q14 near the GJD2 gene in the Rotterdam Study (74), and near the RASGRF1 gene at 15q25 in the TwinsUK cohort (75). Both candidate genes, highly expressed in the retina, provide plausible biological candidates for myopia. GJD2 encodes a neuron-specific protein (connexin36) that is found in retinal photoreceptors, essential in the transmission of rod-mediated visual signals. RASGRF1 is regulated by muscarinic receptors (76) and retinoic acid, both implicated in myopia development in animal models.
The Consortium on Refractive Error and Myopia (CREAM) published an international meta-analysis using spherical equivalent data from over 45 000 participants, which included 37 382 individuals from 27 populations of European ancestry, and 8376 individuals from five Asian cohorts (77). In all, 26 loci were identified at genome-wide significance, including replication of the chromosome 15 regions. Genes identified were involved in neurotransmission (GRIA4), ion transport (KCNQ5, CD55, CHNRG), retinoic acid metabolism (RDH5, RORB, CYP26A1), extracellular matrix remodeling (LAMA2, BMP2) and eye development (SIX4, PRSS56, CHD7). Despite these discoveries, in common with most complex diseases, the significant associations only explained 3–4% of the variation. At the same time, the personal genomics company 23andMe performed an even larger GWAS with almost 26 000 myopic cases and 20 000 controls, using a Cox proportional hazards model of age of onset of myopia as a proxy for severity (a reasonable assumption), and identified 22 significantly associated loci (78). The similarity of results from two different studies using completely different methodologies was truly remarkable: 16 of the 20 novel loci identified by Kiefer et al. were confirmed by CREAM; and of the 22 loci discovered by the CREAM analyses, 14 were replicated by 23andMe and those regions not confirmed had suggestive associations in the other (79).
Further, GWAS meta-analyses have identified RBFOX1 on chromosome 16 as a candidate gene for refractive error susceptibility in European populations (80), and GWAS have identified genetic variants in high myopia studies in Chinese populations (81,82). Future larger GWAS of myopia will provide further evidence of genes, each of smaller effect. Exome sequencing holds promise of identifying genes, particularly in highly affected subjects (83,84), and in families with dominantly inherited high-grade myopia (85,86), though the relevance of these findings to ‘simple’ myopia remains uncertain.
FUTURE DIRECTIONS
A major goal of genomic research is to use genome-wide association findings to develop clinically useful gene-based tests and therapeutic strategies targeted to the disease-related molecular events (87). For AMD, considerable progress has been made in both areas. A SNP risk score combining information from 19 associated loci can distinguish cases and controls reasonably well (area under the receiver operator curve (AUC) = 0.74) (21), suggesting that a SNP score ‘test’ could be used to identify at risk individuals for preventive or preemptive treatment before the onset of disease (88). Ideally, SNP score tests could be used to identify individuals who would benefit most from specific therapies, such as anti-VEFG injections to control neovascularization (89). However, genetic variants contributing to anti-VEGF responsiveness have only been preliminarily identified (90) and further analyses will be needed before pharmacogenetic-based tests can be clinically useful. Additionally, the identification of CFH as a major risk allele for AMD has led to clinical trials investigating the efficacy of anti-complement therapies (91). Other loci associated with AMD could also be targets for novel therapies. The association with lipid pathways suggests that lipid profiles may be clinically useful biomarkers (92); however, these results have not been consistently observed and require further research for confirmation (93). SNP risk scores based on current and new genes associated with glaucoma could also be clinically useful in the future. The identification of novel genes and pathways contributing to glaucoma will also help define disease-specific targets for novel therapeutic approaches. Genomic studies using larger sample sizes, including whole-exome analyses, could lead to the discovery of significant loci for DR. A future area of interest in myopia research is to understand the interaction between associated genes and environmental effects. Through these and other ongoing efforts novel gene-based tests and therapies for common ocular disease can help reduce the global burden of visual impairment.
Conflict of Interest statement. None declared.
FUNDING
The authors acknowledge funding support from NIH/NEI grants EY022302 (L.S.), EY012118 (M.A.P.-V.), EY021453 (J.L.H.), EY022305 (J.L.W.), EY020928 (J.L.W.).
REFERENCES
- 1.Eye Diseases Prevalence Research Group. Causes and prevalence of visual impairment among adults in the United States. Arch. Ophthal. 2004;122:477–485. doi: 10.1001/archopht.122.4.477. [DOI] [PubMed] [Google Scholar]
- 2.Ko F., Vitale S., Chou C.F., Cotch M.F., Saaddine J., Friedman D.S. Prevalence of nonrefractive visual impairment in US adults and associated risk factors, 1999–2002 and 2005–2008. JAMA. 2012;308:2361–2368. doi: 10.1001/jama.2012.85685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manolio T.A. Genomewide association studies and assessment of the risk of disease. N. Engl. J. Med. 2010;363:166–176. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]
- 4.Friedman D.S., O'Colmain B.J., Muñoz B., Tomany S.C., McCarty C., de Jong P.T., Nemesure B., Mitchell P., Kempen J. Eye Diseases Prevalence Research Group. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 5.Clemons T.E., Milton R.C., Klein R., Seddon J.M., Ferris F.L., III Age-Related Eye Disease Study Research Group. Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology. 2005;112:533–539. doi: 10.1016/j.ophtha.2004.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heiba I.M., ELston R.C., Klein B.E., Klein R. Sibling correlations and segregation analysis of age-related maculopathy: the Beaver Dam Eye Study. Genet. Epidemiol. 1994;11:51–67. doi: 10.1002/gepi.1370110106. [DOI] [PubMed] [Google Scholar]
- 7.Klaver C.C., Wolfs R.C., Assink J.J., Van Duijn C.M., Hofman A., de Jong P.T. Genetic risk of age-related maculopathy. Population-based familial aggregation study. Arch. Ophthalmol. 1998;1:1646–1651. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 8.Seddon J.M., Ajani U.A., Mitchell B.D. Familial aggregation of age-related maculopathy. Am. J. Ophthalmol. 1997;123:199–206. doi: 10.1016/s0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 9.Edwards A.O., Ritter R., III, Abel K.J., Manning A., Panhuysen C., Farrer L.A. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;572:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 10.Haines J.L., Hauser M.A., Schmidt S., Scott W.K., Olson L.M., Gallins P., Spencer K.L., Kwan S.Y., Noureddine M., Gilbert J.R., et al. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;572:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 11.Klein R.J., Zeiss C., Chew E.Y., Tsai J.Y., Sackler R.S., Haynes C., Henning A.K., SanGiovanni J.P., Mane S.M., Mayne S.T., et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;572:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold B., Merriam J.E., Zernant J., Hancox L.S., Taiber A.J., Gehrs K., Cramer K., Neel J., Bergeron J., Barile G.R., et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006;36:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yates J.R., Sepp T., Matharu B.K., Khan J.C., Thurlby D.A., Shahid H., Clayton D.G., Hayward C., Morgan J., Wright A.F., et al. Complement C3 variant and the risk of age-related macular degeneration. N. Engl. J. Med. 2007;6:553–561. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 14.Fagerness J.A., Maller J.B., Neale B.M., Reynolds R.C., Daly M.J., Seddon J.M. Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet. 2009;17:100–104. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dewan A., Liu M., Hartman S., Zhang S.S., Liu D.T., Zhao C., Tam P.O., Chan W.M., Lam D.S., Snyder M., et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science. 2006;314:989–992. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 16.Jakobsdottir J., Conley Y.P., Weeks D.E., Mah T.S., Ferrell R.E., Gorin M.B. Susceptibility genes for age-related maculopathy on chromosome 10q26. Am. J. Hum. Genet. 2005;77:389–407. doi: 10.1086/444437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maller J., George S., Purcell S., Fagerness J., Altshuler D., Daly M.J., Seddon J.M. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat. Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 18.Rivera A., Fisher S.A., Fritsche L.G., Keilhauer C.N., Lichtner P., Meitinger T., Weber B.H. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum. Mol. Genet. 2005;2:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Stambolian D., Edwards A.O., Branham K.E., Othman M., Jakobsdottir J., Tosakulwong N., Pericak-Vance M.A., Campochiaro P.A., Klein M.L., et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc. Natl Acad. Sci. USA. 2010;16:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neale B.M., Fagerness J., Reynolds R., Sobrin L., Parker M., Raychaudhuri S., Tan P.L., Oh E.C., Merriam J.E., Souied E., et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC) Proc. Natl Acad. Sci. USA. 2010;107:7395–7400. doi: 10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fritsche L.G., Chen W., Schu M., Yaspan B.L., Yu Y., Thorleifsson G., Zack D.J., Arakawa S., Cipriani V., Ripke S., et al. Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013;45:433–439. doi: 10.1038/ng.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schaumberg D.A., Hankinson S.E., Guo Q., Rimm E., Hunter D.J. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch. Ophthalmol. 2007;125:55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 23.Baird P.N., Robman L.D., Richardson A.J., Dimitrov P.N., Tikellis G., McCarty C.A., Guymer R.H. Gene-environment interaction in progression of AMD: the CFH gene, smoking and exposure to chronic infection. Hum. Mol. Genet. 2008;17:1299–1305. doi: 10.1093/hmg/ddn018. [DOI] [PubMed] [Google Scholar]
- 24.Holliday E.G., Smith A.V., Cornes B.K., Buitendijk G.H., Jensen R.A., Sim X., Aspelund T., Aung T., Baird P.N., Boerwinkle E., et al. Insights into the genetic architecture of early stage age-related macular degeneration: a genome-wide association study meta-analysis. PLoS ONE. 2013;8:e53830. doi: 10.1371/journal.pone.0053830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.National Institute of Diabetes and Digestive and Kidney Diseases. National Diabetes Statistics Fact Sheet: General Information and National Estimates on Diabetes in the United States. Bethesda, MD: U.S. Department of Health and Human Services, National Institute of Health; 2000. Publication No. 02–3892. [Google Scholar]
- 26.Ruta L.M., Magliano D.J., Lemesurier R., Taylor H.R., Zimmet P.Z., Shaw J.E. Prevalence of diabetic retinopathy in Type 2 diabetes in developing and developed countries. Diabet. Med. 2013;30:387–398. doi: 10.1111/dme.12119. [DOI] [PubMed] [Google Scholar]
- 27.Nathan D.M. Long-term complications of diabetes mellitus. N. Engl. J. Med. 1993;328:1676–1685. doi: 10.1056/NEJM199306103282306. [DOI] [PubMed] [Google Scholar]
- 28.Klein R., Klein B.E., Moss S.E., Cruickshanks K.J. The Wisconsin Epidemiologic Study of Diabetic Retinopathy: XVII. The 14-year incidence and progression of diabetic retinopathy and associated risk factors in type 1 diabetes. Ophthalmology. 1998;105:1801–1815. doi: 10.1016/S0161-6420(98)91020-X. [DOI] [PubMed] [Google Scholar]
- 29.Klein R., Klein B.E., Moss S.E., Cruickshanks K.J. Relationship of hyperglycemia to the long-term incidence and progression of diabetic retinopathy. Arch. Intern. Med. 1994;154:2169–2178. [PubMed] [Google Scholar]
- 30.Klein R., Klein B.E., Moss S.E., Linton K.L. The Beaver Dam Eye Study. Retinopathy in adults with newly discovered and previously diagnosed diabetes mellitus. Ophthalmology. 1992;99:58–62. doi: 10.1016/s0161-6420(92)32011-1. [DOI] [PubMed] [Google Scholar]
- 31.Looker H.C., Nelson R.G., Chew E., Klein R., Klein B.E., Knowler W.C., Hanson R.L. Genome-wide linkage analyses to identify loci for diabetic retinopathy. Diabetes. 2007;56:1160–1166. doi: 10.2337/db06-1299. [DOI] [PubMed] [Google Scholar]
- 32.Hietala K., Forsblom C., Summanen P., Groop P.H. Heritability of proliferative diabetic retinopathy. Diabetes. 2008;57:2176–2180. doi: 10.2337/db07-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arar N.H., Freedman B.I., Adler S.G., Iyengar S.K., Chew E.Y., Davis M.D., Satko S.G., Bowden D.W., Duggirala R., Elston R.C., et al. Heritability of the severity of diabetic retinopathy: the FIND-Eye study. Invest. Ophthalmol. Vis. Sci. 2008;49:3839–3845. doi: 10.1167/iovs.07-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brucklacher R.M., Patel K.M., VanGuilder H.D., Bixler G.V., Barber A.J., Antonetti D.A., Lin C.M., LaNoue K.F., Gardner T.W., Bronson S.K., Freeman W.M. Whole genome assessment of the retinal response to diabetes reveals a progressive neurovascular inflammatory response. BMC Med. Genomics. 2008;1:26. doi: 10.1186/1755-8794-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abhary S., Hewitt A.W., Burdon K.P., Craig J.E. A systematic meta-analysis of genetic association studies for diabetic retinopathy. Diabetes. 2009;58:2137–2147. doi: 10.2337/db09-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hirschhorn J.N., Lohmueller K., Byrne E., Hirschhorn K. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 37.Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 38.Sobrin L., Green T., Sim X., Jensen R.A., Tai E.S., Tay W.T., Wang J.J., Mitchell P., Sandholm N., Liu Y., et al. Candidate gene association study for diabetic retinopathy in persons with type 2 diabetes: the Candidate Gene Association Resource (CARe) Invest. Ophthalmol. Vis. Sci. 2011;52:7593–7602. doi: 10.1167/iovs.11-7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong Z., Yang Z., Patel S., Chen H., Gibbs D., Yang X., Hau V.S., Kaminoh Y., Harmon J., Pearson E., et al. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc. Natl Acad. Sci. USA. 2008;105:6998–7003. doi: 10.1073/pnas.0800454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abhary S., Burdon K.P., Casson R.J., Goggin M., Petrovsky N.P., Craig J.E. Association between erythropoietin gene polymorphisms and diabetic retinopathy. Arch. Ophthalmol. 2010;128:102–106. doi: 10.1001/archophthalmol.2009.355. [DOI] [PubMed] [Google Scholar]
- 41.Ciccacci C., Di Fusco D., Cacciotti L., Morganti R., D'Amato C., Novelli G., Sangiuolo F., Spallone V., Borgiani P. TCF7L2 gene polymorphisms and type 2 diabetes: association with diabetic retinopathy and cardiovascular autonomic neuropathy. Acta Diabetol. 2012 doi: 10.1007/s00592-012-0418-x. Jul 28 Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Luo J., Zhao L., Chen A.Y., Zhang X., Zhu J., Zhao J., Quyang H., Luo H., Song Y., Lee J., et al. TCF7L2 variation and proliferative diabetic retinopathy. Diabetes. 2013;62:2613–2617. doi: 10.2337/db12-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buraczynska M., Swatowski A., Markowska-Gosik D., Kuczmaszewska A., Ksiazek A. Transcription factor 7-like 2 (TCF7L2) gene polymorphism and complication/comorbidity profile in type 2 diabetes patients. Diabetes Res. Clin. Pract. 2011;93:390–395. doi: 10.1016/j.diabres.2011.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Fu Y.P., Hallman D.M., Gonzalez V.H., Klein B.E., Klein R., Hayes M.G., Cox N.J., Bell G.I., Hanis C.L. Identification of diabetic retinopathy genes through a genome-wide association study among Mexican-Americans from Starr County, Texas. J. Ophthalmol. 2010 doi: 10.1155/2010/861291. 2010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grassi M.A., Tikhomirov A., Ramalingam S., Below J.E., Cox N.J., Nicolae D.L. Genome-wide meta-analysis for severe diabetic retinopathy. Hum. Mol. Genet. 2011;20:2472–2481. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grassi M.A., Tikhomirov A., Ramalingam S., Lee K.E., Hosseini S.M., Klein B.E., Klein R., Lussier Y.A., Cox N.J., Nicolae D.L. Replication analysis for severe diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 2012;53:2377–2381. doi: 10.1167/iovs.11-8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y.C., Lin J.M., Lin H.J., Chen C.C., Chen S.Y., Tsai C.H., Tsai F.J. Genome-wide association study of diabetic retinopathy in a Taiwanese population. Ophthalmology. 2011;118:642–648. doi: 10.1016/j.ophtha.2010.07.020. [DOI] [PubMed] [Google Scholar]
- 48.Sheu W.H., Kuo J.Z., Lee I.T., Hung Y.J., Lee W.J., Tsai H.Y., Wang J.S., Goodarzi M.O., Klein R., Klein B.E., et al. Genome-wide association study in a Chinese population with diabetic retinopathy. Hum. Mol. Genet. 2013;22:3165–3173. doi: 10.1093/hmg/ddt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Harmon J., Zabrieskie N., Chen Y., Grob S., Williams B., Lee C., Kasuga D., Shaw P.X., Buehler J., Wang N., Zhang K. Using the Utah Population Database to assess familial risk of primary open angle glaucoma. Vision Res. 2010;50:2391–2395. doi: 10.1016/j.visres.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 50.van Koolwijk L.M., Ramdas W.D., Ikram M.K., Jansonius N.M., Pasutto F., Hysi P.G., Macgregor S., Janssen S.F., Hewitt A.W., Viswanathan A.C., et al. Common genetic determinants of intraocular pressure and primary open-angle glaucoma. PLoS Genet. 2012;8:e1002611. doi: 10.1371/journal.pgen.1002611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramdas W.D., van Koolwijk L.M., Ikram M.K., Jansonius N.M., de Jong P.T., Bergen A.A., Isaacs A., Amin N., Aulchenko Y.S., Wolfs R.C., et al. A genome-wide association study of optic disc parameters. PLoS Genet. 2010;6:e1000978. doi: 10.1371/journal.pgen.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macgregor S., Hewitt A.W., Hysi P.G., Ruddle J.B., Medland S.E., Henders A.K., Gordon S.D., Andrew T., McEvoy B., Sanfilippo P.G., et al. Genome-wide association identifies ATOH7 as a major gene determining human optic disc size. Hum. Mol. Genet. 2010;19:2716–2724. doi: 10.1093/hmg/ddq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu Y., Dimasi D.P., Hysi P.G., Hewitt A.W., Burdon K.P., Toh T., Ruddle J.B., Li Y.J., Mitchell P., Healey P.R., et al. Common genetic variants near the Brittle Cornea Syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vitart V., Bencić G., Hayward C., Skunca Herman J., Huffman J., Campbell S., Bućan K., Navarro P., Gunjaca G., Marin J., et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum. Mol. Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 55.Vithana E.N., Aung T., Khor C.C., Cornes B.K., Tay W.T., Sim X., Lavanya R., Wu R., Zheng Y., Hibberd M.L., et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum. Mol. Genet. 2011;20:649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 56.Lu Y., Vitart V., Burdon K.P., Khor C.C., Bykhovskaya Y., Mirshahi A., Hewitt A.W., Koehn D., Hysi P.G., Ramdas W.D., et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet. 2013;45:155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thorleifsson G., Walters G.B., Hewitt A.W., Masson G., Helgason A., DeWan A., Sigurdsson A., Jonasdottir A., Gudjonsson S.A., Magnusson K.P., et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat. Genet. 2010;42:906–909. doi: 10.1038/ng.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiggs J.L., Kang J.H., Yaspan B.L., Mirel D.B., Laurie C., Crenshaw A., Brodeur W., Gogarten S., Olson L.M., Abdrabou W., et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma in Caucasians from the USA. Hum. Mol. Genet. 2011;20:4707–4713. doi: 10.1093/hmg/ddr382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burdon K.P., Macgregor S., Hewitt A.W., Sharma S., Chidlow G., Mills R.A., Danoy P., Casson R., Viswanathan A.C., Liu J.Z., et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat. Genet. 2011;43:574–578. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 60.Wiggs J.L., Yaspan B.L., Hauser M.A., Kang J.H., Allingham R.R., Olson L.M., Abdrabou W., Fan B.J., Wang D.Y., Brodeur W., et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8:e1002654. doi: 10.1371/journal.pgen.1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vithana E.N., Khor C.C., Qiao C., Nongpiur M.E., George R., Chen L.J., Do T., Abu-Amero K., Huang C.K., Low S., et al. Genome-wide association analyses identify three new susceptibility loci for primary angle closure glaucoma. Nat. Genet. 2012;44:1142–1146. doi: 10.1038/ng.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Awadalla M.S., Thapa S.S., Hewitt A.W., Burdon K.P., Craig J.E. Association of variants with primary angle closure glaucoma in two different populations. PLoS ONE. 2013;8:e67903. doi: 10.1371/journal.pone.0067903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Richards A.J., McNinch A., Martin H., Oakhill K., Rai H., Waller S., Treacy B., Whittaker J., Meredith S., Poulson A., et al. Stickler syndrome and the vitreous phenotype: mutations in COL2A1 and COL11A1. Hum. Mutat. 2010;31:E1461–E1471. doi: 10.1002/humu.21257. [DOI] [PubMed] [Google Scholar]
- 64.Rosman M., Zheng Y., Lamoureux E., Saw S.M., Aung T., Tay W.T., Wang J.J., Mitchell P., Tai E.S., Wong T.Y. Review of key findings from the Singapore Malay Eye Study (SiMES-1) Singapore Med. J. 2012;53:82–87. [PubMed] [Google Scholar]
- 65.McBrien N.A. Regulation of scleral metabolism in myopia and the role of transforming growth factor-beta. Exp. Eye Res. 2013 doi: 10.1016/j.exer.2013.01.014. 2013 Feb 8 ePUB ahead of print. [DOI] [PubMed] [Google Scholar]
- 66.Christian P.G., Harkin D.G., Rayner C., Schmid K.L. Comparative effects of posterior eye cup tissues from myopic and hyperopic chick eyes on cultured scleral fibroblasts. Exp. Eye Res. 2013;107:11–20. doi: 10.1016/j.exer.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Thorleifsson G., Magnusson K.P., Sulem P., Walters G.B., Gudbjartsson D.F., Stefansson H., Jonsson T., Jonasdottir A., Jonasdottir A., Stefansdottir G., et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 68.Fan B.J., Pasquale L.R., Rhee D., Li T., Haines J.L., Wiggs J.L. LOXL1 promoter haplotypes are associated with exfoliation syndrome in a U.S. Caucasian population. Invest. Ophthalmol. Vis. Sci. 2011;52:2372–2378. doi: 10.1167/iovs.10-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X., Zhao Y., Gao J., Pawlyk B., Starcher B., Spencer J.A., Yanagisawa H., Zuo J., Li T. Elastic fiber homeostasis requires lysyl oxidase-like 1 protein. Nat. Genet. 2004;36:178–182. doi: 10.1038/ng1297. [DOI] [PubMed] [Google Scholar]
- 70.Schlötzer-Schrehardt U., Pasutto F., Sommer P., Hornstra I., Kruse F.E., Naumann G.O., Reis A., Zenkel M. Genotype-correlated expression of lysyl oxidase-like 1 in ocular tissues of patients with pseudoexfoliation syndrome/glaucoma and normal patients. Am. J. Pathol. 2008;173:1724–1735. doi: 10.2353/ajpath.2008.080535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zenkel M., Krysta A., Pasutto F., Juenemann A., Kruse F.E., Schlötzer-Schrehardt U. Regulation of lysyl oxidase-like 1 (LOXL1) and elastin-related genes by pathogenic factors associated with pseudoexfoliation syndrome. Invest. Ophthalmol. Vis. Sci. 2011;52:8488–8495. doi: 10.1167/iovs.11-8361. [DOI] [PubMed] [Google Scholar]
- 72.Morgan I.G., Ohno-Matsui K., Saw S.M. Myopia. Lancet. 2012;379:1739–1748. doi: 10.1016/S0140-6736(12)60272-4. [DOI] [PubMed] [Google Scholar]
- 73.Nakanishi H., Yamada R., Gotoh N., Hayashi H., Yamashiro K., Shimada N., Ohno-Matsui K., Mochizuki M., Saito M., Iida T., et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;9:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Solouki A.M., Verhoeven V.J., van Duijn C.M., Verkerk A.J., Ikram M.K., Hysi P.G., Despriet D.D., van Koolwijk L.M., Ho L., Ramdas W.D., et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat. Genet. 2010;42:897–901. doi: 10.1038/ng.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hysi P.G., Young T.L., Mackey D.A., Andrew T., Fernández-Medarde A., Solouki A.M., Hewitt A.W., Macgregor S., Vingerling J.R., Li Y.J., et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat. Genet. 2010;42:902–905. doi: 10.1038/ng.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mattingly R.R., Macara I.G. Phosphorylation-dependent activation of the Ras-GRF/CDC25Mm exchange factor by muscarinic receptors and G-protein beta gamma subunits. Nature. 1996;382:268–272. doi: 10.1038/382268a0. [DOI] [PubMed] [Google Scholar]
- 77.Verhoeven V.J., Hysi P.G., Wojciechowski R., Fan Q., Guggenheim J.A., Höhn R., MacGregor S., Hewitt A.W., Nag A., Cheng C.Y., et al. Genome-wide meta-analyses of multi ancestry cohorts identify multiple new susceptibility loci for refractive error and myopia. Nat. Genet. 2013;45:314–318. doi: 10.1038/ng.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kiefer A.K., Tung J.Y., Do C.B., Hinds D.A., Mountain J.L., Francke U., Eriksson N. Genome-wide analysis points to roles for extracellular matrix remodeling, the visual cycle, and neuronal development in myopia. PLoS Genet. 2013;9:e1003299. doi: 10.1371/journal.pgen.1003299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wojciechowski R., Hysi P.G. Focusing in on the complex genetics of Myopia. PLoS Genet. 2013;9:e1003442. doi: 10.1371/journal.pgen.1003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stambolian D., Wojciechowski R., Oexle K., Pirastu M., Li X., Raffel L.J., Cotch M.F., Chew E.Y., Klein B., Klein R., et al. Meta-analysis of genome-wide association studies in five cohorts reveals common variants in RBFOX1, a regulator of tissue-specific splicing, associated with refractive error. Hum. Mol. Genet. 2013;22:2754–2764. doi: 10.1093/hmg/ddt116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li Z., Qu J., Xu X., Zhou X., Zou H., Wang N., Li T., Hu X., Zhao Q., Chen P., et al. A genome-wide association study reveals association between common variants in an intergenic region of 4q25 and high-grade myopia in the Chinese Han population. Hum. Mol. Genet. 2013;20:2861–2868. doi: 10.1093/hmg/ddr169. [DOI] [PubMed] [Google Scholar]
- 82.Shi Y., Gong B., Chen L., Zuo X., Liu X., Tam P.O., Zhou X., Zhao P., Lu F., Qu J., et al. A genome-wide meta-analysis identifies two novel loci associated with high myopia in the Han Chinese population. Hum. Mol. Genet. 2013;22:2325–2333. doi: 10.1093/hmg/ddt066. [DOI] [PubMed] [Google Scholar]
- 83.Shi Y., Li Y., Zhang D., Zhang H., Li Y., Lu F., Liu X., He F., Gong B., Cai L., et al. Exome sequencing identifies ZNF644 mutations in high myopia. PLoS Genet. 2011;7:e1002084. doi: 10.1371/journal.pgen.1002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soler V.J., Tran-Viet K.N., Galiacy S.D., Limviphuvadh V., Klemm T.P., St Germain E., Fournié P.R., Guillaud C., Maurer-Stroh S., Hawthorne F., et al. Study of a US cohort supports the role of ZNF644 and high-grade myopia susceptibility. Mol. Vis. 2012;18:937–944. [PMC free article] [PubMed] [Google Scholar]
- 85.Tran-Viet K.N., Powell C., Barathi V.A., Klemm T., Maurer-Stroh S., Limviphuvadh V., Soler V., Ho C., Yanovitch T., Schneider G., et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am. J. Hum. Genet. 2013;92:820–826. doi: 10.1016/j.ajhg.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aldahmesh M.A., Khan A.O., Alkuraya H., Adly N., Anazi S., Al-Saleh A.A., Mohamed J.Y., Hijazi H., Prabakaran S., Tacke M., et al. Mutations in LRPAP1 are associated with severe myopia in humans. Am. J. Hum. Genet. 2013 doi: 10.1016/j.ajhg.2013.06.002. 2013 Jul 2 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Manolio T.A. Bringing genome-wide association findings into clinical use. Nat. Rev. Genet. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 88.Seddon J.M., Reynolds R., Yu Y., Daly M.J., Rosner B. Risk models for progression to advanced age-related macular degeneration using demographic, environmental, genetic, and ocular factors. Ophthalmology. 2011;118:2203–2211. doi: 10.1016/j.ophtha.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hagstrom S.A., Ying G.S., Pauer G.J., Sturgill-Short G.M., Huang J., Callanan D.G., Kim I.K., Klein M.L., Maguire M.G., Martin D.F., et al. Pharmacogenetics for genes associated with age-related macular degeneration in the Comparison of AMD Treatments Trials (CATT) Ophthalmology. 2013;120:593–599. doi: 10.1016/j.ophtha.2012.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhao L., Grob S., Avery R., Kimura A., Pieramici D., Lee J., Rabena M., Ortiz S., Quach J., Cao G., et al. Common variant in VEGFA and response to anti-VEGF therapy for neovascular age-related macular degeneration. Curr. Mol. Med. 2013;13:929–934. doi: 10.2174/15665240113139990048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Troutbeck R., Al-Qureshi S., Guymer R.H. Therapeutic targeting of the complement system in age-related macular degeneration: a review. Clin. Exp. Ophthalmol. 2012;40:18–26. doi: 10.1111/j.1442-9071.2011.02581.x. [DOI] [PubMed] [Google Scholar]
- 92.Munch I.C., Linneberg A., Larsen M. Precursors of age-related macular degeneration: associations with physical activity, obesity and serum lipids in the Inter99 Eye Study. Invest. Ophthalmol.Vis. Sci. 2013;54:3932–3940. doi: 10.1167/iovs.12-10785. [DOI] [PubMed] [Google Scholar]
- 93.Kabasawa S., Mori K., Horie-Inoue K., Gehlbach P.L., Inoue S., Awata T., Katayama S., Yoneya S. Associations of cigarette smoking but not serum fatty acids with age-related macular degeneration in a Japanese population. Ophthalmology. 2011;118:1082–1088. doi: 10.1016/j.ophtha.2010.10.012. [DOI] [PubMed] [Google Scholar]


