Abstract
GGGGCC repeat expansions in the C9orf72 gene have been identified as a major contributing factor in patients with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Given the overlapping of clinical phenotypes and pathological characteristics between these two diseases and Alzheimer's disease (AD), Parkinson's disease (PD), and essential tremor (ET), we speculated regarding whether C9orf72 repeat expansions also play a major role in these three diseases. Using the repeat-primed polymerase chain reaction method, we screened for C9orf72 in three groups of patients with PD (n = 911), AD (n = 279), and ET (n = 152) in the Chinese Han population. There were no pathogenic repeats (>30 repeats) detected in either the patients or controls (n = 314), which indicated that the pathogenic expansions of C9orf72 might be rare in these three diseases. However, the analysis of the association between the number of repeats (p = 0.001), short/intermediate genotype (short: <7 repeats; intermediate: ≥7 repeats) (odds ratio 1.37 [1.05, 1.79]), intermediate/intermediate genotype (Odds ratio 2.03 [1.17, 3.54]), and PD risks indicated that intermediate repeat alleles could act as contributors to PD. To the best of our knowledge, this study is the first to reveal the correlation between C9orf72 and Chinese PD, AD, or ET patients. Additionally, the results of this study suggest the novel idea that the intermediate repeat allele in C9orf72 is most likely a risk factor for PD.
Keywords: C9orf72, Alzheimer's disease, Parkinson's disease, essential tremor, risk factor
Introduction
A hexanucleotide (GGGGCC) repeat expansion in the first intron of the C9orf72 gene was recently identified as a major contributing factor to the chromosome 9p21-linked diseases amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (Dejesus-Hernandez et al., 2011). As previously reported, the C9orf72 mutation accounts for 23.5–47% of familial ALS/FTD and 4.1–21.0% of sporadic ALS in white populations (Dejesus-Hernandez et al., 2011; Renton et al., 2011; Gijselinck et al., 2012). The pathogenic mechanism of repeat expansions primarily includes interference with the normal expression of the encoded protein or the loss of protein function through the generation of abnormal toxic RNA foci that disrupt normal cellular pathways (Renton et al., 2011; Sha and Boxer, 2012).
There is no doubt that the overlapping presentations of clinical phenotypes, pathological characteristics, and gene mutations exist among Parkinson disease (PD), Alzheimer's disease (AD), and ALS/FTD (Hudson, 1981; Piguet et al., 2011; Arighi et al., 2012; Floris et al., 2012; O'Dowd et al., 2012). First, in the examination of clinical phenotypes, relatives of patients with ALS have an increased risk for developing PD and AD, additionally, some ALS/FTD patients have developed the associated features of Parkinsonism and movement disorders (Hsiung et al., 2012; Takada et al., 2012; Kohli et al., 2013). Second, the presence of TAR DNA-binding protein-43(+) intranuclear inclusions, which are the pathological feature of chromosome 9p21-linked ALS/FTD, have been detected in PD and AD patients (Nakashima-Yasuda et al., 2007; Boeve et al., 2012). Finally, mutations in the microtubule-associated protein tau (MAPT) gene could cause a spectrum of phenotypes which include ALS, Parkinsonism, and cognitive impairment (O'Dowd et al., 2012). Given the considerations above, one question has yet to be addressed. Could the C9orf72 repeat expansions account for other neurodegenerative disorders, such as AD, PD, and essential tremor (ET)?
In C9orf72, Repeat expansions exceeding 30 units have been suggested to be pathological in ALS/FTD patients (Dejesus-Hernandez et al., 2011). Interestingly, pathogenic expansions have also been observed in patients with PD, AD, progressive supranuclear palsy, corticobasal degeneration, and Lewy body dementia (Xi et al., 2012; Cacace et al., 2013; Lesage et al., 2013), which further indicates that the phenotypes that are associated with repeat expansions could include the spectrum of cognitive impairment and movement disorder syndromes. In addition, a previous study has demonstrated that the role of intermediate repeats (7–24 repeat units) is strongly associated with these diseases and the expression of C9orf72. The significantly decreased transcriptional activity of C9orf72 with an increasing number of normal repeats indicates that intermediate repeats may act as predisposing alleles and favors the loss-of-function disease mechanism (Van Der Zee et al., 2013).
In this study, we first assess the prevalence of C9orf72 repeat expansions in a large cohort of Chinese Han patients with AD, PD, or ET to determine whether repeat expansions play a role in these three common disorders. Furthermore, we explore whether repeat expansions of intermediate repeats might be a risk factor for AD, PD, or ET, and/or could affect the age at onset in patients with these three diseases.
Materials and methods
Study samples
Three independent series of patients participated in this study: the first cohort of 911 sporadic PD patients that met the UK brain bank diagnosis criteria (Hughes et al., 1992); the second cohort of 279 sporadic AD patients that met the NINCDS-ADRDA criteria for probable or definite AD (McKhann et al., 1984); and the third cohort of 152 ET patients that met the Washington Heights-Inwood Genetic Study of ET (WHIGET) diagnosis criteria (Louis et al., 1997). All patients were recruited from the outpatient neurology clinics of the Xiangya Hospital, Central South University. In total, 314 healthy Chinese individuals were recruited from the Xiangya Wellness Center as a control group. Informed consents for participation in the study were obtained from all subjects, including patients and controls. This study received prior approval by the institutional review board and the ethics committee of the Xiangya Hospital, Central South University.
Methods
Genomic DNA was isolated from peripheral blood leukocytes using a QIAGEN kit. We screened the presence of the GGGGCC hexanucleotide expansion of C9orf72 using a 2-step polymerase chain reaction protocol. In the first step, we used a previously reported repeat-primed polymerase chain reaction assay to detect the size of the larger expanded alleles (Dejesus-Hernandez et al., 2011). Briefly, DNA samples (50 ng/μ l) were amplified using three primers (MRX-F: 5′FAM-ACAGTACTCGCTGAGGGTGAAC; MRX-R1: 5′CAGGAAACAGCTATGACCGGGCCCGCCCCGACCACGCCCCGGCCCCGGCCCCGG; MRX-M13R: 5′CAAGGAAACAGCTATGACC), and the primers ratio (0.6 μ l of 10 μ M of MRX-F; 0.6 μ l of 10 μ M of MRX-M13R; 0.1 μ l of 10 μ M of MRX-R1) were modified to improve the efficiency of the PCR. Other components of the PCR reaction included the following: 1_μ l of 50_ng/μ l of DNA samples, 2_μ l of 5× Q-solution (QIAGEN Valencia, CA, USA), 0.7 μ l of 100% DMSO (Sigma-Aldrich), 0.36 μ l of 7-deaza-dGTP (New England Biolabs, Ipswich, MA, USA), 0.2 μ l of Roche FastStart Taq DNA polymerase, 0.2 μ l of 10 μ M dNTP and 1 μ l of 10× Buffer (Roche Applied Science, Indianapolis, IN, USA), and 3.24 μ l MQ. The total process was performed using a touchdown thermocycling program. The reaction conditions consisted of 95°C for 5 min, 15 cycles of 95°C for 1 min, 70°C for 1 min, with a decrement of 1°C per cycle, 72°C for 3 min, followed by 25 cycles of 95°C for 1 min, 56°C for 1 min, 72°C for 3 min, 72°C for 60 min, the final temperature was then sustained at 15°C. In the second step, we performed a classical FAM-fluorescent labeled PCR assay to detect the accurate genotype of the non-pathogenic mutation carriers. The fragment length analysis was performed on an ABI 3730×l DNA analyzer and was visualized by the GeneMapper software version 3.2 (Applied Biosystems).
Statistical analysis
A cut-off value of 30 repeats was used to define the pathogenic threshold (Renton et al., 2011). One DNA sample of an ALS patient who was recruited from the Xiangya Hospital was subjected to a repeat-primed polymerase chain reaction, and >30 repeat expansions were detected (unpublished paper), which could verify the reliability and trustworthiness of our experiment. Descriptive statistics were expressed as the mean ± the standard deviation; differences in the distributions of repeat number between the patients and controls were tested using a 2-tailed Mann-Whitney U-test or Kruskal–Wallis H-test, and significance was set at p = 0.05. Considering that the role of ≥7 units in non-pathogenic carriers was strongly correlated with C9orf72 expression (Van Der Zee et al., 2013), all subjects were classified into three genotypes, including S/S, S/I, and I/I (S: short allele <7 units; I: intermediate allele ≥7_units) according to an individual's two repeat alleles. The associations between the number of repeat and disease risk were determined in logistic regression models that were adjusted for the age at onset and gender. Analyses of the associations between the number of repeats and the age at onset were calculated using linear regression models that were adjusted for gender. To adjust p-values for multiple testing, we performed Bonferroni adjustment in logistic regression and linear regression. The statistical analysis was performed using the SPSS program (version 18.0).
Results
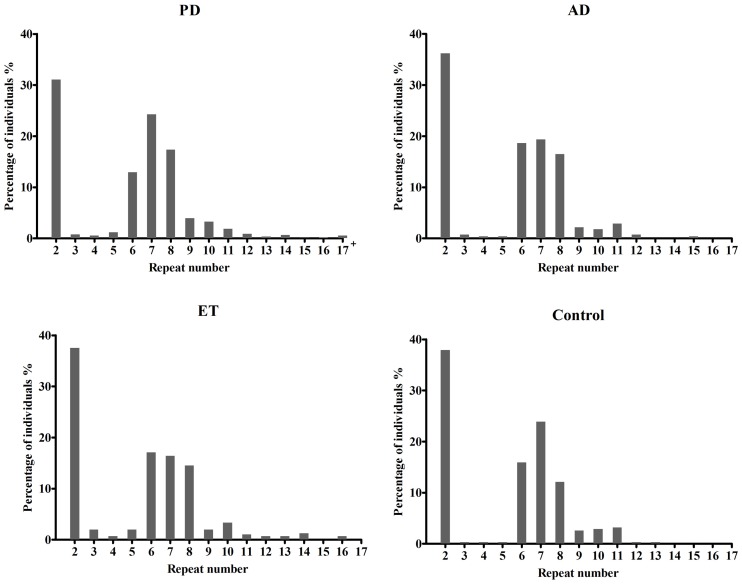
Table 1 presents the demographic information for our study. A total of 1342 patients and 314 healthy controls were successfully subjected to repeat-primed polymerase chain reactions and genotyping. However, no pathological repeat expansion of C9orf72 was detected in either patients or controls. The wide range of repeat expansions in patients was 2–27 units, and the most frequent repeats in all subjects was 2 units, followed by 6, 7, and 8; however, three PD patients harbored marginally larger alleles at 22, 23, and 27 units. In addition, the distributions of repeat numbers in the individuals' larger allele indicated a significant difference in PD (p = 0.01), late onset PD (>50 years) (p = 0.01), and male PD (p = 0.03) when compared with controls. However, no statistical significance was found in AD (p = 0.67), ET (p = 0.13) or their subtypes when compared with control individuals (Figure 1 and Table 2).
Table 1.
Demographic information and average number of repeats among cases and controls.
| Variable | PD | AD | ET | Controls |
|---|---|---|---|---|
| Cases, n | 911 | 279 | 152 | 314 |
| Gender, Male (%) | 533 (58.5) | 167 (59.9) | 89 (58.6) | 158 (50.3) |
| Age | 59.0 ± 11.6 | 72.4 ± 12.0 | 44.7 ± 15.8 | 57.1 ± 13.7 |
| Age at onset | 55.1 ± 11.7 | 70.1 ± 10.0 | 35.4 ± 14.9 | – |
| Repeat number | 5.9 ± 3.2 | 5.4 ± 2.9 | 5.4 ± 3.1 | 5.3 ± 2.9 |
The sample mean ± SD is given for age, age at onset, and repeat number.
Figure 1.
Distributions of repeat number in C9orf72 in Alzheimer's disease, Parkinson disease, essential tremor, and control individuals. There was no significant difference in the distributions of repeat length between AD (p = 0.67), ET (p = 0.13) cases and controls, respectively. However, an evidence of significant distribution was identified between PD cases and controls (p = 0.01); 17+ in PD patients including five PD cases at 18, 19, 22, 23, 27 repeats.
Table 2.
Analysis of the distributions of repeat expansion in cases with different classification.
| EOAD vs. Control | LOAD vs. Control | |||||
|---|---|---|---|---|---|---|
| N | Mean ± SD | P | N | Mean ± SD | P | |
| Cases | 71 | 5.5 ± 2.8 | 0.86 | 208 | 5.4 ± 3.0 | 0.77 |
| Controls | 230 | 5.3 ± 2.9 | 84 | 5.3 ± 2.9 | ||
| Male in AD vs. Control | Female in AD vs. Control | |||||
| Cases | 106 | 5.7 ± 2.9 | 0.36 | 173 | 5.3 ± 2.9 | 0.77 |
| Controls | 158 | 5.2 ± 2.9 | 156 | 5.4 ± 2.8 | ||
| EOPD vs. Control | LOPD vs. Control | |||||
| Cases | 286 | 5.6 ± 2.0 | 0.19 | 603 | 6.0 ± 3.2 | 0.01 |
| Controls | 85 | 5.2 ± 2.7 | 229 | 5.4 ± 2.9 | ||
| Male in PD vs. Control | Female in PD vs. Control | |||||
| Cases | 533 | 5.9 ± 3.0 | 0.03 | 378 | 5.9 ± 3.3 | 0.07 |
| Controls | 158 | 5.3 ± 2.9 | 156 | 5.4 ± 2.8 | ||
| Male in ET vs. Control | Female in ET vs. Control | |||||
| Cases | 89 | 5.5 ± 3.2 | 0.61 | 63 | 5.2 ± 2.9 | 0.57 |
| Controls | 158 | 5.2 ± 2.9 | 156 | 5.4 ± 2.8 | ||
EOAD, early onset AD (≤65 years); LOAD, late onset AD (>65 years); EOPD, early onset PD (≤50 years); LOPD, late onset PD (>50 years).
Given the significant distinction of repeat length in PD and control individuals, 202 PD patients who completed a battery of neuropsychological tests that were recommended by the Movement Disorder Society (MDS) Task Force, were further divided into 70 PD dementia (PD-D), 58 PD mild cognitive impairment (PD-MCI), and 74 PD with no cognitive impairment (PD-NC) according to the MDS Task Force diagnosis criteria(Dubois et al., 2007). However, no significant difference in the distribution of repeats was found among the three subgroups and control individuals (p = 0.98) using Kruskal–Wallis H-test.
In addition, we employed two analytical approaches for association testing, the larger repeat allele as a continuous variable and three genotyping categorical variables based on the individual's short or intermediate alleles. We did not identify any significant evidence of associations between the number of repeat and either the AD or ET risk or the age at onset in patients with PD or ET. Interestingly, we observed a statistically significant result between the I/I genotype and the age at onset (p = 0.007; Regression coefficient 5.50 [0.27, 10.74]) after Bonferroni adjustment for multiple testing, but not in the continuous variable (p = 0.93) (Table 3). Finally, the only significant evidence of association was found between the repeat length and PD risk after using Bonferroni adjustment when considering repeats as a continuous variable (p = 0.001, OR 1.06 [1.01, 1.10]), or when considering three genotypes as a categorical variable (p = 0.0081; S/I: OR 1.37 [1.05, 1.79]; I/I: OR 2.03 [1.17, 3.54]) (Table 4).
Table 3.
Association of allele length with age at onset in patients with PD, AD, and ET.
| Association of allele length with age at onset | ||||||
|---|---|---|---|---|---|---|
| PD (n = 911) | AD (n = 279) | ET (n = 152) | ||||
| Regression coefficient (95% CI) | P | Regression coefficient (95% CI) | P | Regression coefficient (95% CI) | P | |
| Genotype | 0.43 | 0.007 | 0.85 | |||
| S/S | 0.00 (reference) | 0.00 (reference) | 0.00 (reference) | |||
| S/I | 0.02 (−1.60, 1.63) | 1.04 (−1.90, 3.98) | −0.15 (−3.85, 3.55) | |||
| I/I | 1.37 (−1.41, 4.16) | 5.50 (0.27, 10.74) | −0.32 (−7.57, 6.93) | |||
| Repeats on larger allele continuous | ||||||
| 0.20 (−0.48, −0.44) | 0.12 | 0.02 (−0.46, 0.50) | 0.93 | −0.47 (−1.20, 0.25) | 0.20 | |
CI, confidence interval; S, short allele (<7 units); I, intermediate allele (≥7 units).
Regression coefficient and corresponding p results from linear regression models adjusted for gender. After Bonferroni adjustment for multiple testing, p ≤ 0.025 were considered statistically significant (2 tests).
Table 4.
Association of allele length with risk of PD, AD, and ET diseases.
| Association of allele length with risk of disease in comparison with controls (n = 314) | ||||||
|---|---|---|---|---|---|---|
| PD (n = 911) | AD (n = 279) | ET (n = 152) | ||||
| OR (95% CI) | P | OR (95% CI) | P | OR (95% CI) | P | |
| Genotype | 0.0081 | 0.54 | 0.64 | |||
| S/S | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| S/I | 1.37 (1.05, 1.79) | 0.83 (0.55, 1.26) | 0.88 (0.57, 1.37) | |||
| I/I | 2.03 (1.17, 3.54) | 0.91 (0.40, 2.06) | 1.30 (0.56, 3.04) | |||
| Repeats on larger allele continuous | ||||||
| 1.06 (1.01, 1.10) | 0.001 | 0.99 (0.93, 1.05) | 0.69 | 0.96 (0.89, 1.03) | 0.27 | |
OR, odds ratio; CI, confidence interval; S, short allele (<7 units); I, intermediate allele (≥7 units).
Odds ratios and corresponding p-value results from logistic regression models adjusted for age at onset and gender. After Bonferroni adjustment for multiple testing, p ≤ 0.0083 were considered statistically significant (6 tests).
Discussion
Recently, GGGGCC repeat expansions in the C9orf72 gene were identified as major contributing factors for ALS and FTD. However, the preliminary evidence suggested that the C9orf72 mutation rates in patients with clinically diagnosed ALS in China, Japan, Korea, and Taiwan were much lower than that observed in Caucasian populations (Ogaki et al., 2012; Tsai et al., 2012; Zou et al., 2012; Jang et al., 2013), which implied that the number of repeats varied greatly due to different nationalities and ethnicities. Given the clinical heterogeneity with the repeat expansions, we hypothesized that the length of repeats may also account for other neurodegenerative disorders. To investigate the hexanucleotide repeat expansions of C9orf72 on different genetic backgrounds, we screened for C9orf72 in a large group AD, PD, and ET patients with Chinese Han origin. To the best of our knowledge, this study is the first reported investigation of C9orf72 repeat expansions in three cohorts of patients in Asia.
In this study, no pathogenic expansion was observed in either patients or controls, which supported recent data from other independent cohorts. Across these studies, no abnormal repeats were found in 781 patients with PD and 568 patients with AD (Majounie et al., 2012a; Rollinson et al., 2012). The relation between AD, PD, and C9orf72 has been controversial. Several studies have reported that pathogenic repeats were found in 0.7% patients with PD and in less than 1% of clinically diagnosed AD patients (Majounie et al., 2012b; Xi et al., 2012). However, Majounie et al. speculated that the positive results in AD patients might be an incidental rather than a causative finding due to amnesic FTD being misdiagnosed as probable AD (Majounie et al., 2012b).
There are three possible explanations for our negative results. The first possibility is that the hexanucleotide repeat expansions could not cause PD, AD, or ET, and is only specific to ALS/FTD. Although the pathogenic gene mutations were detected in several probable cases of PD or AD, the exact diagnoses of these diseases should be confirmed due to clinical heterogeneity. Another explanation for the result is that the cut-off value of repeat >30 units that is suitable for ALS/FTD is most likely not suitable for AD, PD, or ET patients, and more samples should be included to set a solid cut-off value. Finally, Mok et al. found that the ALS/FTD patients with C9orf72 pathogenic repeats share a similar risk haplotype with Finland, Ireland, Italy, UK, and USA populations. Moreover, an investigation from Japan suggests that the pathogenic expansion is closely tied to the risk haplotype, and the low frequency of the risk haplotype might explain the low frequency of repeat expansions (Konno et al., 2012).
Although no pathogenic expansion was observed in this study, we identified a significant association between the number of intermediate repeats and PD risk, which indicates that the more intermediate repeats, the greater risk of susceptibility to PD. As we know, this study is the first to raise this notion. This notion suggests that the intermediate alleles act as a contributor to PD risk. Although our research to elucidate the disease mechanism of this intermediate repeat remains in its infancy, one previous study has indicated that intermediate repeats could decrease the transcriptional activity of C9orf72 (Van Der Zee et al., 2013). Therefore, how the intermediate alleles act as predisposing alleles and what is the pathogenic pathway of intermediate alleles in PD patients should be addressed in further studies. In addition, this study reported that there was no significant difference in repeat expansions among PDD, PD-MCI, PD-NC patients, and control individuals, which indicated that the number of repeats in C9orf72 might not account for the occurrence and severity of cognitive syndromes in PD patients.
Recently, Kohli et al. (2013) found nine AD patients that carried the C9orf72 mutation with an average disease onset of 77.8 years (all older than 60 years), and speculated that the pathogenic repeat expansions were most likely associated with late onset AD (>65 years). Similarly, in this study, we observed a positive correlation between the I/I genotype and the age at onset, which indicated that AD patients who carried I/I genotypes were susceptible to a higher age at onset. At present, APOE4 is the only major genetic risk factor for the development of late onset AD (Hauser and Ryan, 2013), if the relation above exists, then future research will focus on exploring whether there is an interaction mechanism between these two genes in patients with late onset AD. Finally, there was only a cohort of 106 ET patients that were investigated before our study, and this cohort had the identical result as our study, with no association of normal repeat length with disease risk or with an effect on age at onset, further confirming that GGGGCC repeats did not play a role in patients with ET.
However, there are several limitations in this work. First, an association between the I/I genotype and the age at onset in AD patients might be a false positive observation due to the lack of a statistically significance correlation between the larger alleles (continuous variables) and the age at onset. Therefore, we should recruit more samples to verify the relation between the repeat expansions in C9orf72 and the age at onset in patients with AD. Second, if the relation above was further verified, then we should assess the relation between the C9orf72 repeats and APOE genotypes to explore whether there is an interaction mechanism between them. Third, this study only examined sporadic AD, PD, and ET patients, C9orf72 may play a role in dominant familial forms.
In conclusion, no C9orf72 pathogenic mutations in AD, PD, and ET patients in this study are consistent with previous studies (Majounie et al., 2012b; Dejesus-Hernandez et al., 2013). However, we identified a statistically significant association between the intermediate repeats and PD risk, which implied that intermediate genotypes or alleles might be a risk factor for PD in China. Meanwhile, the lack of association between the intermediate repeats and AD risk has indicated that intermediate genotypes or alleles might play a different role in AD and PD.
Author contributions
Lu Shen, Beisha Tang, and Kun Xia contributed to the conception and organization of our research project; Bin Jiao, Jifeng Guo, Xinxiang Yan, and Lin Zhou contributed to recruiting patients and controls; Bin Jiao, Jifeng Guo, Yaqin Wang, Xiaoyan Liu, Fufeng Zhang, and Yafang Zhou contributed to statistical analysis; Bin Jiao and Jifeng Guo equally contributed to the first draft, and Lu Shen was responsible for the review and critique of our manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to all subjects for their participation in our study. This study was supported by the National Basic Research Program (973 Program) (No. 2011CB510000 to Lu Shen) and the National Natural Science Foundation of China (No. 81171068 to Lu Shen).
References
- Arighi A., Fumagalli G. G., Jacini F., Fenoglio C., Ghezzi L., Pietroboni A. M., et al. (2012). Early onset behavioral variant frontotemporal dementia due to the C9ORF72 hexanucleotide repeat expansion: psychiatric clinical presentations. J. Alzheimers Dis. 31, 447–452 [DOI] [PubMed] [Google Scholar]
- Boeve B. F., Boylan K. B., Graff-Radford N. R., Dejesus-Hernandez M., Knopman D. S., Pedraza O., et al. (2012). Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain 135, 765–783 10.1093/brain/aws004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace R., Van Cauwenberghe C., Bettens K., Gijselinck I., Van Der Zee J., Engelborghs S., et al. (2013). C9orf72 G(4)C(2) repeat expansions in Alzheimer's disease and mild cognitive impairment. Neurobiol. Aging 34, 1712.e1–1712.e7 10.1016/j.neurobiolaging.2012.12.019 [DOI] [PubMed] [Google Scholar]
- Dejesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., et al. (2011). Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejesus-Hernandez M., Rayaprolu S., Soto-Ortolaza A. I., Rutherford N. J., Heckman M. G., Traynor S., et al. (2013). Analysis of the C9orf72 repeat in Parkinson's disease, essential tremor and restless legs syndrome. Parkinsonism Relat. Disord. 19, 198–201 10.1016/j.parkreldis.2012.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B., Burn D., Goetz C., Aarsland D., Brown R. G., Broe G. A., et al. (2007). Diagnostic procedures for Parkinson's disease dementia: recommendations from the movement disorder society task force. Mov. Disord. 22, 2314–2324 10.1002/mds.21844 [DOI] [PubMed] [Google Scholar]
- Floris G., Borghero G., Cannas A., Di Stefano F., Costantino E., Murru M. R., et al. (2012). Frontotemporal dementia with psychosis, parkinsonism, visuo-spatial dysfunction, upper motor neuron involvement associated to expansion of C9ORF72: a peculiar phenotype? J. Neurol. 259, 1749–1751 10.1007/s00415-012-6444-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gijselinck I., Van Langenhove T., Van Der Zee J., Sleegers K., Philtjens S., Kleinberger G., et al. (2012). A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 11, 54–65 10.1016/S1474-4422(11)70261-7 [DOI] [PubMed] [Google Scholar]
- Hauser P. S., Ryan R. O. (2013). Impact of apolipoprotein E on Alzheimer's disease. Curr. Alzheimer Res. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung G. Y., Dejesus-Hernandez M., Feldman H. H., Sengdy P., Bouchard-Kerr P., Dwosh E., et al. (2012). Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain 135, 709–722 10.1093/brain/awr354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson A. J. (1981). Amyotrophic lateral sclerosis and its association with dementia, parkinsonism and other neurological disorders: a review. Brain 104, 217–247 10.1093/brain/104.2.217 [DOI] [PubMed] [Google Scholar]
- Hughes A. J., Daniel S. E., Kilford L., Lees A. J. (1992). Accuracy of clinical diagnosis of idiopathic Parkinson's disease: a clinico-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatr. 55, 181–184 10.1136/jnnp.55.3.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. H., Kwon M. J., Choi W. J., Oh K. W., Koh S. H., Ki C. S., et al. (2013). Analysis of the C9orf72 hexanucleotide repeat expansion in Korean patients with familial and sporadic amyotrophic lateral sclerosis. Neurobiol. Aging 34, 1311.e7–1319.e9 10.1016/j.neurobiolaging.2012.09.004 [DOI] [PubMed] [Google Scholar]
- Kohli M. A., John-Williams K., Rajbhandary R., Naj A., Whitehead P., Hamilton K., et al. (2013). Repeat expansions in the C9ORF72 gene contribute to Alzheimer's disease in Caucasians. Neurobiol. Aging 34, 1519.e5–1519.e12 10.1016/j.neurobiolaging.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno T., Shiga A., Tsujino A., Sugai A., Kato T., Kanai K., et al. (2012). Japanese amyotrophic lateral sclerosis patients with GGGGCC hexanucleotide repeat expansion in C9ORF72. J. Neurol. Neurosurg. Psychiatr. 84, 398–401 10.1136/jnnp-2012-302272 [DOI] [PubMed] [Google Scholar]
- Lesage S., Le Ber I., Condroyer C., Broussolle E., Gabelle A., Thobois S., et al. (2013). C9orf72 repeat expansions are a rare genetic cause of parkinsonism. Brain 136, 385–391 10.1093/brain/aws357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis E. D., Ottman R., Ford B., Pullman S., Martinez M., Fahn S., et al. (1997). The washington heights-inwood genetic study of essential tremor: methodologic issues in essential-tremor research. Neuroepidemiology 16, 124–133 10.1159/000109681 [DOI] [PubMed] [Google Scholar]
- Majounie E., Abramzon Y., Renton A. E., Keller M. F., Traynor B. J., Singleton A. B. (2012a). Large C9orf72 repeat expansions are not a common cause of Parkinson's disease. Neurobiol. Aging 33, 2527.e1–2527.e2 10.1016/j.neurobiolaging.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E., Abramzon Y., Renton A. E., Perry R., Bassett S. S., Pletnikova O., et al. (2012b). Repeat expansion in C9ORF72 in Alzheimer's disease. N. Engl. J. Med. 366, 283–284 10.1056/NEJMc1113592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G., Drachman D., Folstein M., Katzman R., Price D., Stadlan E. M. (1984). Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology 34, 939–944 10.1212/WNL.34.7.939 [DOI] [PubMed] [Google Scholar]
- Nakashima-Yasuda H., Uryu K., Robinson J., Xie S. X., Hurtig H., Duda J. E., et al. (2007). Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 114, 221–229 10.1007/s00401-007-0261-2 [DOI] [PubMed] [Google Scholar]
- O'Dowd S., Curtin D., Waite A. J., Roberts K., Pender N., Reid V., et al. (2012). C9ORF72 expansion in amyotrophic lateral sclerosis/frontotemporal dementia also causes parkinsonism. Mov. Disord. 27, 1072–1074 10.1002/mds.25022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogaki K., Li Y., Atsuta N., Tomiyama H., Funayama M., Watanabe H., et al. (2012). Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol. Aging 33, 2527.e11–2526.e6 10.1016/j.neurobiolaging.2012.05.011 [DOI] [PubMed] [Google Scholar]
- Piguet O., Hornberger M., Mioshi E., Hodges J. R. (2011). Behavioural-variant frontotemporal dementia: diagnosis, clinical staging, and management. Lancet Neurol. 10, 162–172 10.1016/S1474-4422(10)70299-4 [DOI] [PubMed] [Google Scholar]
- Renton A. E., Majounie E., Waite A., Simon-Sanchez J., Rollinson S., Gibbs J. R., et al. (2011). A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollinson S., Halliwell N., Young K., Callister J. B., Toulson G., Gibbons L., et al. (2012). Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiol. Aging 33, 1846.e5–1846.e6 10.1016/j.neurobiolaging.2012.01.109 [DOI] [PubMed] [Google Scholar]
- Sha S. J., Boxer A. (2012). Treatment implications of C9ORF72. Alzheimers Res. Ther. 4, 46 10.1186/alzrt149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada L. T., Pimentel M. L., Dejesus-Hernandez M., Fong J. C., Yokoyama J. S., Karydas A., et al. (2012). Frontotemporal dementia in a Brazilian kindred with the c9orf72 mutation. Arch. Neurol. 69, 1149–1153 10.1001/archneurol.2012.650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C. P., Soong B. W., Tu P. H., Lin K. P., Fuh J. L., Tsai P. C., et al. (2012). A hexanucleotide repeat expansion in C9ORF72 causes familial and sporadic ALS in Taiwan. Neurobiol. Aging 33, 2232.e11–2232.e18 10.1016/j.neurobiolaging.2012.05.002 [DOI] [PubMed] [Google Scholar]
- Van Der Zee J., Gijselinck I., Dillen L., Van Langenhove T., Theuns J., Engelborghs S., et al. (2013). A pan-european study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum. Mutat. 34, 363–373 10.1002/humu.22244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Zinman L., Grinberg Y., Moreno D., Sato C., Bilbao J. M., et al. (2012). Investigation of C9orf72 in 4 neurodegenerative disorders. Arch. Neurol. 69, 1583–1590 10.1001/archneurol.2012.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z. Y., Li X. G., Liu M. S., Cui L. Y. (2012). Screening for C9orf72 repeat expansions in Chinese amyotrophic lateral sclerosis patients. Neurobiol. Aging. 34, 1710.e5–1710.e6 10.1016/j.neurobiolaging.2012.03.014 [DOI] [PubMed] [Google Scholar]