Abstract
Large‐scale longitudinal studies of regional brain volume require reliable quantification using automated segmentation and labeling. However, repeated MR scanning of the same subject, even if using the same scanner and acquisition parameters, does not result in identical images due to small changes in image orientation, changes in prescan parameters, and magnetic field instability. These differences may lead to appreciable changes in estimates of volume for different structures. This study examined scan–rescan reliability of automated segmentation algorithms for measuring several subcortical regions, using both within‐day and across‐day comparison sessions in a group of 23 normal participants. We found that the reliability of volume measures including percent volume difference, percent volume overlap (Dice's coefficient), and intraclass correlation coefficient (ICC), varied substantially across brain regions. Low reliability was observed in some structures such as the amygdala (ICC = 0.6), with higher reliability (ICC = 0.9) for other structures such as the thalamus and caudate. Patterns of reliability across regions were similar for automated segmentation with FSL/FIRST and FreeSurfer (longitudinal stream). Reliability was associated with the volume of the structure, the ratio of volume to surface area for the structure, the magnitude of the interscan interval, and the method of segmentation. Sample size estimates for detecting changes in brain volume for a range of likely effect sizes also differed by region. Thus, longitudinal research requires a careful analysis of sample size and choice of segmentation method combined with a consideration of the brain structure(s) of interest and the magnitude of the anticipated effects. Hum Brain Mapp, 2010. © 2010 Wiley‐Liss, Inc.
Keywords: structural MRI, FreeSurfer, FSL/FIRST, reliability, scan–rescan, automated segmentation
INTRODUCTION
The reliable volumetric assessment of brain structures is critical for clinical neuroscience research. An important and evolving body of literature has focused on using longitudinal within group design. This approach has the advantage of limiting the variance associated with individual differences in brain volumetry and morphometry by using each subject as his or her own control. Longitudinal studies of the volumes of particular brain structures have been used to document brain changes related to aging, disease, treatment regimens, and adverse environmental exposures (Castellanos et al., 2002; Fotenos et al., 2005; Mathalon et al., 2001; Resnick et al., 2003). Manual tracing by experts of magnetic resonance images (MRI) of brain structures has been the standard for brain volume measurement for many studies due to its high inter‐rater reliability for key structures such as the hippocampus and amygdala (∼0.95) (Mervaala et al., 2000; Rojas et al., 2004; Whitwell et al., 2005). However, this labor‐intensive method becomes impractical for studies that require the measurement of many subjects or brain regions or that embody a risk of rater bias. Moreover, variability becomes exacerbated with repeated sessions, which introduces additional sources of variability including changes in subject positioning, variability in prescan and shim settings, and magnetic field drift. This can greatly reduce reliability across scans; e.g., for the hippocampus (∼0.9) and amygdala (∼0.75) (Bartzokis et al., 1993).
Large‐scale quantitative assessment of brain anatomy can only be achieved practicably by using automated brain segmentation algorithms. Despite the prevalence of these techniques, the effects of interscan variability on automated segmentation and labeling algorithms is not well characterized (Wonderlick et al., 2009), and thus, the impact of this variability on the statistical power for discerning differences in brain volumes is largely unknown. Here, we investigate the reliability of automated brain measurement methods using data from normal subjects who were scanned four times: two scans on day 1 and two scans on day 2 occurring 1 week later.
Our main goals were (i) to characterize variability in volume measures of different brain structures commonly studied in clinical neuroscience across repeated anatomical scans, both within and across days and (ii) to determine how variability is influenced by the elapsed time between scans, the size of the brain structure, and the image contrast between neighboring structures. We repeated all analyses using two popular noncommercial programs, FSL/FIRST (FMRIB Integrated Registration and Segmentation Tool, Oxford University, Oxford, UK) and FreeSurfer (Martinos Center for Biomedical Imaging, Harvard‐MIT, Boston). As we were interested in scan–rescan reliability of automated methods, we did not compare the output of these programs with that obtained via manual segmentation, and thus, we did not evaluate the accuracy of these segmentation methods. However, we have recently investigated the accuracy of automated measures of the hippocampal and amygdala volumes compared with the expert manual segmentation (Morey et al., 2009) as have other groups (Barnes et al., 2008; Jatzko et al., 2006; Powell et al., 2008). Finally, because scan–rescan reliability influences experimental power, we estimated the sample size required to detect significant differences in the selected brain structures for a range of likely effect sizes (ES).
METHODS
Subject Data
Twenty‐three healthy subjects (nine females) provided written informed consent for a study approved by the Institutional Review Board of Duke University Medical School. The subjects had an average age of 23.4 (SD = 3.3) and none reported neurologic or psychiatric conditions. Each subject was scanned on two different days. Two scans were conducted 1 h apart on day 1 (scans 1A and 1B) and two scans were conducted 1 h apart at a second session 7–9 days later (scans 2A and 2B). Subjects were removed from the scanner between scans that were conducted on the same day. All anatomical scans were obtained as part of an unrelated functional MRI study that involved an acute tryptophan depletion intervention. Half the subjects had the active intervention prior to scanning on day 1 and the other half had it on day 2. There was no effect of the intervention detected on volumetric results (data not shown). Data were collected between June 2007 and October 2008 with approximately half the subjects scanned in 2007 and the remainder in 2008. All scans were high‐resolution T1‐weighted images with 1‐mm isometric voxels acquired on the same General Electric 3‐Tesla EXCITE system and eight‐channel headcoil using the array spatial sensitivity encoding technique (ASSET) with 3D fast spoiled gradient recall (FSPGR). Image parameters were optimized for contrast between white matter, gray matter, and CSF (TR/TE/flip angle = 7.484 ms/2.984 ms/12°, 256‐mm FOV, 1‐mm slices, 176 slices, 256 × 256 matrix, 1 excitation). Visual inspection of the scans showed no subjective evidence of motion artifact. Interested readers can access the entire dataset in compressed NIFTI format at http://duke.edu/~morey005/ScanRescanData/ along with a readme.txt file that provides information on gender, age, and race/ethnicity for each participant.
Segmentation Methods
Two fully automated segmentation programs, FSL/FIRST (v1.2) (Patenaude, 2007) and FreeSurfer (v4.5) (Fischl et al., 2002), were used to measure the volume of nine brain regions: amygdala, brainstem, hippocampus, lateral ventricles, nucleus accumbens, caudate, putamen, pallidum, and thalamus. Summary results using a prior version of FIRST (v1.0.5) and the FreeSurfer cross‐sectional stream (v4.4) were also calculated. A brief description of segmentation procedures and parameters follows. Further details of these methods are available in Morey et al. ( 2009) and in the documentation provided by the developers of FIRST (http://www.fmrib.ox.ac.uk/fsl/first/index.html) and FreeSurfer (http://surfer.nmr.mgh.harvard.edu/).
Prior to initiating the FIRST processing stream, images from all four time points were coregistered to the image space of the first scan (1A) using 6 DOF rigid transformations (translation, rotation) that were sufficient to achieve alignment between images (1A, 1B, 2A, 2B) from the same individual. The 1A image was used as the base image because of its analogy to the baseline scan in an actual longitudinal study being the most likely candidate for registering subsequent images. The coregistration was necessary for postsegmentation computation of volume overlap. FIRST normalization was then performed with two‐stage affine transformation to the standard space of MNI 152 at 1‐mm resolution. A neck mask was included to improve the registration of our T1 images; this was the only nondefault option selected in our processing stream. The first stage involved a standard 12 degrees‐of‐freedom registration to the template, and the second stage applied 12 degrees‐of‐freedom registration using an MNI 152 subcortical mask to exclude voxels outside the subcortical regions. Next, automated segmentation proceeded via a Bayesian probabilistic approach using shape and appearance models. These models were constructed from a library of manually segmented images, parameterized as surface meshes and then modeled as point distributions. Using the learned models, FIRST searches through linear combinations of shape modes of variation (principal components) to find the most probable shape instance given the observed intensities from the input image. FIRST uses an empirically determined fixed number of modes (iterations) for each structure. Finally, the vertex information or models were transformed to the native space where the boundaries were corrected and volumes (labels) were generated.
Automated segmentation and labeling was also performed by the FreeSurfer longitudinal stream. Prior to initiating longitudinal processing, the data were fully segmented with FreeSurfer's cross‐sectional stream. FreeSurfer utilizes affine transformations and combines information about voxel intensity relative to a probability distribution for tissue classes with information about the spatial relationship of the voxel to the location of neighboring structures obtained from a manually labeled atlas (Fischl et al., 2002). Longitudinal processing began with a mutual coregistration (rigid with six DOF) of all the four time points to create a base image that was not biased by any of the contributing images. The images from each of the four time points were then registered to the base image, and the subcortical segmentation of the base image was used as an initial guess for the segmentation of each time point image in the longitudinal scheme. The subcortical segmentation and parcellation procedure in FreeSurfer involves solving many complex nonlinear optimization problems using iterative methods. Therefore, the results are sensitive to the starting point, in this case the base image. The FreeSurfer developers believe that initializing the processing of a new data set in a longitudinal series with the results of the unbiased template can reduce the random variation in the processing procedure and improve the robustness and sensitivity of the overall analysis. The segmented labels were returned to the base image space using the FreeSurfer library function mri_convert, which applies the inverse transform created during the longitudinal processing. Nearest neighbor resampling was applied to prevent interpolation during the transformation. Individual regions were extracted from the large segmentation volume that contains all the regions of interest.
Statistical Measures
Means and standard deviations of volumes from segmentation using FreeSurfer and FIRST were obtained for each acquisition: 1A, 1B, 2A, and 2B. Separate intraclass correlation coefficients (ICC) were computed to assess the contribution of elapsed time to reliability. The four ICCs included two ICC values for interscan intervals of 1 h (1A vs. 1B, 2A vs. 2B) and two ICC values for interscan intervals of 1 week (1A vs. 2A, 1B vs. 2B). An ICC was also calculated based on volume measures generated from all four scans. Reliability analyses were conducted separately for left and right hemisphere structures except for the brainstem which is a midline structure. The ICC was calculated using two‐way mixed model with measures of absolute agreement (McGraw and Wong, 1996). The mixed model treats subjects as randomly sampled from a larger population and treats the measures from the four scans as a fixed factor being specific to the scanner hardware, software, and acquisition parameters of this study.
Reliability based on the method of segmentation, the interscan interval, and hemisphere was assessed with a repeated‐measures multivariate analysis of variance (MANOVA). Analysis was performed on the dependent variable calculated from the ICC values described earlier using the Fisher r to z transformation, z = 0.5 × [loge(1 + r) − loge(1 − r)]. Factors included method (two levels; FIRST, FreeSurfer), time (two levels; 1 hour interscan, 1 week interscan), and hemisphere (two levels; left, right).
We reasoned that segmentation reliability may be related to the overall volume of the brain structure, to the particular shape, and to the surface area or other unique features of individual structures. Thus, we considered two possible predictors of reliability, volume, and the ratio of volume to surface area. To assess volume as a predictor, the mean volume of structures was parameterized into three groups: (i) small (<2,000 voxels) that included the amygdala, accumbens, and pallidum; (ii) medium‐sized (2,000–4,000 voxels) that included the caudate and hippocampus; and (iii) large (>4,000 voxels) that included the lateral ventricle, brain stem, putamen, and thalamus. Likewise, the ratio of volume to surface area was assessed as a predictor of reliability. Surface area estimation was derived by computing the area of the triangulated mesh of each structure for FIRST and FreeSurfer. The calculated ratios were parameterized into quartiles and the corresponding observations of reliability were allotted into quartiles according to Altman and Bland ( 1994). Repeated measures ANOVA testing was used to assess differences between groups.
To assess the reliability of volume estimates from repeated segmentations, we computed (i) percent volume difference as defined by Eq. (1) and (ii) percent volume overlap or Dice's coefficient as defined in Eq. (2) with repeat scanning. Given labelings L 1 and L 2 from repeat scanning and a function V(L), which takes a label and returns its volume, the percent change in volume ΔV(L 1,L 2) is given by,
| (1) |
For labels with identical volume, ΔV(L1,L2) achieves its optimal value of zero, with increasing values indicating a greater volume difference between the two labelings. Given two different labelings of a structure, L1 and L2, and a function V(L), which takes a label and returns its volume, the percent volume overlap is given by:
| (2) |
For identical labelings, O(L1,L2) achieves its maximum value of 100, with decreasing values indicating less perfect overlap. Note that the overlap between two different labelings will be reduced by slight shifts in the spatial location of one label with respect to another. Percent volume difference and percent volume overlap were computed for the following four separate repeat scan comparisons: 1A vs. 1B, 2A vs. 2B, 1A vs. 2A, 1B vs. 2B.
Means and standard deviations were calculated from volume data summed over the four scans. Cronbach's Alpha, a measure of reliability, and ICC, denoting the ratio of between‐subject variability to total variability, were estimated using SPSS (Release 15.0). Estimated standard deviations and correlation coefficients were used in subsequent power calculations, using a range of likely ES (0.1, 0.2, 0.3, 0.4, 0.5, 0.75, 1.0). Power calculations determined the number of subjects required to detect a given ES with 80% power at an alpha level of 5%. Estimates were calculated using power analysis and sample size (PASS) software [NCSS: Kaysville, UT] (Hintze, 2005).
RESULTS
Sample means and standard deviations of volumes for the nine regions and each of the four scans are summarized in Table I. Scan–rescan ICC values are reported in Table II by region and segmentation method. The scan–rescan reliability was higher for FreeSurfer than FIRST; main effect of method [F(1,17) = 25.8, P < 0.0001]. The data showed greater ICC variation with FIRST (SD = 0.726; variance = 0.528) than FreeSurfer (SD = 0.699; variance = 0.489). The reliability for an interscan interval of 1 h was higher than for an interscan interval of 1 week; main effect of time [F(1,17) = 17.0, P < 0.001]. No difference was found between the reliability of left and right hemisphere segmentation [F(1,17) = 0.31, P > 0.5]. Scan–rescan reliability for FIRST v1.2, which includes improved boundary correction, was higher than for FIRST v1.05 [t(16) = 4.2, P < 0.0001]. Scan–rescan reliability obtained from the longitudinal stream of FreeSurfer was higher than for the cross‐sectional stream [t(16) = 2.7, P < 0.01] (see Supporting Information Table S1).
Table I.
Mean volume by region, scan, and segmentation methoda
| FIRST | FreeSurfer | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1A | 1B | 2A | 2B | 1A | 1B | 2A | 2B | ||
| Accumbens | Mean L | 603 | 596 | 572 | 589 | 557 | 550 | 566 | 572 |
| SD L | 85 | 84 | 108 | 84 | 69 | 66 | 85 | 61 | |
| Mean R | 446 | 458 | 463 | 459 | 506 | 492 | 518 | 513 | |
| SD R | 114 | 112 | 105 | 86 | 79 | 70 | 71 | 64 | |
| Amygdala | Mean L | 1,074 | 1,154 | 1,133 | 1,220 | 1,195 | 1,208 | 1,192 | 1,187 |
| SD L | 263 | 253 | 241 | 216 | 109 | 96 | 104 | 111 | |
| Mean R | 964 | 1,070 | 1,040 | 1,116 | 1,284 | 1,277 | 1,278 | 1,278 | |
| SD R | 238 | 200 | 204 | 255 | 178 | 142 | 145 | 167 | |
| Brain stemb | Mean L | 22,443 | 22,530 | 22,335 | 22,421 | 13,087 | 13,000 | 13,133 | 12,928 |
| SD L | 2,329 | 2,410 | 2,340 | 2,361 | 13,349 | 13,507 | 13,348 | 13,488 | |
| Caudate | Mean L | 3,813 | 3,829 | 3,754 | 3,782 | 3,039 | 3,038 | 3,048 | 3,033 |
| SD L | 524 | 478 | 536 | 462 | 330 | 334 | 332 | 332 | |
| Mean R | 3,969 | 3,994 | 3,996 | 3,982 | 3,108 | 3,114 | 3,134 | 3,115 | |
| SD R | 479 | 479 | 435 | 464 | 349 | 409 | 366 | 367 | |
| Hippocampus | Mean L | 3,956 | 3,982 | 3,976 | 3,977 | 3,257 | 3,278 | 3,259 | 3,266 |
| SD L | 424 | 411 | 416 | 420 | 302 | 302 | 310 | 296 | |
| Mean R | 3,971 | 3,980 | 3,993 | 3,986 | 3,168 | 3,196 | 3,184 | 3,209 | |
| SD R | 366 | 394 | 381 | 377 | 222 | 234 | 232 | 216 | |
| Lat. vent. | Mean L | 6,967 | 7,145 | 7,188 | 7,133 | 3,844 | 3,754 | 3,796 | 3,765 |
| SD L | 2,062 | 2,232 | 2,238 | 2,175 | 1,780 | 1,746 | 1,734 | 1,718 | |
| Mean R | 6,427 | 6,572 | 6,614 | 6,595 | 3,836 | 3,754 | 3,815 | 3,762 | |
| SD R | 2,095 | 2,208 | 2,147 | 2,154 | 2,124 | 2,126 | 2,126 | 2,071 | |
| Pallidum | Mean L | 1,810 | 1,808 | 1,815 | 1,791 | 1,368 | 1,365 | 1,363 | 1,370 |
| SD L | 214 | 230 | 211 | 237 | 149 | 153 | 140 | 138 | |
| Mean R | 1,885 | 1,865 | 1,875 | 1,869 | 1,337 | 1,326 | 1,340 | 1,340 | |
| SD R | 201 | 177 | 174 | 210 | 150 | 158 | 149 | 146 | |
| Putamen | Mean L | 5,588 | 5,770 | 5,769 | 5,789 | 4,838 | 4,820 | 4,825 | 4,838 |
| SD L | 651 | 639 | 587 | 601 | 666 | 677 | 681 | 690 | |
| Mean R | 5,479 | 5,497 | 5,522 | 5,537 | 4,635 | 4,613 | 4,627 | 4,660 | |
| SD R | 690 | 724 | 661 | 708 | 554 | 562 | 568 | 531 | |
| Thalamus | Mean L | 8,830 | 8,844 | 8,830 | 8,854 | 5,630 | 5,634 | 5,654 | 5,630 |
| SD L | 660 | 619 | 646 | 640 | 478 | 533 | 520 | 510 | |
| Mean R | 8,581 | 8,593 | 8,548 | 8,599 | 5,551 | 5,559 | 5,572 | 5,584 | |
| SD R | 640 | 622 | 620 | 619 | 468 | 484 | 500 | 479 | |
The standard deviation (SD) for each scan is a measure of the variance in volume for the group of 23 subjects. Therefore, the magnitude of SD does not necessarily reflect on the accuracy of the mean volume.
The FIRST segmentation of the brainstem includes the fourth ventricle but FreeSurfer does not.
Table II.
Intraclass correlation coefficients for segmentation volumes
| Region (L,R) | FreeSurfer | FIRST | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1A–1B | 2A–2B | 1A–2A | 1B–2B | 4‐scan | 1A–1B | 2A–2B | 1A–2A | 1B–2B | 4‐scan | |
| Accumbens | 0.782 | 0.739 | 0.753 | 0.540 | 0.684 | 0.741 | 0.704 | 0.598 | 0.671 | 0.678 |
| 0.877 | 0.799 | 0.909 | 0.838 | 0.856 | 0.710 | 0.643 | 0.647 | 0.651 | 0.647 | |
| Pallidum | 0.902 | 0.969 | 0.925 | 0.911 | 0.922 | 0.952 | 0.924 | 0.969 | 0.893 | 0.933 |
| 0.895 | 0.948 | 0.902 | 0.885 | 0.910 | 0.922 | 0.879 | 0.814 | 0.912 | 0.889 | |
| Amygdala | 0.889 | 0.873 | 0.843 | 0.806 | 0.866 | 0.728 | 0.712 | 0.791 | 0.679 | 0.749 |
| 0.823 | 0.754 | 0.881 | 0.776 | 0.815 | 0.690 | 0.506 | 0.390 | 0.427 | 0.522 | |
| Caudate | 0.987 | 0.977 | 0.974 | 0.963 | 0.975 | 0.963 | 0.919 | 0.950 | 0.919 | 0.927 |
| 0.968 | 0.984 | 0.979 | 0.969 | 0.975 | 0.864 | 0.978 | 0.834 | 0.944 | 0.895 | |
| Hippocampus | 0.982 | 0.977 | 0.982 | 0.977 | 0.979 | 0.932 | 0.913 | 0.927 | 0.974 | 0.928 |
| 0.924 | 0.952 | 0.924 | 0.934 | 0.935 | 0.809 | 0.863 | 0.822 | 0.886 | 0.855 | |
| Lat. vent. | 0.998 | 0.997 | 0.993 | 0.994 | 0.995 | 0.977 | 0.998 | 0.976 | 0.993 | 0.987 |
| 0.999 | 0.998 | 0.997 | 0.997 | 0.997 | 0.994 | 0.998 | 0.994 | 0.994 | 0.995 | |
| Putamen | 0.972 | 0.984 | 0.972 | 0.974 | 0.973 | 0.287 | 0.970 | 0.293 | 0.952 | 0.613 |
| 0.980 | 0.943 | 0.962 | 0.961 | 0.958 | 0.977 | 0.940 | 0.952 | 0.912 | 0.936 | |
| Thalamus | 0.971 | 0.981 | 0.978 | 0.980 | 0.976 | 0.973 | 0.990 | 0.986 | 0.985 | 0.981 |
| 0.983 | 0.980 | 0.973 | 0.967 | 0.975 | 0.975 | 0.978 | 0.968 | 0.975 | 0.975 | |
| Brain stem | 0.991 | 0.996 | 0.988 | 0.992 | 0.991 | 0.970 | 0.967 | 0.943 | 0.972 | 0.962 |
The scan–rescan reliability of volume measures differed across regions regardless of the segmentation method, main effect for region [F(8, 35) = 81.1, P < 0.0001] (see Table II). High reliability was observed in structures such as the brainstem, lateral ventricle, and thalamus, intermediate reliability in structures such as the putamen, hippocampus, and caudate, and low reliability in structures such as the pallidum, accumbens, and amygdala. Reliability was influenced by both volume [F(2,141) = 75.2, P < 0.0001] and by the ratio of volume to surface area as calculated by FIRST [F(3, 60) = 27.4, P < 0.0001] and FreeSurfer [F(3, 60) = 5.4, P < 0.002]. Thus, structures with large volumes and high volume to surface area ratio had relatively high reliability; whereas structures with small volumes and low volume to surface area ratio had relatively low reliability.
The correlation of amygdala volumes measured in scans 1A and 1B (see Fig. 1) and the hippocampus for scans 1A and 1B (see Fig. 2) illustrate the scan–rescan reliability. The percentage volume difference between repeated scanning is shown in Figure 3 for FIRST and FreeSurfer segmentation. Values for percent volume difference represent the mean of four separate repeat scan comparisons (1A vs. 1B, 2A vs. 2B, 1A vs. 2A, 1B vs. 2B). Examination of volume difference measures illustrates the inconsistency across the four scans (see Fig. 3). As examples, the amygdala and nucleus accumbens showed low consistency; whereas other structures such as hippocampus and thalamus showed generally consistent values.
Figure 1.
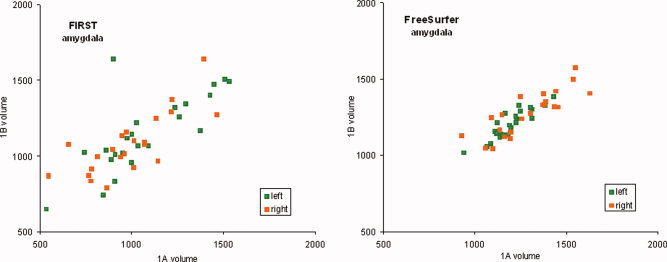
Scatter plots showing correlation between segmented amygdala volumes (mm3) from scan 1A and 1B for FSL/FIRST and FreeSurfer. Left hemisphere volumes are in green and right hemisphere volumes in orange. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 2.
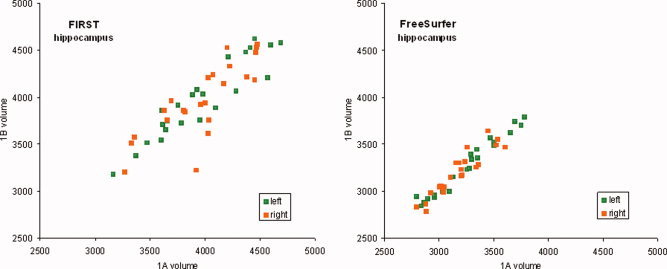
Scatter plots showing correlation between segmented hippocampus volumes (mm3) from scan 1A and 1B for FSL/FIRST and FreeSurfer. Left hemisphere volumes are in green and right hemisphere volumes in orange. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 3.
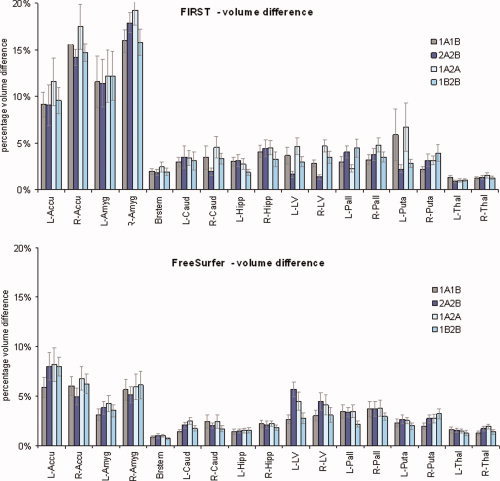
Percent volume difference for scans with a 1‐h and 1‐week interscan interval for nine subcortical brain structures segmented with FIRST and FreeSurfer. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The relationship between volume and particular structures was further assessed to understand the role of contrast between neighboring structures. Comparisons of regions with similar volumes within the medium‐sized group showed the hippocampus, which has poor contrast in the anterior boundary with the amygdala (Pruessner et al., 2000), had a lower ICC than the caudate which has boundaries with generally high contrast [t(15) = 2.3, P < 0.02].
However, the volume difference measure does not capture possible shape variation between segmented regions. Therefore, reliability was further assessed with percent volume overlap shown in Figure 4 for FIRST and FreeSurfer segmentation. The overall mean percent volume overlap across all structures and repeat scanning sessions (see Fig. 4) was better for FIRST (91.5 ± 0.15) than FreeSurfer (87.6 ± 0.29) [t(67) = 9.7, P < 0.0001]. On the other hand, percent volume difference (see Fig. 3) was better for FreeSurfer (3.2 ± 0.03) than FIRST (5.5 ± 0.02) [t(67) = 5.2, P < 0.0001]. This suggests that FreeSurfer segmentations had greater spatial variability over successive repeat scans than FIRST. In contrast to this, FreeSurfer segmentations maintained more reliable volume measures (volume difference) over successive scans than FIRST.
Figure 4.
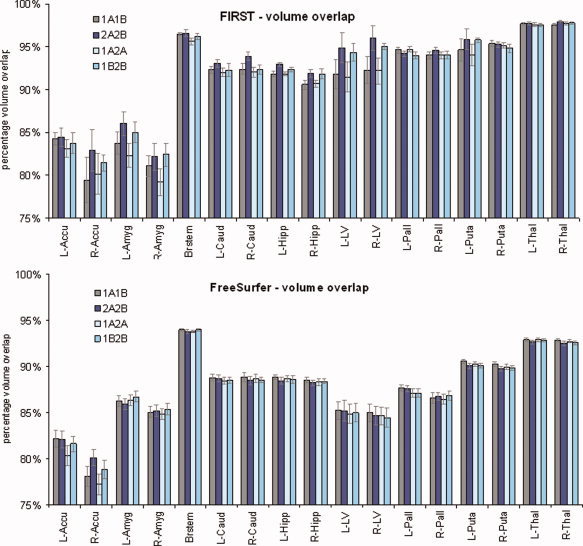
Percent volume overlap for scans with a 1‐h and 1‐week interscan interval for nine subcortical brain structures segmented with FIRST and FreeSurfer. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The images were manually inspected and no gross segmentation errors resulting from image artifact were detected. For structures where a particular correlation between two scanning sessions was appreciably lower than the corresponding pairwise correlations for the same structure, the images were reexamined in the context of the inconsistent segmentation volumes. For instance, FIRST segmentation of the left putamen (see Fig. 5) was dramatically different on the lateral surface for scan 1A (3,777 voxels) compared with scan 1B (7,317 voxels). This resulted in lower intraclass correlations for 1A1B (0.29) and 1A2A (0.29) when compared with 2A2B (0.96) and 1B2B (0.95). Again, there seemed to be no artifact or other overt factor contributing to the discrepancy.
Figure 5.
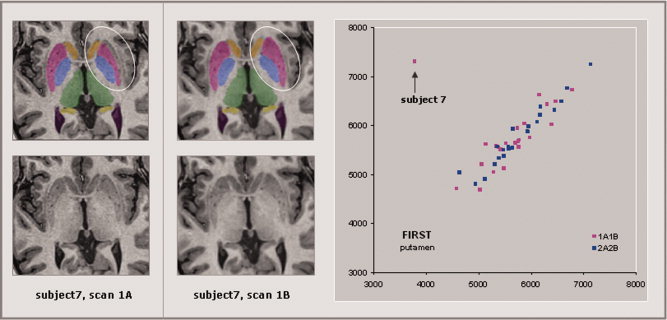
The T1 images and segmentations for subject 7 are shown for scan 1A (left panel) and for scan 1B (center panel). Segmentation of the L‐putamen (circled) is dramatically different on the lateral surface for scan 1A (3,777 voxels) compared to scan 1B (7,317 voxels). This resulted in lower intraclass correlations for 1A1B (0.29) and 1A2A (0.29) as compared to 2A2B (0.96) and 1B2B (0.95). There is no obvious artifact visible in scan 1A that might explain this discrepancy. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Sample Size Estimation
On the basis of variability observed, we estimated the sample size required to achieve 80% power and limit Type 1 error to 5%. The sample size for a range of ES from 0.1 to 1.0 is shown for each of the selected regions and segmentation methods in Figure 6. Estimates showed that structures with ICCs approaching 1 required relatively few subjects (<10) to power studies of longitudinal or repeated measures design for the entire range of ES considered. On the other hand, regions with relatively low ICCs, required few subjects (∼10) for large ES (>0.5), but required much a larger sample (>100) to power studies with small ES (<0.2). For example, to detect a change in amygdala volume corresponding to an ES of 1.0 (∼300 voxels) using FreeSurfer would require eight subjects, whereas detecting a change corresponding to an ES of 0.1 (∼30 voxels) would require more than 500 subjects. By comparison, a region with relatively high ICC, such as the thalamus, would require just 10 subjects to detect a difference with an ES of 0.1 (∼70 voxels); whereas an ES of 1.0 (∼700 voxels) would require just three subjects.
Figure 6.
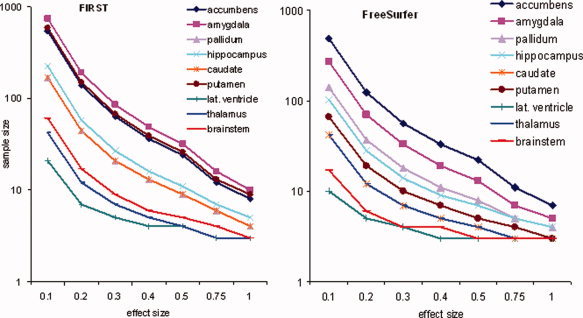
Sample size requirements (y‐axis) for FreeSurfer (left) and FSL/FIRST (right) assuming a within subject design with two observations to achieve 80% power and 5% alpha level are shown for a range of effect sizes (x‐axis) and each of the nine subcortical structures. Note that the sample size is scaled by log10 to enhance visualization of curves at higher effect sizes. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
This study examined the reliability of two fully automatic segmentation and labeling programs for measuring the volumes of subcortical and other brain structures in a group of normal subjects who were repeatedly scanned within a 7–9 day interval. Overall, there was consistent and large‐magnitude scan–rescan variability that was exacerbated for small structures such as the amygdala, accumbens, and pallidum. The choice of software (FreeSurfer or FSL/FIRST) did not strongly influence reliability; however, FIRST produced higher reliability for the small structures measured here. Consistent with the observed main effects, sample size estimates for longitudinal studies were greatest for regions with poor or moderate rescan reliability, particularly when detecting small effects.
We found a difference in reliability between 1 hour and 1 week interscan intervals. (although, for some brain structures, such as the right hippocampus, the 1 hour reliability (2A–2B; ICC = 0.82) was lower than the 1 week reliability (1B–2B; ICC = 0.89). Higher reliability might be expected for shorter interscan intervals due to magnetic field instabilities or drift. Other sources of variance were similar for the 1 hour and 1‐week interscan interval such as the effect of subject repositioning. The Duke scanning site, where these images were obtained, uses rigorous and regular QA procedures (Friedman and Glover, 2006; Keator et al., 2008) that may have diminished some sources of scanner variability.
The goal of our article is to inform longitudinal studies where the selected group is assessed at two different time points and volume data from the two time points are compared. If perfect scan–rescan reliability was achieved then the true change in any structure (e.g., volume) could be measured perfectly in a longitudinal setup. We have approached this problem by examining the special case where the true change of the structure is assumed to be zero and then the measured departure from perfect reliability. The change observed in the selected group of cases can be compared with the longitudinal change in a control group to characterize the effects of the treatment or process (e.g., aging) in question. Although a longitudinal design has the advantage of limiting individual variability with each subject acting as its own control, other sources of variability persist with repeated scanning of the same subject on the same scanner using the same acquisition parameters even with relatively short time duration between scans. These sources of variability include small changes in image orientation, changes in prescan parameters, and instability in the magnetic field.
Previous work examined interscanner reliability, where field strength and manufacturer were varied (Jovicich et al., 2006; Reig et al., 2009; Schnack et al., 2004). Only two studies, to our knowledge, examined rescan reliability (intrascanner) with automated segmentation, with the first study limited to basic tissue class segmentation (GM, WM, CSF) (Agartz et al., 2001). The second study, by Wonderlick et al. ( 2009), reported somewhat similar reliability using FreeSurfer (version 4.0.1) to what we found in this study. For example, similar reliability was obtained in the amygdala (0.85 for Wonderlick et al. vs. 0.87 (left) and 0.82 (right) for this study), caudate (0.99 vs. 0.98 and 0.98), hippocampus (0.96 vs. 0.98 and 0.94), pallidum (0.87 vs. 0.92 and 0.91), putamen (0.95 vs. 0.97 and 0.96), and thalamus (0.97 vs. 0.98 and 0.97). However, these values obtained by Wonderlick et al. were from FreeSurfer 4.0.1 and much higher than those we obtained with the cross‐sectional stream of FreeSurfer (v4.4) as seen in Supporting Information Table S1. There are several factors that may have contributed to these differences including differences in scanner manufacturer, scanner hardware (Siemens 3T TIM Trio in their study vs. GE 3T EXCITE in our study), headcoil (12 channel vs. 8 channel), pulse sequence (MP‐RAGE vs. FSPGR with ASSET), sample size (11 vs. 23), age profile of participants (young subgroup and old subgroup vs. young group), and interscan interval (2 weeks vs. 1 h and 1 week).
We did not undertake an experimental study of the variables that may have contributed to the scan–rescan differences in our brain volumes. One factor that is difficult to control is the precise position of the subject's head within the head coil. A slightly different orientation can result in partial volume effects for different tissue types along the boundary of a brain structure that could change the contrast of the surface boundary with neighboring structures. Such effects are most relevant for boundary voxels, but of lesser consequence for voxels located in the interior of a structure. When the boundary is distinct, meaning there is a minimal overlap in the probability distribution of signal intensity between adjacent structures, this variance has a minor effect on the resulting segmentation. However, when the boundary with a neighboring structure is less distinct, and has a larger overlap in probability distributions of signal intensity, this variance can derail automated segmentation and dramatically change outcome. Reliability may differ across brain structures due to variability in tissue contrast profiles and divergent modeling algorithms (e.g., cortical surface‐based or voxel‐based segmentation methods). A host of other factors specific to the segmentation algorithm and the atlas being used are likely to alter the outcome of segmentation and its vulnerability to MR signal variance in difficult‐to‐segment regions (Shattuck et al., 2008). Thus, depending on its location, a small change in MR signal may lead to a large and sometimes unpredictable difference in the outcome of the segmentation algorithm as suggested by Figure 5. The reliability measurements observed in this sample of young healthy adults are therefore unlikely to limited by the atlases associated with FreeSurfer and FIRST that contain a wide range of demography and pathology. Additional concerns related to the participant sample are covered in the Limitations section that follows.
Rescan reliability was investigated for manual tracing by Bartzokis et al. ( 1993) and showed slightly lower reliability than for automated segmentation for a number of regions such as hippocampus (0.91 for Bartzokis et al. vs. 0.98 (left) and 0.94 (right) for this study with FreeSurfer) and amygdala (0.75 vs. 0.87 and 0.82 for this study with FreeSurfer). Similarly, rescan reliability of intracranial volume with manual tracing (ICC = 0.95) was slightly lower than for intra‐rater (same scan) reliability (ICC = 0.96) (Nandigam et al., 2007). Most volumetric studies that use manual tracing report high intra‐rater reliability even on challenging regions such as the hippocampus and amygdala (ICC = 0.95) (Mervaala et al., 2000; Rojas et al., 2004; Whitwell et al., 2005). When a baseline manual segmentation is performed, fluid registration can be used to attain highly reliable segmentation of repeat scans that is superior to a subsequent manual segmentation (Crum et al., 2001). This approach is especially advantageous when multiple repeat scans are performed longitudinally because it requires manual segmentation of only the first scan. However, the introduction of lesions or rapid degeneration between scans can compromise the fluid registration approach. Thus, fluid registration may be preferred for regions such as the amygdala that are unreliably segmented with the fully automated methods.
The exact sample size recommendations from our power analyses are specific to the hardware, software, segmentation method, pulse sequence, and other parameters used in this study. However, our procedures are typical for academic imaging at many major research institutions, and thus the relative effects across brain regions are likely to generalize. Improvements in multichannel imaging by combining T1, T2, and PD sequences that optimize automated segmentation may improve rescan reliability when compared with single channel acquisition. Such multichannel acquisitions also offer substantial invariance to acquisition parameters (Fischl et al., 2004). Pulse sequences such as high bandwidth multiecho FLASH have a high signal to noise ratio and minimal image distortion from B0 effects have been shown to improve reliability. Wonderlick et al. examined the performance of scan–rescan segmentation with FreeSurfer using recent advances in MR acquisition including high resolution (1 mm isotropic), parallel acquisition (phased array headcoil), and a multiecho T1 weighted sequence using MP‐rage sequence (1.3 × 1.0 × 1.3 mm) for comparison testing (Wonderlick et al., 2009). Even when using these advanced approaches, the effect of MR signal variance on automated segmentation was not eliminated.
Our findings may be specific to FreeSurfer and FIRST—two popular noncommercial software programs used at many research institutions. It is important to emphasize that we did not evaluate the validity or accuracy of the measurements from these two programs. It is possible an accurate measure might result from a given segmentation obtained from a single scan but does not provide information about how consistently it can produce accurate segmentation when the same brain is scanned repeatedly and is generally not assessed in studies of segmentation accuracy. Indeed we show that for certain regions, scan–rescan reliability of automatically segmented brain regions is of concern.
Limitations
The demographic sample of healthy young participants limits the ability to generalize the present findings. Our demographic is unlikely to contain extremes of the population distribution, and one might expect higher scan–rescan reliability in our sample than in a sample representative of a more diverse population. The intriguing point is that despite the limited demographic attributes of this group, the reliability is surprisingly low in some instances and might be even lower in a more diverse sample with respect to demography (e.g., age) or neuropsychiatric pathology. Similarly, our power analyses for estimating sample size are likely to be underestimates of the actual number of subjects required for conducting longitudinal studies in more diverse groups. Studies with a case‐control design are likely to encounter greater variance related to individual differences that are avoided in a longitudinal design where each participant serves as its own control.
CONCLUSIONS
Research based on MR imaging and automated segmentation‐based volumetry requires careful characterization of the reliability and precision of observations and propagation of errors. Initially, small error can introduce substantial variability across repeated observations each feeding into algorithms with multistage signal processing and probabilistic computation. Size and surface contrast features are important factors that influence rescan reliability of regional brain volumes obtained from automated segmentation programs.
Supporting information
Additional Supporting Information may be found in the online version of this article.
Supporting Tables
REFERENCES
- Agartz I, Okuguwa G, Nordstrom M, Greitz D, Magnotta V, Sedvall G ( 2001): Reliability and reproducibility of brain tissue volumetry from segmented MR scans. Eur Arch Psychiatry Clin Neurosci 251: 255–261. [DOI] [PubMed] [Google Scholar]
- Altman DG, Bland JM ( 1994): Quartiles, quintiles, centiles, and other quantiles. BMJ 309: 996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Foster J, Boyes RG, Pepple T, Moore EK, Schott JM, Frost C, Scahill RI, Fox NC ( 2008): A comparison of methods for the automated calculation of volumes and atrophy rates in the hippocampus. Neuroimage 40: 1655–1671. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Mintz J, Marx P, Osborn D, Gutkind D, Chiang F, Phelan CK, Marder SR ( 1993): Reliability of in vivo volume measures of hippocampus and other brain structures using MRI. Magn Reson Imaging 11: 993–1006. [DOI] [PubMed] [Google Scholar]
- Castellanos FX, Lee PP, Sharp W, Jeffries NO, Greenstein DK, Clasen LS, Blumenthal JD, James RS, Ebens CL, Walter JM, et al ( 2002): Developmental trajectories of brain volume abnormalities in children and adolescents with attention‐deficit/hyperactivity disorder. JAMA 288: 1740–1748. [DOI] [PubMed] [Google Scholar]
- Crum WR, Scahill RI, Fox NC ( 2001): Automated hippocampal segmentation by regional fluid registration of serial MRI: Validation and application in Alzheimer's disease. Neuroimage 13: 847–855. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. ( 2002): Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 33: 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Segonne F, Quinn BT, Dale AM ( 2004): Sequence‐independent segmentation of magnetic resonance images. Neuroimage 23 ( Suppl 1): S69–S84. [DOI] [PubMed] [Google Scholar]
- Fotenos AF, Snyder AZ, Girton LE, Morris JC, Buckner RL ( 2005): Normative estimates of cross‐sectional and longitudinal brain volume decline in aging and AD. Neurology 64: 1032–1039. [DOI] [PubMed] [Google Scholar]
- Friedman L, Glover GH ( 2006): Report on a multicenter fMRI quality assurance protocol. J Magn Reson Imaging 23: 827–839. [DOI] [PubMed] [Google Scholar]
- Hintze JL ( 2005): PASS 2005 User's Guide. Kaysville, UT: NCSS. [Google Scholar]
- Jatzko A, Rothenhofer S, Schmitt A, Gaser C, Demirakca T, Weber‐Fahr W, Wessa M, Magnotta V, Braus DF ( 2006): Hippocampal volume in chronic posttraumatic stress disorder (PTSD): MRI study using two different evaluation methods. J Affect Disord 94: 121–126. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, Macfall J, et al. ( 2006): Reliability in multi‐site structural MRI studies: Effects of gradient non‐linearity correction on phantom and human data. Neuroimage 30: 436–443. [DOI] [PubMed] [Google Scholar]
- Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ, et al. ( 2008): A national human neuroimaging collaboratory enabled by the biomedical informatics research network (BIRN). IEEE Trans Inf Technol Biomed 12: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathalon DH, Sullivan EV, Lim KO, Pfefferbaum A ( 2001): Progressive brain volume changes and the clinical course of schizophrenia in men: A longitudinal magnetic resonance imaging study. Arch Gen Psychiatry 58: 148–157. [DOI] [PubMed] [Google Scholar]
- McGraw KO, Wong S ( 1996): Forming inferences about some intraclass correlations coefficients: Correction Psychological Methods. Original in Psychological Methods 1: 30–46. [Google Scholar]
- Mervaala E, Fohr J, Kononen M, Valkonen‐Korhonen M, Vainio P, Partanen K, Partanen J, Tiihonen J, Viinamaki H, Karjalainen AK, et al. ( 2000): Quantitative MRI of the hippocampus and amygdala in severe depression. Psychol Med 30: 117–125. [DOI] [PubMed] [Google Scholar]
- Morey RA, Petty CM, Xu Y, Hayes JP, Wagner HR, Lewis DV, LaBar KS, Styner M, McCarthy G ( 2009): A comparison of automated segmentation and manual tracing for quantifying of hippocampal and amygdala volumes. Neuroimage 45: 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandigam RNK, Chen Y‐W, Gurol ME, Rosand J, Greenberg SM, Smith EE ( 2007): Validation of intracranial area as a surrogate measure of intracranial volume when using clinical MRI. J Neuroimaging 17: 74–77. [DOI] [PubMed] [Google Scholar]
- Patenaude B ( 2007): Bayesian Statistical Models of Shape and Appearance for Subcortical Brain Segmentation, D. Phil. Thesis, Oxford, UK: Oxford University.
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC ( 2008): Registration and machine learning‐based automated segmentation of subcortical and cerebellar brain structures. Neuroimage 39: 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessner JC, Li LM, Serles W, Pruessner M, Collins DL, Kabani N, Lupien S, Evans AC ( 2000): Volumetry of hippocampus and amygdala with high‐resolution MRI and three‐dimensional analysis software: Minimizing the discrepancies between laboratories. Cereb Cortex 10: 433–442. [DOI] [PubMed] [Google Scholar]
- Reig S, Sánchez‐González J, Arango C, Castro J, González‐Pinto A, Ortuño F, Crespo‐Facorro B, Bargalló N, Desco M ( 2009): Assessment of the increase in variability when combining volumetric data from different scanners. Hum Brain Mapp 30: 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick SM, Pham DL, Kraut MA, Zonderman AB, Davatzikos C ( 2003): Longitudinal magnetic resonance imaging studies of older adults: A shrinking brain. J Neurosci 23: 3295–3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas DC, Smith JA, Benkers TL, Camou SL, Reite ML, Rogers SJ ( 2004): Hippocampus and amygdala volumes in parents of children with autistic disorder. Am J Psychiatry 161: 2038–2044. [DOI] [PubMed] [Google Scholar]
- Schnack HG, van Haren NEM, Hulshoff Pol HE, Picchioni M, Weisbrod M, Sauer H, Cannon T, Huttunen M, Murray R, Kahn RS ( 2004): Reliability of brain volumes from multicenter MRI acquisition: A calibration study. Hum Brain Mapp 22: 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW ( 2008): Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage 39: 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Sampson EL, Watt HC, Harvey RJ, Rossor MN, Fox NC ( 2005): A volumetric magnetic resonance imaging study of the amygdala in frontotemporal lobar degeneration and Alzheimer's disease. Dement Geriatr Cogn Disord 20: 238–244. [DOI] [PubMed] [Google Scholar]
- Wonderlick JS, Ziegler DA, Hosseini‐Varnamkhasti P, Locascio JJ, Bakkour A, van der Kouwe A, Triantafyllou C, Corkin S, Dickerson BC ( 2009): Reliability of MRI‐derived cortical and subcortical morphometric measures: Effects of pulse sequence, voxel geometry, and parallel imaging. Neuroimage 44: 1324–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article.
Supporting Tables


