Abstract
Mechanisms of DNA repair and mutagenesis are defined on the basis of relatively few proteins acting on DNA, yet the identities and functions of all proteins required are unknown. Here, we identify the network that underlies mutagenic repair of DNA breaks in stressed Escherichia coli and define functions for much of it. Using a comprehensive screen, we identified a network of ≥93 genes that function in mutation. Most operate upstream of activation of three required stress responses (RpoS, RpoE, and SOS, key network hubs), apparently sensing stress. The results reveal how a network integrates mutagenic repair into the biology of the cell, show specific pathways of environmental sensing, demonstrate the centrality of stress responses, and imply that these responses are attractive as potential drug targets for blocking the evolution of pathogens.
Repair of DNA double-strand breaks (DSBs) by homologous recombination in Escherichia coli is nonmutagenic in unstressed cells but, under stress, switches to a mutagenic mode that is activated by stress responses (1, 2) (Fig. 1A). It is, therefore, a mechanism of stress-induced mutagenesis (SIM): mechanisms in bacterial, yeast, and human cells that promote mutation and potentially accelerate evolution when cells are maladapted to their environment [reviewed (3–6)].
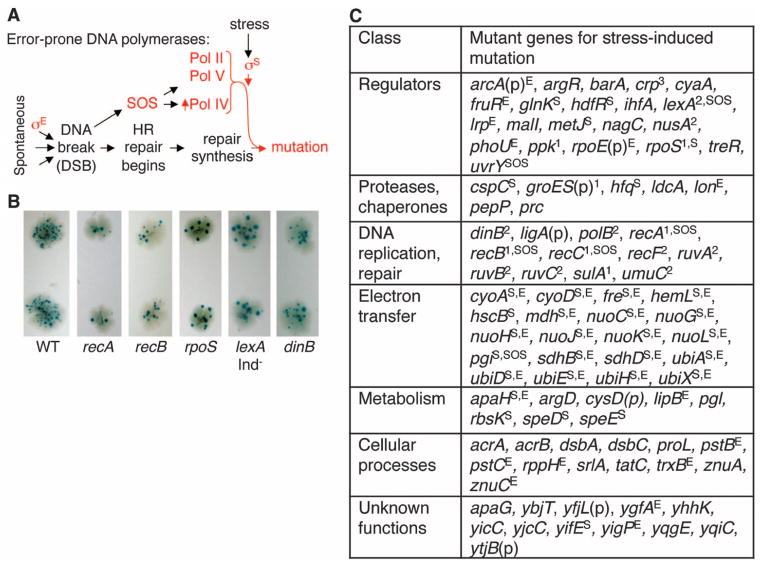
Fig. 1.
(A) Roles of stress responses in mutagenic repair of DNA DSBs by homologous recombination (HR) [reviewed, (7)]. (B) Primary screen for DSB-dependent SIM-deficient mutants. Blue papillae in the white colonies are Lac+ mutant clones formed after prolonged starvation stress (8). (C) Identities of 93 SIM-network genes and results of secondary screens. 1Previously known, found in this screen. 2Previously known, not found in this screen. 3Identified on the basis of genes discovered in this screen. (p), transposon inserted in the promoter (table S12). Superscripts S, E, and SOS indicate decreased σS activity, σE activity, and spontaneous SOS induction, respectively (table S7).
Mutagenic repair of DNA breaks in E. coli requires proteins that mend DSBs by homologous recombination; error-prone DNA polymerases; and activation of the SOS DNA-damage response, the RpoS (σS)–controlled general or starvation stress response, and the RpoE (σE) membrane protein stress response (7). The σE response promotes spontaneous DNA breakage in some DNA regions (8) (Fig. 1A). The SOS response is activated by DSBs and promotes mutation via transcriptional up-regulation of DNA polymerases (Pols) IV and V. However, break repair remains nonmutagenic unless the σS general or starvation response is also activated (1, 2). The σS response licenses the use of Pols IV, II, and V in DSB repair (7) and thus throws the switch to mutagenesis under stress (Fig. 1A) in plasmids (1) and chromosomes of plasmid-free cells (2).
We performed genetic screens for comprehensive discovery of genes required for stress-induced mutagenesis (Methods in supplementary materials and fig. S1). We used a colony–color papillation screen and minitransposon Tn10dCam-insertion mutagenesis, which allows isolation of non-null alleles of essential genes in addition to knock outs (9). Mutants lacking known genes required in DSB-dependent SIM show fewer papillae (Fig. 1B and table S1). A total of 83 genes that facilitate SIM were identified: 76 new and 7 previously known (Fig. 1C). Nine additional previously known genes were not identified, most for reasons that we understand (outlined in table S2), and one was identified by candidate-gene approach (supplementary text S1). We define a network of ≥93 genes (83+9+1) (Fig. 1C and annotations in table S2). All were reconstructed in nonmutagenized cells by using deletion alleles (tables S3 and S4) for nonessential genes and were verified in quantitative SIM assays: 30 showed weak (W, 1.5 to 3); 34 showed moderate (M, 3.1 to 9); and 29 showed strong (S, >9) fold decreases in mutation rate (table S3). In supplementary texts S2 and S3, we estimate total and essential genes likely to have been missed.
We confirmed the mutation-deficient phenotypes of a large representative sample of SIM-deficient mutants using two additional, different assays for DSB-dependent SIM (Fig. 2 and table S5): one (Tet) for frameshift (2) and one (Nal) for base-substitution mutations, both in I-Sce I endonuclease–cleaved chromosomes of starved plasmid-free cells. Both require the σS and SOS stress responses, DSB repair proteins, DNA Pols IV and V, and either starvation or artificial up-regulation of the σS starvation-stress response, all of which characterize DSB-dependent SIM (2) (table S5). Thus, SIM is neither plasmid-specific nor specific to genes selected during the stress [previous concerns (10)]. We confirmed phenotypes of 43 of 52 mutants tested (Fig. 2 and table S5). Those confirmed showed significant correlation of strength of defect between different mutation assays (Lac papillation, Lac quantitative, Tet, and Nal) (tables S1 and S6). Of the remaining nine, all but two displayed sensitivity to SDS-EDTA (tables S1 and S7), a characteristic of cells with membrane stress (σE)–response defects (11). σE functions in SIM by promoting spontaneous DNA breaks and so is not required when DSBs are provided by I-Sce I (8), as is the case in these assays. Thus, the overwhelming majority of genes tested are confirmed as deficient in DSB-dependent base-substitution and frameshift SIM at the three unlinked sites sampled (lac, tet, and gyrA). A summary of SIM-deficient phenotypes of the network mutants in the four mutation assays used is given in table S1.
Fig. 2.
(A to F) Validation of mutants in chromosomal Tet frameshift and Nal base substitution SIM assays. *Significantly SIM-deficient (P values in table S5, two-tailed Student’s t test). Relative mutant frequencies, mutant frequency divided by that of the WT-DSB (I-Sce I–positive) controls assayed in parallel. Means ± SEM (n ≥3 experiments each), for this and all figures.
The 93 SIM genes constitute a functional network. First, protein-protein interaction data (12) show highly significant clustering for the SIM genes (Fig. 3, A and B, and fig. S2). Second, comparison with the Many Microbial Microarrays Database (13) shows that genes in the SIM network are highly significantly coexpressed under various conditions (Fig. 3B), which supports their common function. Highly significant correlations in protein-protein interaction and gene co-expression with the strong class are strengthened by the addition of moderate and weak classes (Fig. 3B) and are also seen in each class individually, with significance increasing from weak to strong (fig. S3).
Fig. 3.
The stress-induced mutation network. (A) Protein-protein interactions: CytoScape 2.8.3 software, “unweighted force-directed layout” (28), links from STRING 9.0 (12). Proteins that promote σS, σE, and SOS activation (Fig. 4), as green, black circle, and red circle, constitute 54% of the network. Downstream of SOS (7), solid red. (B) Coexpression and protein-protein interaction are significantly more clustered than random controls. Gene expression data (13). The 93 SIM genes, (92 × 93)/2 = 4278 pairs, show correlation coefficient distributions (top): bars, entire range; boxes 25th and 75th percentile; red bars, mean. Of 4278 pairs, 3350 show positive correlation coefficient; 928 lie below the zero threshold level. High statistical significance for the strong phenotype (S) genes is increased by addition of moderate (M) and weak (W) (table S3). (Bottom) Significantly more protein-protein interactions for SIM than random genes. Of 4278 pairs, 1320 show positive interaction scores; 2958 pairs do not. P values: sign test of the probability of failure to reject the null hypothesis “number of positively correlated pairs is the same as in the random control.” (C) Allocation of network genes upstream of stress responses (data summarized in tables S1 and S7).
The largest class of genes identified encodes electron transfer chain (ETC) proteins, which function in oxidative phosphorylation (14) (Fig. 1C). These proteins promote mutation by acting upstream of activation of the σS general or starvation stress response during starvation, presumably because they sense stress, as follows.
We tested all network proteins for possible activation of the σS response (Fig. 1C and tables S1 and S7). We identified 31 bona fide σS response–deficient mutants by flow cytometric analysis of the yellow fluorescent protein (YFP) gene expression from the σS-regulated promoter PyiaG (15) (select ETC mutants, Fig. 4A; the rest of the mutants, fig. S4, A and B, and table S8). These mutants showed normal expression of the cyan fluorescent protein (CFP) gene from a Plac promoter-cfp fusion gene (16) (Fig. 4B and fig. S4C), and so do not have reduced transcription overall, but instead are deficient in expression of σS-controlled genes. The decreases in σS-dependent gene expression were not due to delays reaching stationary phase, when σS is induced (Fig. 4C and fig. S5). These proteins might promote SIM by activation of the σS response, acting upstream of σS, sensing and communicating stress.
Fig. 4.
Identification of upstream activators of the σS, σE, and SOS stress responses. Results summarized in table S1. (A) Sample of ETC mutants showing decreased σS activity. See table S8 and fig. S4 for 26 others. (B) No change in transcription from the lac promoter. (C) Mutants enter stationary phase normally (also fig. S5). (D to F) ETC mutants are partially suppressed by up-regulation of σS via deletion of (D) arcB, (E) arcA, or (F) rssB (table S9 and fig. S7). Ratio of mutation rate (bars) and percent mutation restored relative to wild type (WT). (G to I) ΔrpoS is epistatic to ETC mutations in SIM. Double-mutant analyses without (G) or with (H and I) I-Sce I–induced DSBs, showing action in the same pathway. (J) Sample of SIM genes upstream of σE activity [β-galactosidase (β-gal) expression from a σE-regulated promoter, fig. S9, and table S11 for the rest]. (K) Spontaneous SOS induction (21) is reduced in recB, recC, pgi, and uvrY mutants (P = 0.00013, 0.017, 0.0013, and 0.00011, two-tailed Student’s t test). (L) Model: ETC-mediated stress-sensing from starvation to mutation. Described in supplementary text S4. Products of genes identified in screens are in red.
We examined one representative mutant of each protein machine or pathway of the following groups of the ETC: (i) NuoC, NuoG, NuoH, NuoJ, NuoK, and NuoL subunits of NADH: ubiquinone oxidoreductase I (17); (ii) CyoA and CyoD subunits of cytochrome bo′ oxidase (14); and (iii) UbiA, UbiD, UbiE, UbiH, and UbiX, which catalyze biosynthesis of ubiquinone (UQ) (18). First, we find that the SIM deficiency of representative ETC mutants is suppressed, and SIM is partially or mostly restored, by provision of increased σS via arcB, arcA, or rssB mutations (fig. S6), which up-regulate σS levels (19). Mutation is partially restored (Fig. 4, D to F, and table S9), which supports the hypothesis that the ETC proteins act upstream of ArcB, ArcA, and RssB in increasing σS levels (model in Fig. 4L). The incomplete restoration of mutation (Fig. 4, D to F; fig. S7; and table S9) indicates that, in addition to their role upstream of ArcBA and RssB promoting σS accumulation, these proteins also promote SIM via some other route(s).
Second, double-mutant (epistasis) analyses for ETC mutations combined with ΔrpoS show that these genes act in a single SIM-activating pathway with σS. We analyzed double mutants of ΔrpoS with 9 of the 20 ETC mutations (Fig. 4, G to I; fig. S8; and table S10) in standard Lac-mutation assays, as well as in strains with the mutation rate elevated by an I-Sce I endonuclease–generated DSB near lac (1), for greater sensitivity for additive decreases in mutation rate. The decrease in mutation rate for, e.g., the nuoC rpoS double mutant, with or without I-Sce I–induced DSBs, is near that of the rpoS single mutant (Fig. 4, G and H; fig. S8; and table S10), which indicates action in a single pathway. Similar results for cyoD, ubiE, fre, hemL, hscB, mdh, pgi, and sdhD mutants carrying the rpoS mutation (Fig. 4, G to I; fig. S8; and table S10) indicate that rpoS is fully epistatic with these ETC mutants, which shows action in the same pathway. All genetic analyses of σS-upstream genes are summarized in table S1.
A model for the action of the ETC in starvation-sensing and mutation is shown in Fig. 4L (described in supplementary text S4). In it, during starvation, consumption of specific energy sources malate and succinate, during either an internal autophagy-like recycling process or external cannibalism, acts both upstream of σS activation and downstream to promote mutation. Activation of the σS response is a critical hub in the network, supported by a large allocation of network genes (Fig. 3A, green), which demonstrates its importance to mutagenesis. Generalization of the roles of the σS response (7) and supporting network proteins in the chromosomal Tet and Nal assays (Fig. 2 and table S5) indicates that this large network segment (and hub) is generally important to E. coli mutagenesis.
Mutants defective for rpoE, encoding the σE-response transcriptional activator, are sensitive to membrane disruption by SDS with EDTA (11). Forty-four mutants are sensitive to SDS-EDTA (fig. S9 and tables S1 and S7). Thirty-three of these also show decreased transcription from the σE-dependent rpoHP3 promoter (20), which confirms their σE-response deficiency (Fig. 4J, fig. S9, and table S11). Thus, these genes function upstream of activation of the σE response, sensing and transducing the stress signal, which supports the hypothesis that some or all of these may function in SIM via activation of σE. The remaining 11 genes might also encode σE-response activators during SIM, but merely were not confirmed with this particular σE-dependent promoter in these assay conditions (table S11 legend and summarized in tables S1 and S7).
We screened for network mutants defective in the SOS response by sensitivity to ultraviolet (UV) light. Twenty-six show UV sensitivity (table S7 and summary in table S1), which suggests defects in tolerance or repair of DNA damage. Of these, lexA, recA, and recB activate SOS in response to spontaneous DNA breaks (21). We identified three mutants not identified previously as SOS upstream (pgi, recC, and uvrY) by their defects in spontaneous SOS induction using a flow cytometric assay of cells that express the green fluorescent protein (GFP) gene from an SOS-dependent promoter (21) (Fig. 4K). It is possible that more of the UV-sensitive mutants are SOS-induction defective under starvation-stress SIM conditions, which differ from conditions of the GFP assay. All genetic analyses described in the preceding paragraphs are summarized in table S1.
Taken together, our data suggest that as much as 54% of the SIM network proteins (50 out of 93) promote SIM by activating the σS, σE, and/or SOS stress responses (Fig. 3C and tables S1 and S7). Most of the network is devoted to sensing stress and communicating the signal (Fig. 3A).
The fraction of the network not implicated in action upstream of the three stress responses clusters into two groups of coincident gene expression (fig. S10). One group mirrors expression patterns of the σS and SOS groups, whereas the other does not and may represent a separate pathway. Of those genes upstream of stress responses, all are correlated roughly equally for protein-protein interactions and gene coexpression and similarly to all SIM genes (fig. S11).
Our data show that mutagenic DNA break repair (1, 2), previously known to require 16 proteins, is supported by a network of ≥93 proteins, all demonstrated to promote the mutagenesis mechanism. We demonstrated (in the case of the σS controllers) and implicated (for SOS and σE responses) that a total of 50 proteins act upstream of or in the same pathway as these three stress responses: more than half of the network and most of the 77 newly discovered proteins. Stress responses constitute the largest central nodes of the network (Fig. 3A).
DSB-dependent stress-induced mutagenesis produces more than half of spontaneous base substitution and frameshift mutations in starving E. coli (2) and is thus important to evolution. It promotes genetic diversity specifically when cells are maladapted to their environment, that is, when they are stressed. Similar mutagenesis pathways promote antibiotic-induced resistance to ciprofloxacin (antibiotic) in E. coli (6) and bile-induced bile resistance of pathogenic Salmonella (22, 23). Mutagenic DSB repair occurs in yeast (24) and is implicated in mutation hotspots in E. coli (25) and human cancers (26, 27), although whether it is related to stress responses in yeast and human, such as hypoxia-induced SIM (5), is unknown. The identification of stress response regulators as central network hubs suggests to us that these may provide promising candidates as targets for new drugs to inhibit mutagenesis that allows pathogens to adapt in response to antibiotics and the immune system, both stressors. Inhibiting evolution could allow conventional antibiotics and cancer chemotherapies to work without inducing resistance and, perhaps, allow the immune system to overtake pathogens.
Supplementary Material
Acknowledgments
We thank R. Nichols and C. A. Gross for earlier bioinformatic analysis and discussion; S. Gottesman, C. Herman, J. D. Wang, and J. Weissman for discussions; J. Casadesus, S. Gottesman, C. Herman, G. Ira, X. Pan, E. Rogers, T. Silhavy, and J. D. Wang for comments on the manuscript; and D. Satory for help with strains. Bioinformatics supported by NSF 0905536, 1062455, and NIH GM79656 and GM66099 grants (O.L., A.M.L.). R.B.N. was supported by NIH Director’s Pioneer Award DP1-CA174424 (S.M.R.). Supported by NIH grants R01-GM64022 (P.J.H.), R01-GM53158 (S.M.R.) and fellowships F32-GM095267 (R.L.F.) and F32-GM19909 (M.-J.L.). The genes of mutagenic repair of DNA breaks are accessible at http://wiki.porteco.org/tools/simnetwork.
Footnotes
References and Notes
- 1.Ponder RG, Fonville NC, Rosenberg SM. Mol Cell. 2005;19:791. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 2.Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. Proc Natl Acad Sci USA. 2011;108:13659. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galhardo RS, Hastings PJ, Rosenberg SM. Crit Rev Biochem Mol Biol. 2007;42:399. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mittelman D, Wilson JH. Cell Stress Chaperones. 2010;15:463. doi: 10.1007/s12192-010-0205-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindra RS, Crosby ME, Glazer PM. Cancer Metastasis Rev. 2007;26:249. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 6.Cirz RT, et al. PLoS Biol. 2005;3:e176. doi: 10.1371/journal.pbio.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg SM, Shee C, Frisch RL, Hastings PJ. Bioessays. 2012;34:885. doi: 10.1002/bies.201200050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson JL, et al. Mol Microbiol. 2010;77:415. doi: 10.1111/j.1365-2958.2010.07213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleckner N, Bender J, Gottesman S. Methods Enzymol. 1991;204:139. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 10.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Annu Rev Microbiol. 2006;60:477. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 11.Rouvière PE, et al. EMBO J. 1995;14:1032. doi: 10.1002/j.1460-2075.1995.tb07084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szklarczyk D, et al. Nucleic Acids Res. 2011;39(Database issue):D561. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faith JJ, et al. Nucleic Acids Res. 2008;36(Database issue):D866. doi: 10.1093/nar/gkm815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gennis RB, Stewart V. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, et al., editors. Vol. 1. ASM Press; Washington, DC: 1996. pp. 217–261. [Google Scholar]
- 15.Lacour S, Landini P. J Bacteriol. 2004;186:7186. doi: 10.1128/JB.186.21.7186-7195.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 17.Friedrich T. Biochim Biophys Acta. 1998;1364:134. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 18.Meganathan R. FEMS Microbiol Lett. 2001;203:131. doi: 10.1111/j.1574-6968.2001.tb10831.x. [DOI] [PubMed] [Google Scholar]
- 19.Battesti A, Majdalani N, Gottesman S. Annu Rev Microbiol. 2011;65:189. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ades SE, Grigorova IL, Gross CA. J Bacteriol. 2003;185:2512. doi: 10.1128/JB.185.8.2512-2519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pennington JM, Rosenberg SM. Nat Genet. 2007;39:797. doi: 10.1038/ng2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prieto AI, Ramos-Morales F, Casadesús J. Genetics. 2006;174:575. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernández SB, Cota I, Ducret A, Aussel L, Casadesús J. PLoS Genet. 2012;8:e1002459. doi: 10.1371/journal.pgen.1002459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strathern JN, Shafer BK, McGill CB. Genetics. 1995;140:965. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shee C, Gibson JL, Rosenberg SM. Cell Rep. 2012;2:714. doi: 10.1016/j.celrep.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nik-Zainal S, et al. Cell. 2012;149:979. [Google Scholar]
- 27.Roberts SA, et al. Mol Cell. 2012;46:424. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cline MS, et al. Nat Protoc. 2007;2:2366. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.