Abstract
Objectives
To investigate whether selected high-risk MMP7 single nucleotide polymorphisms influence tumor biology or clinical outcomes in patients with clinical early-stage prostate cancer undergoing prostatectomy.
Methods
Two hundred twelve human prostate cancer patients treated with radical prostatectomy were included in the study. Median follow-up was approximately 9.8 years. Genotyping was performed using TaqMan™ technology and custom-designed probes. Three single nucleotide polymorphisms within various regions of the MMP7 gene were assessed with correlation to age at diagnosis, margin status, extracapsular extension, lymph node metastasis, local recurrence and tumor survival in paraffin-embedded prostate tissue specimens from patients with early-stage prostate cancer receiving radical prostatectomy alone.
Results
Rs10895304 was the sole significant polymorphism. The SNP correlated to increased recurrence rates in post-prostatectomy patients (P<0.0094, Log Rank Test). The frequency of the homozygous dominant (A/A) is 74%, the heterozygote (A/G) is 20% and the homozygous recessive (G/G) is 6%. Multivariate analysis (using Chi square analysis) did not detect a confounding relationship between recurrence and age at diagnosis, PSA or Gleason score. None of the other assayed polymorphisms were significant, and no correlations were made to other clinical variables.
Conclusions
The G allele of the rs10895304 polymorphism is predictive of increased local recurrence risk in patients with clinically localized prostate cancer. For this subset of patients, prostatectomy alone may not be adequate for local control. This is a novel and relevant marker that should be evaluated for improved risk stratification of patients who may be candidates for early post-operative radiation therapy to improve local control.
Keywords: polymorphism, MMP7, matrilysin, prostate cancer, prognosis, recurrence
INTRODUCTION
There were an estimated 186,320 prostate cancer cases, and 28,660 prostate cancer deaths in the United States in 20081. Age, race, ethnicity, family history, abnormal digital rectal examination and increased serum PSA levels are all known risk factors. Most screening programs have depended on these factors. However, determining tumor behavior and prognosis is still a great challenge. Standard prognostic factors have included clinical stage, PSA and Gleason score, and additional prognostic information is present in patients who have undergone surgical prostatectomy. They include pathologic staging, margin status, extracapsular extension and seminal vesicle involvement. Though our current prognostic markers are useful, the delineation of pre-treatment markers would be valuable for predicting tumor behavior. Treatment algorithms could be developed using these markers, which would reduce the overtreatment of indolent tumors, and more aggressively treat poor prognostic tumors.
Many promising biomarkers of varied classes have been studied and are in the process of being validated in clinical trials. These include, but are not limited to prostate-specific membrane antigen, early prostate cancer antigen, Chromogranin A, α-methylacyl-CoA racemase, hepsin, enhancer of zeste homolog gene 2, human glandular kallikrein 2, TGF-β1 and Interleukin 62,3. Thus far, there has been varied success with these and other biomarkers in early phase trials. No one biomarker has emerged strong enough to be useful in determining treatment algorithms. However, there is support for the use of panels of validated markers that would as an aggregate serve as prognosticators. Toward this end, the continued search for new and relevant biomarkers is underway3.
Matrix metalloproteinase-7 (MMP-7, PUMP-1, matrin, matrilysin, EC 3.4.24.23) is the smallest member of the large MMP family of Zn2+-dependent extracellular proteases, and has broad substrate specificity with a role in degradation of collagens, elastin, fibronectin, laminin, proteoglycans and vitronectin4–8. It is expressed in higher levels in prostate adenocarcinoma tissues as compared to prostate intraepithelial neoplasia tissues and normal adjacent prostate tissue, and the levels are positively associated with increased Gleason score9. In addition, overexpression of MMP-7 has been associated with tumorigenesis, as well as prostate tumor growth, survival, angiogenesis, invasion and metastasis10–14. Two functional polymorphisms in the MMP-7 promotor region, rs11568818 and rs11568819 are known to modify gene transcription activity, and have been associated with tumorigenesis in multiple tumor types14–18. In addition, several common genetic polymorphisms in the MMP-7 gene, (rs880197, rs10895304 and rs12184413) were evaluated with association to breast cancer risk and prognosis in one or more breast cancer cohorts19. To our knowledge, this is the first analysis of MMP-7 polymorphisms and their association with prostate cancer outcomes and tumor biology. In the present study, we analyze three common genetic polymorphisms in the MMP-7 gene, which were associated with increased risk in the Shanghai Breast Cancer Study19.
METHODS
Study Population
The population consisted of 212 consecutive prostate cancer patients who underwent radical prostatectomy between 1997 and 1999 at Vanderbilt University Medical Center. Ninety-eight percent of the patients were Caucasian with the remainder being 4 African-American patients. The median follow-up for overall survival was 9.1 years (mean 8.3 ± 2.4 years), and for assessment of prostate cancer recurrence was 3.4 years (mean 4.4 ± 3.9 years). The clinical stage was classified according to the American Joint Committee on Cancer TNM staging system20. All patients had histologically confirmed adenocarcinoma. Not all samples were successfully genotyped for each polymorphism. For rs10895304, rs12184413 and rs880197, there were 151, 175 and 162 samples genotyped respectively. This study was approved by the Vanderbilt University Institutional Review Board (IRB No. 030986).
DNA Extraction
Genomic DNA samples were obtained from patient tumor specimens and processed as described previously21,22. Using a standard microtome with disposable blades, 5 μm thick sections of representative areas of normal prostate glands were cut from the paraffin-embedded blocks, stained with hematoxylin-eosin, and examined under a microscrope to verify the absence of prostate cancer. A 5 μm thick section from each patient was used for DNA extraction. The section was deparaffinized with two washes with xylene at room temperature for 30 minutes twice, followed by two washes with 100% ethanol. After the ethanol had completely evaporated, the tissue was completely lysed with proteinase K. Next, the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA) was used to extract and purify the DNA from the tissue according to the manufacturer’s protocol.
SNP Genotyping
The three evaluated SNPs (rs880197, rs10895304 and rs12184413) were genotyped using the TaqMan allelic discrimination assay and the ABI PRISM 7900 Sequence Detection System (Applied Biosystems). TaqMan assays C_7492868_10, C_32018622, and C_32018621_10 were used for the SNPs, respectively. The final volume for each reaction was 5 μl, and included 2.5 μl TaqMan Universal PCR Master Mix, 0.25 μl primers/TaqMan probe mix and 5.0 ng genomic DNA. The PCR profile consisted of an initial denaturation step at 95°C for 10 min, followed by 40 amplification cycles of 92°C for 15 s each, and a subsequent elongation step of 60°C for 1 min. Results were measured with the ABI PRISM 7900HT sequence detector (Applied Biosystem); allele frequencies were determined by ABI SDS software.
Statistical Analysis
Testing of adherence to Hardy-Weinberg equilibrium was performed using a Chi-square goodness of fit test. Relapse-free and overall survival rates were defined as previously described21,22. Biochemical recurrence was defined as a prostate-specific antigen (PSA) detection of > 0.1 ng/ml in at least two consecutive lab draws. Kaplan-Meier survival curves were calculated for each genotype and were compared using the log rank test. Association of the genotypes to various clinicopathologic characteristics was assessed using the Fisher’s exact test.
RESULTS
Patient Characteristics
Table 1 presents demographic, clinical, pathological, recurrence and survival data for the 212 evaluable patients in our study. In order to determine if known prognostic factors were predictive of relapse-free survival, we performed log-rank univariate analyses of these factors. Pre-prostatectomy PSA levels (P<0.045), Gleason score (P<0.001), surgical margin status (P<0.001), extracapsular extension status (P<0.001) and T stage (P<0.001), were all predictive of relapse-free survival. The only one of these factors predictive of overall survival in our study was the Gleason score (P<0.005).
TABLE 1.
| PATIENT DEMOGRAPHICS | ||
|---|---|---|
| FACTORS | Total (N=212) | p-value |
| Race, n(%) | 1.0000 | |
| Black | 4 (1.89) | |
| White | 208 (98.11) | |
| Age at diagnosis (years) | 0.5711 | |
| <=65 | 148 (69.81) | |
| >65 | 64 (30.19) | |
| Pre-prostatectomy PSA (ng/ml) | 0.2925 | |
| Missing | 34 (16.04) | |
| 4–10 | 113 (53.30) | |
| <4 | 22 (10.38) | |
| >10 | 43 (20.28) | |
| Gleason score, n(%) | 0.5079 | |
| 2–6 | 130 (61.32) | |
| 7 | 70 (33.02) | |
| 8–10 | 12 (5.66) | |
| Surgical margin, n(%) | 0.2935 | |
| Missing | 1 (0.47) | |
| Negative | 133 (62.74) | |
| Positive | 78 (36.79) | |
| Extracapsular extension, n(%) | 0.7779 | |
| Missing | 20 (9.43) | |
| Negative | 125 (58.96) | |
| Positive | 67 (31.60) | |
| Disease Stage, n(%) | 0.6483 | |
| Missing | 13 (6.13) | |
| Localized (T1) | 8 (3.77) | |
| Localized (T2) | 134 (63.21) | |
| Localized (T3) | 57 (26.89) | |
Genotyping of MMP7 Polymorphisms and Clinicopathologic Parameters
Table 2 describes our three evaluated single nucleotide polymorphisms in terms of their amino acid transitions, minor allele frequencies in our study population, as well as the gene region as it relates to the promoter start site. Each of the SNPs was analyzed in our patient samples, and gene frequencies were determined. For rs880197, the frequencies of homozygous A/A, heterozygous A/T and homozygous T/T were 15.4%, 22.2% and 62.3% respectively. In the analysis of rs10895304, we found that the frequencies of homozygous A/A, heterozygous A/G and G/G were 70.9%, 24.5% and 4.6% respectively. For rs12184413, the frequencies of homozygous C/C, heterozygous C/T, and homozygous T/T were 76.6%, 18.9% and 4.6% respectively. These frequencies were in Hardy-Weinberg equilibrium (p=0.118).
TABLE 2.
| Polymorphism | Transition | Minor Allele Frequency | Gene Region |
|---|---|---|---|
| rs880197 | A → T | 35% | Promoter (200 kb) |
| rs10895304 | A → G | 20% | 3′FR(−1.2 kb) |
| rs12184413 | C → T | 12% | 3′FR(−1.7 kb) |
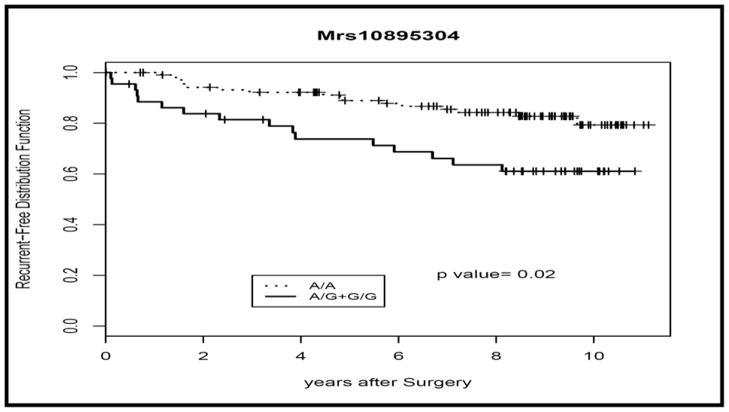
Using chi square analysis, we evaluated the association of each of the SNPs to patient factors including race, patient age at diagnosis, surgical margin status, pre-prostatectomy PSA level, Gleason score, extracapsular extension and disease stage. There was no evidence for association of any of these factors with our evaluated SNPs (data not shown). We used the Kaplan-Meier method to determine the recurrence and survival rates, and log-rank tests to test the difference in these rates between the genotypes of the different polymorphisms. There was no evidence of association of any of the polymorphisms to survival rates. Additionally, there was no statistical significance determined for recurrence rates in the two polymorphisms, rs880197 and rs12184413. However log-rank tests of the genotypes in rs10895304 demonstrated a statistically significant association of this polymorphism with 3 year recurrence-free survival (P= 0.0094, Log-Rank Test). Expression of the G allele of rs10895304 is associated with an increased risk for local recurrence (P=0.02, recessive univariate analysis, Log-rank test). The findings for this polymorphism, rs10895304, are illustrated in Table 3 and Figure 1.
TABLE 3.
| MMP7 (rs10895304) Genotype | ||||||
|---|---|---|---|---|---|---|
| FACTORS | ‘A/A’ (N = 107) | ‘A/G’ (N=37) | ‘G/G’ (N=7) | Missing (N=61) | Total (N=212) | p-value |
| Race, n(%) | 1.0000 | |||||
| Black | 1 (0.93) | 0 (0.00) | 0 (0.00) | 3 (4.92) | 4 (1.89) | |
| White | 106 (99.07) | 37 (100.00) | 7 (100.00) | 58 (95.08) | 208 (98.11) | |
| Age at diagnosis (years) | 0.5711 | |||||
| <=65 | 70 (65.42) | 27 (72.97) | 4 (57.14) | 47 (77.05) | 148 (69.81) | |
| >65 | 37 (34.58) | 10 (27.03) | 3 (42.86) | 14 (22.95) | 64 (30.19) | |
| Pre-prostatectomy PSA (ng/ml) | 0.2925 | |||||
| Missing | 16 (14.95) | 4 (10.81) | 2 (28.57) | 12 (19.67) | 34 (16.04) | |
| 4–10 | 60 (56.07) | 20 (54.05) | 2 (28.57) | 31 (50.82) | 113 (53.30) | |
| <4 | 12 (11.21) | 2 (5.41) | 1 (14.29) | 7 (11.48) | 22 (10.38) | |
| >10 | 19 (17.76) | 11 (29.73) | 2 (28.57) | 11 (18.03) | 43 (20.28) | |
| Gleason score, n(%) | 0.5079 | |||||
| 2–6 | 68 (63.55) | 18 (48.65) | 5 (71.43) | 39 (63.93) | 130 (61.32) | |
| 7 | 33 (30.84) | 16 (43.24) | 2 (28.57) | 19 (31.15) | 70 (33.02) | |
| 8–10 | 6 (5.61) | 3 (8.11) | 0 (0.00) | 3 (4.92) | 12 (5.66) | |
| Surgical margin, n(%) | 0.2935 | |||||
| Missing | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (1.64) | 1 (0.47) | |
| Negative | 73 (68.22) | 20 (54.05) | 5 (71.43) | 35 (57.38) | 133 (62.74) | |
| Positive | 34 (31.78) | 17 (45.95) | 2 (28.57) | 25 (40.98) | 78 (36.79) | |
| Extracapsular extension, n(%) | 0.7779 | |||||
| Missing | 9 (8.41) | 4 (10.81) | 0 (0.00) | 7 (11.48) | 20 (9.43) | |
| Negative | 58 (54.21) | 21 (56.76) | 5 (71.43) | 41 (67.21) | 125 (58.96) | |
| Positive | 40 (37.38) | 12 (32.43) | 2 (28.57) | 13 (21.31) | 67 (31.60) | |
| Disease Stage, n(%) | 0.6483 | |||||
| Missing | 5 (4.67) | 1 (2.70) | 1 (14.29) | 6 (9.84) | 13 (6.13) | |
| Localized (T1) | 5 (4.67) | 3 (8.11) | 0 (0.00) | 0 (0.00) | 8 (3.77) | |
| Localized (T2) | 67 (62.62) | 19 (51.35) | 4 (57.14) | 44 (72.13) | 134 (63.21) | |
| Localized (T3) | 30 (28.04) | 14 (37.84) | 2 (28.57) | 11 (18.03) | 57 (26.89) | |
| Recurrence, n(%) | 0.0094 | |||||
| No | 90 (84.11) | 22 (59.46) | 6 (85.71) | 50 (81.97) | 168 (79.25) | |
| Yes | 17 (15.89) | 15 (40.54) | 1 (14.29) | 11 (18.03) | 44 (20.75) | |
| Overall survival, n(%) | 0.8125 | |||||
| Alive | 85 (79.44) | 30 (81.08) | 5 (71.43) | 55 (90.16) | 175 (82.55) | |
| Dead | 22 (20.56) | 7 (18.92) | 2 (28.57) | 6 (9.84) | 37 (17.45) | |
Figure 1.
COMMENT
In this study, we analyzed three high frequency genetic polymorphisms (rs880197, rs10859304, and rs12184413) to determine if there was evidence for their association with clinicopathologic parameters and survival outcomes in 212 clinically localized prostate cancer patients who underwent radical prostatectomy. We found an association between the rs10859304 and local recurrence. In our patient cohort, the G allele of rs10895304 is associated with increased risk for local recurrence (P=0.0094, Log-rank Test). However there is no evidence for association of any of these polymorphisms with other clinical, histologic or survival outcomes. In our study, pre-prostatectomy PSA levels, Gleason score, surgical margin status, extracapsular extension status and T-stage were all predictive of relapse-free survival. However only Gleason score was predictive of overall survival. It may be that our cohort was underpowered for studies of survival, or that there was not an adequately long enough follow-up period to adequately assess survival.
To our knowledge, no studies have been published on the impact of MMP-7 polymorphisms in prostate cancer. However, there have been multiple studies in other malignancies, which have found positive associations with cancer risk (Li et al, Gyn Onc, 2006; Kubben et al, Br J Can, 2006; Hughes et al, Clin Can Res, 2007; Singh et al, Gyn Onc, 2008; Beeghly-Fadiel et al., Can Res, 2008; Beeghly-Fadiel et al, Int J Can, 2008; Sugimoto et al, J Gastro 2008). The largest of these studies has been a two-stage evaluation of case control study of Chinese women (Beeghly-Fadiel et al., Can Res, 2008; Beeghly-Fadiel et al, Int J Can, 2008), in which women with the recessive genotype (T/T) of rs12184413 had a 30% decreased breast cancer risk over those with the (T/C) and (C/C) genotypes. In vitro analysis revealed an allelic difference in protein binding capacity for the genotypes of this polymorphism. In addition, in silico analysis revealing that this region is rich in CTCF binding sites, which may be involved in transcriptional regulation. Together these findings were consistent with a model in which the homozygous recessive genotype resulted in decreased protein binding, and resultant decreased enzyme activity that could contribute to breast cancer susceptibility. The authors did evaluate the rs10895304, and the homozygotes were significantly associated with an increased breast cancer risk among premenopausal women in the first stage of analysis. Also, the rs880197 polymorphism was associated with a decreased breast cancer risk. However neither of these findings was validated in the second stage of analyses, and neither the rs10895304 or rs880197 polymorphisms were evaluated in vitro for biologic activity.
Our findings of increased local recurrence risk with the G allele of rs10895304 are consistent with a model of increased expression or activity of the MMP-7. MMP-7 is responsible for degrading extracellular matrix (ECM) proteins, such as elastin, E-cadherin, fibronectin, collagens (particular type IV) and proteoglycans (Wilson et al, Int J Biochem cell Biol, 1996; Nelson et al, JCO 2000; Shioma and Okada, Can Met Rev 2003). It is also involved in degrading non-extracellular matrix proteins including tumor necrosis-α precursor, Fas ligand, protumor necrosis factor-α, insulin-like growth factor binding proteins and heparin-binding epidermal growth factor (Wilson et al, Int J Biochem Cell Biol 1996; Egeblad et al, Nat Rev Cancer 2002; Hojilla et al, Br J Can, 2003; Li et al, Exp Biol Med, 2006). The earliest defined role of MMP-7 was for proteolytic break down the physical barriers in the extracellular matrix (Wilson et al, Int J Biochem Cell Biol, 1996). We could hypothesize that rs10895304 is resulting in increased MMP-7 activity, and that increased migration and invasion is promoted by both the degradation of the ECM proteins, as well as the shedding (and inhibition) of E-cadherin and HB-EGF, and release of IGF. Rs10895304 is located ~1.2 kb downstream of the MMP-7 gene, and overlaps with several transcription bindings sites (including: Mammalian transcriptional repressor RBP-J kappa, Myocyte-specific enhancer binding factor and Hepatic Nuclear Factor 1), which may influence transcriptional regulation of the MMP-7 gene. Alternatively, this polymorphism may be in high linkage disequilibrium with another gene region that may be responsible for the increased local recurrence risk.
CONCLUSIONS
In our study, we found an association between the high frequency polymorphism, rs10895304, and local recurrence risk in clinically localized prostate cancer. The G allele of the rs10895304 polymorphism is predictive of increased local recurrence risk in this subset of patients. This represents a marker for tumor aggressiveness, but further in vitro studies would need to be performed to determine if there is an effect on MMP-7 function of this polymorphism. In any case, the subset of patients with clinically localized prostate cancer and the G allele of rs10895304 may have an altered risk-stratification. Perhaps in this group of patients, prostatectomy alone may not be adequate for local control. This is a novel and relevant marker that should be evaluated for improved risk stratification of patients who may be candidates for early post-operative radiation therapy or hormonal therapy to improve local control.
Acknowledgments
Grant Support: This work was supported in part by the DOD grant PC031161 (PI: Bo Lu).
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Parekh DJ, Ankerst DP, Troyer D, Srivastava S, Thompson IM. Biomarkers for prostate cancer detection. J Urol. 2007;178:2252–9. doi: 10.1016/j.juro.2007.08.055. [DOI] [PubMed] [Google Scholar]
- 3.Shariat SF, Karam JA, Margulis V, Karakiewicz PI. New blood-based biomarkers for the diagnosis, staging and prognosis of prostate cancer. BJU Int. 2008;101:675–83. doi: 10.1111/j.1464-410X.2007.07283.x. [DOI] [PubMed] [Google Scholar]
- 4.Miyazaki K, Hattori Y, Umenishi F, Yasumitsu H, Umeda M. Purification and characterization of extracellular matrix-degrading metalloproteinase, matrin (pump-1), secreted from human rectal carcinoma cell line. Cancer Res. 1990;50:7758–64. [PubMed] [Google Scholar]
- 5.Quantin B, Murphy G, Breathnach R. Pump-1 cDNA codes for a protein with characteristics similar to those of classical collagenase family members. Biochemistry. 1989;28:5327–34. doi: 10.1021/bi00439a004. [DOI] [PubMed] [Google Scholar]
- 6.Sellers A, Woessner JF., Jr The extraction of a neutral metalloproteinase from the involuting rat uterus, and its action on cartilage proteoglycan. Biochem J. 1980;189:521–31. doi: 10.1042/bj1890521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson CL, Matrisian LM. Matrilysin: an epithelial matrix metalloproteinase with potentially novel functions. Int J Biochem Cell Biol. 1996;28:123–36. doi: 10.1016/1357-2725(95)00121-2. [DOI] [PubMed] [Google Scholar]
- 8.Woessner JF, Jr, Taplin CJ. Purification and properties of a small latent matrix metalloproteinase of the rat uterus. J Biol Chem. 1988;263:16918–25. [PubMed] [Google Scholar]
- 9.Cardillo MR, Di Silverio F, Gentile V. Quantitative immunohistochemical and in situ hybridization analysis of metalloproteinases in prostate cancer. Anticancer Res. 2006;26:973–82. [PubMed] [Google Scholar]
- 10.Powell WC, Knox JD, Navre M, Grogan TM, Kittelson J, Nagle RB, Bowden GT. Expression of the metalloproteinase matrilysin in DU-145 cells increases their invasive potential in severe combined immunodeficient mice. Cancer Res. 1993;53:417–22. [PubMed] [Google Scholar]
- 11.Knox JD, Wolf C, McDaniel K, Clark V, Loriot M, Bowden GT, Nagle RB. Matrilysin expression in human prostate carcinoma. Mol Carcinog. 1996;15:57–63. doi: 10.1002/(SICI)1098-2744(199601)15:1<57::AID-MC8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto K, Kihira Y, Matuo Y, Usui T. Expression of matrix metalloproteinase-7 and tissue inhibitor of metalloproteinase-1 in human prostate. J Urol. 1998;160:1872–6. [PubMed] [Google Scholar]
- 13.Kuhlmann KF, van Till JW, Boermeester MA, de Reuver PR, Tzvetanova ID, Offerhaus GJ, Ten Kate FJ, Busch OR, van Gulik TM, Gouma DJ, et al. Evaluation of matrix metalloproteinase 7 in plasma and pancreatic juice as a biomarker for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:886–91. doi: 10.1158/1055-9965.EPI-06-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh H, Jain M, Mittal B. MMP-7 (−181A>G) promoter polymorphisms and risk for cervical cancer. Gynecol Oncol. 2008;110:71–5. doi: 10.1016/j.ygyno.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Ghilardi G, Biondi ML, Erario M, Guagnellini E, Scorza R. Colorectal carcinoma susceptibility and metastases are associated with matrix metalloproteinase-7 promoter polymorphisms. Clin Chem. 2003;49:1940–2. doi: 10.1373/clinchem.2003.018911. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Jin X, Kang S, Wang Y, Du H, Zhang J, Guo W, Wang N, Fang S. Polymorphisms in the promoter regions of the matrix metalloproteinases-1, -3, -7, and -9 and the risk of epithelial ovarian cancer in China. Gynecol Oncol. 2006;101:92–6. doi: 10.1016/j.ygyno.2005.09.058. [DOI] [PubMed] [Google Scholar]
- 17.Vairaktaris E, Serefoglou Z, Yapijakis C, Vylliotis A, Nkenke E, Derka S, Vassiliou S, Avgoustidis D, Neukam FW, Patsouris E. High gene expression of matrix metalloproteinase-7 is associated with early stages of oral cancer. Anticancer Res. 2007;27:2493–8. [PubMed] [Google Scholar]
- 18.Zhang J, Jin X, Fang S, Wang R, Li Y, Wang N, Guo W, Wang Y, Wen D, Wei L, et al. The functional polymorphism in the matrix metalloproteinase-7 promoter increases susceptibility to esophageal squamous cell carcinoma, gastric cardiac adenocarcinoma and non-small cell lung carcinoma. Carcinogenesis. 2005;26:1748–53. doi: 10.1093/carcin/bgi144. [DOI] [PubMed] [Google Scholar]
- 19.Beeghly-Fadiel A, Long JR, Gao YT, Li C, Qu S, Cai Q, Zheng Y, Ruan ZX, Levy SE, Deming SL, et al. Common MMP-7 polymorphisms and breast cancer susceptibility: a multistage study of association and functionality. Cancer Res. 2008;68:6453–9. doi: 10.1158/0008-5472.CAN-08-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.American Joint Committee on Cancer. AJCC Cancer Staging Manual. New York: Springer; 2002. Prostate; pp. 309–316. [Google Scholar]
- 21.Browning REt, Li H, Shinohara ET, Cai Q, Chen H, Courtney R, Cao C, Zheng W, Lu B. ATM polymorphism IVS62+60G>A is not associated with disease aggressiveness in prostate cancer. Urology. 2006;67:1320–3. doi: 10.1016/j.urology.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Li HC, Albert JM, Shinohara ET, Cai Q, Freyer A, Cai H, Cao C, Wang Z, Kataoka N, Teng M, et al. E-cadherin promoter polymorphisms are not associated with the aggressiveness of prostate cancer in Caucasian patients. Urol Oncol. 2006;24:496–502. doi: 10.1016/j.urolonc.2006.02.018. [DOI] [PubMed] [Google Scholar]