Abstract
Loss of cardiomyocytes from cardiovascular disease is irreversible and current therapeutic strategies do not redress the loss of myocardium after injury. The discovery that endogenous fibroblasts in the heart can be reprogrammed to cardiomyocyte-like cells after myocardial infarction and heart function is improved subsequently has strong implications in y bringing this treatment paradigm to the clinic. Here we discuss the advances in direct cardiac reprogramming that will potentially act as a springboard in the generation of effective approaches to restoring cardiac function after injury.
Introduction
Cardiovascular diseases claim more lives than any other disease worldwide. The heart's limited ability to repair itself after injuries, such as myocardial infarction, is manifested by scar formation in the heart rather than cardiac muscle regeneration. As a result, individuals experience progressive cardiac dysfunction, contributing to the epidemic of heart failure affecting 23 million worldwide. Although current medical therapies can support a heart with reduced function, there is currently no therapy that could repair damaged cardiac tissue after injury. Much effort has been put into cell replacement strategies in which progenitors, derived either from endogenous cells in the heart, or from exogenous sources, are introduced to the injured heart [1,2]. While these approaches may confer some benefit through paracrine mechanisms that may promote angiogenesis and myocyte survival, they have limited effect on generating new myocardium. Introduction of cardiomyocytes derived from pluripotent stem cells may be able to increase myocardial mass, but more research is needed to overcome issues of purification, delivery, survival, maturation and integration of such cells into the heart.
An alternative strategy that has recently gained momentum involves reprogramming somatic cells from one cell type to another. The scientific foundation for cellular reprogramming was laid in a seminal discovery made over 25 years ago when Davis et al [3] showed that the overexpression of a single transcription factor, MyoD, is sufficient to convert fibroblasts to skeletal muscle in vitro (See Tapacott this issue COGEDE-D-13-00021. Despite the similarities between skeletal and cardiac muscle, a cardiac equivalent of MyoD has yet to be identified. Inspired by the combinatorial effects of multiple transcription factors to reprogram fibroblasts to induced pluripotent stem (iPS) cells [4], we reported that introduction of three members of the core cardiac developmental transcription machinery could induce fibroblasts to adopt a cardiomyocyte-like phenotype in vitro [5]. The same factors function with greater efficiency in vivo [6,7], with addition of other factors [8] or use of microRNAs [9] providing alternative approaches to reprogramming. A similar approach has been used to reprogram fibroblasts to neurons [10]{Yoo, 2011 #17;Ambasudhan, 2011 #22;Son, 2011 #16}, neural stem cells [11–13], hepatocytes (see Suzuki this issue: COGEDE-D-13-00019), hematopoietic cells (see Salci this issue: COGEDE-D-13-00025), and endothelial cells, among other cell types [14–18]. Here we will focus our discussion on reprogramming cardiomyocyte-like cells from fibroblasts. In the first section, we review the development of induced cardiomyocyte-like cells in vitro, and in the second, we highlight its utility in vivo. Finally, we discuss translating cardiac reprogramming from mouse models to human models, as well as potential hurdles for moving cardiac reprogramming into the clinic.
The development of induced cardiomyocyte-like cells
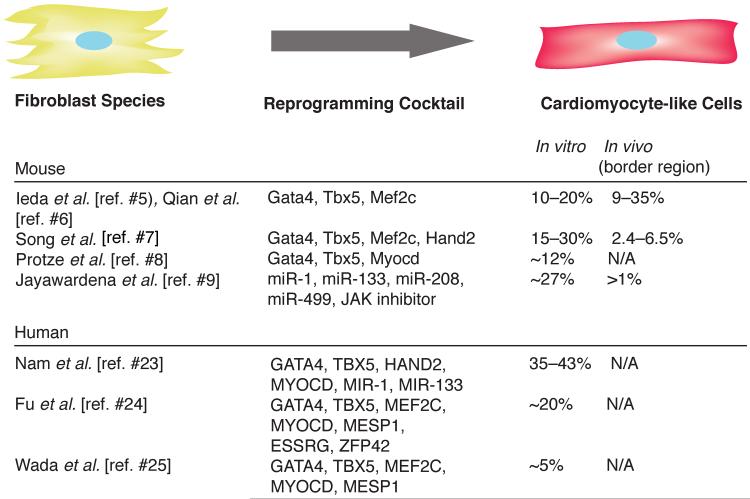
In the first report of cardiac reprogramming, Ieda et al [5] took a candidate approach to identify factors sufficient to convert mouse cardiac and dermal fibroblasts to cardiomyocyte-like cells in vitro. Fourteen developmental cardiac transcription factors were selected and screened for reprogramming using fibroblasts derived from transgenic mice containing EGFP under control of the cardiac-specific α-myosin heavy chain promoter (αMHC-GFP). Few αMHC-GFP+ cells were observed after viral induction of all 14 factors in fibroblasts (<1%). However, serial removal of individual factors demonstrated that a minimum cocktail of three factors, Gata4, Mef2c, and Tbx5 (GMT) was sufficient for generating a greater number of αMHC-GFP+ cells (10–20%). Of the αMHC-GFP+ cells, approximately one- third also expressed cardiac Troponin T (cTnT), 20% developed calcium transients, but only approximately 0.5% of these became more fully reprogrammed with the ability to contract. Despite the low percentage of cells that developed to beating cardiomyocyte-like cells, microarray analyses of the entire αMHC-GFP+ population revealed that the partially reprogrammed population displayed a gene expression profile similar to that of the neonatal cardiomyocytes and the cells showed varying degrees of sarcomere assembly. Furthermore, the reprogramming of these cells was epigenetically stable based on DNA methylation and histone modifications at select loci, and the persistence of phenotypic alteration in the absence of ongoing overexpression of the reprogramming factors. We have termed these cells induced cardiomyocytes (iCMs), which are similar, but clearly distinct, from native cardiomyocytes.
To understand the path of conversion from fibroblasts to iCMs, we utilized fibroblasts from mice harboring an Isl1-Cre or Mesp1-Cre transgene, crossed to a reporter mouse with the reporter activated only upon Cre-recombinase. In this system, if the fibroblasts de-differentiated first into the early mesodermal progenitor (Mesp1+) or cardiac progenitor (Isl1+) state and then differentiated into iCMs, they would be marked by the reporter. The vast majority did not activate Cre-recombinase, suggesting the cells were not obliged to pass through a progenitor state during the conversion. Consistent with this, the electrical activity of the reprogrammed cells was more to a post-natal ventricular cardiomyocyte rather than the relatively immature pluripotent stem cell-derived cardiomyocytes.
The efficiency of in vitro reprogramming can be improved by the addition of a fourth cardiac transcription factor, Hand2 (GHMT) [7], the replacement of Gata4 with the transcription factor Myocd [8], or addition of other developmental transcription factors [19]. There are likely numerous combinations that can activate the cardiac gene program, as the core transcriptional machinery functions in reinforcing feedback loops in which critical transcription factors activate one another and cooperatively interact with each other.
In a screen for candidate cardiac microRNAs for cardiac reprogramming [9], Jayawardena et al found that overexpression of a cocktail of four microRNAs (miRNA), miR-1, miR-133, miR-208, and miR-499, was sufficient to reprogram 1.5–5% of fibroblasts to cardiomyocyte-like cells. Interestingly, when a JAK inhibitor shown to promote cardiac differentiation from partially reprogrammed cells was added to the microRNA cocktail [20], the efficiency of reprogramming improved to 13–27%, and reprogramming can be achieved with just a single miRNA, miR-1. Similar to transcription factor based methods, levels of cardiac mRNA expression increased and Ca2+ oscillations were observed in approximately 1–2% of the transfected cells. Similar to the cardiac transcription factors, these miRNAs make up the core miRNA regulators in the heart [reviewed in 21,22].
The in vitro strategies outlined above can generate cells that are transcriptionally similar to endogenous cardiomyocytes, although there is significant heterogeneity in the degree of sarcomere formation, and only a very small subset are fully reprogrammed and display contractile activity.
Cardiac reprogramming in vivo
To determine if direct reprogramming of resident cardiac fibroblasts, which comprise over 50% of cells in the heart, can improve heart function after injury, cocktails of reprogramming factors were added to the site of injury after myocardial infarction. Mice harboring various lineage markers for the non-myocyte population were subjected to left anterior descending (LAD) ligation, causing fibroblasts to proliferate to form a scar. Retroviral cocktails of GMT [6] or GHMT [7] that infect actively dividing cells, including scar forming fibroblasts, but not non-dividing cells such as cardiomyocytes, were injected into the myocardial border zone after injury. iCMs that were dually positive for cardiac marker and fibroblast lineage tracing markers, were observed in 9–35% of myocytes in the border zone of the infarct, indicating that the reprogrammed iCMs originated from fibroblasts.
iCMs isolated from injured hearts exhibited action potentials, calcium transients, and with varying degrees of maturity, the ability to contract. Evaluation of iCMs by histologic section or single cell isolation at weekly intervals over a four week period revealed progressive changes in sarcomere assembly, with over half ultimately developing well-organized sarcomeres throughout the cell and another 30% displaying sarcomeres through ~2/3 of the cell. A large percentage of in vivo iCMs were more fully reprogrammed (assessed by beating and electrical activity), with up to 50% of reprogrammed cells capable of beating when isolated. In vivo iCMs also electrically coupled with endogenous iCMs and could contract synchronously [6]. Furthermore, significant functional improvements, including cardiac output, ejection fraction, stroke volume, and decreased scar size were observed upon GMT and GHMT treatment, but not after treatment with dsRed control retrovirus. Comparing the efficiency and quality of iCMs in vitro and in vivo, it appears that other unknown factors, including mechanical forces, local signals, and extracellular matrix effects in the in vivo environment may have enhanced the maturity of the iCMs.
A similar approach was taken to determine if the combination of miRNAs and a JAK inhibitor were sufficient for direct reprogramming in vivo [9]. Lentivirus containing the four microRNAs, which unlike retrovirus, infects proliferating and non-proliferating cells, were injected at the site of injury in hearts after LAD ligation. Quantification of cells positive for both fibroblast lineage tracing markers and cTnT revealed that the infarct region contained up to 1% of iCMs. Parameters of heart function, however, were not evaluated in these experiments. It remains unknown if this combination of factors can improve heart function after myocardial infarction.
From mice to humans
An essential step in advancing cardiac reprogramming technology is to translate the knowledge gleaned from studies using the mouse system to human cells. Several recent studies reported the failure of GMT or GMHT to convert fibroblasts directly to cardiomyocyte-like cells, but each described overlapping but distinct combinations of factors that could push human fibroblasts into a more cardiomyocyte-like state [23–25]. Nam et al. reported that a combination of four transcription factors (Gata-4, Hand2, Tbx-5, and Myocd) and two miRNAs (miR-1, and miR-133) were sufficient to reprogram up to 20% of human fibroblasts into cTnT-expressing cells. Similar to in vitro reprogramming reports in mice, calcium transients and increased levels of cardiac-enriched mRNAs could be observed, along with rare beating events. However, not all cardiac genes were upregulated, suggesting the switch from a fibroblast-like phenotype to cardiomyocyte-like phenotype is partial. Similarly, Fu et al. found that adding a nuclear hormone receptor, ESRRG, MESP1, and Myocardin to GMT could induce gene expression shifts in human fibroblasts to a similar extent as GMT in mouse cells in vitro [24]. About 20% of these cells developed calcium transients, and some had action potentials similar to human pluripotent stem cell-derived cardiomyocytes. Wada et al. also found that addition of MESP1 and Myocardin to GMT could reprogram human fibroblasts into cells that displayed many properties of human cardiomyocytes [25]. Although slightly different, human dermal fibroblasts can be reprogrammed using MESP1 and ETS-2 into a cardiac progenitor state that can then differentiate into cardiomyocyte-like cells [26]. Overall, while human cells appear more resistant to reprogramming, various combinations of the core transcriptional machinery are able to shift cells considerably toward the cardiomyocyte state.
Conclusions
The discovery that fibroblasts can be converted to cardiomyocyte-like cells by enforced expression of 3–4 molecular factors in mouse, and 6–7 factors in human cells, reveals a new approach for repairing damaged cardiac tissues. Although cardiomyocyte-like cells generated in the in vitro studies were less than perfect, they created a foundation that ultimately led to a more mature cardiomyocyte-like cell in vivo, capable of improving heart function and reducing scar size after injury. It is clear in the mouse studies that the microenvironment of the heart provided iCMs with unknown cues necessary to take on the hallmarks of a mature cardiomyocyte. Given the experience comparing in vitro vs. in vivo iCMs in mice [6,7,24], it is possible that the current degree of human cell reprogramming will suffice. It will be important to determine if the in vivo environment, possibly in pigs or in non-human primates, will improve reprogramming of human fibroblasts using the current cocktails.
Many important questions and issues in the area of cardiac reprogramming remain. One question is the mechanism underlying reprogramming. It is imperative to identify the DNA targets of the reprogramming factors, their combinatorial function, and the cascade of cellular events they initiate. How transcriptional changes are epigenetically stabilized will also be important to define. Furthermore, it is unknown what role extracellular matrix cues, cell-cell signaling, and tensile forces within the beating heart play during reprogramming. This knowledge of the pathways and cues necessary for reprogramming may be leveraged to further improve cardiac reprogramming.
Another important issue to address from a translational standpoint is safety. In the in vivo studies done in mice, reprogramming factors were delivered virally. Despite a number of viral gene therapy clinical trials underway, the safety of viral vectors remains a concern. Since small molecules have been identified that can replace factors important for reprogramming fibroblasts to induced pluripotent stem cells [27]{Zhu, 2010 #12}, it may be possible to achieve cardiac reprogramming with small molecules as well. Additionally, the safety of partially reprogrammed cells in the heart and the potential disturbances of cardiac rhythm must be evaluated. Further optimization of reprogramming human cells and demonstration of the safety and therapeutic efficacy of this approach should also be performed in large animal models. The paradigm of leveraging cellular reprogramming technology to regenerate tissue by controlling the fate of endogenous support cells in the area of heart injury is an exciting one and may extend to other organs, such as spinal cord, liver, pancreas and brain.
Figure 1. Efficiency of cardiac reprogramming in murine and human cells.
Multiple combinations of transcription factors and miRNAs can induce cardiac reprogramming in mouse and human fibroblasts. To achieve enforced expression, genes were injected into the border region of the infarct.
Acknowledgments
We thank members of the Srivastava laboratory for discussion. E.C.B. was partially supported by the National Science Foundation Predoctoral Fellowship. Work in the laboratory of D.S. was supported by grants from NHLBI/NIH (U01 HL100406, U01 HL098179, R01 HL057181, P01 HL089707), the California Institute for Regenerative Medicine (CIRM), the William Younger Family Foundation, the L.K. Whittier Foundation, and the Eugene Roddenberry Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alexander JM, Bruneau BG. Lessons for cardiac regeneration and repair through development. Trends Mol Med. 2010;16(9):426–434. doi: 10.1016/j.molmed.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cdna converts fibroblasts to myoblasts. Cell. 1987;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142(3):375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485(7400):593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG, Hill JA, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485(7400):599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Protze S, Khattak S, Poulet C, Lindemann D, Tanaka E, Ravens U. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53(3):323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M, Dzau VJ. Microrna-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110(11):1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lujan E, Chanda S, Ahlenius H, Sudhof TC, Wernig M. Direct conversion of mouse fibroblasts to self-renewing, tripotent neural precursor cells. Proc Natl Acad Sci USA. 2012;109(7):2527–2532. doi: 10.1073/pnas.1121003109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl AcadSci USA. 2011;108(19):7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11(1):100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475(7356):386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 15.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475(7356):390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 16.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468(7323):521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 17.Margariti A, Winkler B, Karamariti E, Zampetaki A, Tsai TN, Baban D, Ragoussis J, Huang Y, Han JD, Zeng L, Hu Y, et al. Direct reprogramming of fibroblasts into endothelial cells capable of angiogenesis and reendothelialization in tissue-engineered vessels. Proc Natl Acad Sci USA. 2012;109(34):13793–13798. doi: 10.1073/pnas.1205526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ginsberg M, James D, Ding BS, Nolan D, Geng F, Butler JM, Schachterle W, V R Pulijaal VR, S Mathew S, S T Chasen ST, J Xiang J, et al. Efficient direct reprogramming of mature amniotic cells into endothelial cells by ets factors and tgfbeta suppression. Cell. 2012;151(3):559–575. doi: 10.1016/j.cell.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christoforou N, Chellappan M, Adler AF, Kirkton RD, Wu T, Addis RC, Bursac N, Leong KW. Transcription factors myocd, srf, mesp1 and smarcd3 enhance the cardio-inducing effect of gata4, tbx5, and mef2c during direct cellular reprogramming. PloS one. 2013;8(5):e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13(3):215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 21.Cordes KR, Srivastava D. Microrna regulation of cardiovascular development. Circ Res. 2009;104(6):724–732. doi: 10.1161/CIRCRESAHA.108.192872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu N, Olson EN. Microrna regulatory networks in cardiovascular development. Developmental cell. 2010;18(4):510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nam Y, Song K, Luo X, Daniel E, Lambeth E, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R, Olson EN. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110(14):5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu J-D, Stone NR, Liu L, Spencer CI, Qian L, Hayashi Y, Delgado-Olguin P, Ding S, Bruneau BG, Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. doi: 10.1016/j.stemcr.2013.07.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wada R, Muraoka N, Inagawa K, Yamakawa H, Miyamoto K, Sadahiro T, Umei T, Kaneda R, Suzuki T, Kamiya K, Tohyama S, et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci USA. 2013 doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Islas JF, Liu Y, Weng KC, Robertson MJ, Zhang S, Prejusa A, Harger J, Tikhomirova D, Chopra M, Iyer D, Mercola M, et al. Transcription factors ets2 and mesp1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. 2012;109(32):13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, Ge J, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013 doi: 10.1126/science.1239278. Published online 18 July 2013 [DOI:2010.1126/science.1239278] [DOI] [PubMed] [Google Scholar]