Abstract
The nucleotidyl transferase cGAS, its second messenger product cGAMP and the cGAMP sensor STING, form the basic mechanism of DNA sensing in the cytoplasm of mammalian cells. Several new reports now uncover key structural features associated with DNA recognition by cGAS and the catalytic mechanisms of cGAMP generation. Concurrent studies also reveal unique phosphodiester linkages in endogenous cGAMP that distinguish it from microbial cGAMP and other cyclic-di-nucleotides. Together, these studies provide a new perspective on DNA recognition in the innate immune system.
Introduction
Pattern recognition receptors survey extracellular, endosomal and cytosolic compartments for signs of infection and mount protective immune responses that coordinate host-defense. DNA that gains access to the cytoplasm is perceived as a danger signal that alerts the host to the presence of infectious microorganisms (Atianand and Fitzgerald, 2013). Endogenous DNA that is inappropriately cleared can also accumulate in cytosolic compartments and drive pathological inflammation and autoimmune diseases such as SLE (Systemic lupus erythematosus). Understanding how DNA is detected and how DNA recognition couples to downstream signaling therefore has important implications for infectious and inflammatory diseases. Not surprisingly, elucidation of these pathways has been a focus of considerable attention for many years.
Certain aspects of DNA sensing have been worked out in some detail. DNA activates caspase-1 leading to the maturation of the pro-inflammatory cytokines IL-1β and IL-18. These events are mediated by a DNA-binding protein AIM2, part of a larger PYHIN (Pyrin and HIN200 domain) protein family (Atianand and Fitzgerald, 2013). Although the AIM2 response is important in host-defense, the dominant response to DNA results in transcriptional regulation of type I IFNs. The signal transduction cascade that coordinates this response involves an adapter molecule STING (also known as MPYS, TMEM173, ERIS and MITA) that is localized to the endoplasmic reticulum (Ishikawa and Barber, 2008; Jin et al., 2008; Zhong et al., 2008). STING contains four transmembrane helices and a globular carboxy-terminal domain (CTD) that protrudes into the cytosol. The very C-terminal tail (CTT) of this domain maintains STING in an autoinhibited state (Yin et al., 2012) and facilitates interactions with the IKK related kinase TBK1 in order to facilitate activation of IRF3, an important transducer of IFN gene transcription (Tanaka and Chen, 2012). Putative DNA sensing PRRs were then proposed to act upstream of STING. Indeed, a number of candidate receptors have been linked to varying degrees to STING activation, including an AIM2 related protein IFI16 (Unterholzner and Bowie, 2011) and a DExD/H box helicase called DDX41 (Zhang et al., 2011) amongst others. Insights obtained from studying these molecules have greatly added to our understanding of DNA recognition, but seminal discoveries recently published from the Chen laboratory have been instrumental in elucidating the DNA recognition pathway. Chen and colleagues identified a novel nucleotidyl transferase, cGAS that produces an endogenous second messenger cGAMP (Sun et al., 2013; Wu et al., 2013). In this review, we discuss exciting recent developments regarding the sensing of DNA by cGAS, the enzymatic generation of cGAMP and the mechanisms involved in cGAMP binding to STING.
While mounting evidence indicated STING was an adapter molecule in the DNA sensing pathway, additional studies suggested STING functioned as a direct innate immune sensor of cyclic di-guanylate monophosphate (c-di-GMP) and cyclic-di-adenylate monophosphate (c-di-AMP) (Burdette et al., 2011). These cyclic dinucelotides are conserved signaling molecules produced by bacteria that regulate bacterial motility and biofilm formation. STING binds these small molecules through its CTD. Structural studies indicated that the CTD mediated stable STING-dimerization and that c-di-nucleotides bound at a cleft/groove between two STING monomers (Huang et al., 2012; Ouyang et al., 2012; Parvatiyar et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). Binding of c-di-nucleotides has been proposed to relieve an autoinhibited state, expose the CTT, and stabilize STING dimers (Huang et al., 2012; Shang et al., 2012; Yin et al., 2012). Surprisingly, ligand engagement did not induce significant conformational changes in most of the published STING-c-di-nucleotide structures, which were partially explained by possible conformational changes at the CTT that was not observed in the structures. Clearly major gaps therefore remain in our understanding of STING function and DNA sensing.
Discovery of cGAMP as a second messenger
Using elegant biochemical purification and reconstitution strategies, the Chen lab identified cyclic GMP-AMP (cGAMP) in the cytoplasm of cells exposed to DNA (Wu et al., 2013). cGAMP is an endogenous second messenger structurally similar to cyclic adenosine monophosphate (cAMP). cGAMP is also similar to c-di-GMP and c-di-AMP. Chen and colleagues also used quantitative mass spectrometry and classical protein purification strategies to identify the enzyme responsible for cGAMP generation. Their studies identified E330026A19, an uncharacterized mouse gene with significant structural homology to the catalytic domain of human oligoadenylate synthase (OAS1), which belongs to a large family of nucleotidyl transferases (NTases) including adenylate cyclase (Sun et al., 2013). The authors named this enzyme cGMP-AMP Synthase ‘cGAS’. cGAS binds DNA and catalyzes the synthesis of cGAMP from ATP and GTP in the presence of DNA. cGAMP was shown to bind STING in a manner similar to that of c-di-GMP and activate IRF3, thus reconciling the observations of STING as both an adapter for DNA sensing and a receptor for cyclic di-nucleotides.
Mammalian cGAMP contains unique combinations of phosphodiester linkages
A series of subsequent structural, biophysical and biochemical studies have now advanced our understanding of this fascinating second messenger system. Four independent studies uncovered unique features of the endogenous cGAMP (generated by cGAS) that distinguish it from cGAMP and other c-di-nucleotides of microbial origin (Ablasser et al., 2013; Diner et al., 2013; Gao et al., 2013; Zhang et al., 2013). While the prior studies of Chen and colleagues clearly demonstrated that cGAMP was an endogenous second messenger produced by cGAS in mammalian cells, the exact nature of the phosphodiester linkages between GMP and AMP were not determined, in part because the mass spectrometry approaches used could not unambiguously distinguish these linkages without the availability of all cGAMP isomers as the standard reference (Wu et al., 2013). Surprisingly, using mass spectrometry, enzymatic digestion, NMR analysis, chemical synthesis and structural studies four groups independently found that the cGAMP produced by cGAS contained a phosphodiester linkage between 2′-OH of GMP and 5′-phosphate of AMP and another between 3′-OH of AMP and 5′-phosphate of GMP. This molecule, referred to hereafter as 2′ 3′-cGAMP was found to bind to STING with high affinity and elicit potent IFNβ responses.
Two independent groups arrived at this conclusion while examining IFN responses to cyclic-di-nucleotides and cGAMP in cells expressing a mutant allele of murine STING (R231A), which was previously found to be unresponsive to c-di-nucleotides but normally responsive to DNA (Ablasser et al., 2013; Diner et al., 2013). With the discovery of cGAMP, these groups went on to demonstrate that the R231A mutant was also unresponsive to cGAMP produced by a cGAMP synthase from Vibrio cholera (Davies et al., 2012) and chemically synthesized cGAMP (with conventional 3′-5′ linkages). This observation was puzzling and inconsistent with the hypothesis that cGAMP was critical for driving STING signaling in the DNA sensing pathway. The authors reconciled this paradox by demonstrating that the endogenous cGAMP derived from cGAS had unique phosphodiester linkages different from that of microbial cGAMP or canonical c-di-nucleotides, which all had 3′-5′ linkages. Intriguingly, natural variants of human STING (hSTING) with histidine in place of the normal arginine at position 232 were also identified that were poorly responsive to microbial cGAMP yet responded normally to DNA and cGAS signaling. The presence of the noncanonical 2′-5′ linkage in endogenous cGAMP exerted potent activation of multiple human STING alleles regardless of the residue at position 232. Combined these observations define important differences between the 2′-5′ phosphodiester linkages in endogenous cGAMP and the 3′-5′ linkages found in more conventional c-di-nucleotides with important functional implications for activation of human STING variants.
Through structural determination of cGAS with different nucleotide substrates containing various combination of 2′ or 3′ deoxy nucleotides, the Patel lab found the noncanonical 2′-5′ GpA linkage in the di-nuleotide intermediates as well as the final cyclized product generated by cGAS (Gao et al., 2013). This was then confirmed by reverse-phase HPLC and NMR spectroscopy analysis of di-nucleotides from cGAS catalyzed products both in solution and from the crystals, in comparison with chemically synthesized cGAMP isomers. Employing similar chemical synthesis and biophysical approaches including reverse-phase HPLC, tandem mass spectrometry and circular dichroism, the Chen lab also confirmed the 2′-5′ GpA linkage and 3′-5′ ApG linkage (Zhang et al., 2013).
Using enzymatic digestions Hornung and colleagues expanded their analysis to demonstrate that cGAS first catalyses the synthesis of a linear 2′-5′-linked di-nucleotide, which was then subject to cGAS dependent cyclization in a second step through a 3′-5′ phosphodiester linkage (Ablasser et al., 2013). The Patel group reached the same conclusion using combinations of different nucleotides and in vitro enzymatic reactions (Gao et al., 2013). This 13-membered ring structure therefore defines a novel class of second messenger molecules, extending the family of 2′-5′-linked antiviral biomolecules.
Structural insights into DNA sensing and cGAMP generation by cGAS
Extensive structural studies also add to our understanding of how cGAS functions as a DNA-sensing enzyme (Civril et al., 2013; Gao et al., 2013; Kranzusch et al., 2013). cGAS contains a highly positively charged and poorly conserved N-terminal fragment, followed by a NTase domain that partially overlaps with a C-terminal male abnormal 21 (Mab21) domain. The NTase and Mab21 domains are conserved among different species, and recapitulate the DNA-induced interferon response activity of the full-length protein (Sun et al., 2013). Structures of the cGAS NTase and Mab21 domains reveal a bilobal scaffold, with an N-terminal NTase catalytic core that adopts a mixed α/β fold, and a C-terminal α-helical lobe. This bears significant structural similarity to OAS1, with the exception of a unique zinc-binding motif named “zinc thumb” located between the two lobes of cGAS that was essential for DNA binding.
cGAS binds dsDNA and to some extent also ssDNA, employing a positively charged surface as well as the zinc thumb to interact with the DNA sugar-phosphate backbone (Civril et al., 2013; Gao et al., 2013; Kranzusch et al., 2013). It was reported that at least 16 bp was needed for 2′3′-cGAMP production, and 36 bp and above were necessary for optimal cGAS activity (Gao et al., 2013), which is consistent with another study that demonstrated that two helical turns of dsDNA were required for cGAS binding with an affinity of 87.6 nM (Kranzusch et al., 2013). This is broadly in agreement with previous reports which identified a 45 bp interferon stimulatory DNA (ISD) as the minimal size required to elicit robust interferon production (Stetson and Medzhitov, 2006). Importantly, RNA was shown to be incapable of stimulating cGAS, and this was proposed to be at least partially due to potential steric hindrance between the zinc thumb and the A form structure of dsRNA, which has a different major/minor groove configurations and a wider diameter compared with the B form dsDNA.
Upon binding to dsDNA, a significant conformational change of the NTase catalytic core ensues, involving repositioning of the catalytic residues and widening of the entrance to the catalytic pocket to allow binding of nucleotide substrates. Binding of the GTP and ATP substrates induce small movements of the catalytic residue E211 (mouse) to coordinate two magnesium ions that bind the phosphate groups of the substrates, with the aromatic ring of the nucleotide base stacked over a conserved tyrosine residue. The proposed enzymatic reaction pathway involves a flip-over of the linear intermediate upon formation of the first linkage to allow the second triphosphate access to the catalytic residues (Civril et al., 2013; Gao et al., 2013). A second flipping was proposed upon formation of the final cGAMP product before release from cGAS. This may be facilitated by the charge repulsion between the acidic catalytic residues and the α-phosphate of the di-nucleotide.
Activation of STING by 2′ 3′-cGAMP
Previous structural studies demonstrated that STING binds c-di-GMP through a dimeric configuration (Huang et al., 2012; Ouyang et al., 2012; Shang et al., 2012; Shu et al., 2012; Yin et al., 2012). Surprisingly, most of the reports did not show structural changes in STING upon ligand binding. Recently, Zhang et al demonstrated that 2′3′-cGAMP binds STING with nM affinity whereas c-di-GMP binds ~300 fold weaker, suggesting the former is a much more potent physiological ligand (Zhang et al., 2013). In agreement, the crystal structure of STING in complex with 2′3′-cGAMP shows almost complete enclosure of the ligand at the STING dimer interface with extensive receptor:ligand interactions. This is accomplished by significant ligand-induced conformational changes, including a 20 Å movement of a V-shaped structural element above the binding site, and the formation of a four-stranded β-sheet containing residue R232 (R231 in mouse) as part of the binding pocket. This β-sheet is also call the “lid region”. A similar lid region of the murine STING was also observed in complex with a tricyclic compound 10-carboxymethyl-9-acridanone (CMA) (Cavlar et al., 2013). The function of STING residue R232 (R231 in mouse) in DNA sensing remains controversial. As discussed above murine STING R231A is unresponsive to c-di-GMP but normally responsive to DNA and cGAS (Ablasser et al., 2013; Burdette et al., 2011; Diner et al., 2013). In contrast, Zhang and colleagues showed that the equivalent human STING mutant R232A no longer supports DNA-induced IFN production through cGAS activation (Zhang et al., 2013), which is consistent with the crystal structure that demonstrates direct interaction between residue R232 and the α-phosphates of the 2′3′-cGAMP. In comparison, the available murine STING structures show that residue R231 is either disordered or located outside the binding pocket (Cavlar et al., 2013; Zhang et al., 2013). Because residues in this lid region have high temperature factors compared with the rest of the protein, it is conceivable that the lid region could adopt multiple conformations upon ligand binding. It remains to be determined if murine STING binds 2′3′-cGAMP similar to the human protein at the lid region near residue R231.
cGAS and OAS as nucleic acid-sensing nucleotidyl transferases
cGAS bears significant sequence and structural similarity to the OAS proteins, which are interferon-induced antiviral proteins that catalyze the formation of 2′-5′ oligoadenylate upon stimulation by dsRNA (Baglioni, 1979; Kerr and Brown, 1978). Both are latent NTase enzymes activated through sequence-independent binding of nucleic acids at similar surfaces. The ensuing large conformational changes rearrange the catalytic residues to convert them to competent enzymes. Intriguingly, both cGAS and OAS catalyze the formation of nucleotide products containing 2′-5′ linkages, and both products are implicated in innate immune response to nucleic acids. It is plausible that both cGAS and OAS are members of a larger family of nucleic acid-sensing enzymes with similar scaffolds, which catalyze the formation of second messenger molecules containing noncanonical nucleotide linkages.
Future Directions
Collectively these exciting developments have advanced our understanding of the structure and function of cGAS, clarified the chemical structure of the mammalian cGAMP, and unified the previous observations of STING functions. As the field charges ahead, it is useful to consider what’s next. One of the remaining issues to be resolved is the mysterious requirement of a certain DNA length. The recent structures indicate that the footprint of the cGAS NTase-Mab21 domains on dsDNA is ~10 bp, yet more than 36 bp was required for optimal interferon induction. It is possible that certain oligomerization states of cGAS induced or stabilized by longer dsDNA sizes are required for optimal cGAS activity. This may be analogous to the oligomeric signaling complex of another DNA sensor, the AIM2 inflammasome.
The highly basic N-terminal fragment of cGAS, which was absent in all reported structures, may also play a role in binding DNA of certain lengths, as Sun et al. observed that this fragment also bound to ISD (Sun et al., 2013). Interestingly, the N-terminal fragment may also be involved in intramolecular interactions to maintain cGAS in a latent state, as suggested by Kranzusch and colleagues (Kranzusch et al., 2013). This may be similar to the autoinhibition of other nucleic acid sensor such as RIG-I and AIM2. Such quiescent resting states of these nucleic acid sensors might be critical to prevent spurious activation of these pathways, which could elicit serious tissue damage and manifest in autoimmune and autoinflammatory disorders such as SLE.
The identification of the unique combination of phosphodiester linkages in mammalian cGAMP was a true tour de force. It is however unclear how cGAS catalyzes the combination of 2′-5′ and 3′-5′ linkages that lead to the formation of this unique endogenous cGAMP. Conceivably because the 2′-5′ linkage is uncommon, it may support a longer half-life for the di-nucleotide before cleavage by nucleases. On the other hand, the 2′-5′ phosphodiester bond may encourage depurination of the guanosine which would diminish its stability (Ablasser et al., 2013). More detailed enzymatic analysis will be required to dissect the mechanisms underlying the selective 2′-5′ and 3′-5′ linkages catalyzed by cGAS.
Though the metazoan second messenger 2′3′-cGAMP has been shown to have distinct chemical structure and activities compared with the bacterial second messenger c-di-GMP, what has been learned about the latter might point to future directions for the former. c-di-GMP is synthesized and degraded by diguanylate cyclases (DGCs) and phosphodiesterases (PDEs), respectively (Mills et al., 2011), and acts on diverse receptors containing various protein domains as well as RNA motifs. Could the newly discovered cGAS be the first among several metazoan enzymes that synthesize cGAMP, with each functioning in different cell types or distinct tissue environments? The possibility of cGAMP-specific PDEs is another tantalizing scenario of regulating immune responses to infection and other environment cues.
Finally, a pressing issue remains regarding the role of IFI16 and DDX41 in the broader context of cGAS-cGAMP signaling. It is unclear whether cGAS functions in diverse cell types, or whether each of these different sensors function in different cell types or tissue environments. Furthermore, it remains to be determined whether IFI16 or DDX41 function in concert with or engage directly with the cGAS-cGAMP system to induce interferon responses. For example, DDX41 was previously shown to associate with c-di-GMP with higher affinity than STING (Parvatiyar et al., 2012). It is conceivable that DDX41 could also associate with 2′3′-cGAMP as an accessory receptor of cGAS.
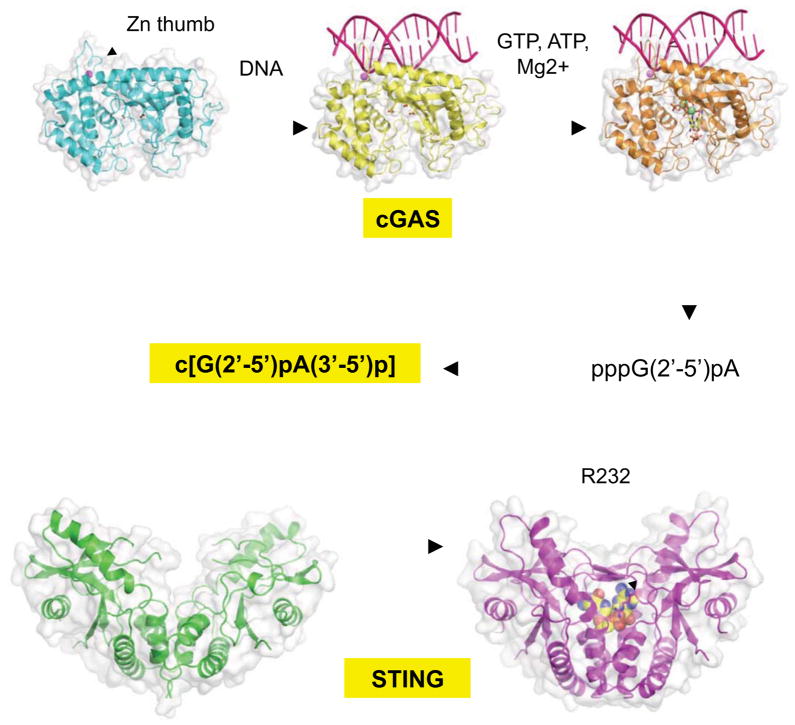
Fig. 1. The cGAS-STING pathway for DNA sensing.
cGAS binds dsDNA using a “zinc thumb” structure and a positively charged surface. DNA-association induces a structural switch that repositions the catalytic residues to bind the GTP and ATP substrates mediated by magnesium ions. The catalysis proceeds from a 2′-5′ GpA linkage to a 3′-5 ApG linkage and forms a cyclic di-nucleotide 2′3′-cGAMP, which then binds and initiates large structural rearrangements in STING. The resulting STING-cGAMP complex may then promote TBK1 and IRF3 activation through its CTT. The zinc and magnesium ions that bind cGAS are colored cyan and green, respectively. The 2′3′-cGAMP is shown as spheres bound to STING.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013;498:380–384. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atianand MK, Fitzgerald KA. Molecular basis of DNA recognition in the immune system. Journal of immunology. 2013;190:1911–1918. doi: 10.4049/jimmunol.1203162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baglioni C. Interferon-induced enzymatic activities and their role in the antriviral state. Cell. 1979;17:255–264. doi: 10.1016/0092-8674(79)90151-x. [DOI] [PubMed] [Google Scholar]
- 4.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavlar T, Deimling T, Ablasser A, Hopfner KP, Hornung V. Species-specific detection of the antiviral small-molecule compound CMA by STING. The EMBO journal. 2013;32:1440–1450. doi: 10.1038/emboj.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Civril F, Deimling T, de Oliveira Mann CC, Ablasser A, Moldt M, Witte G, Hornung V, Hopfner KP. Structural mechanism of cytosolic DNA sensing by cGAS. Nature. 2013;498:332–337. doi: 10.1038/nature12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies BW, Bogard RW, Young TS, Mekalanos JJ. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell. 2012;149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell reports. 2013;3:1355–1361. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, Ascano M, Wu Y, Barchet W, Gaffney BL, Zillinger T, Serganov AA, Liu Y, Jones RA, Hartmann G, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013;153:1094–1107. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang YH, Liu XY, Du XX, Jiang ZF, Su XD. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19:728–730. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 11.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin L, Waterman PM, Jonscher KR, Short CM, Reisdorph NA, Cambier JC. MPYS, a novel membrane tetraspanner, is associated with major histocompatibility complex class II and mediates transduction of apoptotic signals. Mol Cell Biol. 2008;28:5014–5026. doi: 10.1128/MCB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerr IM, Brown RE. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of Human cGAS Reveals a Conserved Family of Second-Messenger Enzymes in Innate Immunity. Cell reports. 2013;3:1362–1368. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mills E, Pultz IS, Kulasekara HD, Miller SI. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cellular microbiology. 2011;13:1122–1129. doi: 10.1111/j.1462-5822.2011.01619.x. [DOI] [PubMed] [Google Scholar]
- 16.Ouyang S, Song X, Wang Y, Ru H, Shaw N, Jiang Y, Niu F, Zhu Y, Qiu W, Parvatiyar K, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36:1073–1086. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paludan SR, Bowie AG. Immune sensing of DNA. Immunity. 2013;38:870–880. doi: 10.1016/j.immuni.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parvatiyar K, Zhang Z, Teles RM, Ouyang S, Jiang Y, Iyer SS, Zaver SA, Schenk M, Zeng S, Zhong W, et al. The helicase DDX41 recognizes the bacterial secondary messengers cyclic di-GMP and cyclic di-AMP to activate a type I interferon immune response. Nature immunology. 2012;13:1155–1161. doi: 10.1038/ni.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shang G, Zhu D, Li N, Zhang J, Zhu C, Lu D, Liu C, Yu Q, Zhao Y, Xu S, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19:725–727. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 20.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19:722–724. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Unterholzner L, Bowie AG. Innate DNA sensing moves to the nucleus. Cell host & microbe. 2011;9:351–353. doi: 10.1016/j.chom.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Wu J, Sun L, Chen X, Du F, Shi H, Chen C, Chen ZJ. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, Wu H. Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING. Mol Cell. 2012;46:735–745. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu YJ. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nature immunology. 2011;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhong B, Yang Y, Li S, Wang YY, Li Y, Diao F, Lei C, He X, Zhang L, Tien P, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]