Abstract
The objective of this study is to evaluate a natural language processing (NLP) algorithm that determines American College of Radiology Breast Imaging Reporting and Data System (BI-RADS) final assessment categories from radiology reports. This HIPAA-compliant study was granted institutional review board approval with waiver of informed consent. This cross-sectional study involved 1,165 breast imaging reports in the electronic medical record (EMR) from a tertiary care academic breast imaging center from 2009. Reports included screening mammography, diagnostic mammography, breast ultrasound, combined diagnostic mammography and breast ultrasound, and breast magnetic resonance imaging studies. Over 220 reports were included from each study type. The recall (sensitivity) and precision (positive predictive value) of a NLP algorithm to collect BI-RADS final assessment categories stated in the report final text was evaluated against a manual human review standard reference. For all breast imaging reports, the NLP algorithm demonstrated a recall of 100.0 % (95 % confidence interval (CI), 99.7, 100.0 %) and a precision of 96.6 % (95 % CI, 95.4, 97.5 %) for correct identification of BI-RADS final assessment categories. The NLP algorithm demonstrated high recall and precision for extraction of BI-RADS final assessment categories from the free text of breast imaging reports. NLP may provide an accurate, scalable data extraction mechanism from reports within EMRs to create databases to track breast imaging performance measures and facilitate optimal breast cancer population management strategies.
Keywords: Breast Imaging Reporting and Data System (BI-RADS), Natural language processing, Imaging informatics, Breast
Introduction
Optimal population management strategies and related research activities for breast cancer require local, regional, and national data repositories of at-risk populations [1]. A key element of such repositories is malignancy risk based upon breast imaging findings, indicated by the American College of Radiology (ACR) Breast Imaging Reporting and Data System (BI-RADS) final assessment category [2]. Large-scale, automated extraction of malignancy risk from radiology reports within the electronic medical record (EMR) may be possible because a standardized lexicon to codify findings in breast imaging studies exists [2]. Use of this lexicon to communicate findings has been broadly adopted because the Mammography Quality Standards Act (MQSA) requires a BI-RADS final assessment category be reported for each mammogram [3].
The MQSA and the ACR Breast Magnetic Resonance Imaging (MRI) Accreditation Program require a medical outcomes audit for quality assurance [3, 4]. The audit determines established quality metrics, such as recall rate and positive predictive value for cancer detection by breast imaging [2, 5]. These metrics are determined using the BI-RADS final assessment categories, which define a study as positive or negative. Studies-assigned BI-RADS assessment categories 0 (need additional imaging evaluation and/or prior imaging for comparison), 4 (suspicious abnormality—biopsy should be considered), and 5 (highly suggestive of malignancy—appropriate action should be taken) are considered positive. Studies-assigned BI-RADS assessment categories 1 (negative), 2 (benign finding), and 3 (probably benign finding) are considered negative based upon imaging. Audits are facilitated by continuously gathering and storing these data in a computer database [5].
BI-RADS categories can be collected at the time of reporting, using structured reporting applications. Alternatively, this information can be extracted from the text of final breast imaging reports manually or in an automated fashion using natural language processing (NLP) algorithms. NLP algorithms can extract meaningful information from free text and have been successfully applied to radiology reports to identify positive findings, recommendations, and tumor status [6–9]. Specific to breast imaging, NLP has been applied to mammography reports to identify findings suspicious for breast cancer [10], correlate findings and their locations [11], determine BI-RADS breast tissue composition [12], and extract multiple other reported attributes [13]. We hypothesized that NLP can accurately extract BI-RADS final assessment categories from radiology reports.
Materials and Methods
Setting
This study was approved by our institutional review board with waiver of informed consent and conducted in compliance with Health Insurance Portability and Accountability Act guidelines. The study setting is a tertiary care academic breast imaging center performing 37,695 screening and diagnostic breast imaging studies in 2009 (25,208 screening mammograms, 6,518 diagnostic mammograms, 4,893 breast ultrasound examinations, and 1,076 breast MRI examinations).
Study Population
The study population was comprised of finalized breast imaging reports within the EMR. Our administrative database from the Radiology Information System (IDXrad v.9.94, GE Healthcare, Burlington, VT) was used to obtain reports. Two hundred fifty breast imaging reports from the time period January 1, 2009 through June 30, 2009 were randomly selected for each type of conventional breast imaging study (screening mammography, diagnostic mammography, breast ultrasound, and combined diagnostic mammography and breast ultrasound performed together). In addition, 250 contrast-enhanced breast MRI reports were randomly selected from July 1, 2009 through December 31, 2009. The conventional breast imaging studies were reported using the Mammography Administration Module (IDXrad, Version 9.94, GE Healthcare, Burlington, VT). This is a breast imaging structured reporting application for creating mammographic and breast ultrasound reports. Negative screening mammogram reports are entered by a transcriptionist, with all other reports entered by a radiologist. The breast MRI studies were reported using speech recognition software (PowerScribe Workstation, Version 4.7, Nuance Communications, Burlington, MA). For all breast imaging reports, the institutional standard practice is to assign a BI-RADS final assessment category to each imaged breast.
Reports were excluded if they contained addenda or if their examination type was misidentified by selection criteria. Reports with addenda were excluded because addenda could modify final assessments but frequently did not include a clear restatement of BI-RADS categories. Report selection criteria did not always yield the desired examination type. For example, reports of breast imaging-guided procedures occasionally were misidentified as screening or diagnostic breast imaging examinations. BI-RADS final assessment categories are typically not assigned to procedures, so these reports were excluded. The study included a minimum of 200 reports per study type to provide 95 % binomial confidence intervals no wider than 5 % at outcomes greater than 85 %. The reports were used to evaluate the recall and precision of data extraction by the NLP algorithm.
Software Tool
BI-RADS Observation Kit (BROK) is an open source, public domain, Java-based information extraction program developed in-house [14]. BROK performs NLP to determine BI-RADS final assessment categories for each imaged breast from text reports. It determines the BI-RADS category and which breast(s) was imaged through regular expression string matches guided by pre-specified report headers and phrases. BROK specifically extracts one BI-RADS final assessment category, stated as a number, for each breast. BROK will also extract a BI-RADS final assessment subcategory in the form of a letter (such as 4a, 4b, or 4c), if a subcategory letter is reported. The NLP algorithm processes breast imaging reports by removing numbers that might be mistaken for BI-RADS final assessment categories. Examples of numbers that are removed are measurements (such as 3 × 4 × 5 cm), locations (such as 3 o'clock), magnetic field strength (such as 3 T), and time intervals (such as 6 months). The NLP algorithm also segments each report into a “body” and “impression.” BROK only extracts data from the report impression, unless report segmentation fails or there is no text within the impression.
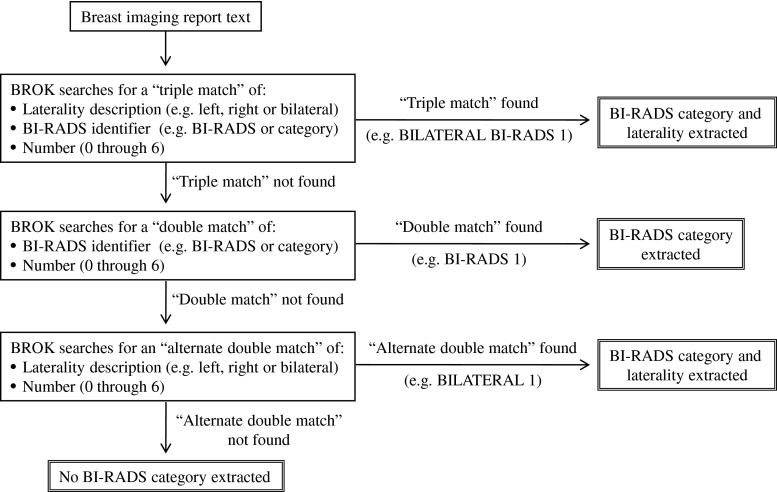
The NLP algorithm searches the report for information about the BI-RADS final assessment category and which breast(s) was assigned that category. If necessary, BROK performs this search through three iterations, as outlined in Fig. 1. BROK also checks the report impression to try to determine if the examination imaged one or both breasts. Once the NLP algorithm has extracted information, it then applies logic to these data so that for each potential breast imaged, BROK outputs: a BI-RADS category number for each breast, “multiple BI-RADS without laterality found,” “no BI-RADS category found,” or “not imaged.” The NLP algorithm also searches for a report addendum. If one is identified, BROK outputs: “addendum” to indicate that the report requires manual human review. BROK was developed using a randomly selected training set of 550 breast MRI reports and 250 breast ultrasound reports taken from the time period January 1, 2009 through June 20, 2009.
Fig. 1.
Iterative information extraction search process for the NLP algorithm
Establishment of a Standard Reference
The performance of the NLP algorithm in creating a database of BI-RADS final assessment categories was compared to a standard reference determined by prospective manual human review of the reports to determine the true BI-RADS categories stated in the final report text. The BI-RADS category could be stated as a number (such as “2”) and/or a standard lexicon phrase (such as “benign”). If there was a discrepancy in the report between the stated number and lexicon phrase, the stated number was taken as the true BI-RADS category.
The report review was performed by a board-certified diagnostic radiologist in a combined breast imaging and imaging informatics fellowship with 6 years of postgraduate experience. The performance of the report review was evaluated by a second physician postdoctoral imaging informatics fellow with 2 years of postgraduate experience. The second physician reviewed a random sample of 60 reports, 12 from each of the five study types, and determined the BI-RADS category for each breast. Agreement between the two reviewers was defined as agreement in BI-RADS categories for both breasts given in each report. There was a high level of agreement, with a ĸ value of 0.988. In the single discordant evaluation, two BI-RADS categories were assigned to one breast. One reviewer did not recognize that two BI-RADS were being reported for that breast. Upon discussion, agreement was reached.
Statistical Analysis
Recall and precision, with 95 % confidence intervals, was determined for the information collected by the NLP algorithm, compared against the standard reference. These metrics were determined for each individual study type and, overall, for all breast imaging studies. Recall, also referred to as sensitivity, measures the proportion of reports from which all the desired information was extracted. Precision, also referred to as positive predictive value, measures the proportion of reports with extracted data that is correct. Successful information extraction required the NLP algorithm to correctly identify which breast(s) was imaged, whether a BI-RADS category was reported for each breast imaged, and the correct BI-RADS category, when it was reported. If the software failed to determine this information for one or both breasts, it was considered a data extraction error for the entire examination. Statistical analysis was performed using SAS version 9.2 (SAS Institute Inc, Cary, NC).
Error analysis was performed to identify the cause of data extraction errors. For the NLP algorithm, reports with outputs of “multiple BI-RADS without laterality found” or “no BI-RADS category found,” for either breast were considered flagged as containing ambiguous information. We determined the percentage of reports with NLP data extraction errors which were flagged by the NLP algorithm. For example, when the NLP algorithm output an incorrect BI-RADS category, rather than identifying the report as containing ambiguous information, this was considered an error which had not been flagged.
Results
Reports
A total number of 1,165 reports were included in the study, consisting of 248 screening mammograms, 227 diagnostic mammograms, 222 breast ultrasound examinations, 234 combined diagnostic mammogram and ultrasound examinations, and 234 breast MRI examinations. Of the 85 reports excluded, 47 contained addendums and 38 had their examination type misidentified by selection criteria. For the 931 conventional breast imaging reports, all but four (all of which were breast ultrasound examinations) contained at least one BI-RADS final assessment category. For the 234 breast MRI reports, all but three contained at least one BI-RADS final assessment category. The conventional breast imaging reports were entered by 23 attending radiologists working with 19 trainees and 3 transcriptionists. The breast MRI examinations were dictated by 13 attending radiologists working with 17 trainees.
NLP Algorithm Performance
For all breast imaging reports, the NLP algorithm demonstrated a recall of 100.0 % (95 % confidence interval (CI), 99.7, 100.0 %) and a precision of 96.6 % (95 % CI, 95.4, 97.5 %) for correct identification of BI-RADS final assessment categories. The NLP algorithm's performance for individual study types is presented in Table 1.
Table 1.
NLP algorithm's performance collecting BI-RADS final assessment categories
| Study type | No. | Recall (95 % confidence interval) | Precision (95 % confidence interval) |
|---|---|---|---|
| Screening mammography | 248 | 100.0 % (98.5, 100.0 %) | 100.0 % (98.5, 100.0 %) |
| Diagnostic mammography | 227 | 100.0 % (98.4, 100.0 %) | 99.6 % (97.6, 100.0 %) |
| Breast ultrasound | 222 | 100.0 % (98.1, 100.0 %) | 88.7 % (83.8, 92.6 %) |
| Combined diagnostic mammography and ultrasound | 234 | 100.0 % (98.4, 100.0 %) | 98.3 % (95.7, 99.5 %) |
| Breast MRI | 234 | 100.0 % (98.4, 100.0 %) | 95.7 % (92.3, 97.9 %) |
| All breast imaging | 1,165 | 100.0 % (99.7, 100.0 %) | 96.6 % (95.4, 97.5 %) |
Error Analysis
NLP failed to extract correct information for 3.4 % (39/1,165) of the breast imaging reports. All 39 errors in data extraction by the NLP algorithm resulted from variations in the content and organization of the report impression with respect to breast(s) imaged and assigned BI-RADS final assessment category. For example, if the report impression did not state which breast(s) was imaged, the NLP algorithm detected a BI-RADS final assessment category in the impression but could not determine to which breast(s) this category applied. The breast(s) imaged was stated in the body of the report, which did not contain a BI-RADS final assessment category. The NLP algorithm did not extract the breast(s) imaged from the body of the report, far away from the statement of the BI-RADS category. This was the cause of 13 of the data extraction errors. Similarly, if the breast(s) imaged was stated in the report impression, but not in proximity to the stated BI-RADS category, the NLP algorithm did not identify which breast was imaged (caused two errors).
Data extraction errors occurred when a report impression stated that only one breast was imaged, but then went on to also discuss the contralateral breast or recommend bilateral breast imaging in the future. The NLP algorithm incorrectly identified the study as a bilateral exam and assigned the BI-RADS final assessment category to both breasts, rather than the single imaged breast (caused eight errors). Errors also occurred with reports where the BI-RADS final assessment category was only stated in words. The NLP algorithm identifies the BI-RADS category as a number and, therefore, failed to extract a BI-RADS category from these reports (caused five errors).
Data extraction errors occurred with reports with two BI-RADS categories assigned to one breast. The NLP algorithm only detected one of these two categories because it is programmed to only detect a single BI-RADS category per breast (caused four errors). An error occurred when the word “category” was misspelled, so that the NLP algorithm did not identify the BI-RADS category (caused one error). When only one breast was imaged and no BI-RADS category was assigned to that breast, the NLP algorithm failed to identify that only one breast was imaged. The algorithm output that both breasts were imaged, but no BI-RADS category, was assigned to either breast (caused six errors).
Flagging Reports with Errors in Data Collection
Of the 39 breast imaging reports with errors in the extracted BI-RADS category, the NLP algorithm flagged 89.7 % (95 % CI, 75.8, 97.1 %) as containing ambiguous information, requiring manual human review. The NLP algorithm's performance flagging errors for individual study types is presented in Table 2.
Table 2.
Proportion of data collection errors flagged by the NLP algorithm
| Study type | NLP % (95 % confidence interval), proportion |
|---|---|
| Screening mammography | Not applicable, 0/0 |
| Diagnostic mammography | 100.0 % (2.5, 100.0 %), 1/1 |
| Breast ultrasound | 96.0 % (79.7, 99.9 %), 24/25 |
| Combined diagnostic mammography and ultrasound | 100.0 % (39.8, 100.0 %), 4/4 |
| Breast MRI | 66.7 % (29.9, 92.5 %), 6/9 |
| All breast imaging | 89.7 % (75.8, 97.1 %), 35/39 |
Discussion
Our study demonstrates high recall and precision for extracting BI-RADS final assessment categories from the free text of breast imaging reports using an open source, public domain NLP tool. NLP may thus provide an accurate automated mechanism to create large-scale data repositories of malignancy risk based on breast imaging findings documented in EMRs. This would enable optimal population management strategies for breast cancer, including quality assurance medical outcomes audits required by the MQSA and the ACR Breast MRI Accreditation Program [3, 4]. The literature suggests that health information technology (HIT) can improve the efficiency and quality of medical care [15]. NLP is an informatics tool that can help to fulfill this promise of HIT. More specifically, NLP may improve and accelerate data collection on a national level with the ACR's National Mammography Database and the National Cancer Institute's Population-based Research Optimizing Screening through Personalized Regimens program [16, 17].
To our knowledge, we are the first to describe a NLP algorithm that extracts BI-RADS final assessment categories from the text of breast imaging reports. A recent publication has described an algorithm that classifies mammography reports by BI-RADS breasts tissue composition, with a very high accuracy of greater than 99.0 % [12]. Overall, this NLP breast composition extractor demonstrated slightly superior performance to our NLP BI-RADS category extractor. This may, in part, be due to the fact that the NLP breast composition extractor determines a single breast composition for each radiology report. Our NLP BI-RADS category extractor identifies a category for each breast, determining both the reported BI-RADS category and the applicable breast(s). This adds a level of complexity to the information extraction process.
A system has also been described in the literature which extracts from breast imaging radiology reports imaging findings and their locations and correlates them [11]. This system demonstrated a recall of 35.7 % and a precision of 91.4 % for breast imaging reports, lower than that of our NLP BI-RADS category extractor. The system for extracting and correlating clinical findings and their locations can localize an imaging finding to a region of the breast, a greater level of detail than our NLP BI-RADS category extractor, which simply determines the breast(s) to which a BI-RADS category is assigned. As a NLP algorithm extracts more granular information, it is likely that achieving high recall and precision becomes more challenging.
In our study, use of the BI-RADS nomenclature fostered NLP's strong performance because the program could specifically search for standardized terminology that was consistently reported. This demonstrates the utility of combining use of an established lexicon with NLP. There is a trend in radiology towards the use of a standardized lexicon for all radiology information resources with the development of RadLex by the Radiological Society of North America [18]. As standardized lexicons are adopted by all radiology disciplines, there will be greater opportunities for NLP to be applied to radiology reports to collect data with a high degree of accuracy. In our study, the errors in data collection by NLP all resulted from nonstandard phrases and formatting. Differences in the performance of NLP between study types (e.g., screening mammography versus breast ultrasound) also resulted from greater variability in report format for certain study types, such as breast ultrasound. If all breast imaging reports were formatted in a more standard manner, information extraction accuracy would be expected to further improve.
Our study was limited by the fact that it was conducted at a single institution to assess the performance of a single NLP algorithm. It is unclear if BROK is representative of other NLP programs. This study focused on the use of a NLP algorithm to determine BI-RADS final assessment category, a well-defined piece of information unique to breast imaging. The accuracy we observed with NLP may not be generalizable, particularly to clinical data created without the use of a standardized lexicon. The NLP we tested is performed retrospectively, not as the examination is reported. NLP will fail to collect data when the information is not clearly reported. This was not a major issue with breast imaging reports because BI-RADS final assessment categories are routine standardized entries. For a variety of radiology subspecialties, NLP would likely be more effective if it was applied at the time of dictation, with the radiologist prompted when key data elements are not found in the report.
Conclusion
NLP may provide an accurate automated approach for large-scale extraction of BI-RADS final assessment categories from the text of radiology reports for a variety of efforts including quality improvement, population management, and related research repositories for breast cancer. Natural language processing has also been applied in breast cancer research to extract information from pathology reports, discover drug treatment patterns, and evaluate health care quality [19–21]. Such applications of NLP will facilitate the creation of breast cancer data repositories that are integrated across multiple disciplines.
Acknowledgments
We would like to thank Drs. E. Francis Cook and E. John Orav for providing guidance for the statistical analysis.
The majority of the contributions of I Ikuta occurred while he was a fellow supported by the National Institutes of Health grant T15LM007092. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
References
- 1.Ballard-Barbash R, Taplin SH, Yankaskas BC, Ernster VL, Rosenberg RD, Carney PA, Barlow WE, Geller BM, Kerlikowske K, Edwards BK, Lynch CF, Urban N, Chrvala CA, Key CR, Poplack SP, Worden JK, Kessler LG. Breast Cancer Surveillance Consortium: a national mammography screening and outcomes database. AJR Am J Roentgenol. 1997;169(4):1001–1008. doi: 10.2214/ajr.169.4.9308451. [DOI] [PubMed] [Google Scholar]
- 2.Breast Imaging Reporting and Data System® (BI-RADS®) 4. Reston: American College of Radiology; 2003. [Google Scholar]
- 3.Mammography Quality Standard Act, 62 Federal Register 559688, 1997
- 4.American College of Radiology Breast Magnetic Resonance Imaging (MRI) Accreditation Program Requirements. Available at http://www.acr.org/~/media/ACR/Documents/Accreditation/BreastMRI/Requirements.pdf. Accessed 10 July 2012
- 5.Sickles EA. Auditing your breast imaging practice: an evidence-based approach. Semin Roentgenol. 2007;42(4):211–217. doi: 10.1053/j.ro.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002;224(1):157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 7.Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, Halpern EF, Thrall JH. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005;234(2):323–329. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 8.Ip IK, Mortele KJ, Prevedello LM, Khorasani R. Repeat abdominal imaging examinations in a tertiary care hospital. Am J Med. 2012;125(2):155–161. doi: 10.1016/j.amjmed.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng LT, Zheng J, Savova GK, Erickson BJ. Discerning tumor status from unstructured MRI reports—completeness of information in existing reports and utility of automated natural language processing. J Digit Imaging. 2010;23(2):119–132. doi: 10.1007/s10278-009-9215-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain NL, Friedman C: Identification of findings suspicious for breast cancer based on natural language processing of mammogram reports. Proc AMIA Annu Fall Symp(829–833), 1997 [PMC free article] [PubMed]
- 11.Sevenster M, van Ommering R, Qian Y. Automatically correlating clinical findings and body locations in radiology reports using MedLEE. J Digit Imaging. 2012;25(2):240–249. doi: 10.1007/s10278-011-9411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Percha B, Nassif H, Lipson J, Burnside E, Rubin D. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc. 2012;19(5):913–916. doi: 10.1136/amiajnl-2011-000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mykowiecka A, Marciniak M, Kupść A. Rule-based information extraction from patients' clinical data. J Biomed Inform. 2009;42(5):923–936. doi: 10.1016/j.jbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 14.How BROK Works. Brigham and Women's Hospital Web site. Available at http://www.brighamandwomens.org/Research/labs/cebi/BROK/default.aspx. Accessed 13 May 2013
- 15.Chaudhry B, Wang J, Wu S, Maglione M, Mojica W, Roth E, Morton SC, Shekelle PG. Systematic review: impact of health information technology on quality, efficiency, and costs of medical care. Ann Intern Med. 2006;144(10):742–752. doi: 10.7326/0003-4819-144-10-200605160-00125. [DOI] [PubMed] [Google Scholar]
- 16.National Mammography Database (NMD). Available at https://nrdr.acr.org/Portal/NMD/Main/page.aspx. Accessed 10 June 2012
- 17.Population-based Research Optimizing Screening through Personalized Regimens (PROSPR). Available at http://appliedresearch.cancer.gov/networks/prospr/. Accessed 10 June 2012
- 18.RadLex. Available at http://www.rsna.org/radlex/. Accessed 10 June 2012
- 19.Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports. Stud Health Technol Inform. 2004;107(Pt 1):565–572. [PubMed] [Google Scholar]
- 20.Savova GK, Olson JE, Murphy SP, Cafourek VL, Couch FJ, Goetz MP, Ingle JN, Suman VJ, Chute CG, Weinshilboum RM. Automated discovery of drug treatment patterns for endocrine therapy of breast cancer within an electronic medical record. J Am Med Inform Assoc 19(e1): e83–e89. doi:10.1136/amiajnl-2011-000295 [DOI] [PMC free article] [PubMed]
- 21.Baldwin KB. Evaluating healthcare quality using natural language processing. J Healthc Qual. 2008;30(4):24–29. doi: 10.1111/j.1945-1474.2008.tb01151.x. [DOI] [PubMed] [Google Scholar]