Abstract
Introduce the notion of cross-sectional relatedness as an informational dependence relation between sentences in the conclusion section of a breast radiology report and sentences in the findings section of the same report. Assess inter-rater agreement of breast radiologists. Develop and evaluate a support vector machine (SVM) classifier for automatically detecting cross-sectional relatedness. A standard reference is manually created from 444 breast radiology reports by the first author. A subset of 37 reports is annotated by five breast radiologists. Inter-rater agreement is computed among their annotations and standard reference. Thirteen numerical features are developed to characterize pairs of sentences; the optimal feature set is sought through forward selection. Inter-rater agreement is F-measure 0.623. SVM classifier has F-measure of 0.699 in the 12-fold cross-validation protocol against standard reference. Report length does not correlate with the classifier’s performance (correlation coefficient = −0.073). SVM classifier has average F-measure of 0.505 against annotations by breast radiologists. Mediocre inter-rater agreement is possibly caused by: (1) definition is insufficiently actionable, (2) fine-grained nature of cross-sectional relatedness on sentence level, instead of, for instance, on paragraph level, and (3) higher-than-average complexity of 37-report sample. SVM classifier performs better against standard reference than against breast radiologists’s annotations. This is supportive of (3). SVM’s performance on standard reference is satisfactory. Since optimal feature set is not breast specific, results may transfer to non-breast anatomies. Applications include a smart report viewing environment and data mining.
Electronic supplementary material
The online version of this article (doi:10.1007/s10278-013-9612-9) contains supplementary material, which is available to authorized users.
Keywords: Radiology reports, Information retrieval, Support vector machine, Text mining, Inter-rater agreement, Textual entailment
Introduction
In breast radiology, the Breast Imaging Reporting and Data System (BI-RADS) [1] mandates that a report encompasses a “clear description of significant finding(s),” and, in a separate section, “final impressions […] based on thorough evaluations of mammographic features of concern.” This organization of information is optimized for the clinical workflow: the findings section state image features that are comprehensible to those skilled in the art of interpreting breast images; the impression or conclusion section answers the clinical question of referring clinicians.
Thus, in BI-RADS-compliant radiology reports, or any report with a similar sectional structure for that matter, there is an informational relation between sentences in the findings section and the sentences in the conclusion section of a report. Sentences can be related in multiple ways. For instance, a conclusion sentence may summarize a finding sentence or correlate it with information from another radiological exam.
In this paper, we introduce the notion of cross-sectional relatedness to the scientific literature: a sentence from the findings section of a radiology report is related to a sentence from the conclusion section of the same report, if the former conveys the same information as or directly elaborates on any of the main phrases or assertions of the latter. Table 1 lists a series of related and nonrelated finding and conclusion sentence pairs.
Table 1.
Pairs of finding and conclusion sentences and their relatedness
| F conveys the same information as C | |
| F | No abnormal findings in right breast |
| C | There are no abnormal findings in right breast |
| F elaborates on the multifocal carcinoma mentioned in C | |
| F | The third lesion may represent a multifocal cancer or metastatic lymph node |
| C | Probably multifocal carcinoma in left breast |
| F elaborates on seroma cavity at site of previous surgery | |
| F | Within the left upper outer quadrant, a 3 × 1.2-cm seroma cavity is present at the previous surgical site |
| C | Breast MRI demonstrates irregular thickened enhancement surrounding a seroma cavity at the site of prior surgery |
| F1 elaborates on abnormality that leads to the diagnosis in C and F2 elaborates on the metastatic lymphadenopathy of C. F3 elaborates on the malignant character mentioned in C | |
| F1 | There is a briskly enhancing nodular lesion in the left breast, measuring 10 × 9 mm |
| F2 | It may represent a multifocal cancer or metastatic lymph node |
| F3 | Kinetic curve of this lesion demonstrated wash-out pattern suggesting malignancy |
| C | Possible metastatic lymphadenopathy |
| F1 and F2 elaborate on assertions in C. F1 discusses a mammogram finding, whereas F2 discusses an ultrasound finding | |
| F1 | There is associated ill-defined increased density with the calcifications suggesting of an infiltrating component |
| F2 | Ultrasound evaluation of the left breast demonstrates an ill-defined hypoechoic mass at the 2 o’clock position |
| C | Malignant left breast mass as detailed above is most compatible with an infiltrating ductal carcinoma with an intraductal component |
| C is not related to any of the sentences F1–F3 since none of these sentences express the same information as C or directly elaborate on any of the main phrases or assertions of C | |
| F1 | There is a tiny cyst at 3 o’clock position in the right breast, and upper outer aspect of the left breast |
| F2 | With dynamic contrast study, no abnormal enhancement is seen in either breast |
| F3 | No abnormal lymph nodes are seen in either axilla |
| C | No MR findings to suggest malignancy |
F, F1, F2, and F3 finding sentences, C conclusion sentence
Understanding the cross-sectional relatedness in a report is key to understanding its information flow and the reasoning pattern of the radiologist who dictated it. This holds for human consumers of reports, such as referring clinicians and radiologists, as well as automated agents that analyze the reports and expose it to downstream consumers.
Such automated agents serve to realize a variety of use cases. For instance, in the radiology workflow, a smart report viewing environment can be devised in which users navigate from sentences in the conclusion section to pertinent sentences or paragraphs in the findings section of the report. This use case can be taken one step further by shuffling the contents of an initial report by grouping together related information from findings and conclusion sections [2]. The technology can also be used for quality control, for instance, for verifying if all critical findings are summarized in the conclusions section of a report [3]. Other applications include automated discovery of relations between image characteristics from the findings section and diagnoses from the conclusion sections by means of data mining techniques.
We define automatic detection of cross-sectional relatedness as a classification problem: given a sentence from a findings section and a sentence from the conclusion section of the same report, are they related? We address this problem with natural language processing methods. General-purpose natural language processing engines [4] have been developed that index medical reports according to linguistic and/or information-theoretic axes [5–8]. The output of these engines has been utilized to solve specific information extraction and classification problems [9–12]. The problem of detecting cross-sectional relatedness in radiology reports has not been addressed in the literature. The topic of detecting entailment between sentences is a topic of continuing interest in the natural language processing community that culminates in the recognizing textual entailment (RTE) challenge series. We relate our work to this effort in the “Discussion” where we also discuss future research directions.
The aims of this paper are as follows:
Standard reference creation and evaluation—develop a standard reference for cross-sectional relatedness in a corpus of breast radiology reports. As this notion is newly introduced to the literature, we also obtain the cross-sectional relatedness annotations of five breast radiologists. We compute inter-rater agreement among the breast radiologists and the standard reference. This analysis serves to analyze the extent to which cross-sectional relatedness is an objective notion, and the quality of the standard reference.
Feature selection and cross-validation—develop a support vector machine (SVM) classifier that automatically detects relatedness between sentences across sections in breast radiology reports. SVMs [13, 14] generally achieve best-in-class results on natural language processing tasks. The domain will be characterized by 13 numerical features based on vector space models and language models [15], which are filtered in a nested cross-validation feature selection protocol with respect to the standard reference.
Comparison against breast radiologists’ annotations—evaluate the SVM classifier against the annotations of the breast radiologists and the standard reference. Depending on the use case, revealing cross-sectional relatedness is potentially most useful in wieldy reports. For this reason, we track the classifier’s performance against report length.
Materials and Methods
Reports
A corpus of 444 de-identified, breast radiology reports was obtained from a radiology department of a US-based university hospital. The reports were automatically split in sections, paragraphs, and sentences by an enhanced sentence boundary detection classifier (C#-port of the OpenNLP library) optimized on our corpus. The section headers were mapped to a fixed set of section types including Clinical history, Findings, and Conclusions. Henceforth, the term finding sentence refers to a sentence in the Clinical history or Findings section; a conclusion sentence refers to a sentence in the Conclusions section.
If s is a finding sentence and s′ a conclusion sentence from the same report, then the pair (s and s′) is an instance. A report with x finding sentences and y conclusion sentences gives rise to x × y instances. An annotation of a report is a subset of the report’s instances. We say that the instances in the annotation of a report are selected. The annotation of a set of reports is simply the union of the annotations of the corpus’ reports.
Breast Radiologists’ Annotation Creation
The corpus of 444 reports was divided into three samples, called S1, S2, and S3 (see Table 2). Care was taken that the second sample, S2, accurately represented the reports of nonsimple cases, so as to avoid that the sample would consist mostly of short and highly similar reports. Three of the reports in S2 had BI-RADS class 0 (“Incomplete; additional imaging required”), seven had BI-RADS class 1 to 3 (benign to probably benign), and ten had BI-RADS class 4 or 5 (probably or most likely malignant); the remaining reports had no BI-RADS classification, which was an artifact of our report acquisition process. Six reports were interpretations of outside exams. Eight reports discussed a combined mammogram–ultrasound exam, ten discussed one or more mammogram exams, and three discussed one or more ultrasound exams; the remaining 16 discussed an MRI exam. Sample S2 was annotated by five breast radiologists.
Table 2.
Details of the three sample sets and the annotations based on them
| Sample | Number of reports | Details |
|---|---|---|
| S1 | 200 | Annotated by first author |
| S2 | 37 | Annotated by five breast radiologists and first author |
| S3 | 207 | Pre-annotated by preliminary SVM classifier, then corrected by first author |
Reports in the corpus contained 9.24 (±5.32) finding sentences and 7.23 (±4.79) conclusion sentences on average. In sample S2, reports had 11.78 (±5.26) finding sentences and 7.19 (±4.79) conclusion sentences on average. Reports in S2 contained significantly more finding sentences than the other reports in the corpus (p < 0.001; Mann–Whitney U test); no significant difference was observed for conclusion sentences (p = 0.671).
A home-grown annotation tool was used for recording related finding–conclusion sentences. In the tool, one conclusion sentence is active at a time (highlighted). The active conclusion sentence can be changed by clicking another conclusion sentence. The annotator indicates that one or more finding sentence s is related to the active conclusion sentence s′ by clicking s, in which case the instance (s, s′) is in the report’s annotation. Conclusion sentences that were selected by the annotator and that were found to be related to at least one finding sentence were highlighted with low intensity, and likewise for finding sentences (see Fig. 1).
Fig. 1.
Annotation tool. In the left pane are (hashed) report identifiers; the middle pane is split in the conclusion section (lower pane, marked “impression”) and the remainder of the report (upper pane)
Annotation guidelines were constructed in an iterative fashion by the first and second author. In each cycle, the current set of annotation criteria were applied to ten fresh reports. Then, annotations were compared and differences discussed. The criteria were updated accordingly. Two iterations yielded the following criteria (italics copied from instructions presented to subjects):
Select by clicking all sentences in the clinical history and/or finding sections that convey the same information as or directly elaborate on any of the main phrases or assertions of the selected impression sentence.
This criterion may not be black and white for all cases. When in doubt, you may want to break the tie by using the following rule as an additional criterion: Select the sentence in question if highlighting it would be clinically useful.
Prior to each session, the participating breast radiologist was trained on the annotation task. This encompassed an introduction to the annotation guidelines and the annotation tool showing two representative reports. The annotations of these reports were discussed with the supervisor of the experiment. On average, annotators used slightly more than an hour to complete the annotation task.
Standard Reference Creation
The standard reference was created by the first author in three phases. In the first phase, reports from sample S1 were annotated in the annotation tool. In this phase, and in this phase only, features were developed and refined by the first author to characterize the instances for the SVM classifier. In the second phase, reports from sample S2 were annotated. Prior to the third phase, an SVM classifier was trained on the annotated reports from S1 and S2 with respect to all features defined. The phased approach ensures that no features were developed during or after exposure to the reports from sample S2, which were annotated earlier by the breast radiologists. In the third phase, reports from sample S3 were pre-annotated by the SVM classifier and then corrected by the first author.
In the standard reference, 8.96 % (1,789/19,972) of the instances in the corpus are selected, and 5.44 % (186/3,419) of the instances in S2 are selected.
Feature Engineering
Each instance (Ι) is described as a tuple 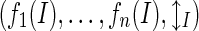 generated from a collection of features (f1, …,fn) and a binary label ℓI. We consider n = 13 features divided over six feature families. In this paper, we only present the six features, spread over four feature families that prove to be optimal. The remaining seven features are described in the first supplemental document.
generated from a collection of features (f1, …,fn) and a binary label ℓI. We consider n = 13 features divided over six feature families. In this paper, we only present the six features, spread over four feature families that prove to be optimal. The remaining seven features are described in the first supplemental document.
We represent a sentence as a bag of its normalized words, excluding stopwords. The normalization step entails removing non-alphanumerical characters and casting alphabetic characters to lower case. We experimented informally with word stems, chunks, bi- and trigrams, as well as extracted concepts from the Systemized Nomenclature of Medicine–Clinical Terms (SNOMED-CT). However, none of these extensions had a positive impact on the performance of the classifier. For the sake of simplicity, we suppressed these extensions.
The features discussed below are rooted in vector space models and language models. We refer the reader to the second supplemental document for a brief introduction to language models. Informally speaking, a language model LC is a probabilistic device that estimates the similarity between a bag of words and a corpus of documents C, such as, for instance, a set of reports. If we have two corpora C and C′, we can combine the language models LC and LC′ into one device LLRC, C′ that estimates the extent to which a given bag of words is more (dis)similar to C than to C′.
Feature Family—Sentence Similarity
We inspect words in the intersection of s and s′ to determine if Ι = (s, s′) should be selected. Each sentence in the report R of s and s′ is considered a separate document. Write dt for the vector of sentence t, where the entry corresponding to word w holds the inverse document frequency of w in the background body defined by R:
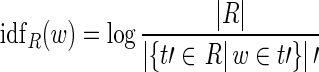 |
if t contains w, and 0 otherwise. In this formula, the nominator denotes the number of sentences in R and the denominator the number of sentences that contain w. No division-by-zero-error can occur, as for every word in R there is sentence containing it.
For two words, idfR(w) > idfR(w ′) indicates that w′ is more frequent in R than w. Inverse document frequency is often combined with a word’s frequency in the document. Being sentences, our documents are very short, which renders frequency a weak parameter.
Let Ι = (s, s′) be an instance with sentences s and s′. Let d and d′ be the vectors that correspond with s and s′, respectively. We compute the similarity between s and s′ by measuring the cosine similarity between their vectors d and d′:
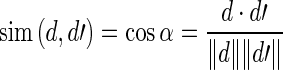 |
where the nominator denotes the dot product of the two vectors and the denominator denotes the product of their Euclidean lengths. Note that α corresponds to the angle between the two document vectors.
Feature Family—Relative Sentence Similarity
If the sentences of Ι = (s, s′) are only weakly similar (that is, sim (s, s′) is low), we may still want to select it, if their similarity is highest among the instances that share a sentence with I. Write Is = {I ′ |I ′ = (t, t ′), t = s} for the set of instances with finding sentence s. Then, we model the relative sentence similarity of Ι with respect to its conclusion sentence as
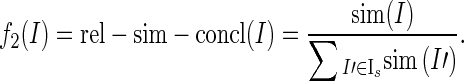 |
Write Js ′ = {I ′ |I ′ = (t, t ′), t ′ = s ′} for the set of instances with conclusion sentence s′. Then, we model the relative sentence similarity of Ι with respect to its finding sentence as
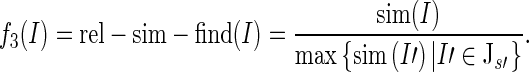 |
Note the asymmetry between rel-sim-concl(Ι) and rel-sim-find(Ι) in terms of their denominators. We experimented with different nominators to model the contextual similarity values; these denominators produced highest scores.
Feature Family—Word Salience
Consider the finding and conclusions sentences from the second group of sentences in Table 1. These sentences have only four words in common: seroma, cavity, at, site, and the. Thus, the sentences are quite dissimilar according to f1. However, the human reader will be alerted by the words seroma and cavity, which are highly salient to him in the sense that if two sentences contain these words, then they are likely to be related. The next feature aims to capture this notion of salience as a numerical value based on language models.
For a given training set of instances O and an annotation A ⊆ O, we obtain the set of potentially salient words by intersecting the sentences in the instances in A:
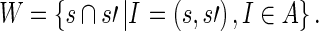 |
We obtain the set of all shared words in the same manner:
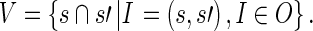 |
The word salience of Ι = (s, s′) in terms of the language models defined by W and V is then given by
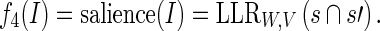 |
Feature Family—A Priori Probability
Some sentences can be excluded from being involved in a selection merely by inspecting their meaning. For instance, the conclusion sentence “findings were discussed with patient and her daughter” is unlikely to have any related finding sentence. On the other hand, the conclusion sentence “new spiculated lesion, with measurement as detailed above” is certainly involved in a selection.
We model the a priori probability that a sentence is involved in a selection. We distinguish between finding and conclusion sentences. Let
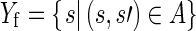 |
be the finding sentences involved in a selection and Nf be the finding sentences in the corpus not contained in Yf.
Then, define:
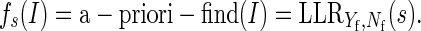 |
We let
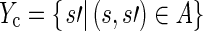 |
be the conclusion sentences involved in a selection. We let Nc be the conclusion sentences in the corpus not contained in Yc. Then, we define the conclusion analogue to the previous feature:
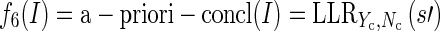 |
Evaluation Metrics
As explained above, the annotation C of a corpus is a set of instances. For two annotations C and C′ of the same corpus, the precision of C with respect to C′ is given by
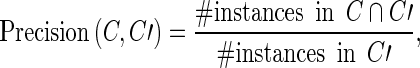 |
whereas the recall of C with respect to C′ is given by
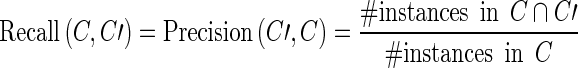 |
The F-measure between C and C′ is the harmonic mean of precision and recall:
 |
The average F-measure of C with respect to annotations D1 …, Dn is the defined as:
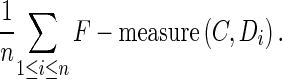 |
The average recall and precision of an annotation is defined with respect to a set of annotations in the same way.
The F-measure between two breast radiologists is the F-measure between their annotations of S2. The F-measure between a breast radiologist and the standard reference is the F-measure between the breast radiologist’s annotation of S2 and the standard reference confined to S2; and similarly for the F-measure between a breast radiologist and the annotation of the SVM classifier of S2.
The average F-measure of a breast radiologist is the average F-measure between his/her annotation of S2 and the annotations of the other breast radiologists and the standard reference confined to S2. The average F-measure of the standard reference is defined similarly with respect to the annotations of S2 of the five breast radiologists. The average F-measure of the SVM classifier is defined as the average F-measure between the annotation of SVM with respect to S2 on the one hand and the standard reference confined to S2 and the annotations of the breast radiologists on the other hand.
We use (Cohen’s) 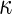 as another measure of inter-rater agreement. F-measure and
as another measure of inter-rater agreement. F-measure and 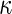 are related in the following way: in domains where the number of true negatives outnumbers the other categories,
are related in the following way: in domains where the number of true negatives outnumbers the other categories, 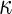 approximates F-measure from below [16].
approximates F-measure from below [16].
Feature Selection and Nested Cross Validation
Overfitting is the phenomenon in pattern recognition that a classifier is biased toward incidental patterns in the training set, which leads to weaker performance on the test set. To avoid overfitting we deploy two strategies.
The first strategy will be to disentangle feature selection from evaluation by means of nested cross validation [17, 18]. In this procedure, we set apart a portion of the standard reference, called the outer fold. Then, we seek the optimal set of features on the remaining reports. Once such a feature set is found an SVM is trained on all reports in the standard reference minus the reports in the outer fold at hand. The resulting SVM is evaluated against the outer fold.
In the feature selection phase, we use the forward selection principle based on feature families, that is, we iteratively include the features from the family that maximally increases F-measure of the SVM classifier in a 12-fold cross validation trial. This cross-validation trial is “nested” within the outer cross-validation process. We stop if none of the remaining feature families improve the performance of the SVM classifier. Conducting feature selection on feature families instead of individual features is our second strategy to combat overfitting.
We experiment with 12 outer folds. In each outer fold, one optimal set of feature families is returned. We label the set of feature families that is returned most frequently as optimal. Then, we run a 12-fold cross-validation protocol on the entire standard reference with respect to the optimal set of feature families. We shall regard the resulting scores as most representative of the SVM’s performance on the standard reference.
Evaluation Protocol
Following the aims of the paper, the evaluation is divided in three steps.
Standard reference creation and evaluation—compute average F-measure of each breast radiologist and average F-measure of the standard reference.
Feature selection and nested cross validation—seek the optimal combination of feature families, following the forward selection principle in a 12-fold nested cross-validation protocol on the standard reference. Evaluate the SVM in an overall 12-fold cross-validation trial. We use 12 folds since sample S2 represents one twelfth of the corpus’ reports and can thus be regarded as one fold in the next step when evaluated against the SVM classifier trained on the annotations of S1 and S3.
Comparison against human annotations—train the SVM classifier on samples S1 and S3 of the standard reference using the optimal combination of feature families. Apply the resulting classifier to S2. Compute the average F-measure of this annotation against the breast radiologists’ annotations and the standard reference.
Results
Annotation Creation and Comparison
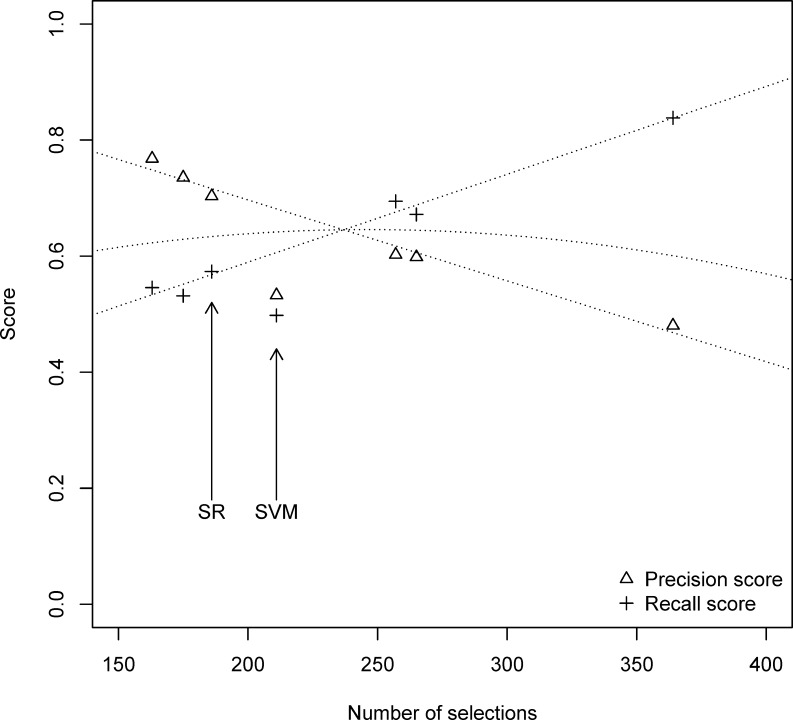
Table 3 compares the breast radiologists’ annotation and the standard reference confined to S2. Overall inter-rater agreement is 0.623 (bottom right cell). The average precision and average recall of the breast radiologists and the standard reference are plotted against the number of selected instances in Fig. 2.
Table 3.
Number of selected instances and evaluation metrics of the breast radiologists and the standard reference on sample S2
| Annotation | Number of selected instances | Average precision | Average recall | Average F-measure | Average 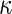
|
|---|---|---|---|---|---|
| BR1 | 163 | 0.768 | 0.546 | 0.628 | 0.606 |
| BR2 | 175 | 0.735 | 0.532 | 0.613 | 0.589 |
| BR3 | 257 | 0.603 | 0.694 | 0.631 | 0.605 |
| BR4 | 364 | 0.480 | 0.838 | 0.605 | 0.574 |
| BR5 | 265 | 0.598 | 0.672 | 0.636 | 0.610 |
| SR | 186 | 0.703 | 0.573 | 0.626 | 0.603 |
| Alla | 235.0 | 0.648 | 0.643 | 0.623 | 0.598 |
aMean scores
Fig. 2.
For each breast radiologist and the standard reference, the number of selections plotted against average precision and average recall. The dotted lines represent the linear regression best fit. The curved dotted lines represent the F-measures associated with the precision and recall linear regressions lines. SR standard reference
Feature Selection and Cross Validation
Table 4 summarizes the feature selection procedure in the nested cross-validation procedure. The set of four feature families, which were detailed above, was selected in nine of the twelve outer folds and is thus considered the (globally) optimal set of feature families. The Structural properties family was selected in three folds; the Domain knowledge family was not selected in any fold. On average, the SVM achieved F-measure 0.702 in the folds in which the globally optimal set of feature families was returned.
Table 4.
For each outer fold, the second column presents the order in which feature families were selected
| Outer fold | Selected feature families | Precision | Recall | F-measure | κ |
|---|---|---|---|---|---|
| 1 | RelS–APr–WSal–Sim–StrP | 0.669 | 0.689 | 0.679 | 0.646 |
| 2 | RelS–APr–WSal–Sim | 0.675 | 0.512 | 0.582 | 0.542 |
| 3 | RelS–APr–WSal–Sim | 0.706 | 0.736 | 0.721 | 0.688 |
| 4 | RelS–APr–WSal–Sim–StrP | 0.645 | 0.630 | 0.639 | 0.609 |
| 5 | RelS–APr–Sim–WSal | 0.843 | 0.705 | 0.768 | 0.744 |
| 6 | RelS–APr–WSal–Sim | 0.634 | 0.712 | 0.671 | 0.636 |
| 7 | RelS–APr–WSal–Sim | 0.776 | 0.735 | 0.755 | 0.732 |
| 8 | RelS–APr–WSal–Sim | 0.722 | 0.698 | 0.710 | 0.679 |
| 9 | RelS–APr–WSal–Sim | 0.702 | 0.677 | 0.689 | 0.663 |
| 10 | RelS–APr–WSal–Sim | 0.757 | 0.680 | 0.717 | 0.692 |
| 11 | RelS–APr–WSal–Sim | 0.700 | 0.712 | 0.706 | 0.672 |
| 12 | RelS–APr–WSal–Sim–StrP | 0.754 | 0.631 | 0.687 | 0.659 |
| {RelS, Apr, WSal, Sim}a | 0.724 | 0.685 | 0.702 | 0.672 | |
| Allb | 0.715 | 0.676 | 0.694 | 0.664 |
For each outer fold, an SVM was trained on all remaining reports with respect to the features found optimal in this fold. The evaluation metrics score the SVM against the reports in the outer fold. RelS relative sentence similarity, APr a priori probability, WSal word salience, Sim sentence similarity, StrP structural properties (see S2)
aPresents the mean of the folds in which the set of feature families was returned that is considered as globally optimal, that is, all folds except 1, 4, and 12
bPresents the mean of all folds
The SVM classifier trained on the optimal set of features has F-measure 0.699 in the overall 12-fold cross-validation trial. Precision is higher than recall (0.711 versus 0.688).
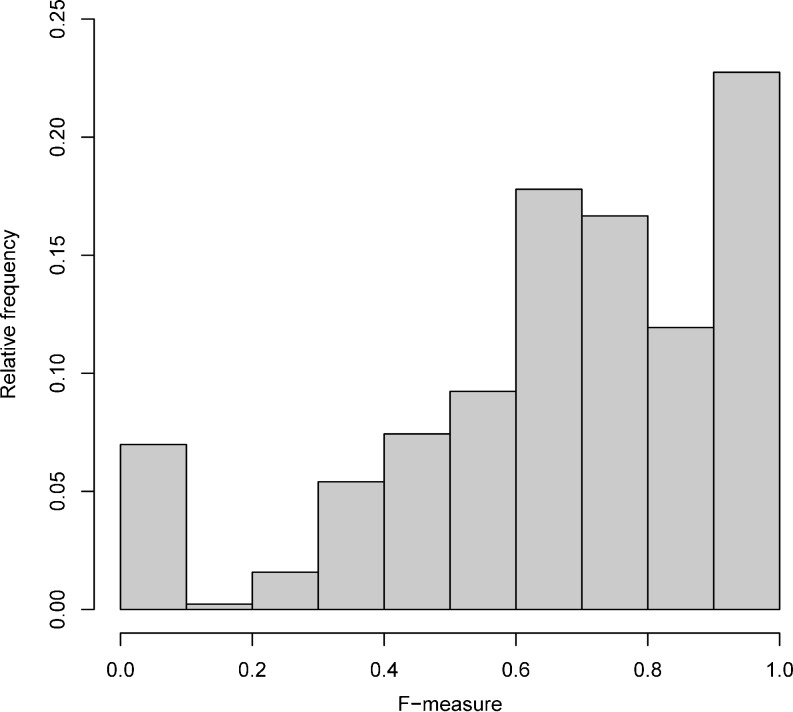
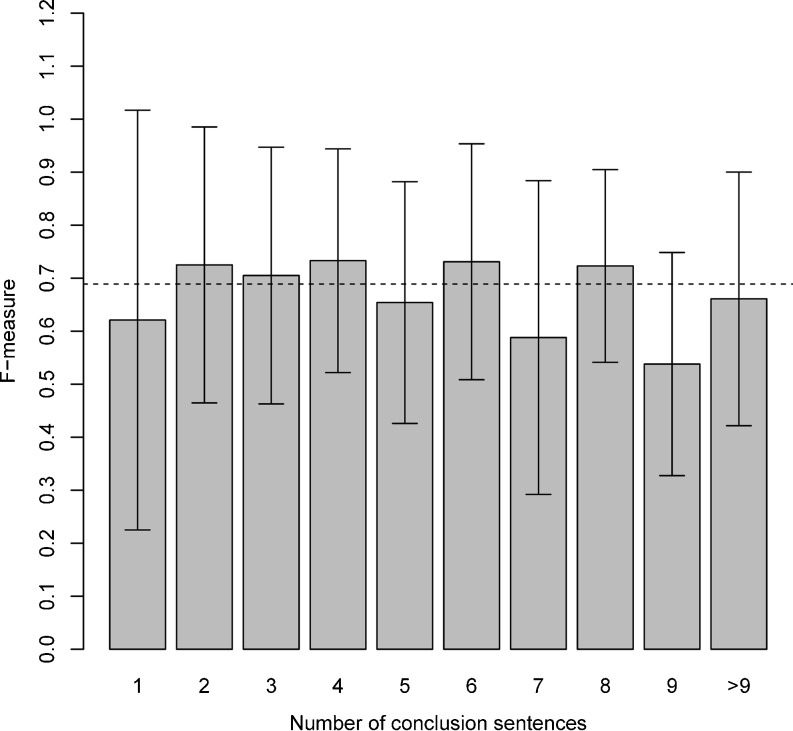
In each fold in the overall trial, we also compute the F-measure between the SVM’s annotation of each individual report and the standard reference’s annotation of that report. Thus we obtain 444 F-measure scores, one for each report. Mean F-measure is 0.698 (±0.268), median is 0.723. Distribution of F-measures is given in Fig. 3. Fig. 4 breaks down the F-measure scores by number of conclusion sentences in the report. The number of instances per report does not correlate with its F-measure (correlation coefficient=−0.073).
Fig. 3.
Distribution of F-measures between standard reference and SVM classifier per report
Fig. 4.
F-measures between standard reference and SVM classifier per report broken down by number of conclusion sentences. Bars represent 1 standard deviation from the mean. Dotted line represents mean F-measure overall
Overall, 6.98 % (31/444) of the reports have F-measure score 0. On these reports, precision and/or recall is 0, meaning that none of the instances selected by the SVM classifier were in the standard reference. Overall, 22.75 % (101/444) of the reports have maximum F-measure of 1, which indicates complete agreement between the SVM classifier and the standard reference.
Comparison Against Human Annotations
Trained on S1 and S3, the SVM makes 211 selections in S2. Table 5 gives the evaluation metrics for the SVM annotation of S2 against the breast radiologists’ annotations and the standard reference. Against the breast radiologists, the SVM has F-measure of 0.491; against the standard reference, it has F-measure of 0.574.
Table 5.
Evaluation metrics of SVM against the breast radiologists and the standard reference confined to S2
| Comparison annotation(s) | Average precision | Average recall | Average F-measure | Average 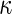
|
|---|---|---|---|---|
| BR1 | 0.490 | 0.607 | 0.542 | 0.517 |
| BR2 | 0.422 | 0.509 | 0.461 | 0.431 |
| BR3 | 0.540 | 0.444 | 0.487 | 0.452 |
| BR4 | 0.664 | 0.385 | 0.487 | 0.445 |
| BR5 | 0.540 | 0.430 | 0.479 | 0.442 |
| BRa | 0.531 | 0.475 | 0.491 | 0.457 |
| SR | 0.540 | 0.613 | 0.574 | 0.549 |
| Allb | 0.533 | 0.498 | 0.505 | 0.473 |
aMean scores against the breast radiologists
bMean scores against the breast radiologists and the standard reference
Average precision and average recall are plotted against the number of selections of SVM in Fig. 2.
Discussion
We address the three aims of the paper in the first three subsections, respectively. The fourth and fifth subsections address related work and directions for future research.
Annotation Creation and Comparison
There is considerable variation between the numbers of selections made by the breast radiologists, ranging from 163 to 364. This may imply that some users interpret the notion of cross-sectional relatedness more liberally than others and/or that they prefer to see more sentences highlighted if the classifier were integrated in a report viewing tool. Such personal preferences can be accounted for by training an ROC curve of SVM classifiers. The SVM classifier that is closest to the user’s preferences can then be selected by setting a slider on a scale from few to many highlights.
Average F-measure of the standard reference is comparable to that of the average F-measures of the breast radiologists. This indicates that the annotation skills of the first author—who is a medical informatician—are comparable to that of breast radiologists.
Generally, inter-rater agreement of 0.8 is considered to be substantial. On our task, the annotators (i.e., the breast radiologists and the first author) have lower agreement, 0.623. We list several root causes:
Strict versus loose interpretation: as observed above with respect to numbers of selections. However, further analysis shows that if we compute F-measure between each annotator and the annotator that is nearest to him/her in terms of number of selected instances, then average F-measure is 0.650. Since the increase with respect to overall inter-rater agreement (0.623) is modest, we posit that this factor has a weak impact on the inter-rater agreement.
Cross-sectional relatedness definition is insufficiently actionable: to mitigate this risk, we framed the annotation task in the context of a workflow enhancement tool. The instruction to select pairs of sentences that would be useful in clinical practice, was intended to contribute to the task’s concreteness. The annotation setting could have been made more concrete by letting the annotation take place behind a PACS workstation simulating the imaging histories of the patients whose reports were annotated. Anecdotal evidence (short chat after experiment) shows that the breast radiologists did not find the task insufficiently clear.
Cross-sectional relatedness between sentences is too fine grained: possibly, higher inter-rater agreement scores are obtained if cross-sectional relatedness between paragraphs is considered instead. In the context of a report viewing tool, such a notion may be equally valuable. There is a concern though, that some radiologists use more paragraphs in their reports than others.
Sample S2 is of higher-than-average complexity. This factor is supported by the observation that the SVM classifier has weaker performance on S2 than on a randomly selected sample of 37 reports, see our discussion below.
Other causes, such as lack of conscientiousness or inherent level of noise, cannot be excluded.
Feature Selection and Nested Cross Validation
The optimal set of features comprised six features. This set was returned in 9 of the 12 outer folds in the nested cross validation trial. The average F-measure of these nine folds was only slightly higher than the F-measure of the SVM in the overall cross validation evaluation (0.702 versus 0.699).
We conclude that report length does not impact the classifier’s performance. This is a positive result: the classifier has the same level of performance on lengthy—and potentially harder to navigate—reports as on short reports. We used report length as a measure for report complexity. An alternative measure would be BI-RADS classification.
Comparison Against Human Annotations
In the 12-fold cross-validation protocol on the standard reference, the SVM classifier has F-measure of 0.699. When trained on samples S1 and S3 and evaluated on sample S2, the SVM classifier has F-measure of 0.574. The difference between these scores may be caused by the fact that the average report complexity of sample S2 is higher than the complexity of the entire corpus. It cannot be caused by an unfavorable proportion of training versus test data. Recall namely that S1 and S3 contain 11 times as many reports as S2. So in terms of number of reports, this evaluation (training on S1 and S3, evaluating on S2) is equivalent to any fold in the 12-fold cross-validation protocol.
On sample S2, the annotation of SVM is in higher agreement with the standard reference than with the breast radiologists’ annotations (see Table 5). This is presumably because of the fact that the standard reference was created by one annotator which guarantees a certain level of consistency but also potentially introduces bias.
Sample S3 was annotated using pre-annotated by an early incarnation of the SVM classifier. Since this mode of annotation was different from the mode of annotation adopted for S1 and S2, this may have caused a bias. This is a limitation of our study.
Related Work
For two fragments of text H (hypothesis) and T (text), H is said to textually entail T if “the meaning of H can be inferred from the meaning of T, as would typically be interpreted by people” [19]. For instance, in the example below [20], T entails H:
T the drugs that slow down or halt Alzheimer’s disease work best the earlier you administer them.
H Alzheimer’s disease is treated using drugs.
Textual entailment is studied in the RTE challenges series organized yearly, starting in 2004. In the RTE challenges, a data set is provided that consists of hypothesis–text pairs. In the first three RTE episodes, focus was on the core task of detecting textual entailment. In later episodes, extensions were explored, such as detecting contradictory relations and, in the two latest episodes (RTE-6 and RTE-7), the main task was defined as follows: “Given a corpus and a set of “candidate” sentences […] from that corpus, RTE systems are required to identify all the sentences from among the candidate sentences that entail a given Hypothesis.”1
In Ref. [21], the authors present the results of their competitive algorithm in the past five RTE challenges. In terms of F-measure, their top scores range from 0.529 (RTE-1) to 0.671 (RTE-3). Top contesters of the first three challenges achieved F-measure scores of 0.580 (RTE-1 [22]), 0.724 (RTE-2 [23]), and 0.766 (RTE-3 [24]). Note that in RTE-2 and RTE-3, a team headed by Hickl ranked higher, but the F-measures of his team’s algorithm were not published [25, 26]. The F-measure score of the winner of RTE-6 (0.480, see Ref. [27]) is substantially lower than the scores of the winners of previous episodes. This may indicate that the complexity of the main task of the first five episodes is lower than that of the two latest episodes. Results of RTE-7 were not available to us, as we did not participate.
Like cross-sectional relatedness, textual entailment is about detecting informational relations in pairs of sentences. A number of differences between our problem and the problems addressed in the RTE challenges must be noted, though:
Cross-sectional relatedness subsumes textual entailment in the sense that if finding sentence F entails conclusion sentence C, then F and C are cross-sectionally related. The converse direction does not hold necessarily.
The fragment pairs of T and H in RTE challenges are sampled from general domains, whereas our standard reference concerns breast radiology reports only. The latter domain is arguably lexically more homogeneous.
The numbers of positive and negative instances are fairly balanced in the RTE challenge, whereas our data is highly unbalanced.
Directions for improvement
Over the years, RTE participants have experimented with various techniques. An early approach [28], brought forward in RTE-1, regards a sentence as a bag of words. It decides that one sentence entails another if the aggregated weight of the words in the intersection of the sentences’ bags of words exceeds a certain threshold (F-measure, 0.628). Advanced features have been used since that model domain knowledge [23, 29] and use background knowledge sources such as WordNet and Wikipedia [30].
Although we have not experimented with WordNet or Wikipedia as background knowledge sources, we are doubtful if they improve performance of the SVM classifier on our task. One of the main use cases for background knowledge is detection of synonymous words or phrases. In the course of our optimizations, we experimented informally with representing each sentence as a bag of SNOMED-CT concepts extracted by MetaMap [31]. This extension did not result in higher outcomes than the bag-of-words approach.
This may not be surprising if we recall that relatedness between sentences is confined by the scope of the report. Since a report is generally written by one radiologist, it is unlikely—and not preferred per BI-RADS—that he uses synonyms to describe the same finding or diagnosis within one report. Other classification problems have been described in the literature in which semantic normalization of narrative content does not improve, and sometimes deteriorates, classification accuracy. We refer the reader to [32] for a good entry point to this literature.
It would be interesting to generalize this research to other areas of radiology. The optimal feature set has no breast-specific features and can be used as a realistic starting point. We have reasons to believe that similar or better results can be obtained on other anatomies. The breast domain is arguably less complex (anatomically and pathologically) than other anatomies, such as, for instance, brain and abdomen/pelvis. Thus, we hypothesize that the terminology base of breast reports is smaller than that of reports of other anatomies. We hypothesize that cross-sectional relatedness detection is harder in domains with a smaller terminology base, since the vector space model has fewer dimensions which may impair its discriminative power.
Instead of describing sentences as bags of words or bags of concepts, it might prove valuable to use semantically richer representations. Such representations could, for instance, impose a hierarchical structure on the sentences’ elements, so that, for instance, it could be determined that “2 o’clock position” is a location, “scattered microcalcifications” is an image finding and that the former is the location of the latter [12]. Domain knowledge, modeled in the form of rules, could then determine that microcalcifications pose an increased risk for malignancy, which can be taken into account when testing relatedness of the sentence with other sentences.
Conclusions
The notion of cross-sectional sentence relatedness in radiology reports was defined and formulated as an information detection problem. We described the development and evaluation of an SVM classifier on this problem. A 444-report standard reference was created; a subset of 37 reports was annotated by five breast radiologists. The SVM classifier has F-measure of 0.699 against the standard reference in a 12-fold cross-validation evaluation. Performance of the SVM classifier is not negatively impacted by the length of the report.
Overall inter-rater agreement on the 37-report subset among the breast radiologists and the first author, who created the standard reference, was mediocre (0.623). Reasons were discussed: (1) definition of cross-sectional relatedness is insufficiently actionable, (2) fine-grained nature of cross-sectional relatedness on sentence level, instead of, for instance, on paragraph level, and/or (3) higher-than-average complexity of 37-report sample.
The optimal feature set of the SVM classifier contains no breast-specific and may thus generalize to other anatomies. We argued that the detection of cross-sectional relatedness may in fact be harder in breast as its lexical base is smaller than that of other anatomies.
Like cross-sectional relatedness, textual entailment is defined in the RTE challenge series as a relation between sentences. Thus, techniques used in these contests may be a source of inspiration for future work.
The ultimate proof of this work lays in its practical utility. Potential outlets include automated report structuring engines that produce mineable data. Since finding sections generally contain image characteristics of findings and conclusion sections diagnostic information, the proposed algorithm can be used to find correlations between image characteristics on the one hand and higher-level information, such as diagnostics information, on the other hand. The annotation task was framed in the context of a smart report reading tool that helps users navigate from sentences in the conclusion section of a report to pertinent information in the rest of the report. This is another potential application of our work.
Electronic Supplementary Material
(DOC 424 kb)
(DOC 452 kb)
Acknowledgments
The authors gratefully acknowledge Yassine Benajiba, Steffen Pauws, and the anonymous referees for valuable comments on an earlier version of this paper.
Footnotes
Contributor Information
Merlijn Sevenster, Phone: +1-914-2620120, FAX: +1-914-9456330, Email: merlijn.sevenster@philips.com.
Yuechen Qian, Email: yuechen.qian@philips.com.
Hiroyuki Abe, Email: habe@radiology.bsd.uchicago.edu.
Johannes Buurman, Email: hans.buurman@philips.com.
References
- 1.Breast Imaging Reporting and Data System Atlas. Reston: American College of Radiology; 2003. [Google Scholar]
- 2.Reiner BI. Customization of medical report data. J Digit Imaging. 2010;23(4):363–73. doi: 10.1007/s10278-010-9307-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gershanik EF, Lacson R, Khorasani R. Critical finding capture in the impression section of radiology reports. AMIA Annu Symp. 2011;2011:465–9. [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman C, Johnson SB. Natural language and text processing in biomedicine. In: Shortliffe EH, Cimino JJ, editors. Biomedical informatics; computer applications in health care and medicine. New York: Springer; 2006. pp. 312–43. [Google Scholar]
- 5.Friedman C, Alderson PO, Austin JHM, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman C, Hripcsak G, Shagina L, Liu H. Representing information in patient reports using natural language processing and the extensible markup language. J Am Med Inform Assoc. 1999;6:76–87. doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreyer KJ, Kalra MK, Maher MM, Hurier AM, Asfaw BA, Schultz T, Halpern EF, Thrall JH. Application of recently developed computer algorithm for automatic classification of unstructured radiology reports: validation study. Radiology. 2005;234(2):323–29. doi: 10.1148/radiol.2341040049. [DOI] [PubMed] [Google Scholar]
- 8.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, Chute CG. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain NL, Knirsch CA, Friedman C, Hripcsak G: Identification of suspected tuberculosis patients based on natural language processing of chest radiograph reports. Proc AMIA Annu Fall Symp, 1996, pp 542–46 [PMC free article] [PubMed]
- 10.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. J Am Med Inform Assoc. 2008;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang PA, Kalra MK, Blake MA, Schultz TJ, Stout M, Halpern EF, Dreyer KJ. Use of Radcube for extraction of finding trends in a large radiology practice. J Digit Imaging. 2009;22(6):629–40. doi: 10.1007/s10278-008-9128-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sevenster M, van Ommering R, Qian Y. Automatically correlating clinical findings and body locations in radiology reports using MedLEE. J Digit Imaging. 2012;25:240–9. doi: 10.1007/s10278-011-9411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang C, Lin C. LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol. 2011;2(3):1–27. doi: 10.1145/1961189.1961199. [DOI] [Google Scholar]
- 14.Kudo T, Matsumoto Y: Chunking with support vector machines. In: Proceedings of the second meeting of the North American Chapter of the Association for Computational Linguistics on Language Technologies, 2001, pp 1–8
- 15.Manning CD, Raghavan P, Schuetze H. Introduction to information retrieval. Cambridge: Cambridge University Press; 2008. [Google Scholar]
- 16.Hripcsak G, Rothschild AS. Agreement, the F-measure, and reliability in information retrieval. J Am Med Inform Assoc. 2005;12(3):296–98. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Varma S, Simon R. Bias in error estimation when using cross-validation for model selection. BMC Bioinforma. 2006;7:91. doi: 10.1186/1471-2105-7-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salzberg SL. On comparing classifiers: pitfalls to avoid and a recommended approach. Data Min Knowl Discov. 1997;1:317–27. doi: 10.1023/A:1009752403260. [DOI] [Google Scholar]
- 19.Dagan I, Glickman O, Magnini B: The PASCAL recognising textual entailment challenge. In Lecture Notes in Computer Science, vol. 3944. Berlin: Springer, 2006, pp 177–190
- 20.Bar-Haim R, Dagan I, Dolan B, Ferro L, Giampiccolo D, Magnini B, Szpektor I. The second PASCAL recognising textual entailment challenge. Proc PASCAL RTE-2 Chall. 2005;3944:177–90. [Google Scholar]
- 21.Pakray P, Bandyopadhyay S, Gelbukh A. Textual entailment using lexical and syntactic similarity. Int J Artif Intell Appl. 2011;2(1):43–58. [Google Scholar]
- 22.Bayer S, Burger J, Ferro L, Henderson J, Yeh A: MITRE’s Submissions to the EU Pascal RTE Challenge. Proc PASCAL RTE-1 Challenge, 2005, pp 41–44
- 23.Tatu M, Iles B, Slavik J, Novischi A, Moldovan D: COGEX at the Second Recognizing Textual Entailment Challenge. Proc. of the PAS-CAL RTE-2 Challenge, 2006
- 24.Tatu M, Moldovan D: COGEX at RTE3. RTE ’07 Proceedings of the ACL-PASCAL Workshop on Textual Entailment and Paraphrasing, 2007, pp 22–27
- 25.Hickl A, Williams J, Bensley J, Roberts K, Rink B, Shi Y: Recognizing textual entailment with LCC’s GROUNDHOG system. Proc. of the PAS-CAL RTE-2 Challenge, 2006
- 26.Hickl A, BensleyJ: A Discourse Commitment-Based Framework for Recognizing Textual Entailment. Proceedings of the Workshop on Textual Entailment and Paraphrasing, 2007
- 27.Li H, Hu Y, Li Z, Wan X, Xiao J: PKUTM participation in TAC2011. Proceeding RTE ’07 Proceedings of the ACL-PASCAL Workshop on Textual Entailment and Paraphrasing, 2010
- 28.Jijkoun V, De Rijke M: Recognizing textual entailment using lexical similarity. Proceedings Pascal 2005 Textual Entailment Challenge Workshop, 2005
- 29.Burnside ES, Davis J, Costa VS, Dutra IDC, Kahn CE, Fine J, Page D: Knowledge discovery from structured mammography reports using inductive logic programming. AMIA Ann Symposium, 2005, pp 96–100 [PMC free article] [PubMed]
- 30.Wang R, Neumann G: Recognizing Textual Entailment Using Sentence Similarity based on Dependency Tree Skeletons. Proceedings of the ACL-PASCAL Workshop on Textual Entailment and Paraphrasing, 2007, pp 36–41
- 31.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. J Am Med Inform Assoc. 2010;17(3):229–36. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldstein I: Automated classification of the narrative of medical reports using natural language processing. University at Albany, State University of New York, 2011
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 424 kb)
(DOC 452 kb)