Abstract
This study aims to determine the most informative mammographic features for breast cancer diagnosis using mutual information (MI) analysis. Our Health Insurance Portability and Accountability Act-approved database consists of 44,397 consecutive structured mammography reports for 20,375 patients collected from 2005 to 2008. The reports include demographic risk factors (age, family and personal history of breast cancer, and use of hormone therapy) and mammographic features from the Breast Imaging Reporting and Data System lexicon. We calculated MI using Shannon’s entropy measure for each feature with respect to the outcome (benign/malignant using a cancer registry match as reference standard). In order to evaluate the validity of the MI rankings of features, we trained and tested naïve Bayes classifiers on the feature with tenfold cross-validation, and measured the predictive ability using area under the ROC curve (AUC). We used a bootstrapping approach to assess the distributional properties of our estimates, and the DeLong method to compare AUC. Based on MI, we found that mass margins and mass shape were the most informative features for breast cancer diagnosis. Calcification morphology, mass density, and calcification distribution provided predictive information for distinguishing benign and malignant breast findings. Breast composition, associated findings, and special cases provided little information in this task. We also found that the rankings of mammographic features with MI and AUC were generally consistent. MI analysis provides a framework to determine the value of different mammographic features in the pursuit of optimal (i.e., accurate and efficient) breast cancer diagnosis.
Keywords: Breast cancer, Mammography, BI-RADS, Decision support, Informatics, Mutual information
Introduction
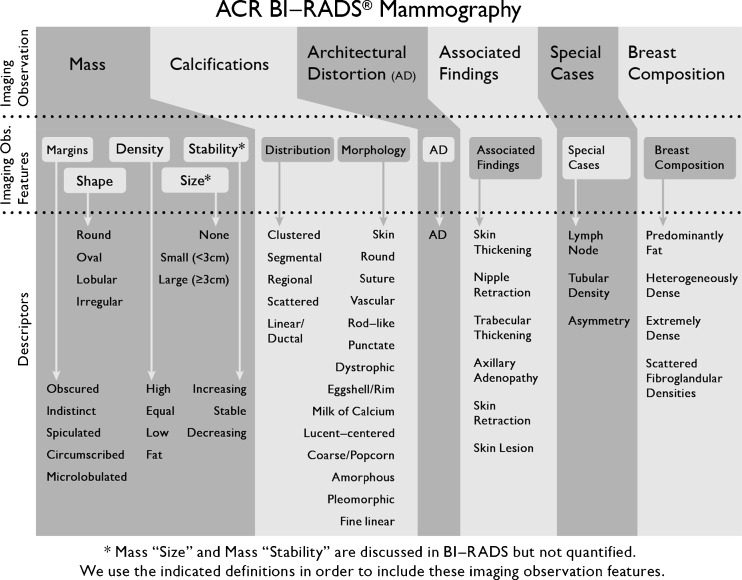
Accurate mammography interpretation depends on careful assessment of predictive mammographic features to estimate the risk of breast cancer and make management recommendations. The Breast Imaging Reporting and Data System (BI-RADS) lexicon standardizes the terminology used to describe mammographic features [1–3] but does not make explicit recommendations as to which features should be prioritized in risk assessment and decision making [4]. In the hierarchical structure of the BI-RADS lexicon (Fig. 1), the imaging observation, representing the type of finding being described, creates the foundation of the lexicon [5]. These imaging observations are then further enriched by imaging observation features under which more granular descriptors convey risks by describing characteristics reflective of pathophysiologic behavior. Our goal is to determine the predictive ability of these imaging observation features in order to help radiologists prioritize their assessment and description of abnormality findings.
Fig. 1.
BI-RADS mammographic features according to our naming conventions for “imaging observation”, “imaging observation features”, and “descriptors”
In the past, evaluation of the predictive ability of mammographic features has not distinguished the imaging observation features and descriptors [4, 6–8], limiting the ability of radiologists to rank and prioritize imaging observation features for decision-making in the clinic. For example, the BI-RADS lexicon provides five descriptors for the imaging observation feature “mass margins”: circumscribed, microlobulated, obscured, indistinct, and spiculated (Fig. 1). Prior literature indicates that spiculated margins had the highest risk of malignancy, and therefore was considered a valuable descriptor. However, this analysis does not establish whether mass margin is the most predictive imaging observation feature (e.g., as compared to mass shape or calcification morphology).
Prior literature has predominantly used two analytical methods to determine the predictive ability of imaging observation features: positive predictive value (PPV) [4, 6] and logistic regression [9–12]. While PPV is a clinically relevant metric for measuring the predictive ability of mammographic features, it has two major shortcomings. First, PPV can only evaluate one binary descriptor at a time, and therefore cannot determine the overall value of an imaging observation feature (e.g., mass margins) including all the descriptors (e.g., circumscribed, microlobulated, obscured, indistinct, and spiculated). This explains why PPV has typically been used to evaluate the BI-RADS lexicon at the descriptor level because the descriptors are most commonly binary. Second, PPV is routinely only available in the setting of biopsy (to establish true positives) and hence the imaging observation feature risk is only estimated in this small subset of patients.
Logistic regression identifies the most important variables based on their coefficients and the correspondent odds ratios under the assumption that the features are independent [10]. It, like PPV, does not allow for a combined measure of risk prediction for imaging observation features with multiple descriptors (e.g., mass margin: circumscribed, microlobulated, obscured, indistinct, and spiculated) [1].
In this study, we use a measure of predictive accuracy for this domain called “Mutual Information” (MI), which is a measure of the information that one variable provides about the other [13, 14]. MI does not exhibit the limitations of PPV and logistic regression because it can quantify not only the relationship between descriptors and breast cancer risk, but also the relationship between imaging observation features (including all descriptors belonging to that feature) and risk. Therefore, MI has the potential to rank imaging observation features for determining the most important in the context of breast cancer diagnosis.
To the best of our knowledge, there is no published study that systematically and comprehensively specifies the relative importance of all of the mammography imaging observation features using data from clinical practice on a series of consecutive patients. The purpose of our study is to quantify and compare the information content using MI inherent in each imaging observation feature included in the BI-RADS lexicon.
Materials and Methods
The institutional review board of the University of Wisconsin Hospital and Clinics exempted this Health Insurance Portability and Accountability Act-compliant retrospective study from requiring informed consent.
Subjects
Our database included all screening and diagnostic mammography examinations collected from full-field digital mammography at the University of Wisconsin Hospital and Clinics from October 1, 2005 to December 30, 2008, in which mammographic findings and demographic risk factors (age, family and personal history of breast cancer, and use of hormone therapy) were described in the BI-RADS format, and were prospectively cataloged by using a structured reporting system (PenRad Technologies, Inc., Buffalo, MN, USA). Mammographic findings were entered by attending radiologists; demographic risk factors were recorded by technologists (Table 1). Eight attending radiologists, who had 7–30 years of experience and specialty in breast imaging practice and met the standards of the Mammography Quality Standards Act, interpreted the mammograms included in the time frame from which we collected clinical data. These mammograms were interpreted in a clinical practice. Screening mammography was interpreted prospectively using single reading and computer-assisted detection (CAD, R2 Technology, Inc., Sunnyvale, CA, USA) by attending radiologists. These interpretations were also done in the context of a teaching hospital; therefore, the majority of these mammograms involved radiology residents and breast imaging fellows.
Table 1.
Demographic risk factors
| Variables | Values |
|---|---|
| Age (years) | <46, 46–50, 51–55, 56–60, 61–65, >65 |
| Hormone therapy | Yes, no |
| Personal history of breast cancer | Yes, no |
| Family history of breast cancera | None, minor, major |
aMinor = non-first-degree family member(s) with a diagnosis of breast cancer, major = one or more first-degree family member(s) with a diagnosis of breast cancer
Imaging Observation Features and the Outcome
We evaluated the relative importance of imaging observation features in the task of differentiating malignant from benign breast abnormalities. We included the following imaging observation features: mass margins, mass shape, mass density, mass size, mass stability, calcification morphology, calcification distribution, architectural distortion, associated findings, and special cases (Fig. 1). All features were estimated by the radiologists, entering them in structured format, as they interpreted these mammography studies in the clinic. We also included breast composition in our analysis since it is an important variable that confers breast cancer risk [15–18] and influences the performance of mammography interpretation [19, 20]. We excluded BI-RADS category [1] in our study since it was a consolidated assessment estimated subjectively from other imaging observation features.
We matched mammographic findings in our database to our University of Wisconsin Cancer Center Registry, which served as our reference standard. The tumor registry achieves high collection accuracy because the reporting of all cancers is mandated by state law, and checked using nationally approved protocols [21]. We considered a finding matched with a registry report of ductal carcinoma in situ or any invasive carcinoma within 1 year as malignant. All other findings shown to be benign by biopsy and those without a registry match within 1 year after the mammogram were considered benign.
Study Design
MI is a basic concept in information theory that quantifies the mutual dependence of two variables, i.e., how much knowing one variable reduces the entropy (uncertainty) of the other [13]. In this study, we measured how much knowing an imaging observation feature reduced the entropy of breast cancer. The mathematical details of MI are discussed in Appendix 1.
In calculating the MI of an imaging observation feature with respect to the outcome (e.g., breast cancer), we calculated the correspondent entropy of the outcome and the conditional entropy given the imaging observation feature. The difference between the two entropy values is the MI of the imaging observation feature. After we obtained the MI of each imaging observation feature with respect to the outcome, we ranked all imaging observation features according to these MI values. A larger MI value for an imaging observation feature indicates that the imaging observation feature provides relatively more information about the outcome. The case of MI value tied ranks is extremely rare since the MI values are expressed with continuous numerical measurements.
In order to evaluate the validity of the MI rankings of imaging observation features, first we calculated probabilities of malignancy for each imaging observation feature with a probabilistic model called a naïve Bayes classifier (NBC) and then, we used these probabilities to construct a receiver operating characteristic (ROC) curve. NBC are known to be equivalent to logistic regression but provide some distinct advantages in terms of simplicity, learning/classification speed, and explanatory capabilities [22, 23].
In this experiment, we used the NBC in the most clinically relevant manner possible. Specifically, we included demographic risk factors (age, family history of breast cancer, personal history of breast cancer, and hormone therapy), which are typically available in clinical practice, in the NBC to measure the pretest probability of disease prior to calculation of the post-test risk when an imaging observation feature became available (Table 1). Therefore, we first trained and tested a NBC on demographic risk factors only without any imaging observation features included (our baseline model), and obtained baseline predictive performance. We then trained and tested a new NBC on these same demographic risk factors plus one specific imaging observation feature. We used the results of the second NBC to construct an ROC curve to measure the predictive accuracy of that given imaging observation feature. In our study, all NBCs were trained and tested with tenfold cross-validation in a software package for machine learning [24] (Weka, version 3.6.4; University of Waikato, Hamilton, New Zealand). We designed the tenfold cross-validation procedure such that all findings from the same patient were included in the same fold. We used the area under an ROC (AUC) as a metric to quantify the predictive performance, based on which we ranked imaging observation features.
Statistical Analysis
To assess the distributional properties of MI estimates, we used a bootstrap methodology [25, 26]. We resampled the actual data set with replacement, and obtained a MI value. We repeated this operation 1,000 times, generated 1,000 estimates of the MI value, and calculated the variance of these estimates. We computed the correspondent 95 % confidence interval (CI) with the variance.
We compared the AUC values associated with NBC of imaging observation features to the baseline predictive performance statistically by using the DeLong method [27]. If we did not find significant differences, we concluded that the feature lacked predictive capability in breast cancer diagnosis. Given our relatively large sample size, and the need to balance statistical and clinical significance, we used p value of 0.001 (two-sided) as the threshold for statistical significance testing. We implemented all statistical analyses in a computing software system (MATLAB, version 2009b; MathWorks, Natick, MA, USA).
Results
Our dataset contains 44,397 consecutively collected mammographic findings, 652 malignant and 43,745 benign, for 20,375 patients. The mean age of the patient population was 54.6 years ± 11.8 (standard deviation). With regard to breast composition, we found 10.8 % predominantly fatty, 43.3 % scattered fibroglandular, 38.7 % heterogeneously dense, and 7.0 % extremely dense; 0.2 % of findings had missing breast composition class. The cancers included 342 masses, 158 calcifications, 43 with combinations of masses and calcifications, 72 false negatives without abnormality findings, and 37 findings categorized as “other” which did not specify whether a finding is a mass or a calcification but had location information.
Our MI analysis reveals that mass margins, mass shape, calcification morphology, mass density, and calcification distribution provided strong information about breast cancer (Table 2). Breast composition, associated findings, and special cases provided little information in distinguishing between malignant and benign findings. Specifically, mass margins provided the most information and special cases supplied the least information in estimating the risk of breast cancer for a given abnormality detected on mammography.
Table 2.
MI of imaging observation features with respect to the outcome (×1,000)
| Imaging observation features | MI (95 % CI) |
|---|---|
| Mass margins | 13.19 (13.11, 13.27) |
| Mass shape | 9.90 (9.83, 9.96) |
| Calcification morphology | 8.29 (8.23, 8.35) |
| Mass density | 8.11 (8.05, 8.17) |
| Calcification distribution | 7.44 (7.38, 7.49) |
| Mass size | 3.58 (3.54, 3.62) |
| Mass stability | 2.50 (2.47, 2.53) |
| Architectural distortion | 1.25 (1.23, 1.28) |
| Breast composition | 0.56 (0.55, 0.57) |
| Associated findings | 0.48 (0.47, 0.49) |
| Special cases | 0.15 (0.15, 0.16) |
By comparing AUC values from our trained NBC, we also found that mass margins achieved the highest predictive performance in distinguishing between malignant and benign findings (Table 3). To demonstrate some specific examples graphically, the AUC value for mass margins which surpassed those for mass density and special cases was shown in Fig. 2. Predictive performance associated with breast composition, associated findings and special cases were the lowest, which agreed with the MI values.
Table 3.
AUC of imaging observation features with respect to the outcome
| Imaging observation features | AUC (95 % CI) | p value (vs. the baseline) |
|---|---|---|
| Mass margins | 0.807 (0.788, 0.828) | <0.001 |
| Mass shape | 0.798 (0.779, 0.820) | <0.001 |
| Calcification distribution | 0.786 (0.764, 0.806) | <0.001 |
| Calcification morphology | 0.785 (0.763, 0.805) | <0.001 |
| Mass density | 0.783 (0.762, 0.804) | <0.001 |
| Mass size | 0.765 (0.742, 0.785) | <0.001 |
| Mass stability | 0.756 (0.736, 0.779) | <0.001 |
| Architectural distortion | 0.747 (0.725, 0.769) | <0.001 |
| Associated findings | 0.743 (0.721, 0.765) | 0.011 |
| Special cases | 0.739 (0.716, 0.761) | 0.232 |
| Breast composition | 0.734 (0.713, 0.757) | 0.423 |
Fig. 2.
ROC curves constructed from the probabilities of the naïve Bayes classifiers for three selected imaging observation features
We also observed that associated findings, special cases, and breast composition did not appear to have the ability to predict whether a mammographic finding was benign or malignant in terms of AUC compared to our baseline model (Table 3). By ranking the imaging observation features in order using MI and AUC, we found that the rankings were consistent for the most part (Table 4).
Table 4.
Rankings from MI and ROC analysis
| Imaging observation features | MI ranks | AUC ranks |
|---|---|---|
| Mass margins | 1 | 1 |
| Mass shape | 2 | 2 |
| Calcification morphology | 3 | 4 |
| Mass density | 4 | 5 |
| Calcification distribution | 5 | 3 |
| Mass size | 6 | 6 |
| Mass stability | 7 | 7 |
| Architectural distortion | 8 | 8 |
| Breast composition | 9 | 9 |
| Associated findings | 10 | 9 |
| Special cases | 11 | 9 |
Discussion
Our results demonstrate that MI has the capability of determining the most informative imaging observation features for breast cancer diagnosis. These results also support and supplement prior literature with regard to the value of these mammographic features. We find that mass margins and mass shape are the most informative, and associated findings and special cases are the least informative features. Moreover, we find that MI provides rankings of imaging observation features, which is reproduced by more conventional approaches to risk ranking including ROC analysis.
MI analysis offers several advantages over conventional feature ranking approaches. First, MI analysis provides a comprehensive methodology for determining the most informative mammographic feature variables, while PPV and regression methods concentrate on rankings of binary variables only (binary descriptors). Second, MI analysis is a straightforward method and is independent of decision algorithms involved, thus reducing computational complexity, while ROC analysis used in this study depends on decision algorithms (naïve Bayes classifier) to generate probabilities of outcomes. Finally, MI analysis measures general statistical dependence between two random variables while traditional correlation coefficient analysis ranks features in order of strength of association with outcomes and is able to find linear dependence only [28, 29].
From a clinical perspective, our results are important for several reasons. First, validating MI as a method that can evaluate the inherent information (the decrease in uncertainty) of an imaging observation feature with regard to the outcome of interest (breast cancer) enables ranking of variable importance. This methodology can rank features which may be useful in helping radiologists order their search pattern or arriving at management decisions when multiple imaging observation features (sometimes conflicting) need to be weighed together. Second, our study reinforces prior literature demonstrating that mass margins and mass shape are the most important imaging observation features to distinguish malignant from benign findings [4, 30–32].
In addition, our results raise the question whether some imaging observation features (e.g., breast composition, associated findings, and special cases) may not contribute substantially to risk estimation for detected mammographic findings. It is interesting that all three of these features do not follow the pattern of more predictive features. Breast composition is a mammography-level feature (rather than an abnormality-level feature) and therefore may predict future risk but not current risk of a mammographic finding. Associated findings and special cases each consist of a list of rarer imaging abnormality findings, a characteristic that may explain the diminished predictive value of these features in our analysis.
Our study provides a more reliable and comprehensive assessment of imaging observation features than most prior studies because we look at the full cohort of consecutive patients seen in a breast imaging clinic with a cancer registry as our reference standard. In contrast, most prior studies used biopsy results as a reference standard thereby only including patients referred for biopsy as the study population [4, 6], which is a small subset of patients evaluated by these imaging observation features. Moreover, MI analysis in our study allows a comprehensive assessment of each imaging observation feature without selecting a single binary descriptor to represent the entire value of the imaging observation feature as has been done in the past [4]. MI is the only well-known technique that can determine how much information (averaged over all the descriptors) each imaging observation feature provides [14].
Based on these observations, we believe that MI analysis may be useful in informing future versions of BI-RADS. The goal of BI-RADS is to standardize mammography practice reporting and is formulated via a data driven process that includes imaging observation features and descriptors that are predictive of benign and malignant disease [3]. It is possible that MI could be used to inform which imaging observation features (and specific descriptors that they contain) should be included in the BI-RADS lexicon as the evidence base in breast imaging grows. Additional research is certainly necessary to determine how robust these rankings are in the clinical setting. Future validation with a multi-institutional trial to confirm these rankings will be important both to demonstrate performance improvement and generalizability. Seamless integration of MI into the clinical workflow (e.g., via structured reporting or PACS software) in order to make MI values available at the time of interpretation and clinical decision making will also be critically important. Nevertheless, our MI analysis appears to be a valuable first step in comprehensively analyzing the value of different imaging observation features on mammography.
There are limitations to our study. First, we calculated MI of each imaging observation feature with respect to the outcome, and did not consider possible effects from other imaging observation features. If there is a strong correlation between two imaging observation features, the contribution of the second imaging observation feature to the estimation of the outcome would be attenuated after the first feature was assessed [33–35]. In clinical practice, radiologists often make clinical decisions based on the information from several imaging observation features simultaneously. A possible line of future research includes looking for a subset of imaging observation features with the highest joint MI result by using multidimensional MI analysis [33–35]. Second, when we used MI analysis to rank imaging observation features on mammography for breast cancer diagnosis, we focused on the predictive accuracy only, and ignored the issues of mortality benefits and cost considerations related to the decision. In the future, it will be important to incorporate utility analysis as a complementary approach to feature ranking [36]. Third, we compared MI values of imaging observation features only in this study. We plan to extend our study of MI analysis to the descriptor level in the future.
Conclusions
Our study demonstrates that MI can be used to efficiently and effectively rank the relative importance of imaging observation features in predicting whether a breast abnormality detected on mammography is malignant. MI analysis may have the potential to improve breast cancer diagnosis by guiding radiologists to the imaging observation feature that is most valuable in discriminating malignant and benign findings on mammography.
Acknowledgments
We thank Elizabeth A. Simcock for figure development and graphic design.
Funding
This work was supported by the National Institutes of Health (grants K07-CA114181, R01-CA127379).
Appendix 1
Originating from Shannon’s information theory [13], MI of a variable X with respect to the outcome Y is defined as the amount by which the uncertainty of Y is decreased with the information X provides. The initial uncertainty of the outcome Y is quantified by entropy H(Y), which is defined (for a discrete outcome) as
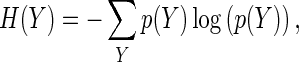 |
where p(Y) is the marginal probability distribution function of Y.
The uncertainty of Y given X, conditional entropy H(Y|X), is defined as
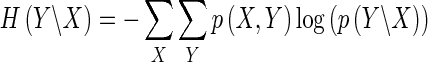 |
where p(X, Y) is the joint probability distribution function of X and Y.
MI can be defined in terms of entropy as 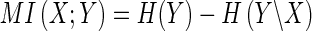 . MI(X; Y) is non-negative, and it is symmetric: MI(X;Y) = MI(Y;X). If X is independent from Y, then H(Y) = H(Y|X), and MI(X;Y) = 0. If base 2 logarithms are used, MI and entropy are in bits. The computation of MI is exemplified below.
. MI(X; Y) is non-negative, and it is symmetric: MI(X;Y) = MI(Y;X). If X is independent from Y, then H(Y) = H(Y|X), and MI(X;Y) = 0. If base 2 logarithms are used, MI and entropy are in bits. The computation of MI is exemplified below.
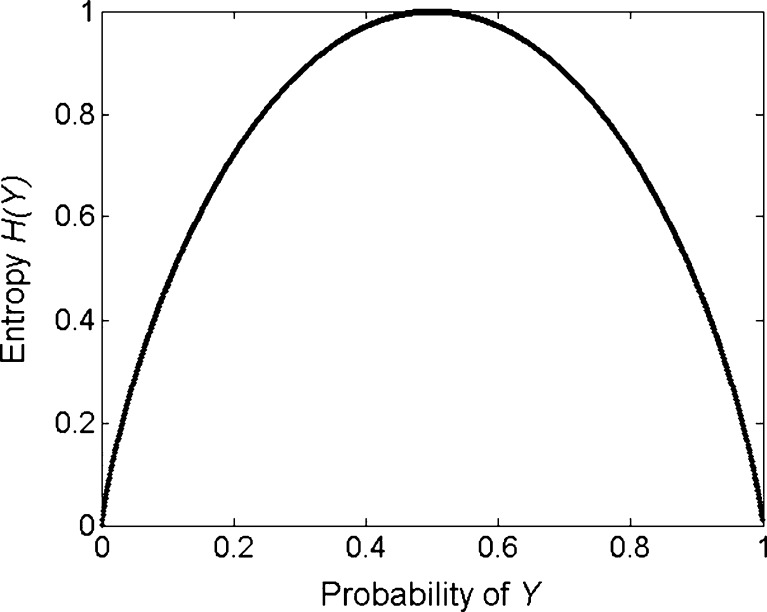
Consider a binary outcome Y with states of malignant and benign. The distribution of Y can be specified by a single probability parameter p0; p(Y = malignant, Y = benign) = (p0, 1 − p0). The entropy associated with Y is maximized when p0 is 0.5 (Fig. 3). The entropy becomes zero when p0 is one or zero since there is no uncertainty for the outcome now. The average uncertainty of Y given knowledge of the feature X (for example, mass margins) is measured by conditional entropy H(Y|X). The difference between initial entropy and conditional entropy represents MI of the variable X with respect to the outcome Y. More details of mutual information and its application to the medical field can be found in other sources [14, 29].
Fig. 3.
Entropy of a binary outcome Y, maximized when the probability of Y is 0.5
References
- 1.American College of Radiology: Breast Imaging Reporting and Data System (BI-RADS) atlas, Reston, Va., 2003
- 2.D'Orsi C, Kopans D. Mammography interpretation: the BI-RADS method. American Family Physician. 1997;55:1548–1550. [PubMed] [Google Scholar]
- 3.Burnside ES, et al. The ACR BI-RADS experience: learning from history. J American College of Radiology. 2009;6:851–860. doi: 10.1016/j.jacr.2009.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liberman L, Abramson A, Squires F, Glassman J, Morris E, Dershaw D. The Breast Imaging Reporting and Data System: positive predictive value of mammographic features and final assessment categories. AJR Am J Roentgenol. 1998;171:35–40. doi: 10.2214/ajr.171.1.9648759. [DOI] [PubMed] [Google Scholar]
- 5.Swets J, Getty D, Pickett R, D'Orsi C, Seltzer S, McNeil B. Enhancing and evaluating diagnostic accuracy. J Medical Decision Making. 1991;11:9–18. doi: 10.1177/0272989X9101100102. [DOI] [PubMed] [Google Scholar]
- 6.Berube M, Curpen B, Ugolini P, Lalonde L, Ouimet-Oliva D. Level of suspicion of a mammographic lesion: use of features defined by BI-RADS lexicon and correlation with large-core breast biopsy. Can Assoc Radiol J. 1998;49:223–228. [PubMed] [Google Scholar]
- 7.Mendez A, Cabanillas F, Echenique M, Malekshamran K, Perez I, Ramos E. Mammographic features and correlation with biopsy findings using 11-gauge stereotactic vacuum-assisted breast biopsy (SVABB) Annals of Oncology. 2003;14:450–454. doi: 10.1093/annonc/mdh088. [DOI] [PubMed] [Google Scholar]
- 8.Venkatesan A, Chu P, Kerlikowske K, Sickles E, Smith-Bindman R. Positive predictive value of specific mammographic findings according to reader and patient variables. Radiology. 2009;250:648–657. doi: 10.1148/radiol.2503080541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayer T, Chhatwal J, Alagoz O, Kahn CE, Jr, Wood R, Burnside ES. Comparison of logistic regression and artificial neural network models in breast cancer risk estimation. RadioGraphics. 2010;30:13–22. doi: 10.1148/rg.301095057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chhatwal J, Alagoz O, Lindstrom MJ, Kahn CE, Jr, Shaffer KA, Burnside ES. A logistic regression model based on the national mammography database format to aid breast cancer diagnosis. AJR Am J Roentgenol. 2009;192:1117–1127. doi: 10.2214/AJR.07.3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform. 2002;35:352–359. doi: 10.1016/S1532-0464(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 12.Tu J. Advantages and disadvantages of using artificial neural networks versus logistic regression for predicting medical outcomes. J Clin Epidemiol. 1996;49:1225–1231. doi: 10.1016/S0895-4356(96)00002-9. [DOI] [PubMed] [Google Scholar]
- 13.Shannon C, Weaver W. The mathematical theory of communication. Urbana, IL: University of Illinois Press; 1949. [Google Scholar]
- 14.Benish W. Mutual information as an index of diagnostic test performance. Methods of Information in Medicine. 2003;42:260–264. [PubMed] [Google Scholar]
- 15.Boyd N, Martin L, Bronskill M, Yaffe M, Duric N, Minkin S. Breast tissue composition and susceptibility to breast cancer. J Natl Cancer Inst. 2010;102:1224–1237. doi: 10.1093/jnci/djq239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyd N, et al. Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol. 2005;6:798–808. doi: 10.1016/S1470-2045(05)70390-9. [DOI] [PubMed] [Google Scholar]
- 17.Martin L, et al. Family history, mammographic density, and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:456–463. doi: 10.1158/1055-9965.EPI-09-0881. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe J. Breast patterns as an index of risk for developing breast cancer. AJR Am J Roentgenol. 1976;126:1130–1137. doi: 10.2214/ajr.126.6.1130. [DOI] [PubMed] [Google Scholar]
- 19.Carney P, et al. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann Intern Med. 2003;138:168–175. doi: 10.7326/0003-4819-138-3-200302040-00008. [DOI] [PubMed] [Google Scholar]
- 20.Mandelson M, et al. Breast density as a predictor of mammographic detection: comparison of interval- and screen-detected cancers. J Natl Cancer Inst. 2000;92:1081–1087. doi: 10.1093/jnci/92.13.1081. [DOI] [PubMed] [Google Scholar]
- 21.Foote M. Wisconsin Cancer Reporting System: a population-based registry. Wisconsin Medical Journal. 1999;98:17–18. [PubMed] [Google Scholar]
- 22.Roos T, Wettig H, Grunwald P, Myllymaki P, Tirri H. On discriminative Bayesian network classifiers and logistic regression. Machine Learning. 2005;59:267–296. [Google Scholar]
- 23.Domingos P, Pazzani M. On the optimality of the simple Bayesian classifier under zero–one loss. Machine Learning. 1997;29:103–130. doi: 10.1023/A:1007413511361. [DOI] [Google Scholar]
- 24.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten I: The Weka data mining software: an update. SIGKDD Explorations(11), 2009
- 25.Efron B, Tibshirani RJ. An Introduction to the bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- 26.Ince R, Mazzoni A, Bartels A, Logothetis N, Panzeri S: A novel test to determine the significance of neural selectivity to single and multiple potentially correlated stimulus features. J Neuroscience Methods, 2011 [DOI] [PubMed]
- 27.DeLong E, DeLong D, Clarke-Pearson D. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. doi: 10.2307/2531595. [DOI] [PubMed] [Google Scholar]
- 28.Guyon I, Elisseeff A. An Introduction to variable and feature selection. J Machine Learning Research. 2003;3:1157–1182. [Google Scholar]
- 29.Tourassi G, Frederick E, Markey M, Floyd C. Application of the mutual information criterion for feature selection in compuet-aided diagnosis. Medical Physics. 2001;28:2394–2402. doi: 10.1118/1.1418724. [DOI] [PubMed] [Google Scholar]
- 30.Winchester D, Winchester D, Hudis C, Norton L. Breast cancer. Heidelberg: Springer; 2007. [Google Scholar]
- 31.Woods R, Oliphant L, Shinki K, Page CD, Shavlik J, Burnside E. Validation of results from knowledge discovery: mass denisty as a predictor of breast cancer. J Digital Imaging. 2010;23:554–561. doi: 10.1007/s10278-009-9235-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woods R, Sisney G, Salkowski L, Shinki K, Lin Y, Burnside E. The mammographic density of a mass is a significant predictor of breast cancer. Radiology. 2011;258:417–425. doi: 10.1148/radiol.10100328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balagani K, Phoha V. On the feature selection criterion based on an approximation of multimensional mutual information. IEEE Trans Pattern Analysis and Machine Intelligence. 2010;32:1342–1343. doi: 10.1109/TPAMI.2010.62. [DOI] [PubMed] [Google Scholar]
- 34.Battiti R. Using mutual information for selecting features in supervised neural net learning. IEEE Trans Neural Networks. 1994;5:537–550. doi: 10.1109/72.298224. [DOI] [PubMed] [Google Scholar]
- 35.Peng H, Long F, Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Analysis and Machine Intelligence. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 36.Sox H, Blatt M, Higgins M, Marton K. Medical decision making. Philadelphia: Butterworth-Heinemann; 1988. [Google Scholar]