Abstract
Genetic resistance is the most effective approach to managing wheat leaf rust. The aim of this study was to characterize seedling and adult plant leaf rust resistance of a world wheat collection. Using controlled inoculation with ten races of Puccinia triticina, 14 seedling resistance genes were determined or postulated to be present in the collection. Lr1, Lr3, Lr10 and Lr20 were the most prevalent genes around the world while Lr9, Lr14b, Lr3ka and/or Lr30 and Lr26 were rare. To confirm some gene postulations, the collection was screened with gene-specific molecular markers for Lr1, Lr10, Lr21 and Lr34. Although possessing the Lr1 and/or Lr10 gene-specific marker, 51 accessions showed unexpected high infection types to P. triticina race BBBD. The collection was tested in the field, where rust resistance ranged from nearly immune or highly resistant with severity of 1 % and resistant host response to highly susceptible with severity of 84 % and susceptible host response. The majority of the accessions possessing the adult plant resistance (APR) gene Lr34 had a maximum rust severity of 0–35 %, similar to or better than accession RL6058, a Thatcher-Lr34 near-isogenic line. Many accessions displayed an immune response or a high level of resistance under field conditions, likely as a result of synergy between APR genes or between APR and seedling resistance genes. However, accessions with three or more seedling resistance genes had an overall lower field severity than those with two or fewer. Immune or highly resistant accessions are potential sources for improvement of leaf rust resistance. In addition, some lines were postulated to have known but unidentified genes/alleles or novel genes, also constituting potentially important sources of novel resistance.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-013-9899-8) contains supplementary material, which is available to authorized users.
Keywords: Triticum aestivum, Puccinia triticina, Lr genes, Gene postulation, APR genes
Introduction
Leaf rust, caused by the fungal pathogen Puccinia triticina Eriks., has been the most prevalent disease in wheat-producing areas (Kolmer 2005). This fungus is adapted to a wide range of environments and it can co-exist with wheat wherever it is grown (Winzeler et al. 2000). It can cause significant yield loss, reaching 15 % or more depending on the crop developmental stage at infection and the susceptibility of the cultivars (Samborski 1985). Genetic resistance has been the most effective method of controlling this disease because it constitutes an environmentally friendly and cost-effective long-term strategy for minimizing yield losses (Pink 2002).
Leaf rust resistance genes named Lr1 to Lr68 have been characterized in bread wheat, durum wheat and diploid wheat species. These genes are located on 20 of the 21 chromosomes of hexaploid wheat (McIntosh et al. 1995, 2007; McCallum et al. 2012). Most Lr genes confer race-specific resistance and follow the gene-for-gene concept leading to a hypersensitive response (HR) or programmed cell death (Flor 1942). Through co-evolution of host and pathogen, most of these genes have been overcome by new pathogen races. Between 1938 and 1964, Australia released many varieties containing single Lr genes, which consequently resulted in an increase in the frequency of corresponding virulent isolates of P. triticina (Park et al. 2001). Gene pyramiding is therefore a viable strategy to provide more durable resistance by slowing down the rate at which single resistance genes are overcome.
Lr seedling resistance genes may be postulated upon inoculation with leaf rust isolates with known avirulence and virulence patterns on wheat differential sets, i.e., cultivars with known Lr genes. Initially established by Loegering et al. (1971) and Browder (1973), gene postulation has been a method of choice for several decades. This method was used to survey leaf rust resistance genes in several collections including a world collection of winter wheat (McVey 1992), American hard red spring wheat (Statler 1984; Oelke and Kolmer 2004), American hard red winter wheat (McVey and Long 1993), American soft red spring wheat (Kolmer 2003; Wamishe and Milus 2004), Mexican wheat cultivars (Singh 1993b; Singh and Rajaram 1991), cultivars from Ethiopia and Germany (Mebrate et al. 2008), Chinese cultivars (Li et al. 2010; Singh et al. 1999), British cultivars (Singh et al. 2001), Argentinean wheat cultivars (Vanzetti et al. 2011) and Eastern, Western and Northern European germplasm (Bartos and Valkoun 1988; Herrera Foessel 2001; Park et al. 2001; Winzeler et al. 2000).
A few Lr genes conferring resistance at the adult plant stage have also been characterized. These genes include Lr12 (McIntosh and Baker 1966), Lr13 (Dyck et al. 1966), Lr22a and b (Dyck 1979), Lr34 (Dyck 1987), Lr35 (Kerber and Dyck 1990), Lr37 (Bariana and McIntosh 1993), Lr46 (Singh et al. 1998), Lr67 (Herrera-Foessel et al. 2011; Hiebert et al. 2010), Lr68 (Herrera-Foessel et al. 2012) and more recently trp-1 and trp-2 (Da-Silva et al. 2012). Genes Lr34, Lr46 and Lr67 provide partial or slow resistance to leaf rust and are considered more durable than seedling resistance genes (Caldwell 1968). The mode of action of these genes is characterized by a longer latent period, a lower infection frequency, smaller uredinia size, a shorter period of sporulation and a lower spore density (Caldwell 1968). Among these genes, Lr34 has not only been durable but it has also been demonstrated to act synergistically with other leaf rust resistance genes (German and Kolmer 1992) and has a pleiotropic effect on other diseases (Singh 1992b; Spielmeyer et al. 2005).
Efficient utilization of genetic resistance relies on an accurate and deep knowledge of the leaf rust resistance genes or gene combinations and of their effectiveness in different environments. This knowledge would undoubtedly raise our understanding of the durability of the genes and assist in pyramiding resistance genes in adapted germplasm. Only four Lr resistance genes have been cloned and sequenced to date: Lr1 (Cloutier et al. 2007), Lr10 (Feuillet et al. 2003), Lr21 (Huang et al. 2003) and Lr34 (Krattinger et al. 2009). Molecular markers have been developed for each of them based on their sequence (Schachermayr et al. 1997; Huang and Gill 2001; Cloutier et al. 2007; Lagudah et al. 2009; Dakouri et al. 2010). Molecular markers for other Lr genes have been developed but these would be linked and not necessarily perfectly diagnostic (for review see McCallum et al. 2012). The goals of the current study were to characterize the seedling and adult plant Lr resistance genes present in a world collection (WC), to evaluate their effectiveness under field conditions and to correlate the results with gene-specific molecular markers in order to validate the postulation and to also identify potential sources of novel resistance and superior gene combinations for use in breeding programs.
Materials and methods
Seedling resistance gene analysis
A total of 275 wheat accessions representing 42 countries were surveyed (Supplementary data Table S1). These lines include wheat cultivars, breeding lines and landraces of hexaploid wheat. Thirty Thatcher near-isogenic lines (NILs) of wheat with known Lr genes constituted the differential set (Table 1).
Table 1.
Seedling infection types observed on the 30 Thatcher near-isogenic lines constituting the differential set upon inoculation with 10 races of Puccinia triticina
| Differential sets | Lr gene | BBBD | MBDS | MGBJ | TJBJ | TDBG | MBRJ | PBDQ | THMJ | TNRJ | TCRJ |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RL 6003 | Lr1 | 0 | 3 | 3 | 3 | 3 | 33+ | 33+ | 3 | 3 | 3 |
| RL 6016 | Lr2a | 0 | 0 | 0 | 3 | 33+ | 0 | 2 | 3 | 3 | 3 |
| RL 6019 | Lr2b | 0 | 0 | 0 | 3 | 3 | 1 | 2 | 3 | 3 | 3 |
| RL 6047 | Lr2c | 1 | 1 | 1 | 3++ | 3 | 0 | 3 | 3 | 3 | 3 |
| RL 6002 | Lr3 | 1− | 3− | 3 | 3 | 3 | 33+ | 3 | 3 | 3 | 3 |
| RL 6010 | Lr9 | 0 | 0 | 0 | 0 | 0 | ; | 0 | 0 | 3 | 0 |
| RL 6005 | Lr16 | 1+ | 2+ | 3− | 3 | 2 | 22+ | ;1 | 3 | 2 | 2 |
| RL 6064 | Lr24 | 0 | 0 | 0 | 3++ | 33− | 0 | 0 | ; | 3 | ; |
| RL 6078 | Lr26 | 1− | 2+ | 1 | 2 | 23 | ; | 2 | 3 | ; | 3 |
| RL 6007 | Lr3Ka | 1 | 2 | 2 | 2 | 2 | 33+ | 2 | 3 | 3 | 3 |
| RL 6053 | Lr11 | 2 | 2+ | 2 | 2+ | 2 | 33+ | 2 | 2 | 3 | 3 |
| RL 6008 | Lr17 | 1 | 3 | 1 | 1+ | 21 | 2 | 3 | ;1 | 0 | ;1 |
| RL 6049 | Lr30 | 2 | 2 | 2 | 2+ | 2 | 33+ | 2 | 3 | 3 | 3 |
| RL 6051 | LrB | 2 | 3 | 2+ | 2+ | 23 | 23 | 3 | 2 | 2 | 2 |
| RL 6042 | Lr3bg | ; | 3 | 3 | 2 | 2 | 2 | 3 | X | X | X |
| RL 6004 | Lr10 | 1− | 3 | 3− | 3 | 33− | 33+ | 3 | 3 | 3 | 3 |
| RL 6013 | Lr14a | 3 | 3 | 3 | 3 | X | 33+ | 2 | 3 | 3 | 3 |
| RL 6006 | Lr14b | 3 | 3 | 3 | 3 | X | 33+ | ;1 | 3 | 0 | 3 |
| RL 6052 | Lr15 | 3− | 3 | 3 | 3 | 33+ | 33+ | 3 | 3 | 2 | 3 |
| RL 6009 | Lr18 | 2 | 2+ | 1 | 2 | 21 | 22+ | 22+ | 22+ | 2 | 23 |
| RL 6040 | Lr19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| RL 6092 | Lr20 | 3 | 3 | 3 | 3 | 2 | 33+ | 3 | 3 | 3 | 3 |
| RL 6043 | Lr21 | 2 | 2+ | 1 | 1 | 2 | ; | 2 | 2 | 0 | 2 |
| RL 6012 | Lr23 | 3− | 3 | 2 | 3 | 33+ | 33+ | 2 | 3 | 0 | 3 |
| RL 6094 | Lr25 | 0 | 0 | 0 | 1 | ; | ; | 3 | 0 | 0 | 2 |
| RL 6079 | Lr28 | 0 | 0 | 3 | 3 | 33+ | 33+ | 33+ | 3 | 3 | 3 |
| RL 6080 | Lr29 | 1 | 1 | 1 | 1 | 13− | 2 | x | ;2 | ; | 2 |
| RL 5497 | Lr32 | 2 | 2 | 1 | 1+ | 2 | 3 | x | x | 0 | 2 |
| RL 6107 | Lr52 | 2 | 2 | 2+ | 1 | 33– | 33+ | ; | ; | 0 | ;1 |
| Thatcher | – | 3 | 3 | 3 | 3++ | 33+ | 33+ | 3 | 3 | 3 | 3 |
Infection types: 0 = no flecks or uredinia, 0; = faint hypersensitive flecks, ; = hypersensitive flecks, 1 = small uredinia with necrosis, 2 = small to medium uredinia with necrosis, 3 = moderate to large size uredinia with/without chlorosis, 4 = very large uredinia without chlorosis, X = mesothetic, a mixture of resistant pustule types, “+” = indicates slightly larger uredinia, “−” = indicates slightly smaller uredinia, infection types (ITs) with two symbols denote a range: e.g., 22+ = indicates a mixture of 2 sizes of uredinia with chlorosis and slightly larger uredinia with chlorosis
The following ten races of the leaf rust pathogen Puccinia triticina were tested on the wheat germplasm and on the differential set: BBBD, MBDS, MGBJ, TJBJ, TDBG, MBRJ, PBDQ, THMJ, TNRJ and TCRJ. The infection type of these races onto the differentials is listed in Table 1.
Five seeds from each accession were planted in 30 × 25 cm fiber flats with 25 accessions in each flat. A mixture of soil, sand and peat moss was used as a growing medium. The fungicide Captan was applied at seeding to prevent fungal infection at germination. The seeded flats were placed in a greenhouse (20–25 °C and 16 h light) and watered as required. Twelve days after planting, i.e., at the two-leaf stage, the seedlings were inoculated with a single race. The inoculation was conducted by spraying the spore solution onto the seedlings using micro-inoculators (University of Minnesota, Minneapolis, USA). The inoculated seedlings were left at ambient environment for 1 h prior to being placed in a humidity chamber for 24 h under conditions of 100 % humidity and 20 °C. After incubation, the flats were returned to the greenhouse and watered daily. Fourteen days after inoculation (DAI), the infection types (ITs) were scored for each race using the 0–4 modified Stakman Scale (Roelfs and Singh 1992). ITs of 3 or higher refer to high infection types (HITs) while ITs of 2 or less were low infection types (LITs). The seedling resistance gene postulation was conducted by comparing the IT patterns of the WC accessions to those of the differential set. Five plants of each accession were scored for their ITs. Seedling tests for lines showing discrepancies between molecular markers and gene postulation were repeated twice for confirmation.
Molecular markers
DNA extraction of the WC was performed as described by Dakouri et al. (2010). Molecular markers specific to Lr1 (Cloutier et al. 2007), Lr10 (Schachermayr et al. 1997), Lr21 (Huang and Gill 2001) and Lr34 (Dakouri et al. 2010) were assessed to confirm the presence or absence of these genes in the WC. Primer names, sequences, PCR conditions and references for each marker are listed (Supplementary data Table S2). For Lr34, the molecular markers caIND11, caSNP4 and caSNP12 were assessed as previously described (Dakouri et al. 2010). Lr1, Lr10 and Lr21 PCR reactions were performed in10 μl aliquots containing 60 ng genomic DNA, 1× PCR buffer, 0.8 mM dNTPs, 1.5 mM MgCl2, 0.4 μM each forward and reverse primers, 0.1 μl of 10× bovine serum albumin (1 mg/mL) and 1 U Taq DNA polymerase. Glenlea, near-isogenic line (NIL) RL6004 (Thatcher-Lr10), McKenzie and NIL RL6058 (Thatcher-Lr34) were used as positive controls for Lr1, Lr10, Lr21 and Lr34, respectively. The PCR products for Lr1, Lr10, Lr21 and Lr34 markers caSNP4 and caSNP12 were resolved on 1.5 % agarose gel while Lr34 marker caIND11 was resolved on an ABI3130xl (Applied Biosystems, Foster City, CA, USA) as previously described (Dakouri et al. 2010).
Field experiments
The world collection was planted in a randomized complete block design (RCBD) with two replications at each of three locations, namely Winnipeg (WPG), Glenlea (GLN) and Portage La Prairie (POR), Manitoba, Canada in 2009, 2010 and 2011. Ten seeds of each accession were planted per hill in WPG and GLN and single 50-cm rows in POR with 25 cm between hills or rows. The wheat cultivars Mckenzie, Thatcher and Thatcher-Lr34 NIL RL6058 were used as checks. Spreader rows of the leaf rust susceptible cultivar Morocco were planted between groups of five rows or hills. Leaf rust epidemics were established by inoculating the entire experiments with a mixture of leaf rust races. In 2009, a mixture of the following races was used: MBDS, MBPS, MBTS, MDNS, MDPS, MFDS, MFNS, MFPS, MLDS, PDBB, TDBG, TDBJ, TFBJ, TFBS, TGBJ, TJBG and TLDS. In 2010, the following races were used: MBPS, MDNS, MDPS, MFDS, MFNS, MFPS, MJDS, MLDS, MNDS, PBDG, PBDQ, TBBJ, TBJS, TDBG, TDBJ, TFBG, TMGJ, TNBQ and TNPS. In 2011, the following races were used: MBPS, MCNS, MDDS, MDNS, MDPS, MDTS, MFDS, MFNS, MFPS, MLDS, PBDQ, PBJQ, TBBG, TBBJ, TDBG, TDBJ, TDGJ, TNBG and TNBJ. Rust severity ratings and reaction types using a modified Cobb scale (Peterson et al. 1948) were collected at maximum rust severity (MRS), which was determined to be when the susceptible check Thatcher showed maximum severity. The host reaction type was evaluated as follows: R, no uredinia present; MR, small uredinia with necrosis and light sporulation; MR-MS, small to medium-size uredinia with moderate sporulation; MS, medium-size uredinia with moderate to intensive sporulation; S, large uredinia with abundant sporulation.
Data analysis
SAS software was used to perform an analysis of variance (ANOVA) using the PROC MIXED model. Each site × year was considered one environment. All factors and their interactions were random effects.
Results
Seedling and APR genes
Gene postulation accompanied by molecular marker data were applied to determine seedling resistance genes in the WC. The ITs of the 30 differential lines to the ten races of P. triticina are shown in Table 1. A total of 14 seedling genes were postulated: Lr1, Lr2a, Lr2c, Lr3, Lr3ka, Lr9, Lr10, Lr14a, Lr14b, Lr15, Lr20, Lr26, Lr28 and Lr30 (Supplementary data Table S3). Of those, Lr20 was postulated in 86 accessions, Lr3 in 62, Lr28 in 19, Lr15 in 16, Lr14a in 12, Lr2c in four and Lr30 and/or Lr3ka in two accessions, while Lr2a, Lr9, Lr14b and Lr26 were each postulated in a single accession (Supplementary data Table S3). Lr2a, Lr2b, Lr11, Lr16, Lr17, Lr25, Lr3bg and LrB were not postulated in any accessions because none of the accessions showed IT patterns similar to their corresponding differential lines. Lr18, Lr19, Lr21, Lr29 and Lr32 had LITs to the ten races and, as such, may not be present in the WC.
Based on this overall gene postulation, the collection was divided into three major groups. Group 1 contained 40 accessions that had HITs to all ten races and thus may not have any of the seedling resistance genes of the differential set. Group 2 contained 11 accessions with ITs that did not match any of the differential lines with known genes, thereby representing unidentified or potentially novel (N) Lr genes. The term unidentified refers to known genes that could not be postulated with our differential set of 30 NILs and race panel of ten but that could be postulated with additional differential lines and/or races. Group 3 contained the remaining 224 lines which may possess one to five seedling resistance genes.
Based on the number of genes, group 3 was further subdivided into four subgroups. Subgroup1 contained 72 accessions postulated to have a single Lr gene. Within this subgroup, 17 and 15 accessions possessed Lr1 and Lr10, respectively. Twenty-four accessions were negative for Lr1 and Lr10 but showed LIT to BBBD and were consequently postulated to have Lr3. Thirteen accessions had a LIT to race TDBG and hence were postulated to have Lr20 alone. Accessions Janetzkis Fruher S and Prospur were postulated to only have Lr28 and Sunbird was postulated to only have Lr9 (Supplementary data Table S3). Subgroup 2 contained 96 accessions that possessed combinations of two seedling resistance genes including Lr1, Lr2c, Lr3, Lr10, Lr14a, Lr15, Lr20, Lr26 and Lr28 as well as unidentified or novel (N) genes. Fifty accessions were postulated to have two of these known genes and 46 accessions had one known gene and one unidentified or novel gene. Subgroup 3 included 44 accessions with three genes, of which six had three known genes while 38 had combinations of known and unidentified or novel genes. Subgroup 4 encompassed 12 accessions with more than three genes including ten accessions with four genes and two with five (Supplementary data Table S3).
Of the postulated or identified genes, Lr1 and Lr10 were the most frequent in North America and Asia, while in South America Lr3, Lr10 and Lr20 were prevalent. In Africa, Lr1, Lr10 and Lr20 were the most frequent. In Europe, Lr1, Lr3, Lr10 and Lr20 were found at higher frequency while in Oceania Lr10 and Lr20 were more frequent (Table 2). Lr1 was more frequent in North America (18/36) followed by Africa (10/25) and South America (7/45). Lr3 was found at higher frequency in South America (16/45) followed by Asia (16/48), and was present in a single accession from Africa. Lr10 occurred at higher frequency in Africa (10/25) followed by Oceania (10/29) and Asia (4/48). Lr20 dominated in African accessions (15/25) followed by South American ones but was found at very low frequency in North American germplasm (3/36) (Table 2).
Table 2.
Geographical distribution of seedling resistance genes in the world collection of wheat
| Continent | No. of accessions | Lr1 | Lr2a | Lr2c | Lr3 | Lr3ka | Lr9 | Lr10 | Lr14a | Lr14b | Lr15 | Lr20 | Lr26 | Lr28 | Lr30 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| North America | 36 | 18 | 1 | 2 | 7 | 2 | 0 | 12 | 3 | 0 | 5 | 3 | 0 | 4 | 2 |
| South America | 45 | 7 | 0 | 1 | 16 | 0 | 0 | 11 | 5 | 0 | 3 | 20 | 0 | 5 | 0 |
| Asia | 48 | 12 | 0 | 0 | 16 | 0 | 0 | 4 | 1 | 1 | 4 | 7 | 1 | 0 | 0 |
| Africa | 25 | 10 | 0 | 0 | 1 | 0 | 0 | 10 | 1 | 0 | 0 | 15 | 0 | 1 | 0 |
| Europe | 65 | 16 | 0 | 0 | 12 | 0 | 0 | 15 | 1 | 0 | 1 | 20 | 0 | 4 | 0 |
| Oceania | 29 | 7 | 0 | 0 | 5 | 0 | 1 | 10 | 1 | 0 | 0 | 11 | 0 | 1 | 0 |
| Unknown | 27 | 4 | 0 | 1 | 5 | 0 | 0 | 8 | 0 | 0 | 3 | 10 | 0 | 4 | 0 |
| Total | 275 | 74 | 1 | 4 | 62 | 2 | 1 | 70 | 12 | 1 | 16 | 86 | 1 | 19 | 2 |
Molecular marker analysis
Gene-specific markers to the seedling resistance genes Lr1, Lr10 and Lr21 were utilized to validate the gene postulation in the WC (Supplementary data Table S3) and to assess their potential as molecular screening tools. The Lr1 and Lr10 markers were present in 74 and 69 accessions, respectively, including 24 accessions that had both markers, while the Lr21 marker was not present in the collection (Supplementary data Table S3). The Lr1 and Lr10 markers validated the gene postulation in the majority of the lines but 51 accessions possessing one and/or the other marker showed unexpected HITs to race BBBD (Supplementary data Table S3). The APR gene Lr34 was assessed in the WC using the three gene-specific markers caSNP4, caIND11 and caSNP12 (Dakouri et al. 2010) and 52 accessions were positive for the Lr34 markers (Supplementary data Table S3).
Field resistance
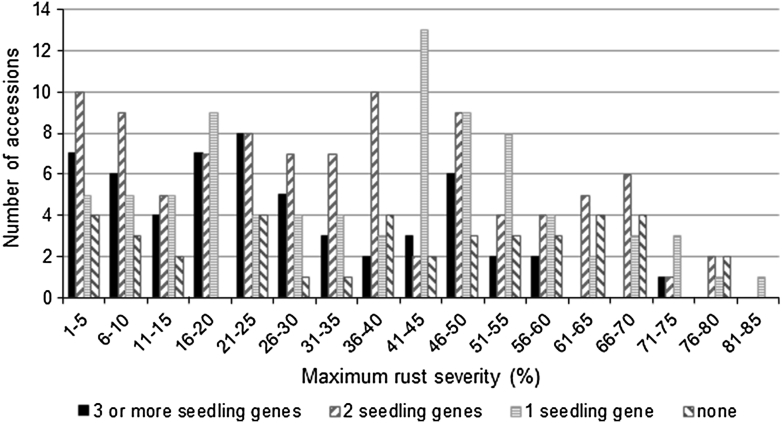
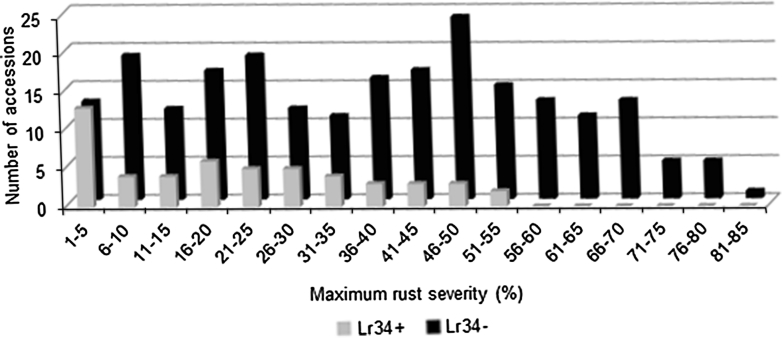
The WC was also evaluated for field resistance at three locations over 3 years. The ANOVA showed significant differences (P < 0.05) between accessions and between environments (Supplementary data Table S4). Significant differences also were observed between accessions with Lr34 and those without Lr34 as well as between accessions within the Lr34+ and Lr34− groups. The averaged maximum rust severity (MRS) and host response (HR) ranged from ~1R to 84S. Overall ~17 % of the accessions were considered highly resistant to leaf rust with MRS <10 %, ~15 % were resistant with MRS ranging from 10 to 20 %, ~28 % were moderately resistant with MRS between 20 and 40 %, 16 % moderately susceptible with MRS of 40–50 %, 20 % susceptible with MRS of 50–70 % and 5 % highly susceptible with MRS >70 %. Host response was not entirely correlated with MRS, and some accessions, for example Albimonte, had very low MRS but large pustules. In general, accessions possessing three or more genes did not have high severity ratings as compared to those having zero, one and two seedling resistance genes (Fig. 1). The average severity rating for the accessions with three or more seedling genes was 26 while it was 33 for those having two seedling genes, 38 for those with a single seedling gene and increased to 40 for the ones having none. Similarly, accessions containing Lr34 were more resistant to leaf rust than those lacking it, regardless of their gene complement (Fig. 2). Of the 52 accessions with Lr34, 41 had an average MRS ranging from 1 to 35 %, which was similar to RL6058, a Thatcher NIL with Lr34. The other Lr34+ accessions showed MRS ranging from 36 to a maximum of 53 %.
Fig. 1.
Frequency distribution of accessions plotted against maximum rust severity (MRS) based on the number of seedling resistance genes. Accessions with three or more genes had lower rust severity than accessions with zero, one or two seedling resistance genes
Fig. 2.
Frequency distribution of accessions plotted against maximum rust severity (MRS) based on the presence or absence of Lr34 shows lower MRS in Lr34+ accessions
Group 1 accessions, i.e., accessions without identified seedling resistance genes, displayed MRS ranging from 2 to 78 %. Seven group 1 accessions were classified as nearly immune or highly resistant with an average MRS ranging from 2 to <10 % and a resistant (R) to moderately susceptible (MS) reaction type (Supplementary data Table S3; Fig. 1). Six group 1 accessions possessed Lr34 and exhibited MRS of 2–35 %. Group 2 with unidentified or N genes showed rust reading ranging from 1R to 76S. Accessions CN99032, Preludio and CN12624 were rated nearly immune or highly resistant, with rust reading 1R, 3RMR and 7RMR, respectively (Supplementary data Table S3). Within group 3, accessions possessing the gene combinations Lr1 + Lr26 and Lr28 + N were particularly highly resistant to leaf rust with MRS <10 %. Accessions containing Lr2c + N, Lr14a + N, Lr20 + N, Lr28 + N, Lr1 + Lr20 + N, Lr1 + Lr14a + N, Lr10 + Lr14a, Lr1 + Lr20 + Lr28 + N and Lr1 + Lr14a + Lr15 + Lr20 + N in combination with Lr34 were highly resistant when compared to other gene combinations.
At the geographical level, the average MRS was the lowest in South America (18 %) and the highest in European (42 %) and Oceanian germplasm (40 %). The majority of South American accessions (21/45) had an MRS from 1–15 %, while the proportion of highly resistant germplasm was much lower in the other continents and the lowest in Africa (1/25) within the same MRS range (Fig. 3). In Africa and North America, the majority of the accessions displayed an MRS ranging from 16 to 35 %. European accessions (23/65) had the highest percentage of susceptible lines with MRS over 50 %.
Fig. 3.
Frequency distribution of accessions plotted against maximum rust severity (MRS) based on their geographical distribution
Discussion
Puccinia triticina, the pathogen causing leaf rust, is present in most of the wheat-growing areas. Genetic resistance remains the most effective, economical and environmentally friendly strategy for controlling the negative impact of this disease. A survey of the seedling and APR genes in a world collection of wheat accessions using molecular markers and gene postulation accompanied by field resistance evaluations at three locations during 3 years is reported.
Seedling and APR genes
Gene postulation has been widely applied as a quick method for hypothesizing the seedling resistance gene composition of germplasm. This method follows the gene-for-gene specificity theory that was first described by Flor (1942). Comparison of infection type profiles of unknown genotypes to various rust races with those of Thatcher near-isogenic lines, each with a single known gene, forms the basis of the gene postulation. However, this method does not enable differentiation between genes that have similar infection type profiles, e.g., Lr1, Lr3 and Lr10. Considering the minimum number of genes required to explain phenotypes (McVey and Long 1993; Singh 1993b), gene postulation and molecular marker analyses of the WC identified 14 known seedling resistance genes present either as single genes or in combinations: Lr1, Lr2a, Lr2c, Lr3, Lr9, Lr10, Lr14a, Lr14b, Lr15, Lr20, Lr26, Lr28 and Lr3ka and/or Lr30.
Lr1, Lr3, Lr10 and Lr20 were the most common genes in the WC, but their distribution across the globe varied. The first three were also the most common in American soft red winter wheat (Roelfs et al. 2000; Wamishe and Milus 2004). Lr20 was the most frequent gene in Ethiopian wheat germplasm (Mebrate et al. 2008) but was found at low frequency in British wheat (Singh et al. 2001) and was rare in Australian germplasm, the American soft red winter wheat and Argentinian germplasm (Singh et al. 2007; Vanzetti et al. 2011; Wamishe and Milus 2004). A high Lr1 frequency was previously reported in American hard red spring wheat, Australian wheat cultivars, Indian and Pakistani wheat, Mexican bread wheat and Chinese cultivars (Oelke and Kolmer 2004; Singh et al. 1999, 2007; Singh and Gupta 1991; Singh and Rajaram 1991) but was rare in Argentinian germplasm (Vanzetti et al. 2011). Lr10 was commonly postulated in international winter wheat nurseries, Indian and Pakistani wheat cultivars, Mexican bread wheat, Brazilian and Argentinian wheat cultivars (McVey 1992; Singh and Gupta 1991; Singh and Rajaram 1991; Vanzetti et al. 2011; Zoldan and Barcellos 2002). Conversely, Lr2a and Lr9 were rare in the WC with one accession each but were common in American soft red winter wheat (Roelfs et al. 2000; Wamishe and Milus 2004) and in cultivars from the UK (Singh et al. 2001). Located on the 1BL.1RS translocation, Lr26 was reported at a high frequency in Chinese wheat germplasm (Singh et al. 1999), Argentinian cultivars (Vanzetti et al. 2011) and British cultivars (Singh et al. 2001) but was present in only a single accession (NING 8331) of the WC. The WC may simply have a low frequency of the rye translocation, a feature we did not evaluate.
Accessions El Gaucho, Bage, AC Minto, Americano 44D and Biggar, postulated to have Lr2c + N, Lr1 + 10 + 20 + N, Lr28 + N, Lr1 + 10 + N and Lr3 + 15 + N, respectively, were previously reported to possess Lr11, Lr3bg, Lr11 + 13 + 21 + 22a, Lr13 + 34 and Lr14a + 13 in the same order (Kolmer 1994; Long and Kolmer 1989; Roelfs 1988). The N gene(s) in these accessions could be known gene(s) not in the differential sets, such as Lr13 or Lr22a, or truly novel gene(s).
Lr19, Lr21, Lr29 and Lr32 displaying LITs to all ten races have not been deployed in wheat varieties (McIntosh et al. 1995; Mebrate et al. 2008; Wamishe and Milus 2004), and so their absence in the WC is not surprising. The same can be said for Lr36 and Lr38. Lr3ka and/or Lr30 were postulated in accessions Katepwa and Burnside but discrimination between the two genes was precluded by the lack of differentiating race(s).
The ten races and the 30 differential lines used in this study permitted gene postulation of 14 seedling resistance genes but these races and differential lines were not sufficient to postulate and determine the identity of all known resistance genes, indicative of some of the limitations of the gene postulation method (Hysing et al. 2006; Mebrate et al. 2008). A total of 96 accessions possessed combinations of known and unidentified (N) seedling resistance genes and 11 accessions were postulated to only have one or more N genes. The unidentified but known genes could be Lr13, Lr27 + Lr31 (Browder 1980; Dyck 1992; Pretorius et al. 1984) or not yet described alleles of known genes (Wamishe and Milus 2004). Allelism tests are appropriate alternatives but they are lengthy to perform (McCallum et al. 2010). Developing molecular markers tightly linked and, ideally, perfectly linked to genes and alleles of interest is the method of choice for determining the identity of known genes (Tanksley et al. 1989).
Variability in the LIT scores (i.e., 0, ;, 1; 2, 22+) is a common indication of heterozygosity for avirulence among P. triticina isolates (Samborski and Dyck 1968; Wamishe and Milus 2004). Temperature sensitivity is another possible cause of LIT score variability such as previously reported for Lr11 and Lr18 (Long and Kolmer 1989; McIntosh et al. 1995). Temperatures as low as 17 °C were used to postulate Lr11 when tested with homozygous avirulent races (Wamishe and Milus 2004). In our study, all races were tested within a temperature range of 18–22 °C, which might explain the lack of detection of Lr11.
Field resistance
To better understand the effect of seedling and APR genes on leaf rust under field conditions, the WC was evaluated in the field at three locations over 3 years. Field data showed that 60 % of the accessions were highly to moderately resistant to leaf rust with MRS of 1–40 %, while 40 % were moderately to highly susceptible with MRS greater than 40 %.
Overall, the rust severity decreased as the number of seedling resistance genes increased, indicating the importance of gene pyramiding in reducing the damage caused by leaf rust including yield loss (Šliková et al. 2004). Gene pyramiding, a process of combining more than one resistance gene in a single genotype, has been a proven strategy for developing long-lasting resistance to plant disease (Singh et al. 2000). This strategy has been exploited for resistance to bacterial blast and bacterial blight in rice (Hittalmani et al. 2000; Huang et al. 1997), powdery mildew and leaf rust in wheat (Liu et al. 2000; Šliková et al. 2004) and stripe rust in barley (Castro et al. 2003). In our study, accessions of the WC contained one to five genes and, in total, 43 gene combinations were observed including 19 two-gene, 19 three-gene, 6 four-gene and 2 five-gene combinations. Accessions possessing three genes or more clearly displayed an overall lower rust severity rating compared to those with zero, one or two genes (Fig. 1).
Subgroup1 is composed of accessions with one seedling resistance gene including Lr1, Lr3, Lr9, Lr10, Lr20 and Lr28 that have already been overcome (Kolmer 1996). Within this subgroup, correlations between the gene content and low MRS were not consistent, which indicates that low levels of MRS in some accessions cannot solely be accounted for by the presence of these genes alone but possibly by the presence of one or more APR genes (Kolmer 1996). Lr20 did not contribute significantly to leaf rust resistance (McIntosh et al. 1995; Wamishe and Milus 2004). The same conclusion may apply to Lr1, Lr3, Lr10 and Lr28.
Subgroup 2 comprised accessions with two genes each. Two-gene combinations also provided variable levels of MRS ranging from 2 to 78 %. Accessions possessing Lr28 + N consistently showed low levels of MRS ranging from 3 to 17 %, indicating the potential of this combination in leaf rust resistance. Accessions El Gaucho and NING 8331with Lr2c + N and Lr1 + Lr26, respectively, were highly resistant with MRS of 2 and 3 % and may also represent good combinations for improved leaf rust resistance.
Despite the fact that most combinations within subgroup 3 provided high to moderate levels of resistance, the gene combination Lr1 + Lr10 + N seems to provide better levels of resistance. The observed resistance level may be solely accounted for by the N gene(s) because Lr1 and Lr10 have been overcome (Kolmer 1996). The only exception was accession Kenya which had a moderately susceptible phenotype accompanied by a HIT to BBBD.
Subgroup 4 with four and five seedling resistance genes was overall more resistant than the other subgroups. The best levels of resistance were observed for combinations with N gene(s), stressing once more the need to further investigate these genes.
Lr34 has been the most important leaf rust resistance gene identified to date because of its race non-specificity, its durability, its synergistic and pleiotropic effects and its positive effect on yield (Dyck 1987, 1991; Dyck et al. 1994; McCallum et al. 2007; McIntosh 1992; Samborski 1985; Singh 1992a, b, 1993a; Singh and Huerta-Espino 1997; Spielmeyer et al. 2005). The majority of Lr34+ accessions (79 %) had high to moderate levels of resistance with MRS ranging from trace to 35 %, similar to RL6058 (i.e., Tc-Lr34 NIL). The remaining eleven Lr34+ accessions (~21 %) showed average MRS ranging from 36 to 52 %. The specific reason(s) for the relatively higher rust severity in these Lr34+ accessions can only be speculated. DNA methylation was reported to play a role in the expression of APR genes in rice, where a correlation was observed between hypermethylation and repression of gene expression (Sha et al. 2005). Recently, Wu et al. (2012) reported the presence of a stripe rust (Yr18 function) inhibitor to Lr34/Yr18/Pm38 in some Chinese landraces. Such inhibitors were not identified in breeding lines or for the Lr34 leaf rust function, but this too suggests the possibility of functional inhibitors to another Lr gene. Population development, gene expression, sequencing and methylation pattern analyses are required to investigate these hypotheses.
The interactions between Lr34 and other leaf rust resistance genes including seedling and APR genes were described in previous reports and also herein. Here, we highlighted gene combinations that have provided excellent levels of resistance in our multi-year, multi-location field trials. The relatively high levels of resistance in Lr34+ accessions with no seedling resistance genes might be explained by the presence of additional APR genes (Li et al. 2010) such as Lr12, Lr13, Lr22a, Lr46, Lr67 and Lr68, or novel APR genes. Synergy between Lr34 and other APR genes was previously reported (Kloppers and Pretorius 1997; Sawhney 1992). Cultivars possessing combinations of Lr34 + Lr12 (e.g., Chinese Spring) and Lr34 + Lr13 (e.g., Roblin) were highly resistant with 5RMR and 10MRMS rust reading, respectively (Dyck 1991, 1992). Accession El Gaucho which had the gene combination Lr34 + Lr2c + N was highly resistant with a severity of 2 % and an RMR reaction type, indicative of the potential synergy between these genes. Seven accessions had the combination of Lr3 + Lr34 with rust rating ranging from 1RMS to 48MSS. Because Lr3 is defeated, additional APR genes are hypothesized in accessions with the lowest MRS. Lr9, previously reported as an important leaf rust resistance gene in soft red winter wheat (Kolmer 2003), was only postulated in Sunbird and, in combination with Lr34, provided only a moderate level of field resistance to leaf rust.
The high levels of resistance in some accessions without Lr34 may be caused by various gene combinations not fully characterized here, including other APR genes. Lr13, which originated from South American germplasm, was commonly found in wheat germplasm worldwide (McIntosh et al. 1995; Singh et al. 1998, 2001). Despite being defeated, Lr13 in combination with other APR genes may provide an acceptable level of field resistance (Kolmer 1992). Lr12 is also frequent in wheat cultivars from Australia, China, North America and South America (Kolmer 2003; McIntosh et al. 1995; Park and McIntosh 1994; Wamishe and Milus 2004). The WC may contain some or all known APR genes as well as novel genes, alone or in combinations. The accession Kanred, for example, had no seedling resistance genes but displayed a low severity of 5 % and a host response (HR) of R in two locations and MS in one, indicating the presence of unidentified genes. The variability in host response (i.e., R in two locations and MS in one) could be due to environmental effects.
The majority of South American accessions had multiple resistance genes, both seedling and APR, combined with unidentified (N) genes, which might explain the low level of rust severity in these accessions. In Europe and Oceania, on the other hand, most accessions had either zero or one seedling gene, thus explaining the high level of rust severity in these accessions. This may indicate intensive breeding programs for leaf rust resistance in South America as compared to the other areas of the world.
Molecular marker analysis
Molecular markers have been invaluable tools in plant breeding, including gene identification and marker-assisted selection. One of the major applications of markers in breeding for leaf rust resistance is to determine the number and identity of the resistance genes in cultivars or germplasm. Markers linked to Lr1, Lr3a, Lr9, Lr10, Lr13, Lr14a, Lr16, Lr17a, Lr19, Lr20, Lr21, Lr22a, Lr23, Lr24, Lr25, Lr26, Lr27, Lr28, Lr29, Lr31, Lr32, Lr34, Lr35, Lr37, Lr38, Lr39, Lr42, Lr46, Lr47, Lr48, Lr49, Lr50, Lr51, Lr52, Lr56, Lr57, Lr58, Lr60, Lr61, Lr63, Lr66 and Lr67 have been published [reviewed in McCallum et al. (2012)] and, since then, markers for Lr68 (Herrera-Foessel et al. 2012) have also been reported. While these markers are linked to Lr genes, they are not all perfectly diagnostic. Their usefulness depends mainly on the genetic background, the tightness of the linkage, whether the gene is from an alien source, the polymorphism level and the robustness of the markers. Gene-specific markers are more accurate but, as we discovered for Lr1 and Lr10, they may not correlate perfectly with the phenotype. This was also observed for Lr1 (Da Silva et al. 2008), although in this case they were not using a gene-specific marker (Feuillet et al. 1995). Our intention was to use the molecular markers to provide validation for the gene postulation and to demonstrate their usefulness as screening tools for breeders and pathologists.
While the majority of the accessions postulated through seedling tests to have Lr1 and/or Lr10 were validated by their respective markers, 51 accessions, although possessing the Lr1 and/or Lr10 gene-specific markers, had HITs to leaf rust race BBBD. Many Lr10 haplotypes have been described but only one was reported to confer resistance (Sela et al. 2012). Such extensive haplotype description has not been done for Lr1. Even though the Lr1 marker is gene-specific, it may not be perfectly diagnostic because the possibility of additional functional and non-functional haplotypes cannot be ruled out. In the case of Lr10, RGA2, a gene closely linked to Lr10, has been shown to be necessary for its function (Loutre et al. 2009). Indeed, some R genes can only act in pairs, but, in addition, NBS-LRR-driven resistance can be mediated through several different resistance mechanisms (Eitas and Dangl 2010). Finally, studies on Lr26 and Pm8 reported their non-functionality in the presence of inhibitor genes (Hanušová et al. 1996; Li et al. 2010). Even though inhibitor genes have not been reported to date for Lr1 and Lr10, this mechanism, the dual gene action and the potential for undescribed functional and non-functional haplotypes may all explain the discrepancies between the marker data and the phenotypic results with BBBD.
Lr21 originated from the diploid wheat species Aegilops tauschii and was first introgressed into wheat cultivar Thatcher (Rowland and Kerber 1974). This gene was not present in the WC, indicating its limited deployment to date in breeding programs.
Our results clearly demonstrate the usefulness of gene-specific markers such as Lr1 (Cloutier et al. 2007), Lr10 (Schachermayr et al. 1997), Lr21 (Huang and Gill 2001) and Lr34 (Krattinger et al. 2009; Lagudah et al 2009; Dakouri et al. 2010) in gene postulation. Gene-specific markers have the potential to be highly diagnostic but they are not always perfect. Extensive haplotyping within the primary and ideally the secondary and tertiary gene pools as well as in evolutionarily related species such as rye, barley and Brachypodium is necessary to provide a comprehensive understanding of the gene(s) and for the development of one or a suite of perfectly diagnostic markers.
APR is conferred by genes that are effective at the adult plant stage. They are mostly race non-specific and provide similar levels of resistance to all leaf rust races (Das et al. 1993; Ohm and Shaner 1976). Using three gene-specific markers (Dakouri et al. 2010), the APR gene Lr34 was identified in 52 of the 275 accessions of the WC. This gene has been widely utilized in breeding for leaf rust resistance and is commonly found in world wheat germplasm (Kolmer et al. 2008), particularly in Asian and South American germplasm (Singh et al. 1999). Molecular markers are especially useful for APR genes because their phenotyping is challenging. APR phenotypes are better characterized under field conditions and the few indoor tests (e.g., cold temperature test) are lengthy (Krattinger et al. 2009). The Lr34 marker caSNP12 was perfectly diagnostic for 700 lines (Dakouri et al. 2010) but it is not ideal as a dominant marker because failed reactions and negative markers are confounded. Marker caIND11 constitutes an excellent alternative, being co-dominant and with a diagnostic rate of 99.7 % (Dakouri et al. 2010).
Conclusions
Efficient exploitation of genetic resistance to leaf rust demands a detailed examination of the occurrence and distribution of both seedling and APR genes. Using molecular markers combined with gene postulation, 14 seedling resistance genes and one APR gene were determined to be present in a world collection of wheat. Additional seedling and APR genes may be present but could not be postulated based on the differential lines, leaf rust races and diagnostic molecular markers. The world collection is a potential reservoir for several novel resistance genes, both seedling and APR. The majority of these accessions would have multiple genes, with some of them providing excellent levels of resistance. However, the actual number of genes contained in each accession can only be determined by genetic analysis. The work presented herein provides further evidence to support the claim that Lr34 is the single most significant and durable gene in breeding programs for leaf rust resistance. The results of this study highlight the potential and limitations of molecular markers for leaf rust gene postulation and the importance of gene pyramiding to provide acceptable long-term resistance to wheat leaf rust with emphasis on the important role of APR genes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Raja Ragupathy, Andrzej Z. Walichnowski, Elsa Reimer, Evelyn Miranda, Braulio Soto-Cerda, Santosh Kumar, Dinushika Thambugala, summer students and the breeding group for help with the field experiments and Pat Seto-Goh for help with seedling experiments and field inoculation. A. D. was supported by the Monsanto’s Beachell-Borlaug International Scholarship Program (MBBISP). This work was supported by an Agriculture and Agri-Food Canada A-Base grant.
References
- Bariana HS, McIntosh RA. Cytogenetic studies in wheat. XV. Location of rust resistance genes in VPM1 and their genetic linkage with other disease resistance genes in chromosome 2A. Genome. 1993;36:476–482. doi: 10.1139/g93-065. [DOI] [PubMed] [Google Scholar]
- Bartos P, Valkoun J. Rust resistance genes in Czechoslovak wheats. Cereal Rusts Powdery Mildews Bull. 1988;16:36–40. [Google Scholar]
- Browder LE. Probable genotype of some Triticum aestivum agent derivatives for reaction to Puccinia recondita f. sp. tritici. Crop Sci. 1973;13:203–206. doi: 10.2135/cropsci1973.0011183X001300020016x. [DOI] [Google Scholar]
- Browder LE. A compendium of information about named genes for low reaction to Puccinia recondita in wheat. Crop Sci. 1980;20:775–779. doi: 10.2135/cropsci1980.0011183X002000060024x. [DOI] [Google Scholar]
- Caldwell RM (1968) Breeding for general and/or specific plant disease resistance. In: Findlay KW, Shepherd KW (eds) Proceedings of the 3rd international wheat genetics symposium. Australian Academy of Science, Canberra, Australia, pp 263–272
- Castro AJ, Chen X, Hayes PM, Johnston M. Pyramiding quantitative trait locus (QTL) alleles determining resistance to barley stripe rust: effects on resistance at the seedling stage. Crop Sci. 2003;43:651–659. doi: 10.2135/cropsci2003.0651. [DOI] [Google Scholar]
- Cloutier S, McCallum BD, Loutre C, Banks TW, Wicker T, Feuillet C, Keller B, Jordan MC. Leaf rust resistance gene Lr1, isolated from bread wheat (Triticum aestivum L.) is a member of the large psr567 gene family. Plant Mol Biol. 2007;65:93–106. doi: 10.1007/s11103-007-9201-8. [DOI] [PubMed] [Google Scholar]
- Da Silva PR, Milach SCK, Sortica VA, Boff T, Brammer SP, Federizzi LC. Validation of molecular markers associated to leaf rust resistance genes in wheat. Pesq Agropec Bras. 2008;43:1357–1363. [Google Scholar]
- Dakouri A, McCallum BD, Walichnowski AZ, Cloutier S. Fine-mapping of the leaf rust Lr34 locus in Triticum aestivum (L.) and characterization of large germplasm collections support the ABC transporter as essential for gene function. Theor Appl Genet. 2010;121:373–384. doi: 10.1007/s00122-010-1316-7. [DOI] [PubMed] [Google Scholar]
- Das MK, Rajaram S, Mundt CC, Kronstad WE, Singh RP. Associations and genetics of three components of slow-rusting in leaf rust in wheat. Euphytica. 1993;68:99–109. doi: 10.1007/BF00024159. [DOI] [Google Scholar]
- Da-Silva PR, Brammer SP, Guerra D, Milach SCK, Barcellos AL, Baggio MI. Monosomic and molecular mapping of adult plant leaf rust resistance genes in the Brazilian wheat cultivar Toropi. Genet Mol Res. 2012;11:2823–2834. doi: 10.4238/2012.August.24.7. [DOI] [PubMed] [Google Scholar]
- Dyck PL. Identification of the gene for adult-plant leaf rust resistance in Thatcher. Can J Plant Sci. 1979;59:499–501. doi: 10.4141/cjps79-078. [DOI] [Google Scholar]
- Dyck PL. The association of a gene for leaf rust resistance with the chromosome 7D suppressor of stem rust resistance in common wheat. Genome. 1987;29:467–469. doi: 10.1139/g87-081. [DOI] [Google Scholar]
- Dyck PL. Genetics of adult plant leaf rust resistance in ‘Chinese Spring’ and ‘Sturdy’ wheats. Crop Sci. 1991;31:309–311. doi: 10.2135/cropsci1991.0011183X003100020016x. [DOI] [Google Scholar]
- Dyck PL. Transfer of a gene for stem rust resistance from Triticum araraticum to hexaploid wheat. Genome. 1992;35:788–792. doi: 10.1139/g92-120. [DOI] [Google Scholar]
- Dyck PL, Samborski DJ, Anderson RG. Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties exchange and Frontana. Can J Genet Cytol. 1966;8:665–671. [Google Scholar]
- Dyck PL, Kerber ER, Aung T. An interchromosomal reciprocal translocation in wheat involving leaf rust resistance gene Lr34. Genome. 1994;37:556–559. doi: 10.1139/g94-079. [DOI] [PubMed] [Google Scholar]
- Eitas TK, Dangl JL. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet C, Messmer M, Schachermayr G, Keller B. Genetic and physical characterization of the Lr1 leaf rust resistance locus in wheat (Triticum aestivum L.) Mol Gen Genet. 1995;248:553–562. doi: 10.1007/BF02423451. [DOI] [PubMed] [Google Scholar]
- Feuillet C, Travella S, Stein N, Albar L, Nublat A, Keller B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. Proc Natl Acad Sci USA. 2003;100:15253–15258. doi: 10.1073/pnas.2435133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. Inheritance of pathogenicity in Melampsora lini. Phytopathology. 1942;32:653–669. [Google Scholar]
- German SE, Kolmer JA. Effect of gene Lr34 in the enhancement of resistance to leaf rust of wheat. Theor Appl Genet. 1992;84:97–105. doi: 10.1007/BF00223987. [DOI] [PubMed] [Google Scholar]
- Hanušová R, Hsam SLK, Bartoš P, Zeller FJ. Suppression of powdery mildew resistance gene Pm8 in Triticum aestivum L. (common wheat) cultivars carrying wheat-rye translocation T1BL·1RS. Heredity. 1996;77:383–387. doi: 10.1038/hdy.1996.157. [DOI] [Google Scholar]
- Herrera Foessel SA (2001) Evaluation of leaf rust (Puccinia triticina) and yellow rust (P. striiformis f. sp. tritici) resistance in 20 Swedish wheats and identification of leaf rust resistance genes. Examensarbete/Minor Field Studies No. 160, Swedish University of Agricultural Sciences
- Herrera-Foessel SA, Lagudah ES, Huerta-Espino J, Hayden MJ, Bariana HS, Singh D, Singh RP. New slow-rusting leaf rust and stripe rust resistance genes Lr67 and Yr46 in wheat are pleiotropic or closely linked. Theor Appl Genet. 2011;122:239–249. doi: 10.1007/s00122-010-1439-x. [DOI] [PubMed] [Google Scholar]
- Herrera-Foessel SA, Singh RP, Huerta-Espino J, Rosewarne GM, Periyannan SK, Viccars L, Calvo-Salazar V, Lan C, Lagudah ES. Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theor Appl Genet. 2012;124:1475–1486. doi: 10.1007/s00122-012-1802-1. [DOI] [PubMed] [Google Scholar]
- Hiebert CW, Thomas JB, McCallum BD, Humphreys DG, DePauw RM, Hayden MJ, Mago R, Schnippenkoetter W, Spielmeyer W. A new gene, Lr67, from the wheat accession PI250413 confers resistance to leaf rust at the adult plant stage. Theor Appl Genet. 2010;121:1083–1091. doi: 10.1007/s00122-010-1373-y. [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet. 2000;100:1121–1128. doi: 10.1007/s001220051395. [DOI] [Google Scholar]
- Huang L, Gill BS. An RGA-like marker detects all known Lr21 leaf rust-resistance gene family members in Aegilops tauschii and wheat. Theor Appl Genet. 2001;103:1007–1013. doi: 10.1007/s001220100701. [DOI] [Google Scholar]
- Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang Q, Kumaravadivel N, Bennett J, Khush GS. Pyramiding of bacterial blight resistance genes in rice: marker-assisted selection using RFLP and PCR. Theor Appl Genet. 1997;95:313–320. doi: 10.1007/s001220050565. [DOI] [Google Scholar]
- Huang L, Brooks SA, Li W, Fellers JP, Trick HN, Gill BS. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics. 2003;164:655–664. doi: 10.1093/genetics/164.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hysing SC, Singh RP, Huerta-Espino J, Merker A, Liljeroth E, Diaz O. Leaf rust (Puccinia triticina) resistance in wheat (Triticum aestivum) cultivars grown in Northern Europe 1992–2002. Hereditas. 2006;143:1–14. doi: 10.1111/j.2005.0018-0661.01917.x. [DOI] [PubMed] [Google Scholar]
- Kerber ER, Dyck PL. Transfer to hexaploid wheat of linked genes for adult-plant leaf rust and seedling stem rust resistance from an amphiploid of Aegilops speltoides × Triticummonococcum. Genome. 1990;33:530–537. doi: 10.1139/g90-079. [DOI] [Google Scholar]
- Kloppers FJ, Pretorius ZA. Effects of combinations amongst genes Lr13, Lr34 and Lr37 on components of resistance in wheat to leaf rust. Plant Pathol. 1997;46:737–750. doi: 10.1046/j.1365-3059.1997.d01-58.x. [DOI] [Google Scholar]
- Kolmer JA. Enhanced leaf rust resistance in wheat conditioned by resistance gene pairs with Lrl3. Euphytica. 1992;61:123–130. doi: 10.1007/BF00026802. [DOI] [Google Scholar]
- Kolmer JA. Genetics of leaf rust resistance in three western Canada spring wheats. Plant Dis. 1994;78:600–602. doi: 10.1094/PD-78-0600. [DOI] [Google Scholar]
- Kolmer JA. Genetics of resistance to wheat leaf rust. Annu Rev Phytopathol. 1996;34:435–455. doi: 10.1146/annurev.phyto.34.1.435. [DOI] [PubMed] [Google Scholar]
- Kolmer JA. Postulation of leaf rust resistance genes in selected soft red winter wheats. Crop Sci. 2003;43:1266–1274. doi: 10.2135/cropsci2003.1266. [DOI] [Google Scholar]
- Kolmer JA. Tracking wheat rust on a continental scale. Curr Opin Plant Biol. 2005;8:441–449. doi: 10.1016/j.pbi.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Kolmer JA, Singh RP, Garvin DF, Viccars L, William HM, Huerta-Espino J, Ogbonnaya FC, Raman H, Orford S, Bariana ES, Lagudah ES. Analysis of the Lr34/Yr18 rust resistance region in wheat germplasm. Crop Sci. 2008;48:1841–1852. doi: 10.2135/cropsci2007.08.0474. [DOI] [Google Scholar]
- Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guedira G, Selter LL, Keller B. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet. 2009;119:889–898. doi: 10.1007/s00122-009-1097-z. [DOI] [PubMed] [Google Scholar]
- Li ZF, Xia XC, He ZH, Li X, Zhang LJ, Wang HY, Meng QF, Yang WX, Li GQ, Liu DQ. Seedling and slow rusting resistance to leaf rust in Chinese wheat cultivars. Plant Dis. 2010;94:45–53. doi: 10.1094/PDIS-94-1-0045. [DOI] [PubMed] [Google Scholar]
- Liu J, Liu D, Tao W, Li W, Wang S, Chen P, Cheng S, Gao D. Molecular marker-facilitated pyramiding of different genes for powdery mildew resistance in wheat. Plant Breed. 2000;119:21–24. doi: 10.1046/j.1439-0523.2000.00431.x. [DOI] [Google Scholar]
- Loegering WQ, McIntosh RA, Burton CH. Computer analysis of disease data to derive hypothetical genotypes for reaction of host varieties to pathogens. Can J Genet Cytol. 1971;13:742–748. [Google Scholar]
- Long DL, Kolmer JA. A North American system of nomenclature for Pucciniarecondita f. sp. tritici. Phytopathology. 1989;79:525–529. doi: 10.1094/Phyto-79-525. [DOI] [Google Scholar]
- Loutre C, Wicker T, Travella S, Galli P, Scofield S, Fahima T, Feuillet C, Keller B. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 2009;60:1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- McCallum BD, Fetch T, Chong J. Cereal rust control in Canada. Aust J Agric Res. 2007;58:639–647. doi: 10.1071/AR06145. [DOI] [Google Scholar]
- McCallum BD, Seto-Goh PL, Xue AG. Physiological specialization of Puccinia triticina in Canada in 2007. Can J Plant Pathol. 2010;32:229–236. doi: 10.1080/07060661.2010.484225. [DOI] [Google Scholar]
- McCallum BD, Hiebert C, Huerta-Espino J, Cloutier S (2012) Wheat leaf rust. In: Sharma I (ed) Disease resistance in wheat, chapter 3, 322 p. CAB International, Wallingford, pp 33–62
- McIntosh RA. Close genetic linkage of genes conferring adult-plant resistance to leaf rust and stripe rust in wheat. Plant Pathol. 1992;41:523–527. doi: 10.1111/j.1365-3059.1992.tb02450.x. [DOI] [Google Scholar]
- McIntosh RA, Baker EP. Chromosome location of mature plant leaf rust resistance in Chinese Spring. Aust J Biol Sci. 1966;19:943–944. [Google Scholar]
- McIntosh RA, Wellings CR, Park RF (1995) Wheat rusts: an atlas of resistance genes. Plant Breeding Institute, University of Sydney, CSIRO, Australia, Published by CSIRO, Australia in conjunction with Kluwer Academic Publishers, London
- McIntosh RA, Devos KM, Dubcovsky J, Rogers WJ, Morris CF, Appels R, Somers J, Anderson OA (2007) Catalogue of gene symbols for wheat: 2007 supplement: http://www.shigen.nig.ac.jp/wheat/komugi/genes/macgene/supplement2007.pdf
- McVey DV. Genes for rust resistance in international winter wheat nurseries XII through VXII. Crop Sci. 1992;32:891–895. doi: 10.2135/cropsci1992.0011183X003200040011x. [DOI] [Google Scholar]
- McVey DV, Long DL. Genes for leaf rust resistance in hard red winter wheat cultivars and parental lines. Crop Sci. 1993;33:1373–1381. doi: 10.2135/cropsci1993.0011183X003300060049x. [DOI] [Google Scholar]
- Mebrate SA, Dehne HW, Pillen K, Oerke EC. Postulation of seedling leaf rust resistance genes in selected Ethiopian and German bread wheat cultivars. Crop Sci. 2008;48:507–516. doi: 10.2135/cropsci2007.03.0173. [DOI] [Google Scholar]
- Oelke LM, Kolmer JA. Characterization of leaf rust resistance in hard red spring wheat cultivars. Plant Dis. 2004;88:1127–1133. doi: 10.1094/PDIS.2004.88.10.1127. [DOI] [PubMed] [Google Scholar]
- Ohm HW, Shaner GE. Three components of slow leaf-rusting at different growth stages in wheat. Phytopathology. 1976;66:1356–1360. doi: 10.1094/Phyto-66-1356. [DOI] [Google Scholar]
- Park RF, McIntosh RA. Studies of single gene adult plant resistances to Puccinia recondita f. sp. tritici in wheat. N Z J Crop Hort Sci. 1994;22:151–158. doi: 10.1080/01140671.1994.9513819. [DOI] [Google Scholar]
- Park RF, Goyeau H, Felsenstein FG, Bartos P, Zeller FJ. Regional phenotypic diversity of Puccinia triticina and wheat host resistance in Western Europe, 1995. Euphytica. 2001;122:113–127. doi: 10.1023/A:1012603500686. [DOI] [Google Scholar]
- Peterson RF, Campbell AB, Hannah AE. A diagrammatic scale for estimating rust intensity of leaves and stems of cereals. Can Res Sec C. 1948;26:496–500. doi: 10.1139/cjr48c-033. [DOI] [Google Scholar]
- Pink DAC. Strategies using genes for non-durable disease resistance. Euphytica. 2002;124:227–236. doi: 10.1023/A:1015638718242. [DOI] [Google Scholar]
- Pretorius ZA, Wilcoxson RD, Long DL, Schafer JF. Detecting leaf rust resistance gene Lr13 in seedlings. Plant Dis. 1984;68:585–586. [Google Scholar]
- Roelfs AP. Resistance to leaf rust and stem rust in wheat. In: Simmonds NW, Rajaram S, editors. Breeding strategies for resistance to the rusts of wheat. Mexico, DF: CIMMYT; 1988. pp. 10–22. [Google Scholar]
- Roelfs AP, Singh RP (1992) In: Hettel GP (ed) Rust diseases of wheat: concepts and methods of disease management. CIMMYT, Mexico, DF
- Roelfs AP, Hughes ME, Long DL (2000) Rust resistance genes in wheat lines and cultivars. U.S. Dep. Agric. Res. Serv. Cereal Dis. Lab. Online, Publication CDL–EP #006. Accessed 7 Nov 2002. http://www.ars.usda.gov/Main/docs.htm?docid=10103
- Rowland GC, Kerber ER. Telocentric mapping in hexaploid wheat of genes for leaf rust resistance and other characters derived from Aegilops squarrosa. Can J Genet Cytol. 1974;16:137–144. [Google Scholar]
- Samborski DJ (1985) Wheat leaf rust. In: Roelfs AP, Bushnell WR (eds) The cereal rusts. Vol. II; diseases, distribution, epidemiology, and control. Academic Press, Orlando, pp 39–59
- Samborski DJ, Dyck PL. Inheritance of virulence in wheat leaf rust on the standard differential wheat varieties. Can J Cytol. 1968;10:24–32. [Google Scholar]
- Sawhney RN. The role of Lr34 in imparting durable resistance to wheat leaf rust through gene interaction. Euphytica. 1992;61:9–12. doi: 10.1007/BF00035541. [DOI] [Google Scholar]
- Schachermayr G, Feuillet C, Keller B. Molecular markers for the detection of the wheat leaf rust resistance gene Lr10 in diverse genetic backgrounds. Mol Breed. 1997;3:65–74. doi: 10.1023/A:1009619905909. [DOI] [Google Scholar]
- Sela H, Spiridon NS, Petrescu AJ, Akerman M, Mandel-Gutfreund Y, Nevo E, Loutre C, Keller B, Schulman AH, Fahima T. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol Plant Pathol. 2012;13:276–287. doi: 10.1111/j.1364-3703.2011.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha AH, Lin XH, Huang JB, Zhang DP. Analysis of DNA methylation related to rice adult plant resistance to bacterial blight based on methylation-sensitive AFLP (MSAP) analysis. Mol Genet Genomics. 2005;273:484–490. doi: 10.1007/s00438-005-1148-3. [DOI] [PubMed] [Google Scholar]
- Singh RP. Association between gene Lr34 for leaf rust resistance and leaf tip necrosis in wheat. Crop Sci. 1992;32:874–878. doi: 10.2135/cropsci1992.0011183X003200040008x. [DOI] [Google Scholar]
- Singh RP. Genetic association of leaf rust resistance gene Lr34 with adult plant resistance to stripe rust in bread wheat. Phytopathology. 1992;82:835–838. doi: 10.1094/Phyto-82-835. [DOI] [Google Scholar]
- Singh RP. Genetic association of gene Bdv1 for tolerance to barley yellow dwarf virus with genes Lr34 and Yr18 for adult plant resistance to rusts in bread wheat. Plant Dis. 1993;77:1103–1106. doi: 10.1094/PD-77-1103. [DOI] [Google Scholar]
- Singh RP. Resistance to leaf rust in 26 Mexican wheat cultivars. Crop Sci. 1993;33:633–637. doi: 10.2135/cropsci1993.0011183X003300030041x. [DOI] [Google Scholar]
- Singh RP, Gupta AK. Genes for leaf rust resistance in Indian and Pakistani wheats tested with Mexican pathotypes of Puccinia recondita f. sp. tritici. Euphytica. 1991;57:27–36. [Google Scholar]
- Singh RP, Huerta-Espino J. Effect of leaf rust resistance gene Lr34 on grain yield and agronomic traits of spring wheat. Crop Sci. 1997;37:390–395. doi: 10.2135/cropsci1997.0011183X003700020014x. [DOI] [Google Scholar]
- Singh RP, Rajaram S. Resistance to Puccinia recondita f. sp. tritici in 50 Mexican bread wheat cultivars. Crop Sci. 1991;31:1472–1479. doi: 10.2135/cropsci1991.0011183X003100060016x. [DOI] [Google Scholar]
- Singh RP, Mujeeb-Kazi A, Huerta-Espino J. Lr46: a gene conferring slow rusting resistance to leaf rust in wheat. Phytopathology. 1998;88:890–894. doi: 10.1094/PHYTO.1998.88.9.890. [DOI] [PubMed] [Google Scholar]
- Singh RP, Chen WQ, He ZH. Leaf rust resistance of spring, facultative, and winter wheat cultivars from China. Plant Dis. 1999;83:644–651. doi: 10.1094/PDIS.1999.83.7.644. [DOI] [PubMed] [Google Scholar]
- Singh RP, Huerta-Espino J, Rajaram S. Achieving near-immunity to leaf and stripe rusts in wheat by combining slow rusting resistance genes. Acta Phytopathol Hung. 2000;35:133–139. [Google Scholar]
- Singh D, Park RF, McIntosh RA. Postulation of leaf (brown) rust resistance genes in 70 wheat cultivars grown in the United Kingdom. Euphytica. 2001;120:205–218. doi: 10.1023/A:1017578217829. [DOI] [Google Scholar]
- Singh D, Park RF, McIntosh RA. Characterisation of wheat leaf rust resistance gene Lr34 in Australian wheats using components of partial resistance and molecular markers. Aust J Agric Res. 2007;58:1106–1114. doi: 10.1071/AR07002. [DOI] [Google Scholar]
- Šliková S, Gregová E, Bartoš P, Hanzalová A, Hudcovicová M, Kraic J. Development of wheat genotypes possessing a combination of leaf rust resistance genes Lr19 and Lr24. Plant Soil Environ. 2004;50:43–438. [Google Scholar]
- Spielmeyer W, McIntosh RA, Kolmer J, Lagudah ES. Powdery mildew resistance and Lr34/Yr18 genes for durable resistance to leaf and stripe rust cosegregate at a locus on the short arm of chromosome 7D of wheat. Theor Appl Genet. 2005;111:731–735. doi: 10.1007/s00122-005-2058-9. [DOI] [PubMed] [Google Scholar]
- Statler GD. Probable genes for leaf rust resistance in several hard red spring wheats. Crop Sci. 1984;24:883–886. doi: 10.2135/cropsci1984.0011183X002400050013x. [DOI] [Google Scholar]
- Tanksley SD, Young ND, Paterson AH, Bonierbale MW. RFLP mapping in plant breeding: new tools for an old science. Bio/Technol. 1989;7:257–264. doi: 10.1038/nbt0389-257. [DOI] [Google Scholar]
- Vanzetti LS, Campos P, Demichelis M, Lombardo LA, Aurelia PR, Vaschetto LM, Bainotti CT, Helguera M (2011) Identification of leaf rust resistance genes in selected Argentinean bread wheat cultivars by gene postulation and molecular markers. Elec J Biotechnol. doi:10.2225/vol14-issue3-fulltext-14
- Wamishe YA, Milus EA. Seedling resistance genes to leaf rust in soft red winter wheat. Plant Dis. 2004;88:136–146. doi: 10.1094/PDIS.2004.88.2.136. [DOI] [PubMed] [Google Scholar]
- Winzeler M, Mesterházy A, Park RF, Bartos P, Csosz M, Goyeau H, Ittu M, Jones E, Loschenberger F, Manninger K, Pasquini M, Richter K, Rubiales D, Schachermayer G, Strzembicka A, Trottet M, Unger O, Vida G, Walther U. Resistance of European winter wheat germplasm to leaf rust. Agronomie. 2000;20:783–792. doi: 10.1051/agro:2000175. [DOI] [Google Scholar]
- Wu L, He H, Yang FP, Xia XC (2012) Characterization of adult plant resistance gene in improved wheat varieties and landraces from China. In: Proceedings of the 13th international cereal rusts and powdery Mildew, Beijing, China, p 114
- Zoldan SM, Barcellos AL. Postulation of genes (Lr) for resistance to leaf rust in Brazilian wheat cultivars. Fitopatol Bras. 2002;27:517–524. doi: 10.1590/S0100-41582002000500012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.