Abstract
Transposable elements (TEs) are DNA fragments that have the ability to move from one chromosomal location to another. The insertion of TEs into gene-rich regions often affects changes in the expression of neighboring genes. Miniature Ping (mPing) is an active miniature inverted-repeat TE discovered in the rice genome. It has been found to show exceptionally active transposition in a few japonica rice varieties, including Gimbozu, where mPing insertion rendered adjacent genes stress-inducible. In the Gimbozu population, it is highly possible that several genes with modified expression profiles are segregating due to the de novo mPing insertions. In our study, we utilized a screening system for detecting de novo mPing insertions in the upstream region of target genes and evaluated the effect of mPing on the stress response of the target genes. Screening for 17 targeted genes revealed five genes with the mPing insertion in their promoters. In most cases, the alteration of gene expression was observed under stress conditions, and there was no change in the expression levels of those five genes under normal conditions. These results indicate that the mPing insertion can be used as a genetic tool to modify an expression pattern of a target gene under stress conditions without changing the expression profiles of those under natural conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s11032-013-9885-1) contains supplementary material, which is available to authorized users.
Keywords: Rice, Transposable element, Transposition, mPing, Gene expression, Reverse genetics
Introduction
Transposable elements (TEs) are DNA fragments that have the ability to move from one chromosomal location to another. Recent genome projects on several species have revealed that TEs are widespread in all organisms, from bacteria to humans, and constitute a significant portion of eukaryotic genomes (Kazazian 2004). They represent over 60 % of the human genome (de Koning et al. 2011), 35 % of the rice genome (Turcotte et al. 2001), and over 85 % of the maize genome (Schnable et al. 2009). Many kinds of TEs have been identified to date, and some of these have increased their copy number up to several thousand copies per genome (Lander et al. 2001; Khan et al. 2011).
The insertion of TEs into gene-rich regions often changes the expression level of neighboring genes through alteration of the DNA sequence. TE insertion can also sometimes disrupt the original cis-elements and occasionally triggers epigenetic silencing, leading to inhibited interaction between the cis-element and the trans-element (Corces and Geyer 1991; Martin et al. 2009). On the other hand, TEs that exhibit promoter activity are able to transcribe the adjacent intergenic sequence, and TE-derived sequences can also provide new regulatory elements for neighboring genes, resulting in these genes showing novel expression patterns (Kloeckener-Gruissem et al. 1992). These TE-derived genetic changes have been broadly used as a tool in reverse genetics. Consequently, TE tagging mutant panels have been established in rice with endogenous retrotranspon Tos17 (Miyao et al. 2003) and exogenous elements, such as Ds and T-DNA (Kolesnik et al. 2004; Jeon et al. 2000).
Miniature Ping (mPing) is the active miniature inverted-repeat transposable element (MITE) discovered in the rice genome (Jiang et al. 2003; Kikuchi et al. 2003; Nakazaki et al. 2003). It is a 430-bp DNA fragment that contains 15-bp terminal inverted repeats and it introduces target site duplications of TAA or TTA, typical characteristics of a tourist-like MITE family. Transposase, which is necessary for the transposition of mPing, is provided from the autonomous elements Ping and Pong. They contain open reading frame (ORF) 1 and ORF2, both of which are needed for the transposition of mPing. ORF1 encodes a protein similar to the DNA-binding domain of the myb transcription factor and ORF2 encodes transposase (Jiang et al. 2003; Yang et al. 2007). mPing, a deletion derivative of Ping, contains the promoter region of ORF1.
The exceptionally active transposition of mPing has been observed in a few japonica rice varieties, including Gimbozu (Naito et al. 2006). In the Gimbozu genome, the copy number of mPing exceeds 1,000 copies, while that of Nipponbare is just 51 copies. Furthermore, mPing increases its copy number by approximately 40 per plant per generation (Naito et al. 2006). A detailed investigation of de novo mPing insertion sites revealed that mPing preferentially inserted in the 1- to- 500-bp upstream regions of genes. The results of this study showed that in fact 9.4 % of de novo insertions of mPing were distributed in the 500-bp upstream regions of genes (Naito et al. 2009). These authors also reported that mPing contained putative stress-responsive cis-elements in its sequence and rendered adjacent genes stress-inducible (Naito et al. 2009).
In the Gimbozu population, it is highly possible that several genes with modified expression profiles caused by the mPing insertion are segregating. Therefore, application of the appropriate evaluation system should allow researchers to use mPing as a genetic tool to modify the expression of a target gene. Several other active transposable elements of rice are suitable tools for gene tagging systems because of their destructive effects on gene function (Miyao et al. 2003; Tsugane et al. 2006; Takagi et al. 2007; Terada et al. 2007). mPing could also be used for the fine-tuning of gene expression. The unique nature of mPing makes it well suited to enrich the genetic resources for conventional rice breeding. In this study, we designed a screening system that detects mPing insertion in upstream regions of the target genes and we observed the screening efficiency. We also evaluated the effect of mPing insertion on the stress response through gene expression analyses under stress conditions.
Materials and methods
Plant material
Oryza sativa ssp. japonica cv. Gimbozu was used. In 2010, 11,520 Gimbozu plants were grown in the experimental field of Kyoto University to establish a DNA pool and a seed pool.
Construction of DNA pool and seed pool
Approximately 2-cm-long leaf blades were harvested from each plant and divided into eight-plant bulked samples. The bulked samples were crushed using a Multi-Beads Shocker (Yasui Kikai, Osaka, Japan) in extraction buffer (100 mM Tris–HCl pH 8.0, 1 M KCl, and 10 mM EDTA; Yamamoto et al. 2007a). After centrifugation, the supernatant was recovered, and an equal volume of isopropyl alcohol was added. Precipitated DNA was recovered by centrifugation, and the pellet was first washed with 75 % ethanol, then dried and dissolved in 1/10 TE (10 mM Tris–HCl pH 8.0 and 1 mM EDTA). A total of 1,440 DNA bulked samples were diluted to 25 ng/μl per sample, and the samples were applied to fifteen 96-well plates for PCR analysis using gene-specific primers.
At plant maturation, we harvested a single panicle from all of the 11,520 plants. Eight panicles of the plants (corresponding to an eight-plant DNA bulked sample) were stored in a bag. We then made 1,440 seed bags (seed pool) corresponding to each of the 1,440 DNA bulked samples (DNA pool).
PCR amplification
PCR primers were designed to amplify the approximately 500-bp target regions [Electronic Supplementary Material (ESM) Table 1]. PCR analyses were performed in a total volume of 5 μl, with 25 ng of DNA. DNA was mixed with 2.5 μl of 2× GoTaq® Green Master Mix (Promega, Madison, WI, USA), 0.5 μl of 2.5 μM primers, and 0.25 μl dimethyl sulfoxide. PCR steps for amplification were as follows: heat denaturation at 94 °C for 3 min, 40 cycles of denaturation at 94 °C for 30 s, annealing at 57 °C for 45 s, and extension at 72 °C for 1.5 min. To select the bulked samples that harbor mPing in the upstream region of the target gene, PCR products were separated by electrophoresis in a 0.8 % agarose gel.
Selection of mPing insertion mutants with DNA pool and seed pool
The DNA pool prepared from 11,520 Gimbozu plants was used for the primary screening of mPing-inserted alleles that have a new mPing insertion in the 1- to 500-bp upstream region from the transcription start site (TSS) of the target genes (Fig. 1). mPing-inserted alleles were detected based on the length difference: PCR products with the insertion were approximately 430 bp longer than the original promoter region. Before the screening, we confirmed that the eight-plant bulk sample was small enough to detect a single heterozygous plant for mPing-inserted allele through PCR (data not shown). Among the 1,440 DNA bulked samples, 17 were omitted from PCR analysis due to the low quality of the DNA.
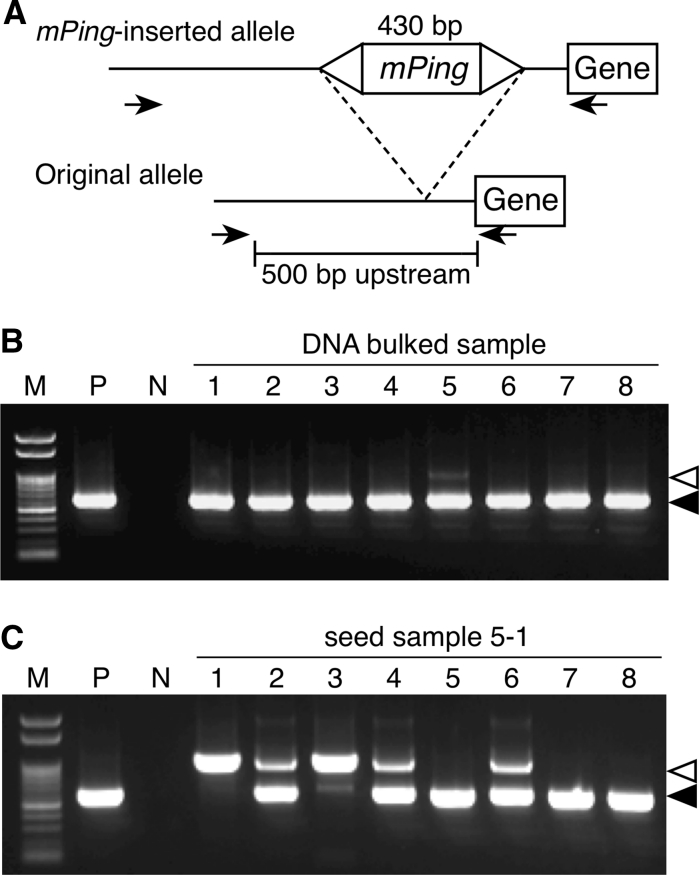
Fig. 1.
Screening scheme of mPing-inserted alleles. A total of 11,520 Gimbozu plants were cultivated and self-fertilized. A single DNA bulked sample and a single seed bag were compiled from eight plants. A set of DNA bulked samples (DNA pool) was used for the primary screening by PCR. PCR was performed using target-specific primers (see Fig. 2a, b). Progenies of eight plants in a seed bag corresponding to a DNA bulked sample with mPing insertion were cultivated, and DNA was then extracted for the secondary screening with target-specific PCR (see Fig. 2c)
In the Gimbozu genome, the number of new insertions of mPing is around 40 per plant per generation (Naito et al. 2006). Based on previous results, we assumed that 10 % of new mPing insertions would be located at the 5′-flanking regions of genes, especially at up to the 500-bp upstream regions from TSS (Naito et al. 2009). There are ≤30,000 protein coding genes (Itoh et al. 2007). Thus, 4/30,000 is a rough estimation of the probability that a gene newly acquires the mPing-inserted promoter per plant per generation. Then, (1–4/30,000)11,520 ≈ 0.22 is the probability that a gene is not harboring the mPing-inserted promoter in the 11,520 plants. This calculation indicates that we could expect at least one mPing-inserted promoter in about 78 % of the rice gene plants in a Gimbozu population of this size.
As shown in Table 1, we selected the stress-related genes as the target genes whose overexpression improved stress tolerance, except salT and wsi18. The upstream region (−500 bp from TSS) of these genes was amplified using specific primer pairs. When we found a DNA bulked sample that exhibited a fragment that was approximately 430 bp longer than the original fragment, we subjected the seed bag corresponding to this DNA bulked sample to further screening. We cultivated 64 plants (8 plants × 8 lines) and examined their genotypes for an mPing-inserted allele. A plant homozygous for an mPing-inserted allele and a plant homozygous for a non-inserted allele were selected as a mPing+ (plus) plant and an mPing− (minus) plant, respectively. Selfed seeds harvested from the mPing+ and mPing− plants were used to raise mPing-inserted lines and non-inserted lines, respectively. The effects of the mPing insertion were evaluated by comparing an mPing-inserted line and a non-inserted line that originated from a single plant.
Table 1.
Target genes
| Gene | Gene function | Acquired abiotic stress tolerance by overexpression | Reference |
|---|---|---|---|
| OsDREB1A | Transcription factor | Salt, drought, and cold tolerance | Ito et al. (2006) |
| DREB1D | Transcription factor | Salt and cold tolerance | Zhang et al. (2009) |
| OsDREB1F | Transcription factor | Salt, drought, and cold tolerance | Wang et al. (2008) |
| ZFP252 | Zinc finger, C2H2-type domain containing protein | Salt and drought tolerance | Xu et al. (2008) |
| ZFP182 | Zinc finger, C2H2-type domain containing protein | Salt tolerance | Huang et al. (2007) |
| SNAC1 | Transcription factor | Salt and drought tolerance | Hu et al. (2006) |
| OsNAC6 | Transcription factor | Salt and drought tolerance | Nakashima et al. (2007) |
| ONAC045 | Transcription factor | Salt and drought tolerance | Zheng et al. (2009) |
| OsLEA3-1 | Late embryogenesis abundant protein | Drought tolerance | Xiao et al. (2007) |
| MYBS3 | Transcription factor | Cold tolerance | Su et al. (2010) |
| OsNHX1 | Na+/H+ antiporter | Salt tolerance | Fukuda et al. (2004) |
| OsGS2 | Glutamine synthetase | Salt tolerance | Hoshida et al. (2000) |
| OsCDPK7 | Calcium-dependent protein kinase | Salt, drought, and cold tolerance | Saijo et al. (2000) |
| SalT | Salt-induced protein | No data | |
| wsi18 | Late embryogenesis abundant protein | No data | |
| OsWRKY11 | Transcription factor | Drought and heat tolerance | Wu et al. (2009) |
| OsbZIP | Transcription factor | Salt and drought tolerance | Xiang et al. (2008) |
Database search of the cis-elements in the upstream region of genes
A database search of the promoter for OsDREB1A, ZFP252, ONAC045, and OsCDPK7 was performed to look for core promoters [TATA box, pyrimidine patch (Y Patch), and regulatory element group (REG)] using the Plant Promoter Database (PPDB; Yamamoto and Obokata 2008). The function feature of the REG was searched by employing the Plant cis-acting Regulatory DNA Elements database (PLACE; Higo et al. 1999). The TATA box and Y Patch are orientation-sensitive factors, while REGs are orientation-insensitive factors that appear upstream of the TATA box (−20 to −400 bp; Yamamoto et al. 2007b). In this analysis, we searched for REGs located up to −800 bp from the TSS because there are no stress-responsive REGs in the region up to −400 bp from the TSS in all target genes.
Stress treatments
Seeds were sterilized with 1,000-fold diluted fungicide (Benlate®; Sumitomo Kagaku, Kogyo, Japan) for 24 h and then immersed in water at 25 °C in the dark. After 72 h of incubation, germinated seeds were cultivated in Kimura B solution in a greenhouse under natural daylight. Seedlings at the three-leaf stage were transferred to a growth chamber that was kept at 25 °C and grown for 2 days under a 14/10-h light/dark photo-cycle before treatment initiation. For cold stress, the seedlings were exposed to 4 °C in the dark for 2 h. For salt stress, the seedlings were transferred to a culture medium containing 250 mM NaCl for 24 h. All of the plants were flash frozen in liquid nitrogen immediately after the respective stress treatment and stored at −80 °C prior to RNA extraction.
Real-time PCR
Total RNA was extracted from the leaves of stress-treated seedlings using TriPure Isolation Reagent (Roche Diagnostics, Indianapolis, IN). The RNA was subsequently treated with Deoxyribonuclease (RT Grade) for Heat Stop (Nippon gene). The DNase-treated RNA was reverse-transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Roche Diagnostics). Real-time PCR was performed using a LightCycler® 1.5 (Roche Diagnostics) real-time instrument with LightCycler® FastStart DNA MasterPLUS SYBR Green I (Roche Diagnostics). Samples were amplified as follows: initial denaturation step at 95 °C for 10 min to activate polymerase, followed by 55 cycles of denaturation at 95 °C for 5 s, annealing at a specific temperature (ESM Table 1) for 8 s, and extension at 72 °C for 12 s. RUBQ (GenBank Accession No. AK121590) was used as an internal reference gene for calculating the relative transcript levels of the target genes. Expression levels of the target genes were determined using three biological replications. The relative expression levels of target genes were then determined in comparison with those of non-inserted lines under the control condition.
Results
Mutation screening of stress tolerance genes with the Gimbozu population
We selected as target genes those stress-related genes whose overexpression could induce high-stress tolerance in rice plants, except for salT and wsi18 (Table 1). mPing-inserted promoters were found in OsDREB1A, ZFP252, ONAC045, and OsCDPK7 (Fig. 2b). For both OsDREB1A and ZFP252, two mPing-inserted promoters were found from two different DNA bulked samples. Sequencing results revealed that two mPing-inserted promoters of OsDREB1A were identical in terms of the insertion site but the directions of the mPing insertion were opposite. This result clearly shows that two mPing-inserted promoters of OsDREB1A resulted from independent insertion events and that there was an insertion hot spot for mPing. Two mPing-inserted promoters of ZFP252 were identical in terms of regards site and direction of mPing; however, those may also have resulted from two independent insertion events that occurred at the same insertion hot spot. We used only one of the aforementioned lines for further analysis. The mPing-inserted promoters were segregated in seed samples (Fig. 2c), and we selected one mPing-inserted promoter homozygous plant and one mPing-non-inserted promoter homozygous plant from each segregating line. The progeny lines of those selected plants were named as shown in Table 2.
Fig. 2.
Detection of mPing-inserted alleles by PCR. a Schematic view of the positions of designed primer pairs. Arrows position of site-specific primers. b An example of the primary screening by PCR. mPing insertion was detected in the No. 5 DNA bulked sample. Filled arrowhead, open arrowhead original amplicon and the mPing-inserted amplicon, respectively. c Example of the secondary screening by PCR. Segregation of the mPing insertion allele in the selfed progeny of the No. 1 panicle of the No. 5 seed bag (5–1 plant). M 100-bp DNA ladder, P Nipponbare, N, standard D.W
Table 2.
Five paired lines originated from a single Gimbozu plant that differ in mPing insertion compared to the upstream region of five targeted genes
| Gene | Line name | Insertion site from TSS (bp) | Orientationa of mPing insertion |
|---|---|---|---|
| OsDREB1A | mfDREB+ | −224 | Forward |
| mfDREB− | No | ||
| OsDREB1A | mrDREB+ | −224 | Reverse |
| mrDREB− | No | ||
| ZFP252 | mZFP+ | −446 | Forward |
| mZFP− | No | ||
| ONAC045 | mNAC+ | −263 | Reverse |
| mNAC− | No | ||
| OsCDPK7 | mCDPK+ | −17 | Reverse |
| mCDPK− | No |
aForward means that the sense strand of Ping’s promoter sequence on mPing is forward relative to the target gene; reverse means that the sense strand of Ping’s promoter sequence on mPing is reverse relative to the target gene
Database search of mPing-inserted promoter
The cis-element interacting with the transcription factors is important for the regulation of gene expression. Sequence comparison between the original promoter and the mPing-inserted promoter will help predict and evaluate the effects of mPing insertion on the stress response of genes. Using the PPDB (Yamamoto and Obokata 2008), we conducted a database search of the promoter for OsDREB1A, ZFP252, ONAC045, and OsCDPK7 and estimated that the core promoters consists of the TATA box, Y Patch, and REG. The function feature of REG was searched using PLACE (Higo et al. 1999; Table 3; Fig. 3). In OsDREB1A, the TATA box, Y Patch, and REG were mined in the region −38, −10, and −486 bp from the TSS, respectively. The REG contains two etiolation-responsive cis-elements (ABRELATERD1 and ACGTATERD1), a calcium-responsive cis-element (ABRERATCAL), and three ABA-responsive cis-elements (CACGTGMOTIF, EBOXNNAPA, and MYCCONSENSUSAT). In mfDREB+ and mrDREB+, mPing insertion was located between the TATA box and the REG. In ZFP252, the TATA box (−31 bp) and Y Patch (−17, −39, and −82 bp) were mined. REG was mined at −701 bp from the TSS and contained a gibberellic acid-responsive cis-element (WRKY71OS), two etiolation-responsive cis-elements (ABRELATERD1 and ACGTATERD1), and two auxin- and salicylic acid-responsive cis-elements (ASD1MOTIFCAMV and HEXMOTIFTAH3H4). mZFP+ has a mPing insertion between the TATA box and the REG. In ONAC045, the TATA box, Y Patch, and a REG-containing photosynthesis-relative cis-element (SITEIIATCYTC) were mined at −33, −15, and −85 bp from the TSS, respectively. mNAC+ has an mPing insertion −263 bp from the TSS. In OsCDPK7, instead of the TATA box, a GA element was mined −17 bp from the TSS as a core promoter (Yamamoto et al. 2009). REGs were mined −222 and −460 bp from the TSS and contained two light-responsive cis-elements (CIACADIANLELHC and SITEIIATCYTC) and a calmodulin-related cis-element (CGCGBOXAT), respectively. In mCDPK+, the mPing insertion was located 1 bp downstream of the GA element.
Table 3.
Results of database search of promoter region
| Gene | Core promoter | Site from TSS (bp) | Name of cis-elements in place | Sequence | Function |
|---|---|---|---|---|---|
| OsDREB1A (AK105599) | TATA box | −27 to −38 | |||
| Y Patch | −1 to −10 | ||||
| REG (CCCACGTG) | −479 to −485 | ABRELATERD1 | ACGTG | Response to etiolation | |
| ABRERATCAL | MACGYGB | Response to calcium | |||
| ACGTATERD1 | ACGT | Response to etiolation | |||
| CACGTGMOTIF | CACGTG | G-box | |||
| EBOXBNNAPA | CANNTG | E-box | |||
| MYCCONSENSUSAT | CANNTG | Response to abscisic acid and cold | |||
| ZFP252 (AY219847) | TATA box | −23 to −31 | |||
| Y Patch | −8 to −17 | ||||
| −32 to −39 | |||||
| −74 to −82 | |||||
| REG (CACGTCAC) | −694 to −701 | WRKY71OS | TGAC | Response to gibberellic acid | |
| ABRELATERD1 | ACGTG | Response to etiolation | |||
| ASF1MOTIFCAMV | TGACG | Response to auxin and salicylic acid | |||
| GTGANTG10 | GTGA | Pollen-specific | |||
| ACGTATERD1 | ACGT | Response to etiolation | |||
| HEXMOTIFTAH3H4 | ACGTCA | Rice OBF1-homodimer-binding site | |||
| ONAC045 (AK067922) | TATA | −25 to −33 | |||
| Y Patch | −7 to −15 | ||||
| REG (TTGTGGGCTTC) | −75 to −85 | SITEIIATCYTC | TGGGCY | Relative to cytochrome, oxidative phosphorylation | |
| OsCDPK7 (AK061881) | GA | −10 to −17 | |||
| REG (CAAGCCCATCA) | −212 to −222 | CIACADIANLELHC | CAANNNNATC | Relative to circadian and response to light | |
| SITEIIATCYTC | TGGGCY | Relative to cytochrome, oxidative phosphorylation | |||
| REG (CTCGCGCGC) | −452 to −460 | CGCGBOXAT | VCGCGB | Response to calmodulin |
Fig. 3.
Distribution of putative core promoters in upstream regions of target genes. Y Patch Pyrimidine patch, REG regulatory element group ABA abscisic acid
Expression changes caused by mPing-inserted promoter under cold and salt stress conditions
To clarify the impact of mPing insertion on the cold and salt response of the neighboring gene expression, we monitored the transcription of OsDREB1A, ZFP252, ONAC045, and OsCDPK7 under cold and salt stress conditions. The expression levels of the target genes are shown in Fig. 4 as relative values to those of non-inserted lines under the control condition.
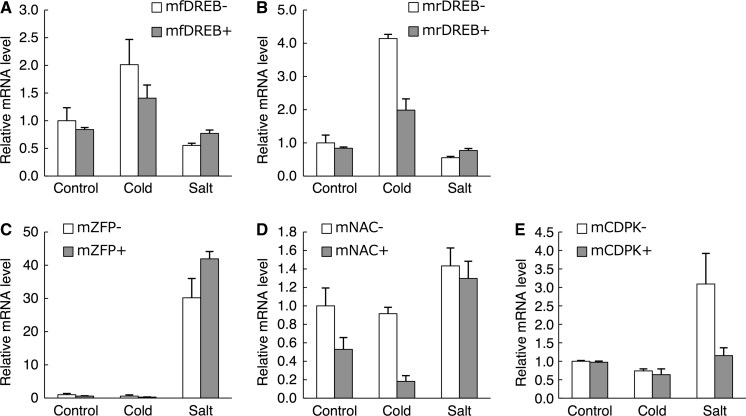
Fig. 4.
Real-time PCR analysis of OsDREB1A (a, b), ZFP252 (c), ONAC045 (d), and OsCDPK7 (e). Total RNA was extracted from mPing-inserted lines and non-inserted lines with no treatment (Control), 4 °C for 2 h (Cold), and 250 mM NaCl for 24 h (Salt). The expression levels of target genes were exhibited as relative values to those of non-inserted lines under the control condition. Data are shown as a the mean ± standard error of three replications
The expression level of OsDREB1A in mfDREB− and mrDREB− was upregulated under cold stress, but it did not change under salt stress (Fig. 4a, b). In mfDREB+ and mrDREB+, the expression level of OsDREB1A was also accelerated only under cold stress. Comparison of mfDREB+ and mrDREB+ revealed that the orientation of mPing did not affect the stress response of OsDREB1A. Under the normal condition, the expression level of OsDREB1A was not affected by the mPing insertion, irrespective of orientation. The database search indicated that mPing insertion between the TATA box and REG did not significantly affect gene expression under the non-stress condition. The REG of OsDREB1A contains a cold-responsive cis-element, such as CACGTGMOTIF and MYCCONSENSUSAT. The degree of the cold response of OsDREB1A in mfDREB+ and mrDREB+ was less than that in mfDREB− and mrDREB−. This ineffective cold-responsive cis-element was caused partly by the increased distance between the REG and TSS due to the mPing insertion. The loss of the REG effect could not be fully recovered by the cis-element within mPing.
In mZFPs, the expression level of ZFP252 was enhanced under salt treatment but not under cold treatment (Fig. 4c). In mZFP+, the expression level of ZFP252 under normal conditions was equal to that of mZFP− as the insertion of mPing did not change the position of the TATA box or the Y Patch (Fig. 3). In mZFP+, the expression level of ZFP252 was also increased only under salt stress, and its expression level was higher than that of mZFP−. The database search indicated that the REG of ZFP252 contained the auxin- and salicylic acid-responsive cis-element ASF1MOTIFCAMV. The auxin- and salicylic acid-responsive gene has been found to also respond to salt stress (Shim et al. 2003; Jain and Khurana 2009). In mZFP+, mPing insertion changes the distance between the REG and the TSS from −694 to −1,124 bp, but mPing insertion provides ASF1MOTIFCAMV as this cis-element is also located in mPing. This compensation of cis-element may one of the reasons why the salt-response in mZFP+ did not change.
In mNAC−, the expression level of ONAC045 was upregulated under salt stress but not under cold stress. With respect to the effects of mPing insertion, the expression level of ONAC045 did not change under the salt stress condition, but that of mNAC+ was lower than that of mNAC− under normal and cold stress conditions (Fig. 4d). As the database search could not mine stress-responsive cis-elements, there must be potential salt-responsive cis-elements in the region unaffected by mPing insertion.
The expression of OsCDPK7 was upregulated under the salt stress condition but not under that of cold stress. mCDPK+ exhibited an expression level equal to that of mCDPK-under normal and cold stress conditions, but its expression level under the salt stress condition decreased with mPing insertion (Fig. 4e). The mPing was inserted without disrupting the GA element and its insertion site is very near the TSS. The effect of the cis-element is usually affected by its orientation when it is located near the TSS. In mCDPK+, the direction of the cold-responsive cis-element on mPing is opposite to that of the TSS, thereby rendering OsCDPK7 cold-inducible. Since mPing insertion reduced the salt response of OsCDPK7, the function of an as yet unidentified salt-responsive cis-element was disturbed by the insertion.
Discussion
In earlier studies (Naito et al. 2006, 2009), we showed that mPing has a high mobility in rice cv. Gimbozu and that it preferentially inserts into the 5′-flanking region of genes that render adjacent genes stress-inducible. In the present study, we conducted a screening analysis of the mPing-inserted promoter with the aim of modifying the expression of stress-inducible genes.
Based on the number of genes in the rice genome and the de novo insertion frequency of mPing, we predicted that screening of 11,520 Gimbozu plants would enable us to find at least one mPing-inserted promoter in about 78 % of the rice genes. Among the 17 genes selected for targeting, we found five genes with an mPing-inserted promoter, which is equivalent to 29 % of the targeted genes. Thus, the number of new insertions of mPing was not 40 per plant per generation, but only ten per plant per generation. In this latter case, approximately 100,000 plants should be cultivated to gain an mPing-inserted promoter in 96 % of the genes. In our previous study, the rate of de novo insertion was estimated by multiplying the number of de novo insertions observed using the Transposon Display technique with 16 primer sets selected from 64 possible primer sets. This different technology could partly explain the large difference in the de novo insertion frequency of mPing between the current and previous studies. We detected two independent mPing insertion events at exactly the same site, indicating the presence of a hot spot targeted by mPing. The large difference in mPing insertion frequency among genes could be one of the main reasons for this inconsistency. Further studies should be conducted to clarify the nature of the hot spot targeted by mPing.
We expected that mPing insertion would render adjacent genes salt and/or cold stress inducible and would increase the response of stress-tolerance genes under stress conditions. The expression analysis of mZFP+, which has a mPing insertion at −446 bp from the TSS of ZFP252, promoted the upregulation of ZFP252 under the salt stress condition. With the exception of mZFP+, the mPing-inserted promoter reduced the original stress response of neighboring genes. Our database search revealed that mPing insertion relocated the stress-responsive core cis-elements, thereby weakening the interaction between trans-elements and cis-elements. These results indicate that it is difficult to strengthen the response of stress-responsive genes by stress-responsive cis-elements in mPing. However, they do clarify that the mPing insertion could make adjacent non-stress-responsive genes stress responsive by providing de novo stress-responsive cis-elements.
Analysis of cis-elements revealed that the mPing insertion did not disrupt core promoter elements such as the TATA box or GA element. In our previous study (Naito et al. 2009), mPing insertions made adjacent genes stress inducible even though their TATA boxes (Os01g0178500, Os02g0582900) were disrupted. Analysis with PPDB and PLACE shows that mPing has the TATA box and GA element in the forward strand and the Y Patch and CA element in the reverse strand (ESM Table 3). Therefore, mPing may act as a core promoter, even if its insertion disrupts the original TATA box.
Our results clearly show that the expression profile of alleles with an mPing-inserted promoter could be altered. mPing strengthens the expression of ZFP252 under salt stress conditions. Since the overexpression of ZFP252 exhibited high salt stress tolerance compared to the wild type (Xu et al. 2008), mZFP+ could be useful in salt tolerance breeding. In addition, some genes negatively regulate stress tolerance (Magnani et al. 2004; Jeon et al. 2010). It should be possible to gain stress-tolerant plants by screening negative-regulating genes that have an mPing insertion in their promoter. Furthermore, it is interesting to note that in most cases the mPing-inserted promoter did not affect the expression level under normal conditions. Transgenic plants constitutively expressing the stress tolerance gene grow well under stress conditions; however, a strong constitutive overexpression plant is likely to exhibit poor growth under normal conditions (Kasuga et al. 1999; Hsieh et al. 2002). The use of a stress-inducible promoter could prevent these problems (Nakashima et al. 2007). Thus, mPing-inserted promoters have the potential to improve stress tolerance without having negative effects on growth or productivity under normal conditions. In terms of Gimbozu, it is also of note that all of the mPing-inserted promoters originated from natural mutation. Thus, it is possible to combine these with a conventional breeding program without any restriction applied to living modified organisms. These advantages of the mPing-inserted promoter are useful in altering the gene expression profiles under several kinds of stress conditions. We look forward to screening the mPing-inserted promoter on a larger scale to extend the collection of genes with mPing-inserted promoters and thereby enable the further selection of genes with modified stress response and use.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from the Ministry of Education, Culture, Sports and Technology of Japan in the form of Grants-in-Aid for Scientific Research, 22380006.
References
- Corces V, Geyer P. Interactions of retrotransposons with the host genome: the case of the gypsy element of Drosophila. Trends Genet. 1991;7:86–90. doi: 10.1016/0168-9525(91)90277-W. [DOI] [PubMed] [Google Scholar]
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, Hirochika H, Tanaka Y. Function, intracellular localization and the importance in salt tolerance of a vacuolar Na(+)/H(+) antiporter from rice. Plant Cell Physiol. 2004;45:146–159. doi: 10.1093/pcp/pch014. [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida H, Tanaka Y, Hibino T, Hayashi Y, Tanaka A, Takabe T. Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol Biol. 2000;43:103–111. doi: 10.1023/A:1006408712416. [DOI] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiol. 2002;129:1086–1094. doi: 10.1104/pp.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Yang X, Wang M, Tang H, Ding L, Shen Y, Zhang H. A novel rice C2H2-type zinc finger protein lacking DLN-box/EAR-motif plays a role in salt tolerance. Biochim Biophys Acta. 2007;1769:220–227. doi: 10.1016/j.bbaexp.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Ito Y, Katsura K, Maruyama K, Taji T, Kobayashi M, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–153. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- Itoh T, Tanaka T, Barrero RA, et al. Curated genome annotation of Oryza sativa ssp. japonica and comparative genome analysis with Arabidopsis thaliana. Genome Res. 2007;17:175–183. doi: 10.1101/gr.5509507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Khurana JP. Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J. 2009;276:3148–3162. doi: 10.1111/j.1742-4658.2009.07033.x. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Lee S, Jung KH, Jun SH, Jeong DH, Lee J, Kim C, Jang S, Yang K, Nam J, An K, Han MJ, Sung RJ, Choi HS, Yu JH, Choi JH, Cho SY, Cha SS, Kim SI, An G. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, Kang NY, Novák O, Ku SJ, Cho C, Lee DJ, Lee EJ, Strnad M, Kim J. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J Biol Chem. 2010;285:23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Bao Z, Zhang X, Hirochika H, Eddy SR, McCouch SR, Wessler SR. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol. 1999;17:287–291. doi: 10.1038/7036. [DOI] [PubMed] [Google Scholar]
- Kazazian HH. Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Khan MF, Yadav BS, Ahmad K, Jaitly AK. Mapping and analysis of the LINE and SINE type of repetitive elements in rice. Bioinformation. 2011;7:276–279. doi: 10.6026/97320630007276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K, Terauchi K, Wada M, Hirano HY. The plant MITE mPing is mobilized in anther culture. Nature. 2003;421:167–170. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- Kloeckener-Gruissem B, Vogel J, Freeling M. The TATA box promoter region of maize Adh1 affects its organ-specific expression. EMBO J. 1992;11:157–166. doi: 10.1002/j.1460-2075.1992.tb05038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S. Establishing an efficient Ac/Ds tagging system in rice: large-scale analysis of Ds flanking sequences. Plant J. 2004;37:301–314. doi: 10.1046/j.1365-313X.2003.01948.x. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Magnani E, Sjölander K, Hake S. From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell. 2004;16:2265–2277. doi: 10.1105/tpc.104.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Troadec C, Boualem A, Rajab M, Fernandez R, Morin H, Pitrat M, Dogimont C, Bendahmane A. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:U1135–U1237. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H. Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell. 2003;15:1771–1780. doi: 10.1105/tpc.012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Cho E, Yang G, Campbell MA, Yano K, Okumoto Y, Tanisaka T, Wessler SR. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran L, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007;51:617–630. doi: 10.1111/j.1365-313X.2007.03168.x. [DOI] [PubMed] [Google Scholar]
- Nakazaki T, Okumoto Y, Horibata A, Yamahira S, Teraishi M, Nishida H, Inoue H, Tanisaka T. Mobilization of a transposon in the rice genome. Nature. 2003;421:170–172. doi: 10.1038/nature01219. [DOI] [PubMed] [Google Scholar]
- Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000;23:319–327. doi: 10.1046/j.1365-313x.2000.00787.x. [DOI] [PubMed] [Google Scholar]
- Schnable P, Ware D, Fulton R, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Shim I, Momose Y, Yamamoto A, Kim D, Usui K. Inhibition of catalase activity by oxidative stress and its relationship to salicylic acid accumulation in plants. Plant Growth Regul. 2003;39:285–292. doi: 10.1023/A:1022861312375. [DOI] [Google Scholar]
- Su C, Wang Y, Hsieh T, Lu C, Tseng T, Yu S. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010;153:145–158. doi: 10.1104/pp.110.153015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Ishikawa N, Maekawa M, Tsugane K, Iida S. Transposon display for active DNA transposons in rice. Genes Genet Syst. 2007;82:109–122. doi: 10.1266/ggs.82.109. [DOI] [PubMed] [Google Scholar]
- Terada R, Johzuka-Hisatomi Y, Saitoh M, Asao H, Iida S. Gene targeting by homologous recombination as a biotechnological tool for rice functional genomics. Plant Physiol. 2007;144:846–856. doi: 10.1104/pp.107.095992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugane K, Maekawa M, Takagi K, Takahara H, Qian Q, Eun CH, Iida S. An active DNA transposon nDart causing leaf variegation and mutable dwarfism and its related elements in rice. Plant J. 2006;45:46–57. doi: 10.1111/j.1365-313X.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- Turcotte K, Srinivasan S, Bureau T. Survey of transposable elements from rice genomic sequences. Plant J. 2001;25:169–179. doi: 10.1046/j.1365-313x.2001.00945.x. [DOI] [PubMed] [Google Scholar]
- Wang Q, Guan Y, Wu Y, Chen H, Chen F, Chu C. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- Wu X, Shiroto Y, Kishitani S, Ito Y, Toriyama K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009;28:21–30. doi: 10.1007/s00299-008-0614-x. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Tang N, Du H, Ye H, Xiong L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008;148:1938–1952. doi: 10.1104/pp.108.128199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 2007;115:35–46. doi: 10.1007/s00122-007-0538-9. [DOI] [PubMed] [Google Scholar]
- Xu D, Huang J, Guo S, Yang X, Bao Y, Tang H, Zhang H. Overexpression of a TFIIIA-type zinc finger protein gene ZFP252 enhances drought and salt tolerance in rice (Oryza sativa L.) FEBS Lett. 2008;582:1037–1043. doi: 10.1016/j.febslet.2008.02.052. [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Obokata J. ppdb: a plant promoter database. Nucleic Acids Res. 2008;36:D977–D981. doi: 10.1093/nar/gkm785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Kitano H, Matsuoka M, Ashikari M. Interaction of two recessive genes, hbd2 and hbd3, induces hybrid breakdown in rice. Theor Appl Genet. 2007;115:187–194. doi: 10.1007/s00122-007-0554-9. [DOI] [PubMed] [Google Scholar]
- Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, Seki M, Shinozaki K, Abe T. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:67. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto YY, Yoshitsugu T, Sakurai T, Seki M, Shinozaki K, Obokata J. Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 2009;60:350–362. doi: 10.1111/j.1365-313X.2009.03958.x. [DOI] [PubMed] [Google Scholar]
- Yang G, Zhang F, Hancock CN, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:10962–10967. doi: 10.1073/pnas.0702080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen C, Jin X, Xiong A, Peng R, Hong Y, Yao Q, Chen J. Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep. 2009;42:486–492. doi: 10.5483/BMBRep.2009.42.8.486. [DOI] [PubMed] [Google Scholar]
- Zheng X, Chen B, Lu G, Han B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem Biophys Res Commun. 2009;379:985–989. doi: 10.1016/j.bbrc.2008.12.163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.