Abstract
Over the last decade, aminoacyl-tRNA synthetases (AARSs) have emerged as a new class of regulatory proteins with widespread functions beyond their classic role in protein synthesis. The functional expansion concurs with the incorporation of new domains and motifs to AARSs and coincides with the emergence of the multi-synthetase complex (MSC) during the course of eukaryotic evolution. Notably, the new domains in AARSs are often found to be structurally disordered or to be linked to the enzyme cores via unstructured linkers. Further bioinformatic analysis performed here classifies the 20 human cytoplasmic AARSs into 3 groups based on their propensities for structural disorder. The analysis also suggests that, while the assembly of the MSC mainly involves ordered structural domains, structurally disordered regions play an important role in activating and expanding the regulatory functions of AARSs.
An ordered protein structure has a defined 3D architecture composed of secondary structurally elements, such as α-helices and β-strands, arranged in a certain manner. In contrast, disordered proteins or regions lack a well-defined structure and exist as dynamic conformational ensembles. Although structural disorders are common in various proteomes, their frequency increases with increasing complexity of the organisms. For example, Ward et al predicted that long (>30 residues) disordered segments occur in 2.0 % of archaeal, 4.2 % of eubacterial and 33.0 % of eukaryotic proteins (Ward, et al., 2004). Genome-scale bioinformatics analyses have shown that structural disorder is most common in proteins involved in signal transduction, transcription and translation regulation due to the ability of structural disorder to offer the malleability and adaptability required in signaling and regulation (Tantos, et al., 2012). A prominent example is the p53 protein, which interacts with many other proteins to carry out its signal transduction function. Most of these interactions are mediated by regions on p53 that are intrinsically disordered, for example, the C-terminus, which, upon interaction with different partners, adopts completely different structures (Oldfield, et al., 2008). In contrast, catalysis is often played by a globular protein, where a well-defined conformation or active site geometry is a pre-requisite for enzymatic function.
Aminoacyl-tRNA synthetases (AARSs) are multifunctional proteins possessing both enzymatic and regulatory functions. Members of the AARS family catalyze the first reaction in protein synthesis, that is, to ligate an amino acid to the 3’-end of its cognate tRNA by using the energy released from hydrolyzing ATP. Importantly, it is the AARS-catalyzed aminoacylation reaction that has established the rules of the genetic code by pairing each amino acid with a tRNA harboring the cognate anticodon trinucleotide. Therefore, tRNA synthetase is considered to be one of the most ancient protein families and is essential in all three domains of life (Woese, et al., 2000). While preserving this enzymatic role, eukaryotic AARSs have been shown to develop other functions during the course of evolution. Human cytoplasmic tRNA synthetases, in particular, regulate diverse functions in different pathways including angiogenesis, inflammation, development and tumorigenesis. Thus, AARSs have emerged as a new class of regulatory proteins with widespread functions beyond their classic role in protein synthesis. The goal of this paper is to propose that structural disorder plays an important role in expanding the ‘functionome’ of AARSs.
New domains of AARSs and the MSC
Comparing the protein sequences of eukaryotic cytoplasmic AARSs with their bacterial and archaeal counterparts, it is immediately obvious that eukaryotic AARSs are generally larger. The size increase is mainly due to the acquisition of new domains or motifs at either N- or C-terminus, and the acquisition continues during the evolution of higher eukaryotes (Guo, et al., 2010). Albeit some exception, these new domains are in general dispensable for the aminoacylation function of the synthetases and, instead, are intimately associated with developing regulatory functions of AARSs.
Also associated with the acquisition of new domains is the emergence of a multi-synthetase complex (MSC) in eukaryotes. A miniature MSC, composed of MetRS, GluRS and a nonsynthetase protein Arc1p, appears in basal eukaryotes such as Saccharomyces cerevisiae. From Drosophila to mammals, the MSC contains 9 AARS components (LysRS, ArgRS, GlnRS, AspRS, MetRS, IleRS, LeuRS and a bifunctional GluProRS) and 3 accessory proteins (p43/AIMP1, p38/AIMP2 and p18/AIMP3). A separate complex exists through the association of ValRS and elongation factor-1H (Motorin Yu, et al., 1987). The AARSs that are not contained in these two complexes are mostly freestanding. Formation of the MSC has been proposed to have a functional dualism: both facilitating protein synthesis by direct channeling of aminoacylated tRNA to the ribosome (Sivaram and Deutscher, 1990) and serving as a reservoir of various regulatory functions associated with its synthetase and non-synthetase components (Lee, et al., 2004; Ray, et al., 2007). Not surprisingly, acquisition of the new domains is also critical for the assembly of the MSC (Rho, et al., 1999).
New domains are often associated with or near structural disorders
Most of the structural information on AARSs has been derived from crystal structure analyses. From those analyses, it is clear that the new domains and motifs of AARSs are often disordered or linked to the conserved enzymatic core via a disordered linker. The first example is found in human TyrRS. Despite tremendous efforts, the protein could not be crystallized without chopping off a C-terminal extension added to TyrRS from insects to human. Structure of the enzyme core – mini-TyrRS – was solved at 1.18 Å, which marks the highest resolution achieved for a tRNA synthetase to this day (Yang, et al., 2002). The C-terminal extension itself, named EMAP-II-like domain for its high homology to a human cytokine – endothelial monocyte activating polypeptide II – that turns out to be a proteolytic product of MSC p43/AIMP1, can also be crystallized and exhibits a globular structure (Yang, et al., 2003). Therefore, a conformational flexibility might be introduced by the way that the EMAP-II-like domain is joined to the core enzyme that makes crystallization of the full-length protein difficult. Indeed, 22 residues (D343-I364) in between the enzyme core and the EMAP-II-like domain were disordered in the crystal structure of mini-TyrRS (Yang, et al., 2002).
TrpRS is the closest homolog of TyrRS and provides another example for a disordered linker in between the enzyme core and an appended domain. From fish to humans, TrpRS possesses a N-terminal extension named WHEP domain. Crystals were able to grow with the full-length human TrpRS but the helix-turn-helix WHEP domain was only resolved in one of two subunits of the dimeric protein structure (Yang, et al., 2003). The WHEP domain was asymmetrically resolved because of the half-of-the-sites binding of Trp-AMP in the same subunit to help engage the WHEP domain. However, even in the subunit where the WHEP domain was resolved, 21 residues (D61-E81) in the linker region between the WHEP domain and the enzyme core were disordered.
Structural disordering has also been observed within the appended domains of AARSs. Human LysRS contains a eukaryote-specific N-terminal extension. NMR studies in solution revealed that the extension is mostly unstructured (Liu, et al., 2012). Consistently, LysRS can be crystallized only when this extension was removed (Guo, et al., 2008). As another example, the C-terminal vertebrate-specific UNE-S domain is completely disordered in the crystal structure of human SerRS (Xu, et al., 2012). The N-terminal WHEP domains of GlyRS and HisRS were also missing from the electron density maps, and at least for GlyRS, the WHEP domain is disordered regardless of the space group that the protein is crystallized and whether or not in complex with Gly-AMP or other ligands (Guo, et al., 2009).
Classification of human tRNA synthetases based on structural disorders
When a domain is disordered in the crystal structure, it is not immediately obvious whether the domain is intrinsically unstructured or the disorder is a result of a flexible linker. In this case, other complementary methods are needed for clarification. For example, although the WHEP domain in human HisRS is completely disordered in the crystal structure (Xu, et al., 2012), it exhibits the expected helix-turn-helix conformation in solution as detected by NMR (PDB 1X59). This suggests that the WHEP domain is essentially structured however adopts multiple conformations relative to the enzyme core and therefore is missing from the electron density map.
Theoretically, this ambiguity can also be clarified by bioinformatics approaches, as structural disorder is an intrinsic property encoded by the amino acid sequences of a protein. Amino acids can be grouped into three classes: order-promoting residues C, W, Y, I, F, V, L and probably H, T and N; disorder-promoting residues E, P, Q, S, R, K, M and probably D; and neural residues A and G (Radivojac, et al., 2007). Clearly, bulky hydrophobic residues promote order, whereas polar and charged residues have the opposite effect. Other distinct properties of disordered regions, such as richness in proline residues and low sequence complexity, have also been identified and incorporated into various algorithms to predict the presence of intrinsic disorders in proteins.
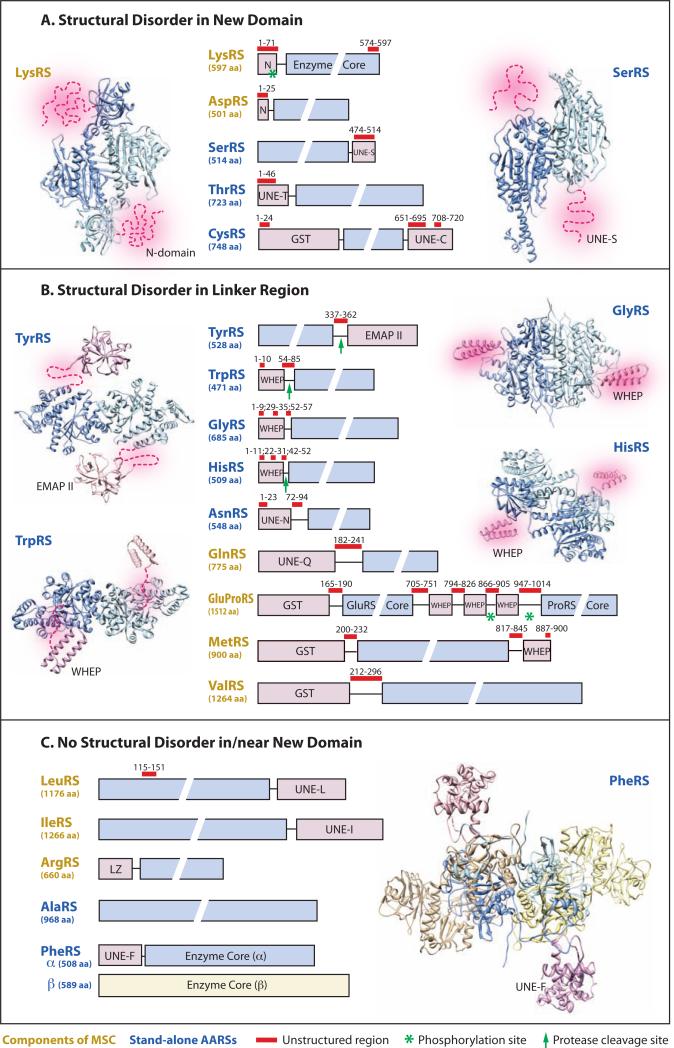
To systematically analyze structural disorders in human cytoplasmic tRNA synthetases, a prediction server metaPrDOS (http://prdos.hgc.jp/meta) based on a meta approach that integrates the results of seven different predication methods was used in this study (Ishida and Kinoshita, 2008). Remarkably, the results are consistent with what we have learned from crystal and NMR structural analysis (Figure 1). For example, with 5% false positive rate, the server predicted that 26 residues (A337-E362) in the linker region between the TyrRS enzyme core and the EMAP-II-like domain are intrinsically disordered. The server also predicted that 32 residues (A54-V85) in the linker region between the TrpRS enzyme core and the WHEP domain, the entire N-terminal extension (M1-V71) of LysRS and the C-terminal UNE-S domain (A474-A514) of SerRS to be structurally disordered. The WHEP domains of GlyRS and HisRS are predicted to be partially disordered, each having two internal stretches of ordered region corresponding to the two helices of the helix-turn-helix motif. Yet, not all new domains are disordered or near a disordered linker. Based on the prediction, the 20 cytoplasmic human tRNA synthetases can be categorized into three groups: A) disordered in new domains; B) disordered in linker regions; C) have no significant structural disorder associated with new domains or linkers (Figure 1). The classification and further analyses provide insights on the role of structural disorder in AARSs.
Figure 1.
Classification of human cytoplasmic AARSs based on their propensities for structural disorder. Significant structural disorders predicted by metaPrDOS are labeled as red bars on top of the domain structures of each AARS. A significant structural disorder refers to a stretch of disorder that is longer than 20 residues, or a group of shorter disordered stretches that are near each other, such as those seen in the WHEP domain of GlyRS and HisRS. Arrows and asterisks indicate protease cleavage and phosphorylation sites, respectively. Structures of human AARSs obtained by crystal structure or NMR analyses are shown to validate the structural disorder prediction results (LysRS: PDB 3BJU; SerRS: PDB 3VBB; TrpRS: PDB 1R6T; GlyRS: PDB 2ZT5; HisRS: PDB 4G84; PheRS: 3L4G). TyrRS structure is modeled based on the crystal structures of mini-TyrRS (PDB 1N3L) and the EMAP-II-like domain (PDB 1NTG). WHEP domains in GlyRS and HisRS, and in one subunit of TrpRS were disordered in their respective crystal structures and are also created by modeling.
Structural disorders in new domains
Five AARSs (i.e. LysRS, AspRS, SerRS, ThrRS and CysRS) exhibit significant structural disorders that are exclusively associated with their appended domains (Figure 1A). Majority of the new domains within this group, including the N-terminal extensions of LysRS, AspRS and ThrRS and the C-terminal extension of CysRS, started to appear in basal eukaryotes and were documented to exhibit tRNA binding or recognition capacities (Francin, et al., 2002). As exemplified by the N-terminal extension of LysRS (see below), such capacities are likely to be expanded or diverted to mediate interactions with other partners in higher eukaryotes.
Among the 5 AARSs of this group, LysRS and AspRS are components of MSC. While new domains are often involved in MSC assembly, the N-terminal extensions of LysRS and AspRS are the exceptions: both LysRS and AspRS interact with p38/AIMP2 in the MSC via their enzyme cores (Ofir-Birin, et al., 2013; Robinson, et al., 2000). The structurally disordered N-terminal extension of LysRS, instead, interacts with other protein partners both inside and outside the MSC. Inside the MSC, it interacts with p38 MAP kinase to become phosphorylated at Thr52, which triggers its release from MSC. The released LysRS is translocated to the plasma membrane to interact with the transmembrane region of 67LR laminin receptor to enhance laminin-induced cancer cell migration (Kim, et al., 2012). Although the exact binding site of 67LR on LysRS is not yet defined, the interaction involves the N-terminal region of LysRS that includes the N-terminal extension.
SerRS provided the first example illustrating that the regulatory function of a tRNA synthetase could be essential (Kawahara and Stainier, 2009). Three independent forward genetics studies in zebrafish established that SerRS plays a critical role in vascular development, and this role is independent of aminoacylation but dependent on the presence the UNE-S domain appended to vertebrate SerRS from fish to humans (Xu, et al., 2012). UNE-S contains a robust nuclear localization signal sequence that directs SerRS into the nucleus to regulate VEGF expression. Presumably, UNE-S interacts with a specific importin for mediating the nuclear import and the flexible conformation of the disordered UNE-S is key for this interaction. In fact, a point mutation (F383V) in SerRS linked to abnormal vasculature disrupts SerRS nuclear localization by sequestering the UNE-S domain (Xu, et al., 2012).
Structural disorders in the linker region
Significant structural disordering is also found in 10 other AARSs (i.e. GlyRS, HisRS, TyrRS, TrpRS, GlnRS, AsnRS, MetRS, ValRS and GluProRS). Compared to the unstructured N-terminal extension of LysRS and UNE-S domain of SerRS in the last group, the appended domains within this group have a preformed structure, but are linked to the enzyme cores via structurally disordered linkers of various lengths (Figure 1B).
The unstructured linkers are used as cleavage sites for proteolysis-based activation of the regulatory functions of TyrRS and TrpRS. Human TyrRS can be secreted and its regulatory functions outside the cell are self-inhibited until the EMAP-II-like domain is removed through proteolysis. While the EMAP-II-like domain itself exhibits cytokine functions similar to that of EMAP-II, mini-TyrRS manifests IL-8-like pro-inflammatory and pro-angiogenic cytokine activities through binding to chemokine receptor CXCR1 and CXCR2 (Vo, et al., 2011; Wakasugi and Schimmel, 1999). Apparently, linking the two functional entities with a flexible linker facilitates the activation due to the increased sensitivity of a flexible linker to protease cleavage.
TrpRS is highly upregulated by interferon-γ. The upregulation promotes both secretion and nuclear localization of TrpRS. When secreted, TrpRS possesses anti-angiogenic activities by inhibiting VE-cadherin to form cell-cell junction of endothelial cells (Tzima, et al., 2005). Similar to TyrRS, the anti-angiogenic activity of TrpRS is self-inhibitory until the WHEP domain is removed to expose the TrpRS active site for binding to VE-cadherin through its protruding Trp side chains (Zhou, et al., 2010). Here again, the activation of the anti-angiogenic activity of TrpRS is associated with proteolysis cleavage at a structurally disordered linker between the enzyme core and an appended domain.
The disordered linkers also harbor sites for posttranslational modification, as exemplified in the bi-functional GluProRS. The protein is linked together via 5 long disordered regions (36-, 47-, 33-, 40- and 68-residue, respectively) that joins the N-terminal GST domain, the GluRS core, the first, second and third WHEP domains and the C-terminal ProRS core (Figure 1B). Upon interferon-γ stimulation, the protein is phosphorylated at Ser886 between the second and the third WHEP domains, and at Ser999 between the third WHEP domain and the C-terminal ProRS. The phosphorylation events triggers the release of GluProRS from the MSC and the subsequent assembly of the “GAIT” (γ-interferon-activated inhibitor of translation) complex that includes GluProRS and three other proteins (ribosomal protein L13a, NS1-associated protein-1, and glyceraldehyde-3-phosphate dehydrogenase), resulting in translational silencing of VEGF expression (Arif, et al., 2009).
Intriguingly, GlnRS and MetRS are also components of the MSC, and ValRS exists in association with elongation factor-1H (Motorin Yu, et al., 1987). Each appended domain in GlnRS, MetRS and ValRS are linked to their respective enzyme core via a long and disordered linker (Figure 1B). Possibly, these linkers are also targeted for posttranslational modifications to trigger the dissociation of the synthetases from the MSC (or from the elongation factor) to participate in other regulatory processes.
A disordered region of 23 residues (R72-N94) in between the eukaryote-specific N-terminal domain (UNE-N) and the human AsnRS core is predicted by metaPrDOS. Similar structural disordering in a parasitic AsnRS (Brugia malayi) was revealed by NMR analysis (Crepin, et al., 2011). Interestingly, AsnRS is highly expressed in Brugia malayi, produced at least 10 times more than any other AARSs, and is secreted with the ability to bind IL-8 receptors, CXCR1 and CXCR2, and to activate their down-stream pathways (Ramirez, et al., 2006). The IL-8-like activity of the parasitic AsnRS resides in the UNE-N domain containing a β-hairpin-α-helix motif also seen in other CXC cytokines, such as IL-8 and SDF-1(Kron, et al., 2012). Although, in this case, the UNE-N domain does not need to be cleaved off from the synthetase core to exhibit cytokine activities, the flexibility of the disordered linker may still be important for ensuring the conformational independence of UNE-N. Interestingly, human AsnRS does not act on CXCR1 and CXCR2 (Ramirez, et al., 2006), but activates another chemokine receptor CCR3 (Howard, et al., 2002).
The flexible linkers are also implicated in diseases. The WHEP domain of HisRS is the main epitope recognized by HisRS autoantibodies in myositis-interstitial lung disease (Levine, et al., 2007). The disordered linker between the WHEP domain and the enzyme core harbors a granzyme B cleavage site (45LGPG48). The cleavage itself and/or the conformational freedom of the WHEP domain have been implicated in the initiation of the autoimmune disease (Levine, et al., 2007). GlyRS also has a WHEP domain that is flexibly linked to the enzyme core and its autoantibodies are associated with polymyositis and dermatomyositis complicated by interstitial lung disease (Stojanov, et al., 1996). In addition, GlyRS is implicated in Charcot-Marie-Tooth disease through genetic mutations. As demonstrated for one Charcot-Marie-Tooth disease-causing mutation (G526R), the conformation of the WHEP domain is dramatically affected in the mutant versus WT GlyRS (He, et al., 2011).
No structural disorder
The appended domains of PheRS, ArgRS, LeuRS and IleRS do not exhibit significant structural disorder (Figure 1C). And as expected, no significant structural disorder is found in AlaRS, which is the only AARS that did not acquire an appended domain during the course of eukaryotic evolution. The lack of structural disordering seems to correlate with fewer numbers of regulatory functions. In fact, only one prominent regulatory function has been reported for this group of AARSs, which is the critical role of LeuRS as a leucine sensor for the mTOR-signaling pathway. Both human and yeast LeuRS activates TORC1 signaling through a leucine-dependent interaction with Rag GTPase (Bonfils, et al., 2012; Han, et al., 2012). However, the interaction is mediated through different sites on human versus yeast LeuRS, and the appended domain in human LeuRS (UNE-L) is not responsible for the interaction.
PheRS is the only human cytoplasmic tRNA synthetase encoded by two genes, forming a (αβ)2 heterotetrameric architecture. Eukaryotic PheRS-α is longer than its prokaryotic orthologs with a N-terminal extension UNE-F, however eukaryotic PheRS-β is shorter than its prokaryotic counterparts by the loss of the anticodon-binding domain B8. The structure of the N-terminal extension of the α-subunit was well resolved in the crystal structure of human PheRS to fold into three continuous DNA-binding fold domains and was predicted by structural modeling to interact with the D, T loops and the anticodon stem of the cognate tRNA. Consistently, truncation of the DNA-binding fold domains resulted in complete loss of tRNA aminoacylation while the activation of phenylalanine was not affected. Therefore, the role of UNE-F in PheRS-α is to compensate the loss of B8 domain in PheRS-β and is distinct from regulatory functions discussed here.
It is worth noting that ArgRS, LeuRS and IleRS are components of the MSC, and the UNE-L domain of LeuRS interacts with both the leucine zipper domain of ArgRS and the UNE-I domain of IleRS in the MSC (Ling, et al., 2005; Rho, et al., 1999). Other appended domains important for the MSC assembly such as the GST domains of MetRS and GluProRS also have preformed structures. In contrast, the disordered appended domain of LysRS and AspRS are not involved in MSC assembly. Therefore, it seems that the assembly of MSC mainly involves appended domains that are well structured, while the unstructured regions within the MSC play an important role in regulating the release of AARSs from the MSC and in mediating their regulatory functions outside the MSC.
Functional significance of structural disorders in AARSs
From the above analysis, the role of structural disordering in developing the regulatory functions of AARSs can be summarized as below:
Regulate the release from MSC
As exemplified by GluProRS and LysRS, release of the tRNA synthetase components from its cellular ‘depot’ (MSC) is regulated by posttranslational modifications (i.e. phosphorylation) on structurally disordered regions. Although phosphorylation can also act on side chains within structured regions of tRNA synthetases (Kwon, et al., 2011; Ofir-Birin, et al., 2013), a disordered region can more easily fold onto the modifying enzyme to facilitate substrate binding and to provide more efficient regulations (Iakoucheva, et al., 2004).
Regulate the activation of regulatory functions
As for the same reason that structurally disordered regions provide better substrate display for posttranslational modifications, the disordered regions provide enhanced susceptibility for proteolysis cleavage. As shown by the examples of TyrRS and TrpRS, such cleavage is critical for the activation of otherwise masked regulatory functions of AARSs. In addition, the conformational advantage of an internally disordered region could dominant over distinct sequence specificities associated with different proteases to provide a unifying mechanism that converges various signaling pathways into the activation of a regulatory function of AARS. Indeed, proteases with different sequence specificities can cleave off the EMAP-II-like domain to activate the regulatory functions of TyrRS (Yang, et al., 2007).
Ensure conformational independence of the new domains and potentially coordinate regulation with translation
In addition to their sensitivity to proteolysis-based regulations, a flexible linker allows the connecting domains to have the conformational freedom to recruit their own binding partners. In the context of AARS where a new regulatory domain is attached to an essential component of the translation machinery, this provides a potential coordination between regulation and translation. The concept, in some sense, is relevant to the regulatory function of TrpRS in the nucleus (Sajish, et al., 2012). In this case, the WHEP domain of TrpRS binds to DNA-PK and PARP-1 and bridges the two proteins for p53 activation. However, if Trp-AMP is bound to the TrpRS enzyme core, it induces a large conformational change of the WHEP domain that abolishes its ability to bind to DNA-PK and PARP-1. Therefore, the flexible linker not only allows the WHEP domain to freely interact with the large-sized DNA-PK and PARP-1 to activate p53 when the enzyme core is not in use, but also makes it possible for Trp-AMP, an aminoacylation reaction intermediate, to induce a large-scale interdomain conformational change that controls the nuclear function of TrpRS.
Mediate multiple interactions with high specificity and low affinity
The N-terminal extension of human LysRS demonstrates the adaptability of a disordered region to different interaction partners from tRNA to MAP kinase and to 67LR laminin receptor. With multiple interaction partners, specificity is a question. Interaction with a disordered partner has the advantage of high specificity that is rendered by the induced-fit mechanism. For example, tRNA interaction induces part of the N-terminal extension of LysRS (S19-E45) to adopt a helical structure that aligns positively charged residues on one side of the helix to enhance the specificity (Guo, et al., 2010; Liu, et al., 2012).
The high specificity interaction with a disordered partner is coupled with low affinity resulted from large entropy penalty paid for the disorder-to-order transition (Wright and Dyson, 2009). The combination of high specificity with low affinity is widely exploited in regulatory processes and is required for cargo transportation. This could be the reason that the UNE-S domain of SerRS and the C-terminal region of LysRS (K574-V597), both of which harbor a nuclear localization signal sequence, are completely disordered.
Concluding remarks
Although the importance of intrinsic disorder has been broadly recognized in cell signaling and regulatory processes (Ward, et al., 2004), its role in expanding the ‘functionome’ of tRNA synthetases has not been recognized. It is clear that structural disordering is key to every aspect of the functional expansion, and majority of the AARS regulatory functions involve disordered structures in one way or another. Evolutionarily speaking, because of their smaller numbers of structural constraints, intrinsically disordered regions are capable of fast development of new functions (Rezaei-Ghaleh, et al., 2012). This explains, at least in part, the association of structural disordering with the ‘new’ functions of AARS and, in turn, suggests that identifying and understanding structurally disordered regions will guide the discovery of more regulatory functions of AARS.
Acknowledgements
I would like to thank the reviewers of this Perspective for their constructive and insightful comments. The work was supported by a grant from the National Institute of Health (GM 088278).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arif A, Jia J, Mukhopadhyay R, Willard B, Kinter M, Fox PL. Two-site phosphorylation of EPRS coordinates multimodal regulation of noncanonical translational control activity. Mol Cell. 2009;35:164–180. doi: 10.1016/j.molcel.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfils G, Jaquenoud M, Bontron S, Ostrowicz C, Ungermann C, De Virgilio C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol Cell. 2012;46:105–110. doi: 10.1016/j.molcel.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Crepin T, Peterson F, Haertlein M, Jensen D, Wang C, Cusack S, Kron M. A hybrid structural model of the complete Brugia malayi cytoplasmic asparaginyl-tRNA synthetase. J Mol Biol. 2011;405:1056–1069. doi: 10.1016/j.jmb.2010.11.049. [DOI] [PubMed] [Google Scholar]
- Francin M, Kaminska M, Kerjan P, Mirande M. The N-terminal domain of mammalian Lysyl-tRNA synthetase is a functional tRNA-binding domain. J Biol Chem. 2002;277:1762–1769. doi: 10.1074/jbc.M109759200. [DOI] [PubMed] [Google Scholar]
- Guo M, Ignatov M, Musier-Forsyth K, Schimmel P, Yang XL. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: Implications for multisynthetase complex formation. Proc Natl Acad Sci U S A. 2008;105:2331–2336. doi: 10.1073/pnas.0712072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Schimmel P, Yang XL. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584:434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo RT, Chong YE, Guo M, Yang XL. Crystal structures and biochemical analyses suggest a unique mechanism and role for human glycyl-tRNA synthetase in Ap4A homeostasis. J Biol Chem. 2009;284:28968–28976. doi: 10.1074/jbc.M109.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JM, Jeong SJ, Park MC, Kim G, Kwon NH, Kim HK, Ha SH, Ryu SH, Kim S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell. 2012;149:410–424. doi: 10.1016/j.cell.2012.02.044. [DOI] [PubMed] [Google Scholar]
- He W, Zhang HM, Chong YE, Guo M, Marshall AG, Yang XL. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc Natl Acad Sci U S A. 2011;108:12307–12312. doi: 10.1073/pnas.1104293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard OM, Dong HF, Yang D, Raben N, Nagaraju K, Rosen A, Casciola-Rosen L, Hartlein M, Kron M, Yang D, et al. Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J Exp Med. 2002;196:781–791. doi: 10.1084/jem.20020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- Kawahara A, Stainier DY. Noncanonical activity of seryl-transfer RNA synthetase and vascular development. Trends Cardiovasc Med. 2009;19:179–182. doi: 10.1016/j.tcm.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DG, Choi JW, Lee JY, Kim H, Oh YS, Lee JW, Tak YK, Song JM, Razin E, Yun SH, et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 2012;26:4142–4159. doi: 10.1096/fj.12-207639. [DOI] [PubMed] [Google Scholar]
- Kron MA, Wang C, Vodanovic-Jankovic S, Howard OM, Kuhn LA. Interleukin-8-like activity in a filarial asparaginyl-tRNA synthetase. Molecular and biochemical parasitology. 2012;185:66–69. doi: 10.1016/j.molbiopara.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Kwon NH, Kang T, Lee JY, Kim HH, Kim HR, Hong J, Oh YS, Han JM, Ku MJ, Lee SY, et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc Natl Acad Sci U S A. 2011;108:19635–19640. doi: 10.1073/pnas.1103922108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Cho BH, Park SG, Kim S. Aminoacyl-tRNA synthetase complexes: beyond translation. J Cell Sci. 2004;117:3725–3734. doi: 10.1242/jcs.01342. [DOI] [PubMed] [Google Scholar]
- Levine SM, Raben N, Xie D, Askin FB, Tuder R, Mullins M, Rosen A, Casciola-Rosen LA. Novel conformation of histidyl-transfer RNA synthetase in the lung: the target tissue in Jo-1 autoantibody-associated myositis. Arthritis Rheum. 2007;56:2729–2739. doi: 10.1002/art.22790. [DOI] [PubMed] [Google Scholar]
- Ling C, Yao YN, Zheng YG, Wei H, Wang L, Wu XF, Wang ED. The C-terminal appended domain of human cytosolic leucyl-tRNA synthetase is indispensable in its interaction with arginyl-tRNA synthetase in the multi-tRNA synthetase complex. J Biol Chem. 2005;280:34755–34763. doi: 10.1074/jbc.M413511200. [DOI] [PubMed] [Google Scholar]
- Liu S, Decker A, Howell M, Caperelli C, Tsang P. (1)H, (13)C and (15)N resonance assignment of the N-terminal domain of human lysyl aminoacyl tRNA synthetase. Biomolecular NMR assignments. 2012 doi: 10.1007/s12104-012-9430-x. [DOI] [PubMed] [Google Scholar]
- Motorin Yu A, Wolfson AD, Orlovsky AF, Gladilin KL. Purification of valyl-tRNA synthetase high-molecular-mass complex from rabbit liver. FEBS Lett. 1987;220:363–365. doi: 10.1016/0014-5793(87)80847-5. [DOI] [PubMed] [Google Scholar]
- Ofir-Birin Y, Fang P, Bennett SP, Zhang HM, Wang J, Rachmin I, Shapiro R, Song J, Dagan A, Pozo J, et al. Structural Switch of Lysyl-tRNA Synthetase between Translation and Transcription. Mol Cell. 2013;49:30–42. doi: 10.1016/j.molcel.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC genomics. 2008;9(Suppl 1):S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophysical journal. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez BL, Howard OM, Dong HF, Edamatsu T, Gao P, Hartlein M, Kron M. Brugia malayi asparaginyl-transfer RNA synthetase induces chemotaxis of human leukocytes and activates G-protein-coupled receptors CXCR1 and CXCR2. The Journal of infectious diseases. 2006;193:1164–1171. doi: 10.1086/501369. [DOI] [PubMed] [Google Scholar]
- Ray PS, Arif A, Fox PL. Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci. 2007;32:158–164. doi: 10.1016/j.tibs.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Rezaei-Ghaleh N, Blackledge M, Zweckstetter M. Intrinsically disordered proteins: from sequence and conformational properties toward drug discovery. Chembiochem : a European journal of chemical biology. 2012;13:930–950. doi: 10.1002/cbic.201200093. [DOI] [PubMed] [Google Scholar]
- Rho SB, Kim MJ, Lee JS, Seol W, Motegi H, Kim S, Shiba K. Genetic dissection of protein-protein interactions in multi-tRNA synthetase complex. Proc Natl Acad Sci U S A. 1999;96:4488–4493. doi: 10.1073/pnas.96.8.4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JC, Kerjan P, Mirande M. Macromolecular assemblage of aminoacyl-tRNA synthetases: quantitative analysis of protein-protein interactions and mechanism of complex assembly. J Mol Biol. 2000;304:983–994. doi: 10.1006/jmbi.2000.4242. [DOI] [PubMed] [Google Scholar]
- Sajish M, Zhou Q, Kishi S, Valdez DM, Jr., Kapoor M, Guo M, Lee S, Kim S, Yang XL, Schimmel P. Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-gamma and p53 signaling. Nature chemical biology. 2012;8:547–554. doi: 10.1038/nchembio.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaram P, Deutscher MP. Existence of two forms of rat liver arginyl-tRNA synthetase suggests channeling of aminoacyl-tRNA for protein synthesis. Proc Natl Acad Sci U S A. 1990;87:3665–3669. doi: 10.1073/pnas.87.10.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojanov L, Satoh M, Hirakata M, Reeves WH. Correlation of antisynthetase antibody levels with disease course in a patient with interstitial lung disease and elevated muscle enzymes. Journal of clinical rheumatology : practical reports on rheumatic & musculoskeletal diseases. 1996;2:89–95. doi: 10.1097/00124743-199604000-00006. [DOI] [PubMed] [Google Scholar]
- Tantos A, Han KH, Tompa P. Intrinsic disorder in cell signaling and gene transcription. Molecular and cellular endocrinology. 2012;348:457–465. doi: 10.1016/j.mce.2011.07.015. [DOI] [PubMed] [Google Scholar]
- Tzima E, Reader JS, Irani-Tehrani M, Ewalt KL, Schwartz MA, Schimmel P. VE-cadherin links tRNA synthetase cytokine to anti-angiogenic function. J Biol Chem. 2005;280:2405–2408. doi: 10.1074/jbc.C400431200. [DOI] [PubMed] [Google Scholar]
- Vo MN, Yang XL, Schimmel P. Dissociating quaternary structure regulates cell-signaling functions of a secreted human tRNA synthetase. J Biol Chem. 2011;286:11563–11568. doi: 10.1074/jbc.C110.213876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Schimmel P. Two distinct cytokines released from a human aminoacyl-tRNA synthetase [see comments]. Science. 1999;284:147–151. doi: 10.1126/science.284.5411.147. [DOI] [PubMed] [Google Scholar]
- Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Woese CR, Olsen GJ, Ibba M, Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol Mol Biol Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Shi Y, Zhang HM, Swindell EC, Marshall AG, Guo M, Kishi S, Yang XL. Unique domain appended to vertebrate tRNA synthetase is essential for vascular development. Nature communications. 2012;3:681. doi: 10.1038/ncomms1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei Z, Zhou JJ, Ye F, Lo WS, Wang F, Lau CF, Wu J, Nangle LA, Chiang KP, et al. Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure. 2012;20:1470–1477. doi: 10.1016/j.str.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X-L, Liu J, Skene RJ, McRee DE, Schimmel P. Crystal structure of an EMAP-II-like cytokine released from a human tRNA cytokine. Helvetica Chimica Acta. 2003;86:1246–1257. [Google Scholar]
- Yang X-L, Skene RJ, McRee DE, Schimmel P. Crystal structure of a human aminoacyl-tRNA synthetase cytokine. Proc Natl Acad Sci U S A. 2002;99:15369–15374. doi: 10.1073/pnas.242611799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Kapoor M, Otero FJ, Slike BM, Tsuruta H, Frausto R, Bates A, Ewalt KL, Cheresh DA, Schimmel P. Gain-of-function mutational activation of human tRNA synthetase procytokine. Chem Biol. 2007;14:1323–1333. doi: 10.1016/j.chembiol.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc Natl Acad Sci U S A. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Kapoor M, Guo M, Belani R, Xu X, Kiosses WB, Hanan M, Park C, Armour E, Do MH, et al. Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat Struct Mol Biol. 2010;17:57–61. doi: 10.1038/nsmb.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]