Abstract
The HPV infection in men (HIM) study examines the natural history of genital HPV infection in men. Genotyping methods used in this study identify 37 α-HPV types; however, the viral type could not be identified in approximately 22% of male genital specimens that were HPV PCR positive. Our aim was to genotype HPV-unclassified specimens by sequencing PGMY09/11, GP5+/6+ or FAP59/64 PCR products. Using this approach we were able to detect 86 unique HPV types among 508 of 931 specimens analyzed. We report for the first time the presence of a broad range of α-, β- and γ-HPV at the male genitals.
Keywords: Human papillomavirus, Genera, Viral prevalence, Men, Genital, Penile
Introduction
Human papillomaviruses (HPV) are primarily transmitted by sexual contact and infection by these viruses is strongly associated with the development of cancers of the cervix, vagina, and vulva in women, cancer of the penis in men, in addition to cancers of the anus and oropharynx in both genders (IARC, 2007). To date more than 150 different HPV types have been described. Forty α-HPV types have been shown to infect the anogenital tract of which 13 are classified as high-risk or oncogenic according to the World Health Organization (IARC, 2007).
HPV taxonomy relies on L1 late gene sequence variability: new viral types are defined when the L1 sequence differs by at least 10% from the closest known type. The majority of HPV types groups in three different genera: α-papillomavirus, predominantly isolated from mucosal and genital lesions, and β- and γ-papillomavirus mostly detected in cutaneous specimens (Bernard et al., 2010). Most studies of genital HPV infection in men have examined only α-HPV types; the distribution of other HPV types present at the male genitals remains unknown. In the HIM study (Giuliano et al., 2008), over 66% of men tested positive for HPV at their first study visit; however, the viral type could not be identified in nearly 22% of these men (e.g., HPV PCR positive, no genotype specified) (Akogbe et al., 2012). Understanding the natural history of HPV infection in men is essential to better understand the transmission of these viruses and to study HPV related disease. In the present study, we aimed to sequence HPV-unclassified genital specimens using three different generic HPV primer sets to identify α-, β- and γ-HPV infections in a well-defined cohort of adult men.
Results
A total of 931 specimens from the external genitalia of 649 men were included in this study: 348 specimens from 244 men in the USA, 338 specimens from 233 men in Brazil, and 245 specimens from 172 men in Mexico. These genital specimens were collected from a range of visits, through the end of 2008, which corresponds to the HIM study baseline through 3 years of follow up: 118, 279, 210, 147, 114, 52, 8, and 3 samples from visits 1, 2, 3, 4, 5, 6, 7, and 8, respectively.
Among all 931 specimens, 393 were characterized by sequencing PGMY09/11 PCR products. Next, nested PCR with GP5+/6+ primers was able to amplify 34 samples; and finally the FAP59/64 amplified 398 additional samples. Overall, 106 (11.4%) were HPV negative using any of the three primers sets (13.5%, 9% and 12% of the specimens from the USA, Brazil and Mexico, respectively), indicating that a fraction of unclassified HPV detection most likely represents spurious PCR products. Among the PCR-positive specimens using either primers PGMY09/11 or FAP59/64, HPV could not be genotyped in 133 (14.3%) of specimens since BlastN analysis indicated high similarity to partial HPV sequences in GeneBank as yet without type designation. Additionally, in 182 specimens (19.6%) direct sequencing of the FAP59/64 amplicons was unsuccessful due to overlapping peak patterns most possibly due to the presence of more than one HPV type.
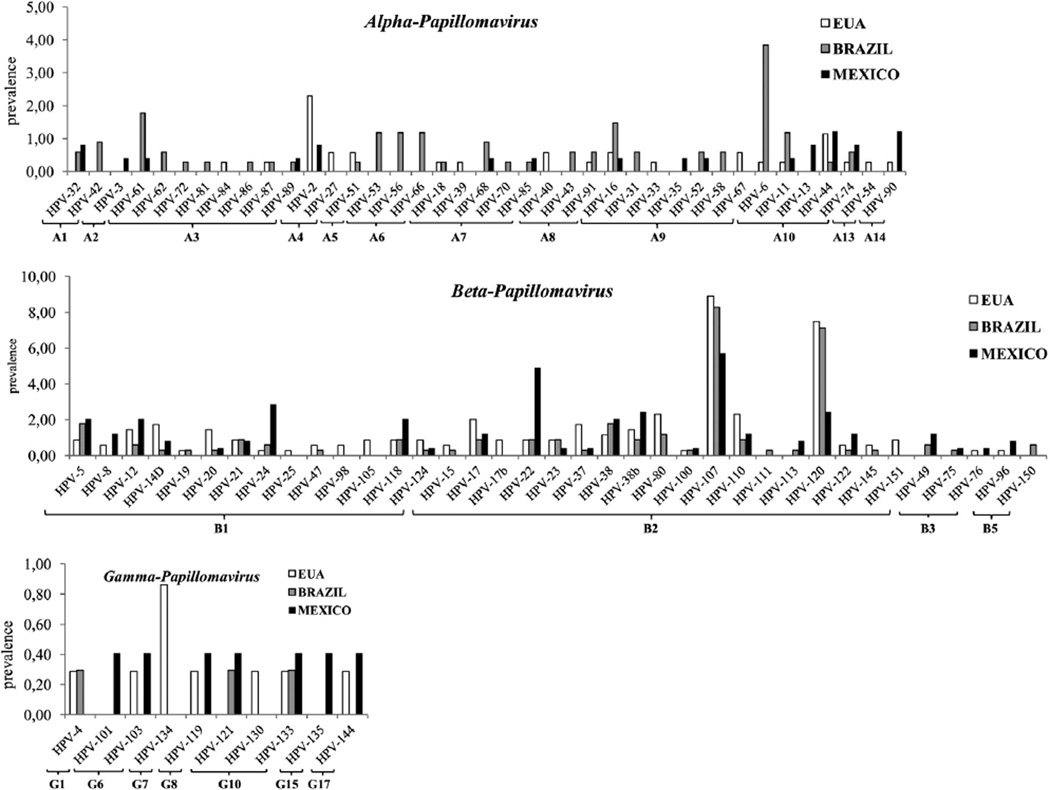
We were able to successfully type 508 (54.6%) previously unclassified specimens, comprising 86 different HPV types: 13.7% α-HPV, 38.7% β-HPV, and 2% γ-HPV. Alpha-HPVs were detected in 9.5% (33/348), 21.3% (72/338) and 9.4% (23/245) of the specimens from the USA, Brazil and Mexico, respectively. Some divergence was observed concerning HPV type frequency across populations; for instance, HPV-6 was more commonly detected among Brazilian samples (13/338; 3.8%), whereas HPV-2 was detected in eight of 348 samples from the USA (2.3%) (Fig. 1). Most samples analyzed contained β-HPV: 44.8% (n = 156), 32.5% (n = 110) and 38.7% (n = 95) of the specimens from the USA, Brazil and Mexico, respectively. HPV β-2 species was the most frequent and diverse group: we detected 18 different HPV genotypes from this species of which the more common were HPV-107 and HPV-120. HPV-22 was more frequent among samples from Mexico (12/245; 4.9%). In a small proportion of the samples, we identified HPV types of three different γ-HPV species; only HPV-133 was detected in all three populations. HPV-147 which has not been taxonomically classified thus far (http://pave.niaid.nih.gov/#home) was detected in one sample from each country.
Fig. 1.
HPV type frequency by country of α-, β-, and γ-HPVs in unclassified samples of the HIM study. The y-axis indicates frequency of individual HPV types. HPV species are indicated in the x-axis.
Among the 649 men included in this study, we analyzed two or more specimens from 199 individuals. DNA of the same HPV type was detected in only 57 men, in more than one visit (Table 1). HPV-107 and HPV-120 were the most common types detected at more than one study visit.
Table 1.
Number of HPV persistent detection by country in unclassified samples of the HIM study.
| Genus | Specie | Type HPV | EUA | Brazil | Mexico | Total |
|---|---|---|---|---|---|---|
| α-HPV | α6 | HPV-56 | 1 | 1 | ||
| α4 | HPV-2 | 1 | 1 | |||
| α10 | HPV-6 | 1 | 1 | |||
| HPV-44 | 1 | 1 | ||||
| β-HPV | β1 | HPV-5 | 1 | 1 | 2 | |
| HPV-14D | 1 | 1 | ||||
| HPV-20 | 1 | 1 | ||||
| HPV-24 | 1 | 1 | ||||
| HPV-47 | 1 | 1 | ||||
| HPV-105 | 1 | 1 | ||||
| HPV-118 | 1 | 1 | 1 | 3 | ||
| β1 | HPV-22 | 3 | 3 | |||
| HPV-23 | 1 | 1 | ||||
| HPV-37 | 2 | 2 | ||||
| HPV-38b | 1 | 1 | 2 | 4 | ||
| HPV-80 | 1 | 1 | 2 | |||
| HPV-107 | 3 | 6 | 2 | 11 | ||
| HPV-110 | 1 | 1 | 2 | |||
| HPV-120 | 5 | 6 | 1 | 12 | ||
| Uncharacterized | 1 | 4 | 1 | 6 | ||
| Total | 20 | 23 | 14 | 57 |
Discussion
Using a PCR-sequencing procedure, we were able to show that over 11% of unclassified HPV infections (i.e., PCR-positive but genotyping-negative) were truly HPV negative and that 55% of such infections were positive for a broad distribution of α-, β- and γ-HPV types. These unclassified HPV infections could result from non-specific amplification of genes unrelated to HPV or represent viral types different from the 37 α-HPVs possibly genotyped by the Linear Array HPV genotyping test.
We detected α-HPVs in 128 samples, 80 of which contained HPV types that should have been identified with the linear array genotyping test. These false-negative results are likely due to low viral copy numbers which were detected on a different PCR reaction followed by sequencing, a more sensitive technical approach. Interestingly, by sequencing PGMY09/11 and GP5+/6+ PCR products we were able to identify a wide range of β-HPV types in the samples analyzed, although both primer sets were primarily designed to detect α-HPVs. However, a high proportion of β-HPV specimens were further characterized and identified by sequence analysis of FAP59/64 PCR products. Additionally, all γ-HPV specimens were discovered using FAP59/64 primers (data not shown). Overall, we detected 37 different HPV types from four β-HPV species. Some differences were observed regarding the prevalence of individual types among the three geographical locations.
We observed in 14% of the samples high sequence identity with HPVs not fully characterized, most of which consisted of partial FAP HPV sequences previously detected in the skin of healthy individuals, dialysis patients and renal transplant recipients (Antonsson et al., 2000). The detection of 133 putative new HPV types in human skin samples supports that β- and γ-HPVs are more diverse than α-HPV (Forslund, 2007). Furthermore, sequences with overlapping peak patterns indicate the presence of more than one HPV type, as was observed in 19.5% of the samples analyzed in this study. This could be overcome by sequencing multiple clones or by using the Luminex technology (Gheit et al., 2007). In fact, the latter system was employed in the analysis of 17 unclassified HPV penile samples from the HIM study and showed that most specimens contained more than three HPV cutaneous types (T. Gheit and M. Tommasino, personal communication). These results suggest that an even greater amount of genital samples from the HIM study may actually contain DNA from cutaneous HPV types. These include α-HPV positive samples that were both characterized using the Linear Array genotyping test and PCR-sequencing. Multiple infection by β-HPVs are commonly detected in cutaneous tumors and normal skin samples while using FAP59/64 primers set (Ekström et al., 2011; Forslund et al., 1999).
Former surveys of penile HPV prevalence mainly focused on high-risk genital α-HPV infections (IARC, 2007). Our data points to a broader distribution of β- and γ-HPVs than was previously recognized, as these viral types were primarily described in skin samples and for this reason we may not exclude the possibility that a high number of cutaneous HPV also circulate in the anogenital area of women. Recently, HPV types from both genera were also detected in condyloma samples (Johansson et al., in press). Further, a wide spectrum of known and novel HPV types that phylogenetically cluster into the β- and γ-HPV genera were identified in the oral cavity (Bottalico et al., 2011). Nevertheless, it seems that only α-HPVs infections are associated to oral cancer development (Paolini et al., 2013). In addition to certain types of sexual behavior that may increase the risk of β-HPV infection in the anogenital region, direct hand to penis contact may favor viral transmission. Further, the presence of cutaneous HPV DNA in the penis could reflect deposition of virions released from other body sites with productive infections.
Persistence of high-risk α-HPV types constitutes the most important factor for the development of cervical cancer (IARC, 2007). In this study, DNA of the same β-HPV type was detected in more than one visit in 47 of 250 (18.8%) of individuals with at least one specimen harboring viral types from this branch. Data in the literature concerning persistence of β-HPV is limited. Still, it was described that 43% of skin infections by cutaneous HPVs persisted for 6 years in healthy individuals and renal transplant recipients (Hazard et al., 2007). Interestingly, a lack of association between HPV persistence and presence of warts was observed.
We report for the first time the presence of a broad range of novel and candidate β- and γ-HPV at the male genitals. The implication of β-HPV persistence for the pathogenesis of cancer is unknown as yet and requires further investigation.
Materials and methods
Study population
The HPV infection in men (HIM) study is a multinational, prospective study of HPV natural history in men recruited from three different populations (São Paulo, Brazil; Cuernavaca, Mexico; Tampa, Florida). Over 4000 men ages 18–70 years were enrolled in the study and were followed every 6 months for up to 4 years. Details of the HIM study are described elsewhere (Giuliano et al., 2008, 2011). This analysis is restricted to 931 samples from 649 men who had at least one unclassified genital HPV specimen collected between March 2005 and December 2008. No exclusions were made for men with external genital lesions. This study was approved by the ethics committees of the hospitals and institutions involved. All participants gave written informed consent.
Genital specimen collection and HPV detection
At each study visit, genital specimens were obtained from the coronal sulcus, glans penis, penile shaft, and scrotum using Dacron (Digene, Gaithersburg, MD, USA) swabs, and combined into one sample. HPV DNA was extracted using the QIAGen Media Kit (QIAGen, Valencia, CA, USA) and PCR (PGMY09/11) was performed. Specimens were genotyped using the linear array method (Roche Molecular Diagnostics, Alameda, CA, USA) which identifies 37 α-HPVs commonly detected in the cervix (Gravitt et al., 1998). Specimens that tested PCR-positive and genotyping-negative were considered unclassified infections, and underwent additional typing.
Typing of unclassified samples
Purified DNA was genotyped by direct sequencing of the PCR amplimers generated by PGMY09/11 primers or cloning of the amplicons followed by sequencing. Next, 1 µl of PGMY09/11 negative products was used in a nested PCR using GP5+/6+ primers (de Roda Husman et al., 1995) and positive samples were cloned and sequenced. Finally, nested PGMY09/11-GP5+/6+ PCR negative samples were submitted to a new PCR using FAP59/64 primers and amplimers were analyzed by direct sequencing (Forslund et al., 1999). All PCRs were performed using AmpliTaq Gold polymerase (Perkin-Elmer, Foster City, CA, USA). Before sequencing, PCR products were purified using EXO SAP-IT (GE Healthcare, Buckinghamshire, UK). Sequencing reactions were performed in an ABI 3130XL Genetic Analyzer (AB Applied Bio-systems, CA, USA) using the BigDye® Terminator v3.1 Cycle Sequencing kit (AB Applied Biosystems, CA, USA). Sequence identity was determined by comparison with the BlastN database: sequences with scores higher than 90% within at least 200 bp were conclusively typed.
Acknowledgments
We are thankful to Tarik Gueit and Massimo Tommasino for discussions and HPV Luminex analysis. HIM study group: Brazil: Elimar Gomes, Elisa Brito, Filomena Cernicchiaro, Rubens Matsuo, Vera Souza, Ricardo Cintra, Ricardo Cunha, Birgit Fietzek, Raquel Hessel, Viviane Relvas, Fernanda Silva, Juliana Antunes, Graças Ribeiro, Roberta Bocalon, Rosária Otero, Rossana Terreri, Sandra Araujo, Meire Ishibashi, the CRT–DST/AIDS Nursing team; Mexico: Jorge Salmerón, Manuel Quiteria, Aurelio Cruz, Pilar Hernandez, Griselda Diaz Garcia, Oscar Rojas Juarez, Rossane del Carmen Gonzales Sosa, Rene de Jesus Alvear Vazquez; USA: Martha Abrahamsen, Christine Gage, Kathy Eyring, Nadia Lambermont, Emily Jolles, Kayoko Kay, Kim Isaacs, Andrea Leto, Dan’elle Smith, Kyle Wolf, Anthony Bilotto, Abidemi Ajidahun, Michael Blackmer, Michael O’Keefe, Bradley Sirak, Alan G. Nyitray, Beibei Lu, Mary Papenfuss, Hui-Yi Lin; HIM study Co-Investigator: Ray Viscidi.
Role of the funding source
Financial support: Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [Grant numbers 10/15282-3 to L.S. and 08/57889-1 to L.L.V.]; Conselho Nacional de Desenvolvimento Científico e Tencnológico (CNPq) [Grant number 573799/2008-3 to L.L.V.] and National Institute of Health (NIH) [Grant number RO1 CA098803 to A.R.G.].
References
- Akogbe GO, Ajidahun A, Sirak B, Anic GM, Papenfuss MR, Fulp WJ, Lin HY, Abrahamsen M, Villa LL, Lazcano-Ponce E, Quiterio M, Smith D, Schabath MB, Salmeron J, Giuliano AR. Race and prevalence of human papillomavirus infection among men residing in Brazil, Mexico and the United States. Int. J. Cancer. 2012;131(3):E282–E291. doi: 10.1002/ijc.27397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Forslund O, Ekberg H, Sterner G, Hansson BG. The ubiquity and impressive genomic diversity of human skin papillomaviruses suggest a commensalic nature of these viruses. J. Virol. 2000;74(24):11636–11641. doi: 10.1128/jvi.74.24.11636-11641.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard HU, Burk RD, Chen Z, van Doorslaer K, zur Hausen H, de Villiers EM. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401(1):70–79. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottalico D, Chen Z, Dunne A, Ostoloza J, McKinney S, Sun C, Schlecht NF, Fatahzadeh M, Herrero R, Schiffman M, Burk RD. The oral cavity contains abundant known and novel human papillomaviruses from the Betapapillomavirus and Gammapapillomavirus genera. J. Infect. Dis. 2011;204(5):787–792. doi: 10.1093/infdis/jir383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekström J, Bzhalava D, Svenback D, Forslund O, Dillner J. High throughput sequencing reveals diversity of Human Papillomaviruses in cutaneous lesions. Int. J. Cancer. 2011;129(11):2643–2650. doi: 10.1002/ijc.26204. [DOI] [PubMed] [Google Scholar]
- Forslund O. Genetic diversity of cutaneous human papillomaviruses. J. Gen. Virol. 2007;88(10):2662–2669. doi: 10.1099/vir.0.82911-0. [DOI] [PubMed] [Google Scholar]
- Forslund O, Antonsson A, Nordin P, Stenquist B, Hansson BG. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 1999;80(9):2437–2443. doi: 10.1099/0022-1317-80-9-2437. [DOI] [PubMed] [Google Scholar]
- Gheit T, Billoud G, de Koning MN, Gemignani F, Forslund O, Sylla BS, Vaccarella S, Franceschi S, Landi S, Quint WG, Canzian F, Tommasino M. Development of a sensitive and specific multiplex PCR method combined with DNA microarray primer extension to detect Betapapillomavirus types. J. Clin. Microbiol. 2007;45(8):2537–2544. doi: 10.1128/JCM.00747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano AR, Lazcano-Ponce E, Villa LL, Flores R, Salmeron J, Lee JH, Papenfuss MR, Abrahamsen M, Jolles E, Nielson CM, Baggio ML, Silva R, Quiterio M. The human papillomavirus infection in men study: human papillomavirus prevalence and type distribution among men residing in Brazil, Mexico, and the United States. Cancer Epidemiol. Biomarkers Prev. 2008;17(8):2036–2043. doi: 10.1158/1055-9965.EPI-08-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano AR, Lee JH, Fulp W, Villa LL, Lazcano E, Papenfuss MR, Abrahamsen M, Salmeron J, Anic GM, Rollison DE, Smith D. Incidence and clearance of genital human papillomavirus infection in men (HIM): a cohort study. Lancet. 2011;377(9769):932–940. doi: 10.1016/S0140-6736(10)62342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 1998;36(10):3020–3027. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazard K, Karlsson A, Andersson K, Ekberg H, Dillner J, Forslund O. Cutaneous human papillomaviruses persist on healthy skin. J. Invest. Dermatol. 2007;127(1):116–119. doi: 10.1038/sj.jid.5700570. [DOI] [PubMed] [Google Scholar]
- IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Smokeless Tobacco Products, 90. Lyon, Inc.689; 2007. [Google Scholar]
- Johansson H, Bzhalava D, Ekstrom J, Hultin E, Dillner J, Forslund O. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology. 440(1):1–7. doi: 10.1016/j.virol.2013.01.023. [DOI] [PubMed] [Google Scholar]
- de Roda Husman AM, Walboomers JM, van den Brule AJ, Meijer CJ, Snijders PJ. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 1995;76(4):1057–1062. doi: 10.1099/0022-1317-76-4-1057. [DOI] [PubMed] [Google Scholar]
- Paolini F, Rizzo C, Sperduti I, Pichi B, Mafera B, Rahimi SS, Vigili MG, Venuti A. Both mucosal and cutaneous papillomaviruses are in the oral cavity but only alpha genus seems to be associated with cancer. J. Clin. Virol. 2013;56(1):72–76. doi: 10.1016/j.jcv.2012.09.016. [DOI] [PubMed] [Google Scholar]