Abstract
Nonalcoholic Fatty Liver Disease (NAFLD) is an obesity-related condition affecting over 50% of individuals in some populations and is expected to become the number one cause of liver disease worldwide by 2020. Common, robustly associated genetic variants in/near five genes were identified for hepatic steatosis, a quantifiable component of NAFLD, in European-ancestry individuals. Here we tested whether these variants were associated with hepatic steatosis in African and/or Hispanic Americans and fine-mapped the observed association signals. We measured hepatic steatosis using computed tomography in five African-American (n=3124) and one Hispanic-American (n=849) cohorts. All analyses controlled for variation in age, age2, gender, alcoholic drinks, and population substructure. Heritability of hepatic steatosis was estimated in three cohorts. Variants in/near PNPLA3, NCAN, LYPLAL1, GCKR, and PPP1R3B were tested for association with hepatic steatosis using a regression framework in each cohort and meta-analyzed. Fine-mapping across African-American cohorts was conducted using meta-analysis. African- and Hispanic-American cohorts were 33.9/37.5% male, with average age of 58.6/42.6 years and body mass index of 31.8/28.9kg/m2, respectively. Hepatic steatosis was 0.20–0.34 heritable in African-and Hispanic-American families (p<0.02 in each cohort). Variants in or near PNPLA3, NCAN, GCKR, PPP1R3B in African Americans and PNPLA3 and PPP1R3B in Hispanic Americans were significantly associated with hepatic steatosis; however, allele frequency and effect size varied across ancestries. Fine-mapping in African Americans highlighted missense variants at PNPLA3 and GCKR and redefined the association region at LYPLAL1.
Conclusions
We show for the first time that multiple genetic variants are associated with hepatic steatosis across ancestries and explain a substantial proportion of the genetic predisposition in African and Hispanic Americans. Missense variants in PNPLA3 and GCKR are likely functional across multiple ancestries.
Keywords: liver steatosis, single nucleotide polymorphisms, obesity, meta-analysis, genetic variance
The prevalence of nonalcoholic fatty liver disease (NAFLD) has increased with the rise in obesity and is predicted to become the leading cause of liver disease in the world by 2020. (1) NAFLD is a spectrum of diseases that includes liver steatosis (fat), nonalcoholic steatohepatitis (NASH; fat and inflammation) and fibrosis/cirrhosis (scarring). (2) NAFLD is correlated with central obesity, high levels of triglycerides (TG), low levels of high-density lipoprotein (HDL) cholesterol, high blood pressure, impaired glucose tolerance (3, 4), and cardiovascular disease. (5, 6) Consequently, the mortality related to both liver and non-liver causes of death is predicted to reach epidemic proportions by the end of the decade. Unfortunately, few medical treatments presently exist for NAFLD. A better understanding of the pathophysiology of NAFLD will improve the diagnosis, management, treatment, and ultimately prevention of NAFLD.
Human genetic studies have provided new insights into NAFLD. Computed tomography (CT) can be used to reliably quantitate hepatic steatosis non-invasively in population based samples. CT measured hepatic steatosis has been found to be heritable (h2=0.26–0.27) in European-ancestry cohorts (7) and shown to predict histological steatosis. (8–11) Using a genome-wide association study (GWAS) in 7176 individuals of European ancestry, the Genetics of Obesity-related Liver Disease (GOLD) Consortium identified genetic variants that reproducibly associated with hepatic steatosis in or near the genes PNPLA3, LYPLAL1, PPP1R3B, NCAN, and GCKR. (7) Interestingly, the prevalence of NAFLD and metabolic disease varies amongst ancestries.(12, 13) In particular, Hispanic Americans have the highest prevalence of NAFLD whereas African Americans have the lowest. (14, 15) Some of this variation may be influenced by genetics. (16) Since the correlation between single nucleotide polymorphisms (SNPs), known as linkage disequilibrium (LD), or the non-random association of alleles, varies amongst ancestries it remains to be determined which susceptibility variants are shared or divergent across ancestries.
In the present study, we first determined the proportion of observed variation in hepatic steatosis which can be attributed to genetic factors (heritability) in African- and Hispanic-American families. We then aimed to replicate, for the first time, the effects of multiple variants in or near PNPLA3, GCKR, NCAN, PPP1R3B, and LYPLAL1, which were robustly associated with hepatic steatosis in European-ancestry individuals, and summarized the effects by performing a meta-analysis. Finally, using GWAS data we fine-mapped the association signal at these loci in African Americans in an effort to identify the putative causal variant(s).
EXPERIMENTAL PROCEDURES
Ethics Statement
All work performed in this manuscript was approved by local institutional review boards or equivalent committees.
Study Design
A total of four cohorts were included in the current study: the Jackson Heart Study (JHS (17); both the JHS and JHS-ARIC (Atherosclerosis Risk in Communities Study; samples of African Americans), the Insulin Resistance Atherosclerosis Family Study (IRASFS (18); African and Hispanic Americans), Genetic Epidemiology Network of Arteriopathy (GENOA (19); African Americans), and Family Heart Study (FamHS (20); African Americans) (Appendix Table 1).
Outcome Variable and Covariates
In each cohort, hepatic steatosis was measured by CT-scanning using a standardized protocol. Either a phantom (JHS, GENOA, and FamHS) or spleen density (IRASFS) was used to adjust attenuation values as part of quality control. Details of the protocol can be found in Appendix Table 2. Age, gender, and alcohol intake was self-reported.
Genotyping
Details of the genotyping methods, quality control, and imputation procedures used in each participating cohorts are shown in the Appendix Tables 3 and 4. Briefly, the genotypes of index variants from European-ancestry populations in patatin-like phospholipase domain containing 3 (PNPLA3; rs738409), protein phosphatase 1, regulatory subunit 3B (PPP1R3B; rs4240624), neurocan (NCAN; rs2228603), glucokinase regulator (GCKR; rs780094), and lysophospholipase-like 1 (LYPLAL1; rs12137855) were determined by array coupled with imputation (Affymetrix Human Chip v6·0: JHS, JHS-ARIC and GENOA; Illumina 1M-Duo v3.0: FamHS) or de novo (Sequenom: IRASFS) genotyping.
Statistical Analysis
Heritability determination
Cohort-specific estimates of heritability for hepatic steatosis were obtained in both African (IRASFS, GENOA, and FamHS) and Hispanic Americans (IRASFS) families. While the amount of alcohol consumption was generally low across samples, it was non-negligible; therefore, each study adjusted hepatic steatosis values for the reported number of drinks per week. Estimates of population substructure were obtained from principal components-derived from genome-wide data in African Americans and ancestry informative markers in Hispanic Americans. In each cohort, hepatic steatosis was inverse normally transformed to reduce the impact of outliers and deviations from normality and thus standardize the phenotype in preparation for meta-analysis. Hepatic steatosis was adjusted for age, age2, gender, alcohol intake, and population substructure. Thus, heritability calculations reflect the “residual” heritability after controlling for covariates. Heritability estimates were computed using a variance component procedure implemented in SOLAR. (21)
Association analyses
In each cohort, hepatic steatosis was inverse normally transformed. Cohort-specific association analyses were performed in family-based samples accounting for family structure using either a random effects model (IRASFS; SOLAR (21)) or a mixed linear effects model (GENOA and FamHS, R) (Appendix Table 5). Association analyses in JHS were carried out using regression. Analyses were adjusted for age, age2, gender, alcohol intake, and principal component estimates of ancestry. Meta-analysis by race and overall was performed using the inverse variance weighted method as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal/). For significance, a meta-analysis P-value less than 0.01 (Bonferroni correction for five tests) was considered as significant while values less than 0.10 with consistent direction of effect were considered trending.
To facilitate fine-mapping of these five loci, genotyped and imputed data were extracted from GWAS datasets (JHS, JHS-ARIC, GENOA and FamHS) (Appendix Table 5). The region of interest for each locus was defined using a LD threshold of r2>0.1 from the index variant in the European-ancestry meta-analysis as estimated from the HapMap European (CEU) population. These regions are most likely to harbor causal variants in LD with the index variant reported in the original study in those of European ancestry. (7) Variants from individual cohorts were included in the meta-analysis if they had a minor allele frequency ≥1% and imputation quality of r2≥0.3. Furthermore, SNPs needed to be present in three out of four cohorts to be included in analyses. Meta-analysis was performed using the inverse variance weighted method as implemented in METAL (http://www.sph.umich.edu/csg/abecasis/metal/).
RESULTS
The study sample characteristics are shown in Table 1. In general, subjects were middle-aged, although subjects from IRASFS were younger (mean age ~43 versus 53–70 years in other cohorts). Despite their younger age, the mean CT liver attenuation of Hispanic Americans trended towards being lower – indicating increased hepatic steatosis – than subjects of African ancestry. The alcohol consumption was generally low across cohorts.
Table 1.
Characteristics of study populations
| EA meta- analysis |
AA cohorts | IRASFS HA |
|||||
|---|---|---|---|---|---|---|---|
| FamHS | GENOA | IRASFS | JHS | JHS-ARIC | |||
| Demographics | |||||||
| N (% male) | 7,227 (45.4) | 622 (34.2) | 555 (24.9) | 364 (38.1) | 1,180 (38.9) | 403 (27.5) | 849 (37.5) |
| Mean age (SD), years | 62.8 (6.3) | 53.4 (10.8) | 69.0 (7.85) | 43.6 (13.7) | 55.5 (9.9) | 69.9 (5.0) | 42.6 (13.9) |
| Median drinks per week (P25, P75)* | 0–3 | 0 (0, 1) | 0 (0,0) | 0.23 (0, 2.8) | 0.9 (0.1, 3.9) | 0.6 (0.1, 2.0) | 0.23 (0, 2.8) |
| Never drinkers (%) | 2,436 (37.7) | 349 (68.3) | 430 (77.5) | 170 (46.7) | 611 (51.8) | 128 (31.7) | 398 (47.0) |
| Mean body mass index (SD), kg/m2 | 27.5 (0.6) | 32.7 (7.4) | 32.7 (7.3) | 29.9 (6.6) | 32.1 (6.5) | 30.7 (6.0) | 28.9 (6.1) |
| CT attenuation | |||||||
| Reference | Spleen andPhantom HD | Phantom HD | Phantom HD | Spleen | Phantom HD | Phantom HD | Spleen |
| Median hepatic steatosis, HU (P25, P75)* | 60–68 | 61.6 (56.3, 65.6) | 60.2 (56.0, 65.7) | 57.5 (52.8, 61.1) | 60.1 (54.8, 64.5) | 61.4 (55.7, 65.5) | 55.5 (45.4, 60.5) |
EA: European Ancestry; AA: African American; HA: Hispanic American; FamHS: Family Heart Study; GENOA: Genetic Epidemiology Network of Arteriopathy; IRASFS: Insulin Resistance Atherosclerosis Family Study; JHS: Jackson Heart Study; ARIC: Atherosclerosis Risk in Communities; SD: Standard Deviation; CT: Computed Tomography
Range of median values among EA studies; SD: standard deviation; P25, P75: 25th and 75th percentiles; Phantom HD: high density external hydroxyapetite CT control; HU: Hounsfield Units
Estimates of heritability of hepatic steatosis are shown in Table 2. Two of the African American cohorts showed slightly higher heritability (0.30±0.08 and 0.34±0.10 in GENOA and FamHS, respectively), while the African- and Hispanic-American subjects from IRASFS showed lower heritability (0.22±0.12 and 0.20±0.07, respectively). These data indicate that liver fat in adult subjects is moderately heritable across the cohorts in the current study and justifies exploring the contribution of genetic variants.
Table 2.
Heritability estimates for hepatic steatosis in African-American and Hispanic-American (HA) cohorts
| Cohort | N subjects |
N families |
Ascertainment | Age range (years) |
Heritability | SE | P-value |
|---|---|---|---|---|---|---|---|
| African Americans | |||||||
| FamHS | 622 | 214 | families of hypertensive probands | 30 – 83 | 0.34 | 0.10 | 1.90E-04 |
| GENOA | 555 | 410 | sibs of hypertensive probands | 40 – 98 | 0.30 | 0.08 | 4.00E-03 |
| IRASFS | 364 | 41 | families of T2D, IGT and NGT probands | 18 – 80 | 0.22 | 0.12 | 1.10E-02 |
| Hispanic Americans | |||||||
| IRASFS | 849 | 86 | families of T2D, IGT and NGT probands | 18 – 81 | 0.20 | 0.07 | 7.00E-05 |
FamHS: Family Heart Study; GENOA: Genetic Epidemiology Network of Arteriopathy; IRASFS: Insulin Resistance Atherosclerosis Family Study; T2D: Type 2 Diabetes; IGT: Impaired Glucose Tolerance; NGT: Normal Glucose Tolerance
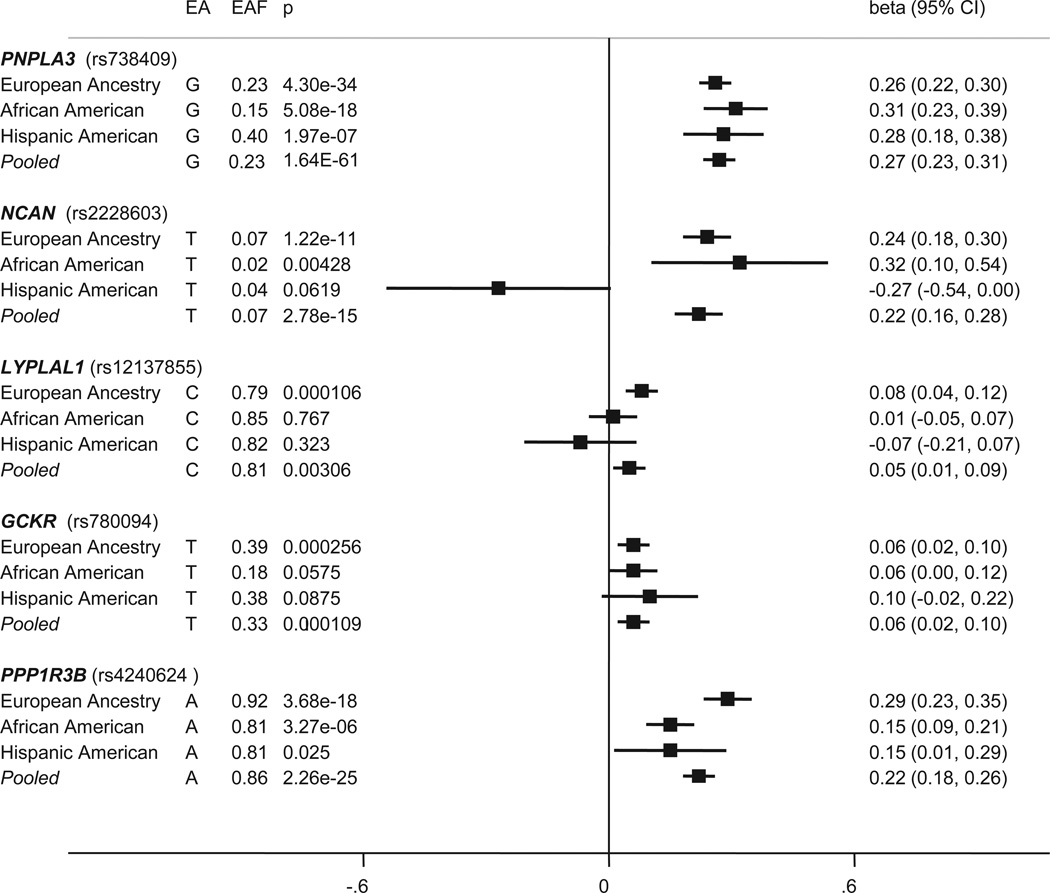
The summary of association evidence for loci replicated from the previously published meta-analysis of European-ancestry in African and Hispanic American individuals is shown in Table 3 and Figures 1 and 2 (cohort-specific results for African Americans are shown in Appendix Figure 1 and Appendix Table 6). Using the criterion P-value<0.01 (Bonferroni correction for five tests), three of the variants - PNPLA3 (rs738409), PPP1R3B (rs4240624), and NCAN (rs2228603) - were significant in the African American meta-analysis, with allele frequencies and effect direction consistent with the European-ancestry results. PNPLA3 (rs738409) also displayed significant association in the Hispanic-American study; however, the other two loci trended toward significance and the association with NCAN (rs2228603) was in the opposite direction, suggesting that, if real, it would have a small protective effect in Hispanic Americans, contrary to its fat-increasing effect in other ancestry groups. This was the only locus to have a significant P-value of heterogeneity across ancestries (data not shown). GCKR (rs780094) displayed trending evidence of association (P-value<0.10), and although it did not reach significance in the African or Hispanic American analyses, the direction and magnitude of the effect of the T allele was consistent with the European-ancestry findings, indicating a general consistency across racial groups. LYPLAL1 (rs12137855) was not significant in either African or Hispanic Americans and despite comparable allele frequencies.
Table 3.
Association summary statistics for the five hepatic steatosis loci
| Locus | EA meta-analysis N=7,176 |
AA meta-analysis N=3,124* |
IRASFS HA N=839 |
ALL meta-analysis N=11,139* |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP ID | Chr (Position) |
Nearest Gene |
EA | EAF | Beta (SE) |
P-value | EAF | Beta (SE) |
P-value | EAF | Beta (SE) |
P value | EAF | Beta (SE) |
P-value |
| rs738409 | 22 (42656060) | PNPLA3 | G | 0.23 | 0.26 (0.02) | 4.30E-34 | 0.15 | 0.31 (0.04) | 5.08E-18 | 0.40 | 0.28 (0.05) | 1.97E-07 | 0.23 | 0.27 (0.02) | 1.64E-61 |
| rs2228603* | 19 (19190924) | NCAN | T | 0.07 | 0.24 (0.03) | 1.22E-11 | 0.02 | 0.32 (0.11) | 4.28E-03 | 0.04 | −0.27 (0.14) | 0.062 | 0.07 | 0.22 (0.03) | 2.78E-15 |
| rs12137855 | 1 (217515001) | LYPLAL1 | C | 0.79 | 0.08 (0.02) | 1.06E-04 | 0.85 | −0.01 (0.03) | 0.767 | 0.82 | −0.07 (0.07) | 0.323 | 0.81 | 0.05 (0.02) | 3.06E-03 |
| rs780094 | 2 (27594741) | GCKR | T | 0.39 | 0.06 (0.02) | 2.56E-04 | 0.18 | 0.06 (0.03) | 0.058 | 0.38 | 0.10 (0.06) | 0.088 | 0.33 | 0.06 (0.02) | 1.09E-04 |
| rs4240624 | 8 (9221641) | PPP1R3B | A | 0.92 | 0.29 (0.03) | 3.68E-18 | 0.81 | 0.15 (0.03) | 3.27E-06 | 0.81 | 0.15 (0.07) | 0.025 | 0.86 | 0.22 (0.02) | 2.26E-25 |
EA: European Ancestry; AA: African American; IRASFS: Insulin Resistance Atherosclerosis Family Study; HA: Hispanic American; Chr: chromosome; Position (Mb): Position in megabases using build 35; EA: effect allele; EAF: effect allele frequency; Beta: regression coefficient for the coded allele on hepatic steatosis (measured using liver attenuation controlled for scan penetrance) per one standard deviation increase in rank equivalent to 1 standard deviation increase in normally transformed trait value; SE: standard error of the beta; P-value: P-value of association
rs2228603: N=1,947 for AA meta-analysis and N=9,965 for all meta-analysis.
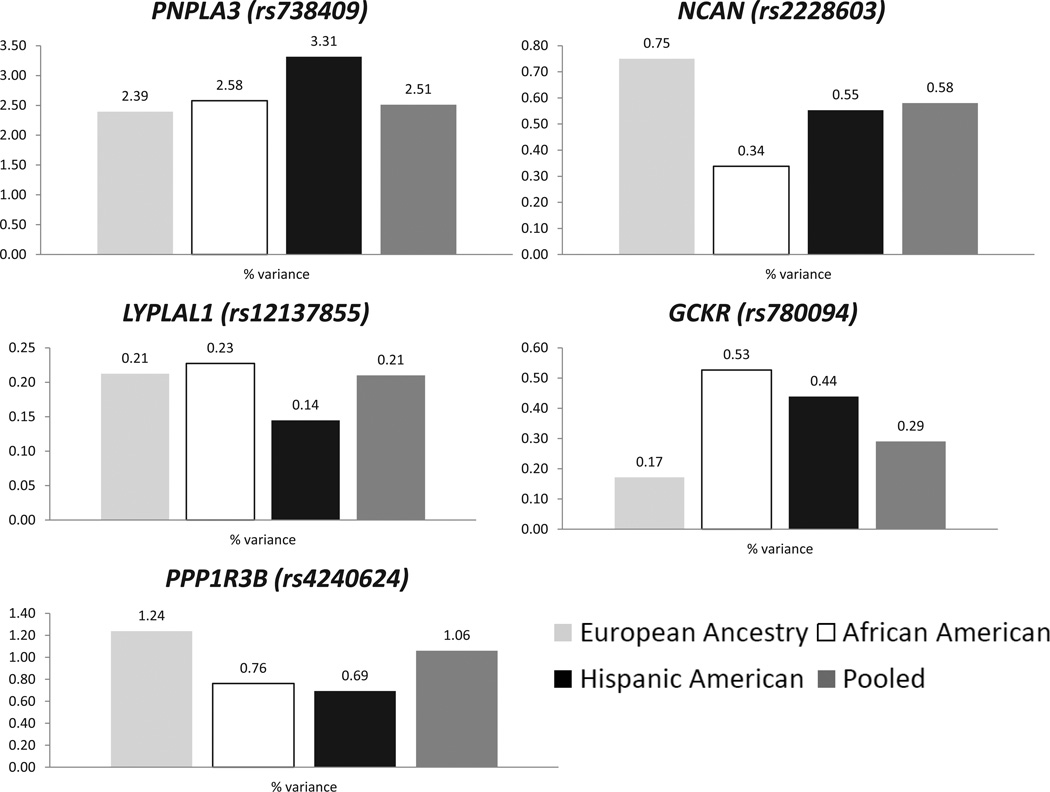
The total variances explained across the five loci were: 4.76%, 3.63%, and 5.64%, in individuals of European Ancestry, African Americans, and Hispanic Americans, and 4.56% of variance in all individuals combined.
Figure 1. Forest plot of the effect for hepatic steatosis index variants in European-Ancestry, African-American, and Hispanic-American populations.
For each race and a pooled analysis, the effect allele (EA) and frequency (EAF) are listed with the corresponding p-value (p), beta value (beta) and 95% confidence interval (95% CI) for association with hepatic steatosis adjusting for age, age2, gender, alcoholic drinks, and population substructure. Values greater than zero indicate an increase in the amount of hepatic steatosis. The solid vertical line represents no effect (beta=0).
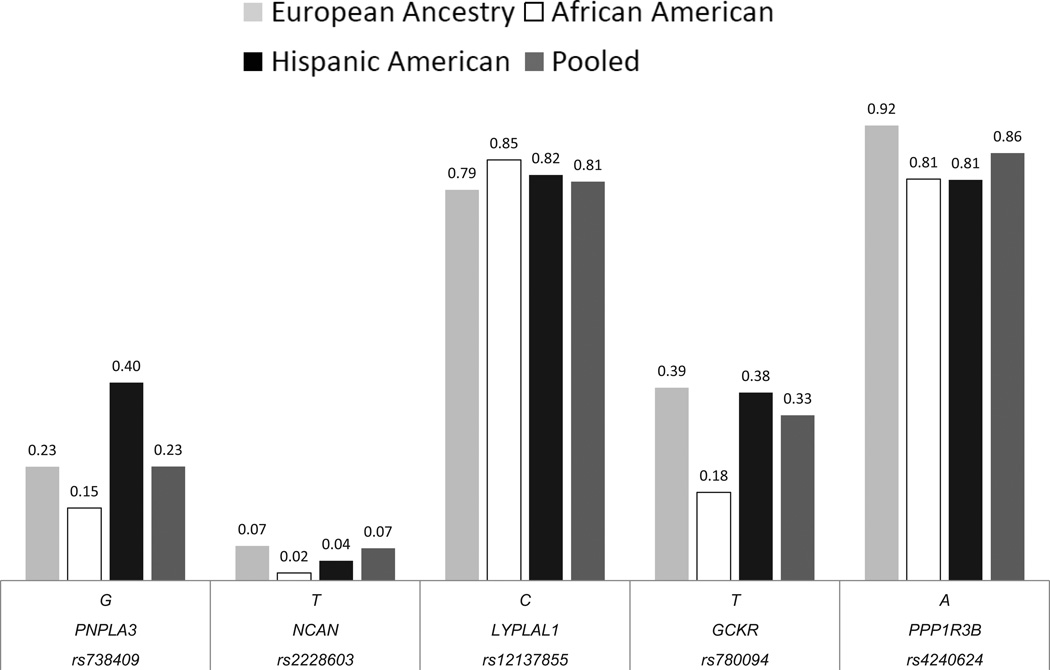
Figure 2. Allele frequency distribution for hepatic steatosis index variants in European-Ancestry, African-American, and Hispanic-American populations.
For each index variant, the effect allele is listed with a corresponding bar denoting the frequency observed in European-Ancestry (gray), African-American (white), and Hispanic-American (black) populations as well as in the pooled analysis (dark gray).
Meta-analysis of all available data across racial groups bears out these trends supporting a consistent role for variants near PNPLA3, PPP1R3B, and NCAN with diminished evidence of association with GCKR and LYPLAL1. However, the effect of the GCKR variant remains consistent, but modest, across all racial groups and the lack of significance may be more an issue of power. In contrast, the results indicate lack of evidence for association of the variant near LYPLAL1 in African and Hispanic Americans.
The variance explained varied by locus and ancestry (Figure 3). In particular, PNPLA3 (rs738409), explained the most variance in Hispanic Americans (3.31%), less in African Americans (2.58%) and the least in European-ancestry populations (2.39%); this could be attributed to differences in the allele frequency of the fatty liver promoting variant (G) as the effect size of this allele was roughly equivalent across ancestries. PPP1R3B (rs4240624), in contrast, had an effect size in European-ancestry populations that was twice that in the other ancestries whereas its frequency was roughly the same across the three groups. As a consequence this variant explained a higher proportion of the variance in European-ancestry individuals (1.24%) than African and Hispanic Americans (0.76% and 0.69%, respectively). GCKR (rs780094) had the same effect across ancestries but its frequency in African Americans was half of that in European-ancestry individuals and Hispanic Americans which were about equal (Figure 2). The LYPLAL1 (rs12137855) association signal was not significant in African and Hispanic Americans making the slight differences in the actual association signals across ancestries difficult to interpret; further evaluation of this locus in Hispanic Americans by fine mapping is warranted. Finally, NCAN (rs2228603) had a strong effect in European-ancestry individuals and African Americans but possibly goes in the opposite direction in Hispanic Americans (Figure 1). The total variances explained across the five loci were: 4.76%, 3.63%, and 5.64%, in individuals of European Ancestry, African Americans, and Hispanic Americans, and 4.56% of variance in all individuals combined.
Figure 3. Weighted average of the variance explained by each SNP for European-Ancestry, African-American, and Hispanic-American populations.
For each index variant, the corresponding bars represent the proportion of variance explained in European-Ancestry (gray), African-American (white), and Hispanic-American (black) populations as well as in the pooled analysis (dark gray).
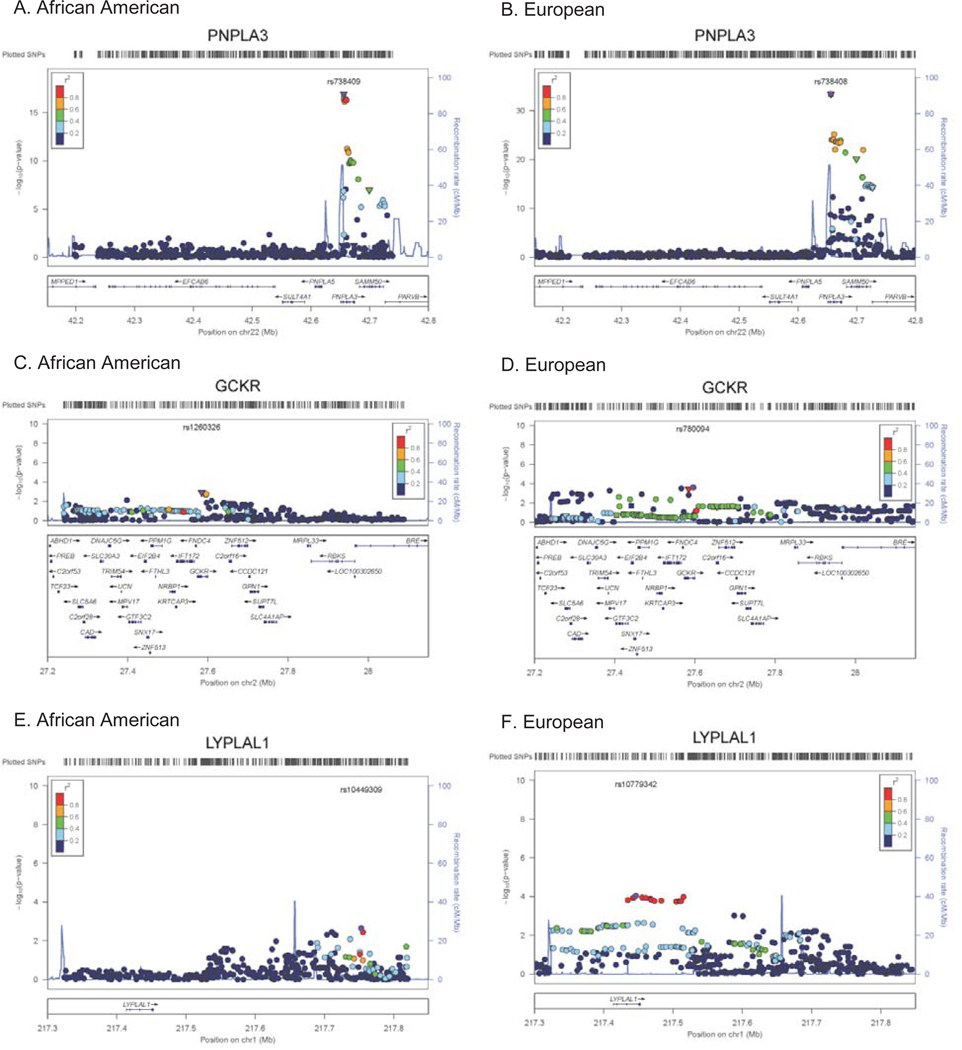
Fine-mapping of these loci in African Americans revealed that the strongest associating variants at PNPLA3 and GCKR were missense variants (rs738409, Figure 4A and 4B and rs1260326, Figure 4C and 4D, respectively). Analysis of the PPP1R3B locus revealed a similar pattern of association as seen in European-ancestry individuals (Appendix Figure 2A and 2B). The association signal at LYPLAL1 in African Americans (rs10449309, P-value=0.00223) appears to be distinct from that observed in European-ancestry populations and is separated by a recombination hotspot (Figure 4E and 4F). Results from fine-mapping of the NCAN (Appendix Figure 2C and 2D) locus revealed a differential pattern of LD with no additional variants showing stronger association.
Figure 4. Regional plots of loci robustly assoicated with NAFLD in European-ancestry population and fine-mapped in the African-American population.
A) PNPLA3 in African Americans, B) PNPLA3 in European Americans, C) GCKR in African Americans, D) GCKR in European Americans, E) LYPLAL1 in African Americans, F) LYPLAL1 in European Americans. The variant most robustly associated is denoted in purple annotated by SNP ID with additional genotyped and imputed SNPs passing study-specific quality controls. SNPs are plotted with their meta-analysis p-values as a function of position (hg19). Shape of the data point is indicative of function, i.e. non-synonymous (▼) and no annotation (●), and color indicates LD (r2) with the previously identified variant taken from HapMap (red, r2=0.8–1.0; yellow, r2=0.6–0.8; green, r2=0.4–0.6; cyan, r2=0.2–0.4, and blue, r2<0.2). Estimated recombination rates (HapMap) reflect the local LD structure. Gene annotations were taken from the University of California Santa Cruz genome browser.
DISCUSSION
Here we explored the genetic underpinnings of NAFLD across ancestries. The prevalence of this condition has been shown to vary with individuals of Hispanic ancestry having more NAFLD than individuals of European ancestry who have higher prevalence of NAFLD than individuals of African ancestry. The underlying causes of these differences remain to be determined. Here we test whether previously identified genetic variants contributing to hepatic steatosis, a quantifiable component of NAFLD, yield similar effects in different racial populations.
We found evidence that hepatic steatosis was 20–34% heritable in African- and Hispanic-Americans families after accounting for variation associated with age, age2, gender, alcohol intake, and population substructure. This was similar to what has been reported in European-ancestry populations in previous studies of the same quantitative component of NAFLD (7), in children of Hispanic ancestry using hepatic steatosis quantified from liver biopsies or magnetic resonance scans (22), and in the IRASFS in which the heritability of liver density without control for scan penetrance (spleen) yielded modestly greater heritability estimates among African-Americans (h2=0.20) and Hispanic-Americans families (h2=0.35). (23) These results reinforce that development of NAFLD is partially genetically influenced.
We also tested whether specific variants that associate with hepatic steatosis in European-ancestry individuals associated with this phenotype in African and Hispanic Americans. We found that variants in or near PNPLA3 (rs738409), PPP1R3B (rs4240624), and NCAN (rs2228603) were significantly associated in African Americans and PNPLA3 (rs738409) in Hispanic Americans. Interestingly, the effect size of PNPLA3 (rs738409) was similar across the ancestries but the frequency of the effect allele (G) varied so that the variance explained in Hispanic Americans was higher than in European-ancestry individuals than in African Americans and consistent with the observed prevalence of NAFLD. This trend has been previously noted.(16) However, this was not a universal trend across the variants. PPP1R3B (rs4240624) had an effect size in European-ancestry individuals that was twice that in the other ancestries whereas its frequency was roughly the same across the three groups. As a consequence, this variant explains a higher proportion of the variance in European- than African- than Hispanic-ancestry individuals. The association signal was in the same region in European-ancestry and African-American individuals suggesting that a difference in LD across populations is not likely to explain this difference in effect. This suggests other factors may be important in determining the effect of this variant across ancestries (see below). The effect of GCKR (rs780094) was similar across ancestries but its frequency in African Americans was half of that in European-ancestry and Hispanic-American individuals (which were about equal). LYPLAL1 (rs12137855) had, if anything, a small, or arguably no effect in African and Hispanic Americans. This may be, in part, due to the signal not being present or being different (see fine-mapping) in African and Hispanic Americans; further evaluation of this locus in Hispanic Americans by fine-mapping is warranted. Finally, NCAN (rs2228603) has a strong effect in European-ancestry individuals and African Americans; however the direction of effect of the alleles is in the opposite direction in Hispanic Americans. Whether this observation is due to differences in LD, power, or interactions with other genes and/or environments in Hispanic Americans compared with those of other ancestries remains to be elucidated. Since the fatty liver promoting allele is low frequency in all ancestries and particularly rare in African Americans, sequencing may be required to find the causal variant(s) at this locus.
We took advantage of LD differences between ancestries to fine-map the five NAFLD associated loci in individuals of African ancestry. We found that at PNPLA3 (rs738409) and GCKR (rs1260326), among the strongest associated variants in African Americans, were missense variants that have been shown to have functional consequences. (24, 25) Recent reports propose that PNPLA3 helps to break down triglycerides in the liver. (26) Interestingly, mice lacking PNPLA3 do not develop fatty liver (27) whereas overexpression of the mutant (but not wild-type) PNPLA3 in mouse liver does lead to steatosis (24) suggesting that the variant in PNPLA3 may exert its effects via a gain of function. GCKR sequesters glucose kinase (GK) in the nucleus and is also a competitive inhibitor of GK preventing it from phosphorylating glucose. (25, 28) Phosphorylated glucose is a precursor to formation of glycerol and of fatty acids, both of which are needed for formation of triglycerides. The rs1260326 variant eliminated GCKR activity and results in disinhibition of GK leading to glucose to triglyceride shifts. (25, 28) Therefore, our data combined with the above findings suggests that these associated variants may be functional across ancestries.
The differences in the variance explained by PNPLA3 (rs738409) across ancestries may be due, in part, to differences in allele frequency but may also be due to gene-environment interactions. In FamHS it has been found that PNPLA3 (rs738409) has an interaction with visceral adipose tissue and gender having more of an effect in women and in those with higher levels of visceral fat. (29) In the IRASFS, African Americans had lower visceral adipose tissue than Hispanic Americans which may, in part, account for the lower effect of PNPLA3 on hepatic steatosis.(30) Further, in a study of 127 children and adolescents, researchers found that the ratio of n-6 to n-3 polyunsaturated fatty acids (PUFAs) interacted with the GG gentoype at rs738409 in PNPLA3 promoting hepatic steatosis. This suggests a nonadditive increase in hepatic steatosis in individuals with the GG gentoype at rs738409 when consuming higher n-6 versus n-3 PUFAs. (31) Whether this or other dietary interactions account for some of the effects of PNPLA3 (rs738409) on hepatic steatosis across ancestries remains to be determined.
We further fine-mapped the signal at LYPLAL1 and found that the strongest association signal in individuals of African ancestry (rs10449309) may not be the same as in European-ancestry populations (rs10779342). Indeed, the strongest association signal was distal of the nearby recombination interval in African Americans whereas in European-ancestry individuals it was proximal to this landmark. However, there could be two signals near LYPLAL1 in European-ancestry populations, one of which corresponding to the signal seen in African Americans. This signal may be better defined with increased European-ancestry sample sizes in future studies. Alternatively, it may also be better defined through functional experiments.
A limitation of our study is that it uses cross-sectional cohort data that may not accurately depict genetic risk over time which has been shown to be cumulative and larger than in cross-sectional designs.(32) The analyses presented herein mirror those of the original report (7) with covariates that are not hypothesized to be in the causal pathway between the SNP and development of NAFLD. It remains to be determined whether the effects of our variants on variation in NAFLD may be mediated through changes in insulin resistance, obesity, serum lipid levels, or other mechanisms. Formal mendelian randomization experiments can be carried out in the future to help elucidate these possibilities. Additionally, Hispanic Americans were only represented by one study and will require replication in more cohorts to verify effects with increased power. Finally, this was a study of hepatic steatosis and the effects of these variants on developing nonalcoholic steatohepatitis and/or fibrosis remain to be determined.
This is the largest study to date assessing the effects of genetic variants across ancestries for hepatic steatosis, a quantifiable component of NAFLD. This report confirms the effects of variants previously identified in European-ancestry populations in African and Hispanic populations. In addition, fine-mapping efforts in the African American population have identified a novel association region in LYPLAL1 and implicated a missense variants in GCKR (rs1260326) and PNPLA3 (rs738409) in NAFLD pathology. These results demonstrate how cross ancestry comparisons of association results can help fine-map association signals. This is the first study to show heterogeneity of genetic effects across ancestries suggesting that other elements besides genotype may affect the expressivity of these variants. Clinically, our findings shows that genetic predisposition to disease can vary across ancestries and in this way may contribute to health disparities between groups. Further, by characterizing the mechanisms by which these genetic variants act across ancestries we may be able to identify the genes and biological pathways that they act through so that we can target these to prevent or treat this disease more effectively in the future. We have also identified the likely causal variants that predispose to NAFLD across ancestries at two loci which may be used as part of a cumulative genetic NAFLD risk marker panel in the future. Finally, although we show that development of NAFLD can be genetically influenced, much of the variation in the development of the trait is not genetic and thus leaves much hope and promise for being able to abrogate the disease in the future by changing environmental influences.
Supplementary Material
Acknowledgments
Financial Support:
Support for the Jackson Heart Study was provided by National Heart Lung and Blood Institute and the National Center on Minority Health and Health Disparities grants N01-HC95170, N01-HC95171 and N01-HC95172. Support for the Insulin Resistance Atherosclerosis Family Study was provided by the National Heart, Lung and Blood Institute grants 5R01HL060944, 5R01HL061019, 5R01HL060919, 5R01HL060894, and 5R01HL061210. Support for the Genetic Epidemiology Network of Arteriopathy was provided by the National Institutes of Health, grant numbers HL085571, HL087660, and HL100245 from National Heart, Lung, Blood Institute. Support for Family Heart Study was provided by the National Heart, Lung and Blood Institute grant 5R01HL08770003. IRASFS genotyping was carried out with funds from the Department of Internal Medicine at the University of Michigan. Analysis was partially supported by the Mid-Atlantic Nutrition Obesity Research Center (P30 DK072488) from the National Institute of Diabetes and Digestive and Kidney Diseases. In addition, we would like to acknowledge the National Institute of Diabetes and Digestive and Kidney Diseases (R00DK081350, NDP), the American Diabetes Association Mentor-Based Postdoctoral Fellowship Program (7-07-MN-08, RH), the National Institute of Diabetes and Digestive and Kidney Diseases (K23 DK080145 EKS and BK), the Doris Duke Charitable Foundation (Grant 2012067, EKS and BK), and the Department of Internal Medicine at the University of Michigan (EKS and BK).
Abbreviations
- NAFLD
Nonalcoholic Fatty Liver Disease
- PNPLA3
Patatin-like Phospholipase Domain Containing 3
- NCAN
Neurocan
- LYPLAL1
Lysophospholipase-like 1
- GCKR
Glucokinase Regulator
- PPP1R3B
Protein Phosphatase 1, Regulatory Subunit 3B
- NASH
Nonalcoholic Steatohepatitis
- TG
Triglycerides
- HDL
High-density Lipoprotein
- CT
Computed Tomography
- h2
Heritability
- GWAS
Genome-wide Association Study
- GOLD
Genetics of Obesity-related Liver Disease
- SNPs
Single Nucleotide Polymorphisms
- LD
Linkage Disequilibrium
- JHS
Jackson Heart Study
- JHS-ARIC
Jackson Heart Study-Atherosclerosis Family Study
- IRASFS
Insulin Resistance Atherosclerosis Family Study
- GENOA
Genetic Epidemiology Network of Arteriopathy
- FamHS
Family Heart Study
- SOLAR
Sequential Oligogenic Linkage Analysis Routines
- CEU
CEPH (Utah residents with ancestry from northern and western Europe)
- PUFAs
polyunsaturated fatty acids
Contributor Information
Nicholette D Palmer, Email: nallred@wfubmc.edu.
Solomon K Musani, Email: smusani@umc.edu.
Laura M Yerges-Armstrong, Email: lyerges@medicine.umaryland.edu.
Mary F Feitosa, Email: mfeitosa@wustl.edu.
Lawrence F Bielak, Email: lfbielak@umich.edu.
Ruben Hernaez, Email: rhernaez@jhsph.edu.
Bratati Kahali, Email: bratati@med.umich.edu.
J Jeffrey Carr, Email: jcarr@wakehealth.edu.
Tamara B Harris, Email: harrista@nia.nih.gov.
Min A Jhun, Email: minajhun@umich.edu.
Sharon LR Kardia, Email: skardia@umich.edu.
Carl D Langefeld, Email: clangefe@wakehealth.edu.
Thomas H Mosley, Jr, Email: tmosley@umc.edu.
Jill M Norris, Email: Jill.Norris@ucdenver.edu.
Albert V Smith, Email: albert@hjarta.is.
Herman A Taylor, Email: htaylor@umc.edu.
Lynne E Wagenknecht, Email: lwgnkcht@wakehealth.edu.
Jiankang Liu, Email: jliu@umc.edu.
Ingrid B Borecki, Email: iborecki@wustl.edu.
Patricia A Peyser, Email: ppeyser@umich.edu.
Elizabeth K Speliotes, Email: espeliot@umich.edu.
REFERENCES
- 1.Bellentani S, Marino M. Epidemiology and natural history of non-alcoholic fatty liver disease (NAFLD) Ann Hepatol. 2009;8(Suppl 1):S4–S8. [PubMed] [Google Scholar]
- 2.Harrison SA, Neuschwander-Tetri BA. Nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Clin Liver Dis. 2004;8:861–879. doi: 10.1016/j.cld.2004.06.008. ix. [DOI] [PubMed] [Google Scholar]
- 3.Speliotes EK, Massaro JM, Hoffmann U, Vasan RS, Meigs JB, Sahani DV, Hirschhorn JN, et al. Fatty liver is associated with dyslipidemia and dysglycemia independent of visceral fat: the Framingham Heart Study. Hepatology. 2010;51:1979–1987. doi: 10.1002/hep.23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stepanova M, Younossi ZM. Independent Association Between Nonalcoholic Fatty Liver Disease and Cardiovascular Disease in the US Population. Clin Gastroenterol Hepatol. 2012;10:646–650. doi: 10.1016/j.cgh.2011.12.039. [DOI] [PubMed] [Google Scholar]
- 5.Adams LA, Lymp JF, St Sauver J, Sanderson SO, Lindor KD, Feldstein A, Angulo P. The natural history of nonalcoholic fatty liver disease: a population-based cohort study. Gastroenterology. 2005;129:113–121. doi: 10.1053/j.gastro.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 6.Hamaguchi M, Kojima T, Itoh Y, Harano Y, Fujii K, Nakajima T, Kato T, et al. The severity of ultrasonographic findings in nonalcoholic fatty liver disease reflects the metabolic syndrome and visceral fat accumulation. Am J Gastroenterol. 2007;102:2708–2715. doi: 10.1111/j.1572-0241.2007.01526.x. [DOI] [PubMed] [Google Scholar]
- 7.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, et al. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7:e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limanond P, Raman SS, Lassman C, Sayre J, Ghobrial RM, Busuttil RW, Saab S, et al. Macrovesicular hepatic steatosis in living related liver donors: correlation between CT and histologic findings. Radiology. 2004;230:276–280. doi: 10.1148/radiol.2301021176. [DOI] [PubMed] [Google Scholar]
- 9.Kodama Y, Ng CS, Wu TT, Ayers GD, Curley SA, Abdalla EK, Vauthey JN, et al. Comparison of CT methods for determining the fat content of the liver. AJR Am J Roentgenol. 2007;188:1307–1312. doi: 10.2214/AJR.06.0992. [DOI] [PubMed] [Google Scholar]
- 10.Bohte AE, van Werven JR, Bipat S, Stoker J. The diagnostic accuracy of US, CT, MRI and 1H-MRS for the evaluation of hepatic steatosis compared with liver biopsy: a meta-analysis. Eur Radiol. 2011;21:87–97. doi: 10.1007/s00330-010-1905-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernaez R, Lazo M, Bonekamp S, Kamel I, Brancati FL, Guallar E, Clark JM. Diagnostic accuracy and reliability of ultrasonography for the detection of fatty liver: a meta-analysis. Hepatology. 2011;54:1082–1090. doi: 10.1002/hep.24452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Browning JD, Szczepaniak LS, Dobbins R, Nuremberg P, Horton JD, Cohen JC, Grundy SM, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40:1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 13.Fan JG, Zhu J, Li XJ, Chen L, Li L, Dai F, Li F, et al. Prevalence of and risk factors for fatty liver in a general population of Shanghai, China. J Hepatol. 2005;43:508–514. doi: 10.1016/j.jhep.2005.02.042. [DOI] [PubMed] [Google Scholar]
- 14.Mohanty SR, Troy TN, Huo D, O'Brien BL, Jensen DM, Hart J. Influence of ethnicity on histological differences in non-alcoholic fatty liver disease. J Hepatol. 2009;50:797–804. doi: 10.1016/j.jhep.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuqua SR, Wyatt SB, Andrew ME, Sarpong DF, Henderson FR, Cunningham MF, Taylor HA., Jr Recruiting African-American research participation in the Jackson Heart Study: methods, response rates, and sample description. Ethn Dis. 2005;15:S6-18-29. [PubMed] [Google Scholar]
- 18.Henkin L, Bergman RN, Bowden DW, Ellsworth DL, Haffner SM, Langefeld CD, Mitchell BD, et al. Genetic epidemiology of insulin resistance and visceral adiposity. The IRAS Family Study design and methods. Ann Epidemiol. 2003;13:211–217. doi: 10.1016/s1047-2797(02)00412-x. [DOI] [PubMed] [Google Scholar]
- 19.Investigators TF. Multi-center genetic study of hypertension: The Family Blood Pressure Program (FBPP) Hypertension. 2002;39:3–9. doi: 10.1161/hy1201.100415. [DOI] [PubMed] [Google Scholar]
- 20.Higgins M, Province M, Heiss G, Eckfeldt J, Ellison RC, Folsom AR, Rao DC, et al. NHLBI Family Heart Study: objectives and design. Am J Epidemiol. 1996;143:1219–1228. doi: 10.1093/oxfordjournals.aje.a008709. [DOI] [PubMed] [Google Scholar]
- 21.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwimmer JB, Celedon MA, Lavine JE, Salem R, Campbell N, Schork NJ, Shiehmorteza M, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136:1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagenknecht LE, Scherzinger AL, Stamm ER, Hanley AJ, Norris JM, Chen YD, Bryer-Ash M, et al. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring) 2009;17:1240–1246. doi: 10.1038/oby.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285:6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beer NL, Tribble ND, McCulloch LJ, Roos C, Johnson PR, Orho-Melander M, Gloyn AL. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18:4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang Y, Cohen JC, Hobbs HH. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J Biol Chem. 2011;286:37085–37093. doi: 10.1074/jbc.M111.290114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basantani MK, Sitnick MT, Cai L, Brenner DS, Gardner NP, Li JZ, Schoiswohl G, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52:318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rees MG, Wincovitch S, Schultz J, Waterstradt R, Beer NL, Baltrusch S, Collins FS, et al. Cellular characterisation of the GCKR P446L variant associated with type 2 diabetes risk. Diabetologia. 2012;55:114–122. doi: 10.1007/s00125-011-2348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Graff M, North KE, Franceschini N, Reiner AP, Feitosa M, Carr JJ, Gordon-Larsen P, et al. PNPLA3 gene-by-visceral adipose tissue volume interaction and the pathogenesis of fatty liver disease: The NHLBI Family Heart Study. Int J Obes (Lond) 2012 doi: 10.1038/ijo.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagenknecht LE, Palmer ND, Bowden DW, Rotter JI, Norris JM, Ziegler J, Chen YD, et al. Association of PNPLA3 with non-alcoholic fatty liver disease in a minority cohort: the Insulin Resistance Atherosclerosis Family Study. Liver Int. 2011;31:412–416. doi: 10.1111/j.1478-3231.2010.02444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santoro N, Savoye M, Kim G, Marotto K, Shaw MM, Pierpont B, Caprio S. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS One. 2012;7:e37827. doi: 10.1371/journal.pone.0037827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.