Abstract
Multiple Myeloma (MM) is a complex disease driven by numerous genetic and epigenetic alterations. Comprehensive oncogenomic analysis indicates the presence of many highly recurrent and highly focal amplifications/deletions in the MM genome. Integrated oncogenomic analyses of human MM have identified candidates resident within regions of amplification/deletions predicted to be involved in MM pathogenesis and progression. The biological behavior and clinical outcome in MM is dependent on these molecular determinants, which are also attractive therapeutic targets. The data obtained from extensive analysis of patient samples with annotated clinical outcome have now provided insight into molecular mechanism of disease behaviour, help develop sensitive prognostic models, identified novel therapeutic targets provided the framework for the development of molecularly-based therapies and eventually help develop individualized therapy to improve outcome whith reduced toxicity.
Introduction
Multiple myeloma (MM) is characterized by a significant heterogeneity at multiple levels, i.e., clinical presentation, biologic characteristics, response to treatment, and clinical outcome. The current data supports the hypothesis that this heterogeneity is mainly related to molecular characteristics of the tumor clone. Although genetic changes are the hallmark of cancer cell, in many hematologic neoplasms, these changes are very limited (like in chronic myeloid leukemia, and most acute leukemias). In contrast, solid tumors usually present a wide variety of chromosomal and genomic rearrangements. Myeloma is probably in between these two extreme genetic landscapes. Actually, karyotypes in MM are usually complex, with both quantitative (chromosome number) and qualitative (chromosome structure) abnormalities.1–5 However, despite this complexity, several recurrent changes are observed, including hyperdiploidy,5 loss of chromosome 13,6–9 and specific translocations like t(11;14)(q13;q32),10–12 t(4;14)(p16;q32),13–16 or t(14;16)(q23;q32).17,18 The objective of this review is to look at how the currently available genomic data can help build a pathogenetic and prognostic models that could be used for patient management.
Pathogenesis
Although complex karyotypic recurrent changes have been described in myeloma, the karyotypic oncogenetic classification, mainly based on hyperdiploidy and 14q32 translocations19 is only partially confirmed by molecular studies. However, now myeloma has been evaluated at the transcriptional level, using gene expression profiling by several studies. In this approach, CD138+ purified malignant plasma cells are used to extract RNAs and hybridized on an array to evaluate expression of genes representative of the whole transcribed genome. Using unsupervised bioinformatic methods, tumors are then classified according to gene expression profile similarities. One of the first analysis compared gene expression profiling in cohorts of patients with monoclonal gammopathy of undetermined significance (MGUS) and MM demonstrating sequential genetic changes from normal plasma cells to malignant PCs in the process providing clues to molecular basis for malignant transformation as well as potential therapeutic targets (Figure 1).52 The first molecular classification in 2002 clustered genes according to similarities with either MGUS or human myeloma cell lines20, and identified 4 classes of MM, according to their similarities to MGUS or MM cell line profiles. In 2003, another study showed that the most relevant profiles were related to Ig gene expression.21 More recently, using un-supervised analyses, three reports identified subgroups mostly driven by chromosomal aberrations. The first report identified 8 different subgroups, mainly based on the cyclin D gene expression, and on the different 14q32 recurrent translocations.19 This molecular classification has been refined in 2006, identifying seven subclasses of myeloma.22 In this pathogenetic model, the first class is defined by the translocation t(4;14), identified by overexpression of the MMSET and/or FGFR3 genes. The second class is defined by upregulation of one of the MAF genes, related to the translocations t(14;16) or t(14;20). Cases with CCND1 or CCND3 upregulation (due to the translocations t(11;14) or t(6;14)) clustered in two different groups, named CD1 and CD2. CD2 group was characterized by CD20 expression. The fifth group was characterized by hyperdiploidy. The last two groups were characterized by a low incidence of bone disease, according to a low DKK1 expression, whereas the last group was characterized by high expression of genes involved in proliferation. This molecular classification has been partially confirmed and further refined by a recent study by the HOVON group.23 Although the “low bone disease” group was not confirmed, three other sub-groups were identified: one group enriched by “myeloid” genes (that could be related to plasma cell sorting problems), one group characterized by overexpression of cancer testis antigen genes, and finally a group defined by overexpression of positive regulators of the NFκB pathway.
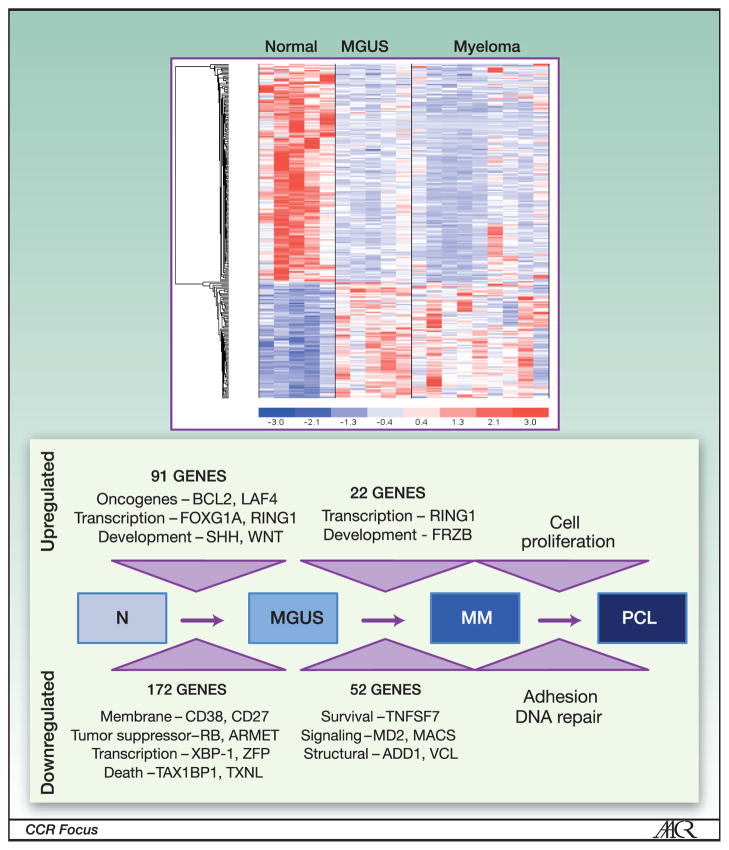
Figure 1. Progression to Myeloma - Gene Expression Analysis.
High-throughput gene expression profile identifies distinct pattern in myeloma and MGUS compared to normal plasma cells (above). Analysis of the expression profile data identifies a multistep model of progression from normalplasma cells to MGUS cells, and to multiple myeloma (MM) PCL- plasma cell leukemia. Figure adapted from Davies et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003;102:4504-4511. © the American Society of Hematology.
DNA-based techniques such as array-CGH.24,25 have identified the role of NFκB pathway in myeloma. In two separate studies, it has been shown that the NFκB pathway can be activated, either by deletions of NFκB inhibitors (such as TRAF3 or CYLD), or by activation of NFκB activators (such as NIK or CD40). Other studies based on the analysis of copy number changes by high-density SNP-array has identified other levels of molecular heterogeneity.26,27,49,50 These studies identified genomic heterogeneity within the hyperdiploid group driven by the presence of either chromosome 1q gain and/or chromosome 11 gain, chromosome 13 loss or chromosome 5 gain conferring significant outcome difference. Furthermore these studies demonstrated that integration of copy number changes and gene expression values allowed to convert genomic heterogeneity into identification of potential cancer target genes. Subclasses of hyperdiploid multiple myeloma patients with clinical and biological associations were also characterized by gene expression profiling.28 Initial attempts at understanding the genesis of these genomic heterogeneity resides in uncontrolled recombination mechanisms which may become potential target to understand the biology as well as develop effective therapeutic strategy.29
To summarize the tremendous work performed on molecular description of myeloma, a huge heterogeneity is present, that so far prevents the identification of specific definitive entities to dissect myeloma into different disease sub-groups. However, it is interesting to note that the emerging classifications are essentially based on recurrent chromosomal changes. This situation is highly reminiscent of the non-Hodgkin lymphomas. In this latter tumors, their classification is based on chromosomal changes specific of each entities, such as t(11;14) for mantle cell lymphoma, t(8;14) for Burkitt’s lymphoma, or t(14;18) for follicular lymphoma. Further work is needed to confirm such a classification.
Prognosis
If molecular studies have so far not provided a definite myeloma subclassification into specific diseases correlating with biology or clinical behavior, they have definitely contributed to identification of several prognostic factors that significantly influence the patient outcome. By conventional cytogenetics recurrent chromosomal changes have been identified and correlated with clinical outcome (Table 1). The abnormalities such as t(4;14), t(14;16), part or whole chromosome 13 deletion as well as loss of 17p13 carry a poor prognosis in patients undergoing high-dose therapy; while hyperdiploidy and t(11;14) translocations are associated with better outcome. The significance of chromosome 13 deletion remains enigmatic as it is also observed in patients with MGUS with unclear relationship to its transformation to myeloma.
Table 1.
Recurrent cytogenetic changes in myeloma
| Common cytogenetic alterations | |
|---|---|
| Hyperdiploidy: | 50–60% |
| t(4:14): | 15% |
| t(11;14): | 20% |
| t(14;16): | 3% |
| t(14;20): | 1% |
| del; 13 or 13q: | 45% |
| del 17p : | 8% |
| Recently identified alterations | |
| 1q+ : | 35% |
| 1p−: | 30% |
| 5q+: | 50% |
| 12p−: | 10% |
As myeloma cells have low proloiferative index, prognostic significance of genetic abnormalities is analyzed by interphase fluorescence in situ hybridization (FISH). Among the specific 14q32 translocations discussed previously, t(4;14) is definitely the most important one from a clinical point of view. Number of studies have confirmed that patients who display this translocation (15% of the patients) have a specifically poor prognosis.30–35 Interestingly, these patients may require a specific therapeutic approach to include the novel agents such as proteasome inhibitor or immunomodulatory agents. Previously reported del13 is not considered to predict poor outcome by itself. The most important chromosomal change for prognosis is del(17p). Present in 8–10% of the patients, this deletion is associated with a remarkably short survival, irrespective of the therapy utilized.34,36,37 The molecular target of this deletion could be TP53, but no clear biological evidence supports this hypothesis, and mutations are observed in only a subset of patients with del(17p).38 Finally, several reports have shown that gains of chromosome 1q (observed in one third of the patients) also confer a poor prognosis.39,40 This abnormality is typically a secondary event, not specific of myeloma, acquired during evolution.
The prognostic significance of molecular changes have been analyzed by high-throughput microarray profiling techniques, focused on copy number alteration using either SNP array or gene expression profiling. These techniques are potentially more powerful since they analyze the whole genome.
An analysis of genome-wide copy number alterations (CNA) in 192 newly-diagnosed uniformly-treated patients with MM using high-density SNP arrays suggested a global genomic instability in MM.43 One of three distinct patterns of CNAs are present in 98% cases: loss or gain of the chromosome, loss or gain of a whole arm of the chromosome, and/or interstitial losses or gains. Analysis of the most frequent lesions (>10%) have identified two main groups: the first group encompasses almost exclusively (with the exception of chromosome 11) either gain or loss of entire chromosomes or interstitial gain or loss of the flanking centromeric regions. This group includes gains of chromosomes 3, 5, 7, 9, 11, 15, 18, 19, 21 and loss of chromosomes 13, 22 and X (in female cases). The second group is characterized by genetic lesions that affect gain or loss of sub-chromosomal material, including amplification of 1q and 6p as well as deletion of 1p, 6q, 8p, 12p, 14q, 16p, 16q and 20p. The analysis of prognostic significance of CNAs in MM identified amplifications of 1q and deletions of 1p, 12p, 14q, 16q, and 22q to be associated with poor prognosis, while amplifications of chromosomes 5, 9, 11, 15, and 19 conferred a superior outcome. A multivariate analysis identified a prognostic model that includes amp(1q23.3), amp(5q31.3), and del(12p13.31) as the most powerful independent adverse markers (P < .0001) and the prognostic significance of the model has been validated in an independent cohort of 273 patients with MM. These findings therefore demonstrate the feasibility of molecular karyotyping using SNP profiling to predict outcome in MM. This prognostic model has to be confirmed in independent series. Recurrent cytogenetic changes in myeloma are listed in Table 150
Two large studies have evaluated prognostic significance of gene expression profiling to identify poor-risk patient populations. One of the 2 models, the UAMS 70-gene model, has 30% of the informative genes mapped to chromosome 1.41 In the other model, the IFM 15-gene model, high-risk patients were enriched in genes controlling proliferation and chromosomal instability, whereas low-risk patients were enriched in hyperdiploid karyotypes.42 It is interesting to note that the two models do not have a single common gene, reflecting mainly the redundancy in the genes and pathways that control growth, proliferation and survival besides differences in platforms used for the microarray analyses and/or differences in the treatment used to define the patient population. However, in an attempt to validate the techniques, the IFM 15-gene set was shown to be powerful in the UAMS population, but with a lower significance.42 Interestingly, both sets identified patients with a short survival, but none of them identified very good-risk patients, probably because of a short follow-up. An international large-scale effort is needed to fully validate a uniform set of genes predictive of outcome irrespective of the treatment used to make gene expression profiling a routine in clinical practice. Moreover as CpG methylation affects gene expression and thus may be relevant to pathogenesis and behavior of myeloma cells, a genome-wide methylation profile have been analyzed using microarray. In a recent study methylation patterns, especially hypomethylation, was capable of distinguishing nonmalignant from malignant plasma cells.51 In fact differential methylation was also evident at transition of MGUS cells to MM cells. Interestingly, genes involved in cell-cell signaling and cell adhesion were remethylated in cells from plasma cell leukemia stage suggesting development of independence from the interaction with bone marrow microenvironment cells.
Recently two transcriptome modifiers have been investigated in myeloma. Alternate splicing is an important post translational change that alters specificity of gene function. Dysregulated alternative splicing has been reported in myeloma with effect on overall clinical outcome.60 MicroRNA, are small noncoding RNA molecules that regulate multiple target genes through cleavage of targeted transcripts and by inducing translational inhibition. Differential expression of number of microRNAs have been described in myeloma and MGUS compared to normal plasma cells.53 In one study miRs -21, -106b-25 and -181a/b were overexpressed in MM and MGUS with respect to normal PCs, while miRs-32, and -17-92, were exclusively over expressed in MM compared to MGUS. Two target genes of over-expressed miRs, SOCS-1 and p300-CBP, were identified as having influence on myeloma pathogenesis. Down-regulation of miRs 15a/-16 present on Chromosome 13 has also been described with potential effect on MM cell proliferation,54 however its relation with chromosome 13 or 13q34 deletion is not established.55 Some relation between miR expression pattern and molecular and genetic subgroups in myeloma have been described56,57; e.g. overexpression of miR-let 7e, -125-5p and -99b located at 19q13.33 in patients with t(4;14) translocation56 and miRs -1 and -133a in t(14;16) MM is reported.57 Combined mRNA and miR profiling has identified microRNA/mRNA regulatory network with early evidence of differential expression in high-risk disease.58 miRs -192, -194 and -215 downregulated in subset of MM patients is correlated with transcriptional activation by p53 and modulation of MDM2 expression suggesting these miRNAs as positive regulators of p53 with important role in MM development.59 Unsupervised clustering analysis of microRNA expression profile data also identifies groups with different survival outcome recognizing critical microRNAs as modulators of gene expression and signaling pathways and provides potential novel microRNA and gene targets in MM to both understand biological behavior and for therapeutic application.61
Therapeutic implications
So far, treatment options were especially driven by age and/or physiological conditions. In patients under 65 years of age, the standard of care is usually a short induction followed by a high-dose melphalan with stem cell rescue. For older patient or for ones with co-mobidities, long-term treatment with a combination is usually chosen. However, the availability and understanding of genomic data has significantly contributed to therapy of myeloma. First both in vitro and in vivo models have been developed to characterize MM cell-bone marrow stromal cell (BMSC) interactions, as well as signaling pathways controlling growth, survival, drug resistance, or migration within the BM milieu. These studies have delineated the signaling cascades and molecular mechanisms and identified that MM cell growth is mediated via ERK/MAPK, survival via JAK/STAT, drug resistance via PI3-K/Akt, and migration via PKC dependent signaling cascades. These systems have been used to identify potential novel therapeutic targets, as well as validate novel targeted therapies. The genomic studies especially the gene expression profiling confirmed the significance of these targets as well as the role and effects of agents directed at these targets. These studies led to the development and FDA approval of bortezomib, a proteasome inhibitor and thalidomide and Lenalidomide, an immunomodulator in MM. Other representative novel agents include the newer proteasome inhibitors carfilzomib and NPI-0052; multitargeted kinase inhibitor PKC-412; histone deacetylase (HDAC) inhibitors; heat shock protein 90 (hsp90) inhibitor, telomerase inhibitors; small molecule inhibitors against Akt (perifosine); cyclin-dependent kinase inhibitors; as well as anti-CD40, anti-CD56, and anti-CD138. The promising targets, agents and stage of their clinical development is listed in Table 2. Secondly, the genomic studies have begun to identify new targets e.g dickoff-1 (DKK-1) which have been validated in vitro and in vivo with translation to clinical studies. Thirdly, these studies have informed the development of combination therapies based on targeting of dual apoptotic pathways or different mechanisms of action or overcoming resistance to one of the agents. The list of such combinations is described in table 2.
Table 2.
Novel target in myeloma, agents and stage of ongoing clinical trials
| Cell surface targets | |||
|---|---|---|---|
| Target | Agent | Clinical Study Phase | Single Agent(S)/Combination(C) |
| FGF3 | Dasatinib | I/II | S |
| FGF, PDGF | (mAb)TKI258 | I | S |
| CD38 | mAb | I | S |
| CD40 | SGN-40 (mAb) | I/II | S, C (Lenalidomide) |
| HCD122 (mAb) | I | S | |
| CD56 | huN901-DM1 (C-mAb) | I | S |
| CS1 | HuLuc63 (mAb) | II/III | S, C (Lenalidomide, bortezomib) |
| CD138 | BT062 (mAb-DM4) | I | S |
| RANKL | AMG162 (mAb) | I/II | S |
| MUC1 | AR20.5 (mAb) | I/II | S |
| BAFFR | LY2127399 (mAb) | I/II | S |
| CD52 | Alemtuzumab (mAb) | II | S |
| TRAIL | Apo2L/TRAIL (Apo2 ligand) | I | S |
| Mapatumumab | I/II | S | |
| IGF1/R | IGF1R CP-571 (mAb) | I | S |
| EM164 (mAb) | I | S | |
| IL6/R | CNTO328 (mAb) | II/III | S, C (bortezomib) |
| Altizumab (mAb) | III | S | |
| VEGF/R | Bevacizumab (mAb) | II | S |
| SU5416 | II | S | |
| Zactima (ZD6474) | II | S | |
| DKK-1 | BHQ-880 | I/II | S |
| Activin A | I/II | S | |
| KIR | IPH101 | I/II | S |
| CXCR3 | AMD3100 | II | C (bortezomib) |
| Intracytoplasmic and/or nuclear targets | |||
| CDK | Alvocidib (NSC649890) | I | S |
| CDK and GSK3β | AT7519M | I/II | S, C (bortezomib) |
| IKK | RTA402 | I | S |
| Akt | perofosine | III | C (bortezomib) |
| HDAC | panabinostat | III | C (bortezomib) |
| Vorinostat | II/III | C (bortezomib) | |
| Romidopsin | II/III | C ( bortezomib) | |
| Farnesyltransferase | Tipifarnib (R115777) | II | S, C (bortezomib) |
| HSP90 | KOS953 | II | C ( bortezomib) |
| AUY922 | II | C (bortezomib) | |
| IPI504 | I/II | C (bortezomib) | |
| Proteasome | Carfilzomib | II/III | S, C (lenalidomide) |
| NPI-0052 | I | S | |
| MLN9708 | I | S | |
| Mitochondria | GCS-100 | I/II | C |
| mTOR | CCI-779 II | II | C (bortezomib) |
| RAD001 | II | C (lenalidomide, bortezomib) | |
| INK128 | II | S | |
| PKC | Enzastaurin | I/II | S, C (bortezomib) |
| Telomerease | GRN163L | I/II | S, C (bortezomib) |
Data collected from National Cancer Institute Clinical Trials website, Multiple
Myeloma Research Foundation website and the International Myeloma Foundation website. mAb, monoclonal antibody.
Finally, attempt to use genetic data in treatment selection has been proposed based on patients’ myeloma cell genetic characteristics, e.g. for patients displaying the t(4;14). A few studies did show that t(4;14)-patients may benefit from the use of bortezomib, either as induction therapy, or as long-term treatment.44–46 In some of these studies, the long-term use of bortezomib totally overcame the poor prognosis associated with t(4;14).44,45 For other high-risk parameters such as del(17p), or gene expression-defined high-risk disease, no specific treatment has so far demonstrated beneficial effect. Another important question would be to define a standard of care for very good-risk patients. However, these patients are not yet clearly identified, and long-term analyses are needed to try to define these patients, and then to possibly propose less toxic approaches for these patients. Finally, a major objective for individualized therapeutic approaches would be to define what is the best frontline or subsequent line combination for a specific patient. This objective requires genomic studies performed in well-defined populations of patients, treated with a specific combination (such as bortezomib-dexamethasone, or lenalidomide-dexamethasone), with a primary endpoint based on progression free survival. Several studies are currently ongoing.
Sequencing
Recently 2 reports have presented data on sequencing in myeloma. The first study utilized massively parallel whole genome paired end sequencing on 2 myeloma patient samples collected 6 months apart and identified 29 somatic rearrangements, including three that were present only in the second sample.47 One of these was on chromosome 13. Breakpoint sequencing revealed a 64.9Kb homozygous (no wild-type read pairs found) deletion involving the first two exons of the RB1 gene. No reads spanning this breakpoint were found in the matching sample taken six months earlier. A second much larger effort in 29 patients (22 whole genomes and 17 whole exomes) used 30X coverage deep sequencing identified number of unique recurrent biologically important mutations involving histone methyltransferases, transcription factor IRF4, BRAF, genes involved in protein translation, and surprisingly genes involved in blood coagulation. 48 These results early sequencing efforts provide important insight into the pathogenesis of disease progression .as well as confirms the potential of whole genome sequencing to inform biology of the disease that may affect the therapeutic approach in future.
Conclusion
To conclude, all the reported studies so far show that myeloma is characterized by a wide molecular heterogeneity. The next steps will reside in developing a combination of several molecular approaches (Figure 2), including copy number change analyses, gene expression profiling, massive parallel sequencing, miRNA analyses and epigenetic changes survey in large uniformly treated patient cohorts, in order to get a clear landscape of the molecular changes, and their impact in myeloma classification, prognosis, and ultimately therapeutic management.
Figure 2. High-throughput genomic analysis spanning all regulatory checkpoints.
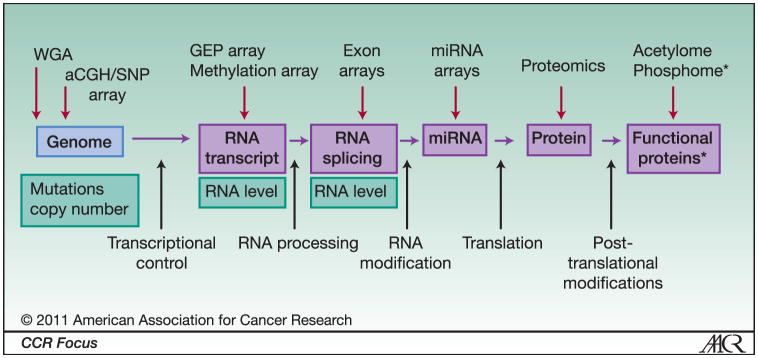
Genomic information is translated through various processes including post-translational protein modification (middle row). Abnormalities at these various levels potentially play a role in development of malignant transformation and behavior of the cancer cell (bottom row). Various high -throughput genomic analysis methods and arrays spanning all regulatory checkpoints are available to identify these various genomic abnormalities to develop an integrated approach that will lead to understanding of the molecular pathogenesis of cancer, identification of novel targets and therapies, development of personalized medicine, and predictive models for outcome. * protein modification such as phosphorylation, acetylation, ubiqiitination, sumoylation etc.
Acknowledgments
This study was supported in part by the National Institutes of Health Grant PO1 CA155258-01 (NCM and HAL), RO1-124929, P50-100007, PO1-78378, and Dept. of Veterans Affairs Merit Review Awards (NCM); and by Intergroupe Francophone du Myélome; and the French Institute National du Cancer Grant R09076NN (HAL).
References
- 1.Dewald GW, Kyle RA, Hicks GA, Greipp PR. The clinical significance of cytogenetic studies in 100 patients with multiple myeloma, plasma cell leukemia, or amyloidosis. Blood. 1985;66:380–90. [PubMed] [Google Scholar]
- 2.Sawyer JR, Waldron JA, Jagannath S, Barlogie B. Cytogenetic finding in 200 patients with multiple myeloma. Cancer Genet Cytogenet. 1995;82:41–9. doi: 10.1016/0165-4608(94)00284-i. [DOI] [PubMed] [Google Scholar]
- 3.Laï JL, Zandecki M, Mary JY, et al. Improved cytogenetics in multiple myeloma: a study of 151 patients including 117 patients at diagnosis. Blood. 1995;85:2490–7. [PubMed] [Google Scholar]
- 4.Calasanz MJ, Cigudosa JC, Odero MD, et al. Cytogenetic analysis of 280 patients with muliple myeloma and related disorders: Primary breakpoints and clinical correlations. Genes Chromosom Cancer. 1997;18:84–93. [PubMed] [Google Scholar]
- 5.Smadja NV, Bastard C, Brigaudeau C, Leroux D, Fruchart C. Hypodiploidy is a major prognostic factor in multiple myeloma. Blood. 2001;98:2229–38. doi: 10.1182/blood.v98.7.2229. [DOI] [PubMed] [Google Scholar]
- 6.Zojer N, Konigsberg R, Ackermann J, et al. Deletion of 13q14 remains an independent adverse prognostic variable in multiple myeloma despite its frequent detection by interphase fluorescence in situ hybridization. Blood. 2000;95:1925–30. [PubMed] [Google Scholar]
- 7.Desikan R, Barlogie B, Sawyer J, et al. Results of high-dose therapy for 1000 patients with multiple myeloma: durable complete remissions and superior survival in the absence of chromosome 13 abnormalities. Blood. 2000;95:4008–10. [PubMed] [Google Scholar]
- 8.Facon T, Avet-Loiseau H, Guillerm G, et al. Chromosome 13 abnormalities identified by FISH analysis and serum beta2-microglobulin produce a powerful myeloma staging system for patients receiving high-dose therapy. Blood. 2001;97:1566–71. doi: 10.1182/blood.v97.6.1566. [DOI] [PubMed] [Google Scholar]
- 9.Fonseca R, Harrington D, Oken MM, et al. Biological and prognostic significance of interphase fluorescence in situ hybridization detection of chromosome 13 abnormalities (delta13) in multiple myeloma: An eastern cooperative oncology group study. Cancer Res. 2002;62:715–20. [PubMed] [Google Scholar]
- 10.Chesi M, Bergsagel PL, Brents LA, et al. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 1996;88:674–81. [PubMed] [Google Scholar]
- 11.Fonseca R, Blood EA, Oken MM, et al. Myeloma and the t(11;14)(q13;q32): evidence for a biologically defined unique subset of patients. Blood. 2002;99:3735–41. doi: 10.1182/blood.v99.10.3735. [DOI] [PubMed] [Google Scholar]
- 12.Garand R, Avet-Loiseau H, Accard F, Moreau P, Harousseau JL, Bataille R. t(11;14) and t(4;14) translocations correlated with mature lymphoplasmacytoid and immature morphology, respectively, in multiple myeloma. Leukemia. 2003;17:2032–2035. doi: 10.1038/sj.leu.2403091. [DOI] [PubMed] [Google Scholar]
- 13.Chesi M, Nardini E, Brents LA, et al. Frequent Translocation t(4;14)(p16.3;q32.3) in multiple myeloma is associated with increased expression and activating mutations of fibroblast growth factor receptor 3. Nat Genet. 1997;16:260–4. doi: 10.1038/ng0797-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chesi M, Nardini E, Lim RSC, et al. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34. [PubMed] [Google Scholar]
- 15.Li Z, Zhu YX, Plowright EE, et al. The myeloma-associated oncogene fibroblast growth factor receptor 3 is transforming in hematopoietic cells. Blood. 2001;97:2413–9. doi: 10.1182/blood.v97.8.2413. [DOI] [PubMed] [Google Scholar]
- 16.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J. A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–6. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 17.Chesi M, Bergsagel PL, Shonukan OO, et al. Frequent dysregulation of the c-maf proto-oncogene at 16q23 by translocation to an Ig locus in multiple myeloma. Blood. 1998;91:4457–63. [PubMed] [Google Scholar]
- 18.Hurt EM, Wiestner A, Rosenwald A, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–9. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 19.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhan F, Hardin J, Kordsmeier B, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–57. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 21.Magrangeas F, Nasser V, Avet-Loiseau H, et al. Gene expression profiling of multiple myeloma reveals molecular portraits in relation to the pathogenesis of the disease. Blood. 2003;101:4998–5006. doi: 10.1182/blood-2002-11-3385. [DOI] [PubMed] [Google Scholar]
- 22.Zhan F, Huang Y, Colla S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broyl A, Hose D, Lokhorst H, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010 Jun; doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 24.Annunziata CM, Davis RE, Demchenko Y, et al. Frequent engagement of the classical and alternative NFκB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12 :115–30. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keats JJ, Fonseca R, Chesi M, et al. Promiscuous mutations activate the noncanonical NFκB pathway in multiple myeloma. Cancer Cell. 2007;12 :131–44. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrasco DR, Tonon G, Huang Y, et al. High-resolution genomic profiles define distinct clinicopathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–25. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 27.Avet-Loiseau H, Li C, Magrangeas F, et al. Prognostic significance of copy- number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chng WJ, Kumar S, Vanwier S, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–9. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 29.Shammas MA, Shmookler Reis RJ, et al. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113:2290–7. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang H, Sloan S, Li D, et al. The t(4;14) is associated with poor prognosis in myeloma patients undergoing autologous stem cell transplant. Br J Haematol. 2004;125:64–8. doi: 10.1111/j.1365-2141.2004.04867.x. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez NC, Castellanos MV, Martin ML, et al. Prognostic and biological implications of genetic abnormalities in multiple myeloma undergoing autologous stem cell transplantation: t(4;14) is the most relevant adverse prognostic factor, whereas RB deletion as a unique abnormality is not associated with adverse prognosis. Leukemia. 2007;21:143–50. doi: 10.1038/sj.leu.2404413. [DOI] [PubMed] [Google Scholar]
- 32.Gertz MA, Lacy MQ, Dispenzieri A, et al. Clinical implications of t(11;14)(q13;q32), t(4;14)(p16.3;q32), and -17p13 in myeloma patients treated with high-dose therapy. Blood. 2005;106:2837–40. doi: 10.1182/blood-2005-04-1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreau P, Attal M, Garban F, et al. Heterogeneity of t(4;14) in multiple myeloma. Long-term follow-up of 100 cases treated with tandem transplantation in IFM99 trials. Leukemia. 2007;21:2020–4. doi: 10.1038/sj.leu.2404832. [DOI] [PubMed] [Google Scholar]
- 34.Avet-Loiseau H, Attal M, Moreau P, et al. Genetic abnormalities and survival in multiple myeloma: the experience of the Intergroupe Francophone du Myélome. Blood. 2007;109:3489–95. doi: 10.1182/blood-2006-08-040410. [DOI] [PubMed] [Google Scholar]
- 35.Fonseca R, Blood E, Rue M, et al. Clinical and biologic implications of recurrent genomic aberrations in myeloma. Blood. 2003;101:4569–75. doi: 10.1182/blood-2002-10-3017. [DOI] [PubMed] [Google Scholar]
- 36.Drach J, Ackermann J, Fritz E, et al. Presence of a p53 gene deletion in patients with multiple myeloma predicts for short survival after conventional-dose chemotherapy. Blood. 1998;92:802–9. [PubMed] [Google Scholar]
- 37.Chang H, Qi C, Yi QL, et al. p53 gene deletion detected by fluorescence in situ hybridization is an adverse prognostic factor for patients with multiple myeloma following autologous stem cell transplantation. Blood. 2005;105:358–60. doi: 10.1182/blood-2004-04-1363. [DOI] [PubMed] [Google Scholar]
- 38.Lode L, Eveillard M, Trichet V, et al. Mutations in TP53 are exclusively associated with del(17p) in multiple myeloma. Haematologica. 2010 doi: 10.3324/haematol.2010.023697. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanamura I, Stewart JP, Huang Y, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–32. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fonseca R, Van Wier SA, Chng WJ, et al. Prognostic value of chromosome 1q21 gain by fluorescent in situ hybridization and increase CKS1B expression in myeloma. Leukemia. 2006;20:2034–40. doi: 10.1038/sj.leu.2404403. [DOI] [PubMed] [Google Scholar]
- 41.Shaughnessy JD, Zhan F, Burrington BE, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–84. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 42.Decaux O, Lode L, Magrangeas F, et al. Prediction of survival in multiple myeloma based on gene-expression profiles revealed cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients. J Clin Oncol. 2008;26:4798–805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 43.Avet-Loiseau H, Li C, Magrangeas F, et al. Prognostic significance of copy- number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585–90. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.San Miguel JF, Schlag R, Khuageva NK, et al. Bortezomib plus melphalan and prednisone for initial treatment of multiple myeloma. N Engl J Med. 2008;359:906–17. doi: 10.1056/NEJMoa0801479. [DOI] [PubMed] [Google Scholar]
- 45.Pineda-Roman M, Zangari M, Haessler J, et al. Sustained complete remissions in multiple myeloma linked to bortezomib in total therapy 3: comparison with total therapy 2. Br J Haematol. 2008;140:625–34. doi: 10.1111/j.1365-2141.2007.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avet-Loiseau H, Leleu X, Roussel M, et al. Bortezomib plus dexamethasone induction improves outcome of patients with t(4;14) myeloma but not outcome of patients with del(17p) J Clin Oncol. 2010 doi: 10.1200/JCO.2010.28.3945. in press. [DOI] [PubMed] [Google Scholar]
- 47.Munshi NC, Avet-Loiseau H, Stephens P, et al. Whole Genome Paired End Sequencing Identifies Genomic Evolution in Myeloma. Blood. 2008;112:2846a. [Google Scholar]
- 48.Golub T. First impressions of the multiple myeloma genome. Proceedings of AACR. 2010:SY30–03. [Google Scholar]
- 49.Fonseca R, Bergsagel PL, Drach J, et al. International Myeloma Working Group. International Myeloma Working Group molecular classification of multiple myeloma: spotlight review. Leukemia. 2009 Dec;23(12):2210–21. doi: 10.1038/leu.2009.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walker BA, Leone PE, Chiecchio L, Dickens NJ, Jenner MW, Boyd KD, Johnson DC, Gonzalez D, Dagrada GP, Protheroe RK, Konn ZJ, Stockley DM, Gregory WM, Davies FE, Ross FM, Morgan GJ. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010 Oct 14;116(15):e56–65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 51.Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, Davies FE, Ross FM, Morgan GJ. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011 Jan 13;117(2):553–62. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- 52.Davies FE, Dring AM, Li C, Rawstron AC, Shammas MA, O’Connor SM, et al. Insights into the multistep transformation of MGUS to myeloma using microarray expression analysis. Blood. 2003 Dec 15;102(13):4504–11. doi: 10.1182/blood-2003-01-0016. [DOI] [PubMed] [Google Scholar]
- 53.Pichiorri F, Suh SS, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, Munker R, Volinia S, Boccadoro M, Garzon R, Palumbo A, Aqeilan RI, Croce CM. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc Natl Acad Sci U S A. 2008;105(35):12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corthals SL, Jongen-Lavrencic M, de Knegt Y, Peeters JK, Beverloo HB, Lokhorst HM, Sonneveld P. Micro-RNA-15a and micro-RNA-16 expression and chromosome 13 deletions in multiple myeloma. Leuk Res. 2010 May;34(5):677–81. doi: 10.1016/j.leukres.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 56.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009 Dec 10;114(25):e20–6. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 57.Gutiérrez NC, Sarasquete ME, Misiewicz-Krzeminska I, Delgado M, De Las Rivas J, et al. Deregulation of microRNA expression in the different genetic subtypes of multiple myeloma and correlation with gene expression profiling. Leukemia. 2010 Mar;24(3):629–37. doi: 10.1038/leu.2009.274. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, Cartron MA, van Rhee F, Nair B, Waheed S, Pineda-Roman M, Alsayed Y, Anaissie E, Shaughnessy JD., Jr High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc Natl Acad Sci U S A. 2010 Apr 27;107(17):7904–9. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM, Jr, Hofmainster C, Alder H, Garofalo M, Di Leva G, Volinia S, Lin HJ, Perrotti D, Kuehl M, Aqeilan RI, Palumbo A, Croce CM. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell. 2010 Oct 19;18(4):367–81. doi: 10.1016/j.ccr.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Munshi NC, Li C, Minvielle S, Amin SB, Moreau P, Magrangeas F, et al. Alternate Splicing Is a Frequent Event and Impacts Clinical Outcome in Myeloma: A High-Density Exon Array Analysis of Uniformly Treated Newly-Diagnosed Myeloma Patients. Blood. 2008;112:498a. [Google Scholar]
- 61.Adamia S, AvetLoiseau H, Amin SD, Tai Y, Treon SP, Moreau P, et al. MicroRna Expression Profile Identifies Distinct Clinically Relevant Sub-Groups in Multiple Myeloma: Novel Prognostic Markers and Potential Targets for Therapy. Blood. 2008;112:96a. [Google Scholar]