Abstract
The endosomal sorting complex required for transport-III (ESCRT-III) proteins are essential for budding of some enveloped viruses, the formation of intraluminal vesicles at the endosome and for the abscission step of cytokinesis. ESCRT-III proteins form polymers that constrict membrane tubes, leading to fission. We have used electron cryomicroscopy (cryo-EM) to determine the molecular organization of pleiomorphic ESCRT-III CHMP2A-CHMP3 polymers. The three-dimensional reconstruction at 22 Å resolution reveals a helical organization of filaments of CHMP molecules organized in a head-to-tail fashion. Protease susceptibility experiments indicate that polymerization is achieved via conformational changes that increase the protomer stability. Combinatorial siRNA knockdown experiments indicate that CHMP3 contributes synergistically to HIV-1 budding, and the CHMP3 contribution is ∼10-fold more pronounced in concert with CHMP2A than with CHMP2B. This is consistent with SPR affinity measurements that suggest sequential CHMP4B-CHMP3-CHMP2A recruitment while showing that both CHMP2A and CHMP2B interact with CHMP4B, in agreement with their redundant functions in HIV-1 budding. Our data thus indicate that the CHMP2A-CHMP3 polymer observed in vitro contributes to HIV-1 budding by assembling on CHMP4B polymers.
Keywords: CHMP2A, CHMP2B, CHMP3, CHMP4, ESCRT, ESCRT-III, HIV-1, budding
Introduction
Endosomal sorting complexes required for transport (ESCRT) were first discovered in yeast, where they catalyze membrane protein sorting into intraluminal vesicles at the endosomal membrane. This process creates multivesicular bodies, which ultimately fuse with the vacuole/lysosome and thereby target the proteins for lysosomal degradation (Henne et al., 2011). The ESCRT system is highly conserved in all eukaryotes and comprises five different complexes that are assembled sequentially (Saksena et al., 2007). ESCRT-0 recognizes ubiquitylated cargo (Ren and Hurley, 2010), ESCRT-I and -II have a role in vesicle formation; and ESCRT-III constitutes the fission machinery (Boura et al., 2012; Wollert and Hurley, 2010), with support from the VPS4 complex, which also recycles ESCRT-III from cellular membranes (Babst et al., 2011; Baumgartel et al., 2011; Shestakova et al., 2010; Saksena et al., 2007) Selected ESCRT proteins are recruited during budding of some enveloped viruses including HIV-1 (Sundquist and Krausslich, 2012; Martin-Serrano and Neil, 2011; Usami et al., 2009; Morita and Sundquist, 2004) and during cytokinesis of eukaryotes (Carlton et al., 2012; Guizetti and Gerlich, 2010; McDonald and Martin-Serrano, 2009). Even Crenarchaea, which lack endosomes, employ ESCRT-III- and VPS4-like proteins for cell division (Samson et al., 2008)(Ghazi-Tabatabai et al., 2009; Lindas et al., 2008). All known ESCRT-mediated processes employ ESCRT-III and VPS4, consistent with their requirement for a final common membrane fission step (Guizetti and Gerlich, 2012; Peel et al., 2011; Hurley and Hanson, 2010).
S. cerevisiae express six ESCRT-III proteins, which form at least 2 subcomplexes composed of Vps20-Snf7 and Vps2-Vps24 (Babst et al., 2002); Vps20 recruits Snf7, which in turn recruits Vps24 followed by Vps2, to form a large ESCRT-III core complex that can deform membranes in vitro (Saksena et al., 2009; Teis et al., 2008). The four core yeast ESCRT-III proteins have been shown to trigger vesicle fission in vitro from giant unilamellar vesicle (GUV) membranes, with Vps20 and Snf7 playing central roles in fission in the in vitro system, and Vps24 rendering fission more efficient (Wollert and Hurley, 2010). Mammalian cells express 12 ESCRT-III proteins, typically termed CHMP (CHarged Multivesicular body Protein), which have the potential to form analogous core complexes composed of CHMP6 (Vps20), a member of the CHMP4 (A, B, or C) family (Snf7), CHMP3 (Vps24), and a member of the CHMP2 (A or B) family (Vps2). Other ESCRT-III family members (CHMP1A, B, CHMP5, CHMP7, and IST1) appear to play regulatory roles both in yeast and higher eukaryotes (Babst et al., 2011; Henne et al., 2011; Peel et al., 2011; Hurley and Hanson, 2010).
ESCRT-III members share a common N-terminal core domain architecture (Bajorek et al., 2009; Xiao et al., 2009; Muziol et al., 2006). The C-terminus folds back onto the core in an auto-inhibited cytosolic form, and CHMP activation entails its displacement (Bajorek et al., 2009; Lata et al., 2008a; Shim et al., 2007; Zamborlini et al., 2006; Lin et al., 2005). Activation is thought to favor membrane binding and polymerization (Peel et al., 2011). Only CHMP4 and CHMP2 polymers have thus far been observed in vivo, which form either CHMP4-containing plasma membrane tubes upon co-expression with dominantly inhibitory VPS4B protein (Hanson et al., 2008) or plasma membrane tubes decorated with helically arranged CHMP2B (Bodon et al., 2011). Helical structures of a larger diameter that apparently contain CHMP2A have been imaged by cryo electron tomography at the midbody of HeLa cells (Guizetti et al., 2011). In vitro, several types of polymers have been observed in the absence of membranes, including two or three stranded filaments of yeast Vps24 (Ghazi-Tabatabai et al., 2008), circular arrays of CHMP4B (Pires et al., 2009) and tubular polymers of IST1, CHMP1A (Bajorek et al., 2009) and CHMP2A/CHMP3 complexes (Lata et al., 2008b). In particular, the yeast Vps24 and Vps2 proteins interact preferentially, and the analogous human CHMP2A and CHMP3 can co-assemble into regular straight or cone-shaped tubular helical structures in vitro (Babst et al., 2002), (Bajorek et al., 2009; Lata et al., 2008b). Based upon the observation that some of these tubes are closed at one end, a model for ESCRT-III-driven neck closure has been proposed in which ESCRT-III spiraling filaments could catalyze membrane fission by spiraling closed and drawing the opposing membranes together (Fabrikant et al., 2009; Lata et al., 2008b).
Although nearly all ESCRT-III members play pivotal roles during cell division (Morita et al., 2010), siRNA depletion experiments indicate that HIV-1 budding has a strong requirement for a CHMP4 family member (particularly CHMP4B) and a CHMP2 family member (either A or B), but a much weaker requirement for CHMP3 (Morita et al., 2011). The VPS4 complex is also essential for budding (Kieffer et al., 2008; Strack et al., 2003; von Schwedler et al., 2003) and functions as a component of the fission machinery in higher eukaryotes (Baumgartel et al., 2011; Jouvenet et al., 2011). During HIV-1 budding, CHMP4 assembles first within the neck of a budding virion where it can apparently recruit either CHMP2A or B by direct interaction leading to virion release (Morita et al., 2011). Thus, in contrast to the yeast system, where single rounds of vesicle budding can occur in the absence of the Vps2p (CHMP2) (Wollert and Hurley, 2010), mammalian CHMP2 isoforms appear to play a key role in the mammalian fission process during HIV-1 budding (Morita et al., 2011).
Here we present a helical reconstruction of the ESCRT-III CHMP2A-CHMP3 polymer by cryo-EM. The reconstruction reveals a linear arrangement of CHMP proteins into filaments that interact laterally to produce the tube-like structures. CHMP2A-CHMP3 polymerization increased the thermostability and protease resistance of the subunits as compared to the monomeric proteins, suggesting that polymerization generates free energy that might be employed for membrane remodeling. The physiological importance of the CHMP2A-CHMP3 combination is supported by observation that CHMP3 acts synergistically with CHMP2A and, to a lesser extent, with CHMP2B during HIV-1 release. We find that CHMP4B binds ∼16 fold more tightly to CHMP3 than CHMP2A, supporting the idea that recruitment at the HIV-1 budding site normally follows the order CHMP4B:CHMP3:CHMP2A.
Results
Cryo-EM analyses and 2D classification of CHMP2A-CHMP3 polymers
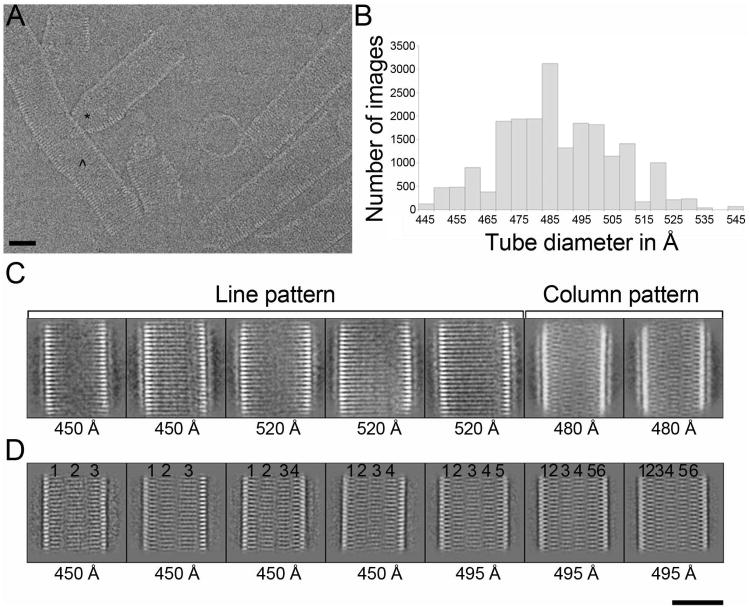
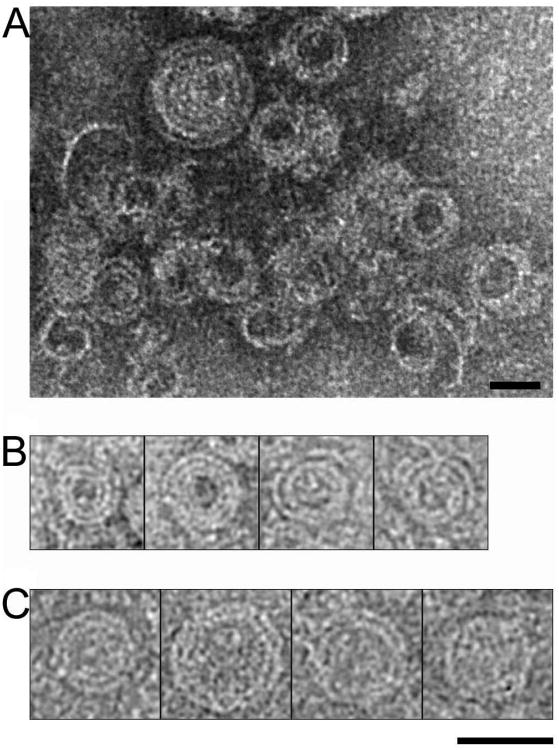
Tubular polymers composed of MBP-CHMP2AΔC (residues 9-161) and CHMP3 (residues 1-222) were assembled as described (Lata et al., 2008b) and MBP was then removed by proteolysis to optimize data quality (Figure S1). Although MBP cleavage from CHMP2AΔC induced aggregation of CHMP2AΔC-CHMP3 tubes, single tubes could still be imaged by cryo-EM (Figure 1A). The CHMP2AΔC-CHMP3 tubes are pleiomorphic with regard to: 1) length, 2) diameter and 3) straight, curved or cone-shaped walls (Figure 1A). We collected three datasets, two of which were from tubes in which MBP was completely removed from CHMP2AΔC (collected on a FEI Tecnai G2 Polara microscope) and one that was collected on tubes in which the MBP was partially removed, (FEI CM200 microscope). Tube images were cut into short segments and classified by a reference-free approach, resulting in a large number of two-dimensional (2D) class averages of tubes with variable diameters between 445 and 545 Å (Figure 1B). These 2D class averages could be sorted into two general groups that exhibited two different types of internal striations. One group showed continuous parallel transversal striations, (termed the “line pattern”) (Figure 1C), while the second group does not have continuous striations. Instead, these tubes exhibit vertical, column-like segments that contain similar striations that are not continuous between segments (Figures 1C and D). The column-like pattern predominated in two of the three data sets. Because the line-pattern classes were only frequent in the sample in which the MBP moiety had been partially removed, we compared the 2D class averages obtained after reference-free classification of MBP un-cleaved and cleaved tubes derived from the same purification batch. Both classifications produced class averages that had the same column-pattern, implying that MBP removal had no significant effect on the structural organization of the tubes (Figure S2). This experiment confirmed that the line- and column-patterns observed in the 2D-classes arise from genuinely different types of CHMP2AΔC-CHMP3 tubes.
Figure 1. Structural diversity of CHMP2AΔC-CHMP3 co-polymers.
(A) Cryo-EM raw image of CHMP2AΔC-CHMP3 tubes after removal of MBP from CHMP2AΔC reveals structural heterogeneity; (*) highlights a cone shaped structure and (^) highlights a tube with continuous variation in dimensions.
(B) Histogram of the number of segmented tube images showing each diameter from one dataset after 2D classification by multivariate statistical analysis.
(C) Representative 2D class averages obtained after 2D classification of one of the three datasets. Brackets delineate classes with typical line and column patterns.
(D) Representative class averages for a second dataset (corresponding to the histogram shown in B). The numbers on top of each image mark the position of the different columns seen in each class. The number of columns increased with the diameter of the tubes, but could also vary for classes having the same diameter. Diameters of each class are indicated on each individual image. Scale bars in A, C and D are 400 Å.
CHMP2AΔC and CHMP3 can form helical polymers of different symmetries
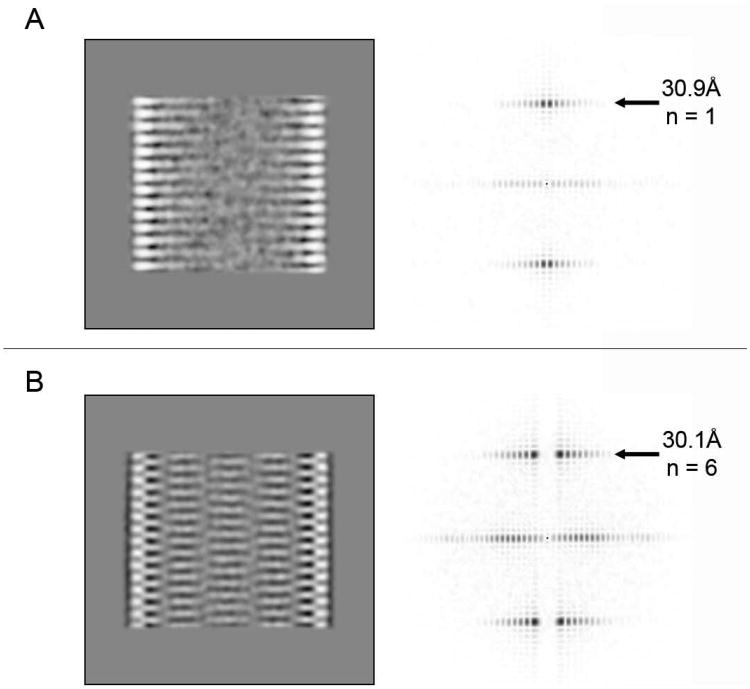
2D Fourier transformations of classes displaying the line-like and column-like patterns show layer lines at ∼30.9 and 30.1 Å, respectively. The small difference in their heights is most likely due to the fact that the images were taken on different electron microscopes at different magnifications (Figures 2A and B). However, the horizontal position of the maximum amplitude of the layer line in both Fourier transformations differed and corresponded to a Bessel function of the order n=1 for the line-pattern and n=6 for the column-pattern. To characterize these differences further, we set up a systematic test of helical symmetry parameters to probe which pitch of a putative helix is the most consistent with the 2D class average (Desfosses et al., 2011). The correlation between the class average and the 2D projection of a 3D model revealed the maximum correlation for a pitch of 30.9 Å in case of the line-pattern (Figure S3A). This suggests a 1-start helix with a pitch of 30.9 Å consistent with the Bessel order n=1 calculated from the 2D Fourier transform. Following the same approach, the search for the best pitch for the 5 column-like pattern class average (Figure 2B) showed a maximum correlation at 181 Å and lower correlation peaks for pitches, in multiples of 30 Å (Figure S4A). The correctness of the model was validated by calculating an isosurface of the 3D model with a 181-Å-pitch and by comparing its 2D projections shown at different out-of-plane angles with the best matching reference-free 2D class averages (Figures S4B and C). Thus, the 5 column-like pattern class average (Figure 2B) can be best described by a 6-start helix having a 181-Å-pitch. Other 2D class averages having different diameters and different column numbers (Figure 1D) can also be explained by an n-start helix (n is an integer) model having an n-times 30-Å-pitch (data not shown). In addition, the 2D projections calculated at different out-of-plane angles of the 3D model of the 1-start helix match the ab-initio reference-free 2D class averages representing the line-pattern (Figures S3B and C). In summary, CHMP2AΔC and CHMP3 can form tubes with different helical packing described by an n-start helix with a n*30 Å pitch (1 ≤ n ≤ 7 for the data analyzed in this study).
Figure 2. 2D Fourier transform provides evidence for differences in helical packing.
(A) 2D class averages representing the line-pattern and (B) the column pattern, and their corresponding sum of the amplitude spectra of all class members (120 and 242 members in A and B, respectively). The heights of the first layer line and the measured Bessel order (n) are indicated.
3D reconstruction of CHMP2A-CHMP3 tubes
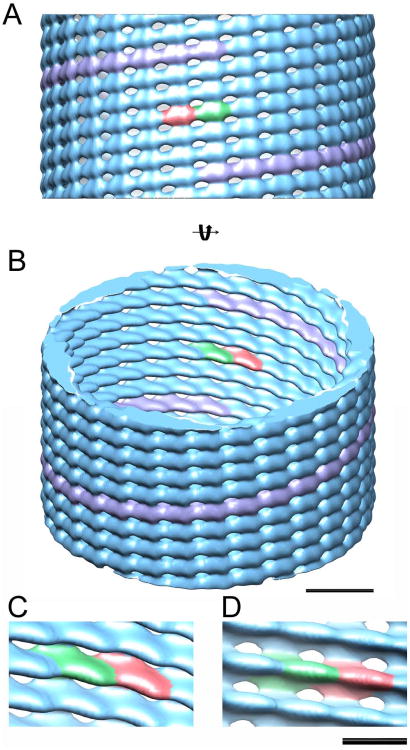
Cyclic symmetry of the order n around the helical axis of the tubular structures is further corroborated by the presence of either inversion or mirror symmetry in some 2D class averages (Figure S4D). We therefore used the Iterative Helical Real Space Reconstruction (IHRSR) algorithm (Egelman, 2007) to calculate a 3D reconstruction on the most populated class (mean diameter of 485 Å), which is best described by a 6-start helix with a 181-Å-pitch (Figure S4B). Upon testing various cyclic symmetries, only C6 produced reliable results. IHRSR converged to a final solution having an axial rise per subunit z of 6.03 Å and an azimuthal angle φ of 11.9° for a resolution of 22 Å at a Fourier shell correlation (FSC) of 0.5 (Figure S5A). The in plane 2D projection of the 3D reconstruction reveals the same column pattern as in Figure 2B and its corresponding 2D Fourier transformation confirms a single layer line at 30 Å (Figure S5B). The 3D reconstruction presents a 6-stranded helical structure with each filament being separated by ∼ 30 Å and made up of 30.2 asymmetric units per turn. The asymmetric unit is elongated in shape (Figure 3, red and green densities and Figure S5C) and on the inside of the structure, clear grooves separate each strand (Figure 3B-D). There are also lower density zones between strands (appearing as holes at the threshold used in the isosurface representation of Figure 3). The long axis of the asymmetric unit is tilted by ∼ 45° with respect to the tube axis such that lateral interactions between units produce a tube width of ∼40 Å as well as the external 40 Å dimension of the asymmetric unit (Figure 3C). The surface making up the lateral interactions between filaments is substantially smaller than that of linear inter-monomer interactions (Figures 3C and D). Following the same procedure, a 3D reconstruction was calculated for tubes with a 460 Å diameter (Figure S6A). This revealed a 5-start helical structure with a pitch of 152 Å for 25.3 subunits per turn. Notably, the overall architecture and unit cell shape are similar to those of the 485 Å diameter tube reconstruction (Figure S6B).
Figure 3. 3D reconstruction of CHMP2A-CHMP3 tubes.
(column pattern, n=6) (A) Isosurface representation (blue) of the 3D reconstruction of a tube having a 485 Å average diameter. The raise of 1 helical turn is indicated in pink and two helically related asymmetric units are colored green and red, respectively. (B) Same isosurface as shown in (A) rotated by 30° provides insight into the tube. Scale bar is 100 Å.
(C-D) Close-up of the linear monomer-monomer interaction and the lateral inter-filament contacts. C is the same view as B and D the same as A, but looking from the inside. Scale bar is 50 Å
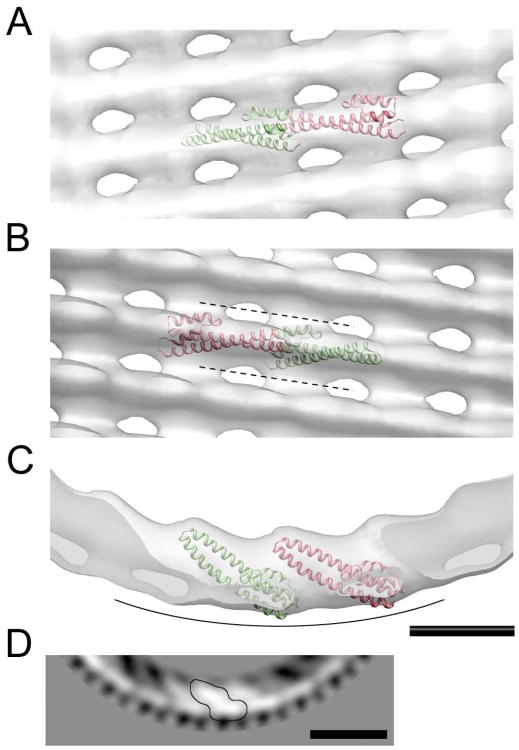
Despite the intermediate resolution of the cryo-EM reconstruction, the CHMP3 monomer containing helices 1 to 4 could be docked inside the map. Both hands of the cryo-EM map were tested and could not be discriminated at the current resolution. However, both solutions support the conclusion that a CHMP3 monomer with its 70-Å-long helical hairpin could be positioned diagonally with the tip of the helical hairpin pointing towards the inside of the tube (Figure S7A and B). One reasonable possibility is that the CHMP filament consists of head to tail interactions of successive CHMP2A and CHMP3 monomers (Figures 4A-C). Even though the asymmetric unit appears slightly too large for a single monomer at this resolution (Figure 3), it is not big enough to fit a CHMP dimer model (Lata et al., 2008b) without steric clashes. Thus the interpretation of the density of the asymmetric unit (Figure 4D) might be better explained with a different CHMP conformer model (even though the core monomer (helices 1-4) lacks ∼ 20 amino acids). Despite the fact that attempts to calculate a 3-D reconstruction from the data representing the 1-start helix model (Figure S3) did not converge to a reliable model, probably due to the limited data available for analysis, we propose that the conformation of the CHMP filament is the same.
Figure 4. Docking of the CHMP3 structure (helices 1 to 4) into the cryo-EM map. (A – D).
Zoomed views on the docked monomers inside the cryo-EM map. The cryo-EM density is shown as an isosurface in transparent gray color. Adjacent CHMP3 monomers are shown in green and red ribbons. (A) Front view. (B) Same orientation as shown in (A) viewed from the inside; the dotted lines mark the positions of grooves between consecutive strands. (C) Top view, the curved line indicates the membrane-binding surface. (D) Part of a gray-scaled 2D section from the cryo-EM map viewed along the helical axis (corresponding to the orientation in C). One helical asymmetric unit is highlighted (contoured line). Scale bars are respectively 50 Å in panels A to C and 100 Å in panel D.
Molecular architecture of CHMP2A polymers
CHMP2AΔC assembled into coil-like structures as determined by negative-stain electron microscopy (Figure 5A). Some of these filaments almost close at one end by successively narrowing the coil diameter as viewed from the top (Figures 5B and C). The CHMP2A coils are ∼ 30 Å in width, which is similar to the CHMP filaments observed in the CHMP2A-CHMP3 polymer, the CHMP2B membrane tubes (Bodon et al., 2011), and in CHMP4B polymers (Pires et al., 2009). This suggests that CHMP2A, CHMP2B, CHMP3 and CHMP4B employ similar structural principles to form linear filaments of constant width.
Figure 5. Conserved filament organization of ESCRT-III.
(A) Negative stained image of CHMP2AΔC polymers. (B) Gallery of spiral structures formed by CHMP2AΔC obtained by negative staining electron microscopy and (C) cryo-EM. CHMP2AΔC forms 30 Å-thick strands, which are similar to the width of CHMP2B and CHMP2AΔC-CHMP3 filaments. Scale bars are 400 Å.
ESCRT-III CHMP2A-CHMP3 stability
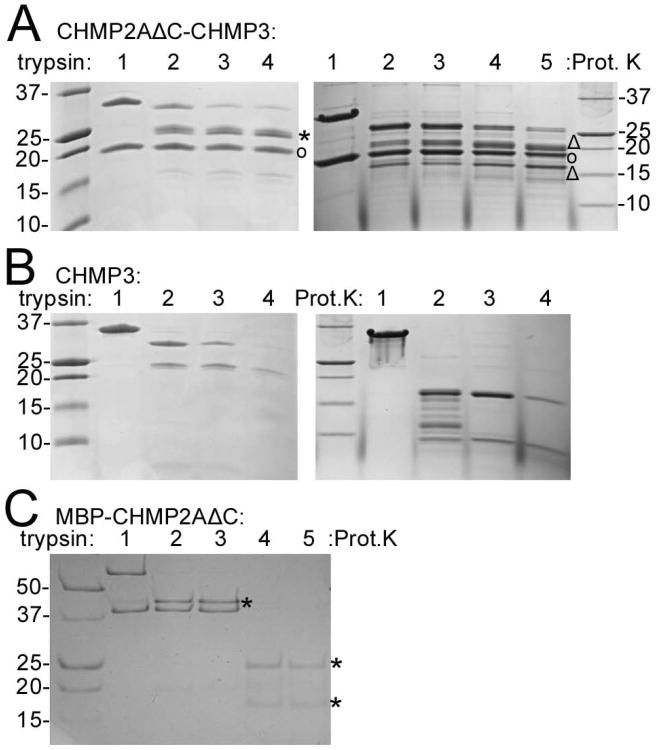
Trypsin treatment of CHMP2AΔC-CHMP3 polymers did not reduce the size of CHMP2AΔC significantly as judged from the SDS-PAGE analysis (Figure 6A, left panel, circle), while full length CHMP3 proteolyzed to a fragment migrating at ∼ 25 kDa (Figure 6A, left panel, asterisk), which contained residues 7 to 183 as determined by mass spectrometry. Proteinase K digestion of CHMP2AΔC-CHMP3 tubes reduced the size of CHMP3 further to produce fragments corresponding to residues 2 to 168 (Figure 6A, right panel, triangle) and residues 8 to 160, (Figure 6A, right panel, triangle) while CHMP2AΔC stayed largely resistant (Figure 6A, right panel, circle). In contrast, monomeric CHMP3 and MBP-CHMP2AΔC were degraded by trypsin and proteinase K (Figures 6B and C), indicating that polymer formation rendered CHMP3 and CHMP2AΔC protease resistant. Trypsin-treated CHMP2AΔC-CHMP3 polymers were indistinguishable from wild type polymers, suggesting that the core of the tubular polymer comprises CHMP2A and CHMP3 helices 1 to 4 plus the helix 4/5 linker. Circular dichroism thermostability measurements showed a melting temperature (TM) of ∼64°C for MBP-CHMP2A (MBP has a TM of 55°C), a TM of 60°C for CHMP3 and an increase of the TM to 70°C for the CHMP2AΔC-CHMP3 tubular polymer. Taken together, these observations indicate that polymer formation renders CHMP2A and CHMP3 more stable.
Figure 6. CHMP2AΔC-CHMP3 polymerization into tubes renders CHMP2A and CHMP3 protease resistant.
(A) Trypsin and proteinase K proteolysis of CHMP2AΔC-CHMP3 tubes. Right panel: lane 1, no trypsin; lanes 2-4 trypsin: CHMP2AΔC-CHMP3 tube ratios of 1:70; 1:35 and 1:20 (w/w), respectively. Right panel: lane 1, no proteinse K; lanes 2-5, proteinase K ratios of 1:500, 1:250, 1:100 and 1:50 (w/w), respectively. (CHMP2AΔC (°); CHMP3 bands produced by trypsin (*) and Proteinase K (Δ) digestion)
(B) Trypsin and proteinase K proteolysis of CHMP3. Right panel: lane 1, no trypsin; lanes 2-4, trypsin: CHMP3 ratios of 1:70, 1:35 and 1:20 (w/w), respectively.
(C) Trypsin and proteinase K proteolysis of MBP-CHMP2AΔC. Lane 1, no trypsin; lanes 2 and 3 trypsin/MBP-CHMP2AΔC ratios of 1:100 and 1:50 (w/w); lanes 4 and 5, proteinase K/MBP-CHMP2AΔC ratios 1:100 and 1:50 (w/w). (MBP bands after proteolysis (*)).
CHMP3 exerts a synergistic effect on CHMP2A during HIV-1 budding
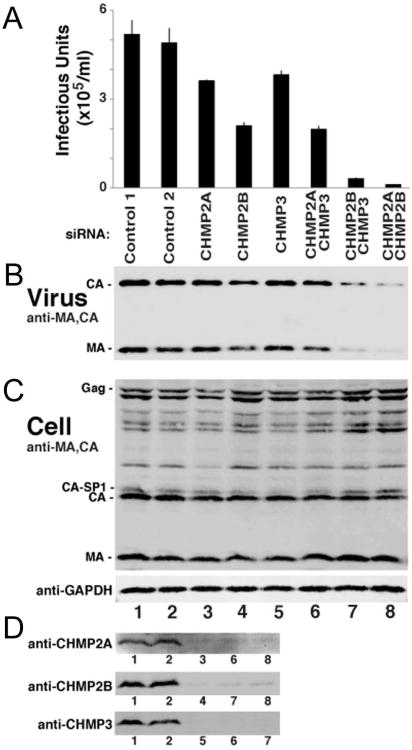
Morita and colleagues have reported that HIV-1 release from host cells requires a CHMP4 family member, together with either CHMP2A or B (Morita et al., 2011). Here we extended the siRNA knockdown study to double knock-downs of CHMP2A-CHMP3 and CHMP2B-CHMP3. As expected from previous results, single knock-downs of CHMP2A, CHMP2B and CHMP3 had only modest effects on virion release and infectivity. Knock-down of CHMP2A and CHMP3 reduced infectious units by a factor of ∼1.4 and CHMP2B knock-down produced a ∼ 2.5-fold reduction (Figures 7A and B). The double knock-down of CHMP2A and CHMP3 reduced infectivity 2.6-fold, while the double knockdown of CHMP2B-CHMP3 reduced infectivity 17-fold and the CHMP2A-CHMP2B double knockdown reduced infectivity 46 fold (Figures 7A and B). Detection of cellular Gag, CA, MA and GAPDH demonstrated similar protein concentrations (Figure 7C) and efficient single and double knock-downs were verified by western blot analyses (Figure 7D). Our data indicate that loss of CHMP3 synergizes slightly with loss of CHMP2A in inhibiting HIV-1 release (co-depletion induced a 2.6-fold reduction versus a 1.4-fold reduction of either CHMP2A or CHMP3 single depletions) but shows much greater synergy with loss of CHMP2B (17-fold reduction, while CHMP2B depletion causes a 2.5-fold and CHMP3 depletion a 1.5-fold reduction on their own). These results show that the function of CHMP2A (left intact in the CHMP2B-CHMP3 co-depletion, 17-fold reduction) is significantly more affected by the absence of CHMP3 than the function of CHMP2B (left intact in the CHMP2A-CHMP3 co-depletion; 2.6-fold reduction). Thus our data suggest that during HIV-1 budding, CHMP3 synergizes more efficiently with CHMP2A than with CHMP2B.
Figure 7. ESCRT-III CHMP3 exerts a synergistic effect on ESCRT-III CHMP2A and CHMP2B during HIV-1 budding.
293T cells expressing HIV-1 were treated either with control siRNAs (lanes 1 and 2) or with siRNAs that target the indicated ESCRT-III CHMP family members (lanes 3-8).
(A) Viral titers, n=6 titer measurements from two independent experiments.
(B) Western blot analyses of virion-associated CA and MA proteins released into the cell culture supernatant.
(C) Western blot analyses of intracellular viral Gag derived proteins (panels B and C, anti-MA and anti-CA Abs) and expression levels of cellular GAPDH (loading control)
(D) Western blot analyses of endogenous cellular ESCRT-III CHMP2A, CHMP2B and CHMP3 protein levels, corresponding to the designated lanes of the upper blots.
ESCRT-III interactions in vitro support their redundant function during HIV-1 budding
It was shown previously that CHMP2A can interact directly with CHMP4B, which is required for HIV-1 budding (Morita et al., 2011). Because CHMP3 depletion synergized with CHMP2 proteins in inhibiting HIV-1 budding, we performed biosensor analyses that extended previous CHMP-CHMP interaction studies (Martin-Serrano et al., 2003; von Schwedler et al., 2003) and analyzed the pairwise interactions of CHMP4B with CHMP3, CHMP2A and CHMP2B. Because some of the binding data could not be satisfactorily analyzed by the Langmuir model, binding curves were recorded until equilibrium was reached and the equilibrium data were evaluated by non-linear least squares fitting to hyperbolic binding models. This analysis revealed a KD of 2.4
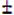 0.2 μM for CHMP2A monomer binding to immobilized CHMP4B monomers (Figure S8A). At low concentrations, CHMP3 monomers bound to CHMP4B monomers with an approximate 16-fold tighter KD1 of 0.15
0.2 μM for CHMP2A monomer binding to immobilized CHMP4B monomers (Figure S8A). At low concentrations, CHMP3 monomers bound to CHMP4B monomers with an approximate 16-fold tighter KD1 of 0.15
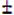 0.04 μM. At higher concentrations, a second KD2 of 69
0.04 μM. At higher concentrations, a second KD2 of 69
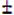 12 μM was observed, which might indicate CHMP3 polymerization upon saturation of the CHMP4B binding sites (Figure S8B). CHMP2A and CHMP3 interacted with a KD of 3.2
12 μM was observed, which might indicate CHMP3 polymerization upon saturation of the CHMP4B binding sites (Figure S8B). CHMP2A and CHMP3 interacted with a KD of 3.2
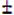 0.2 μM (Figure S8C), which is in agreement with the requirement of ∼ 7 μM concentrations of CHMP2A and CHMP3 for polymer formation (Lata et al., 2008b). Finally, CHMP4B and CHMP2B interacted with a KD of 0.56 μM and CHMP2B-CHMP3 interacted with a KD of 1.4 μM (Figure S8D). These data confirm that CHMP4B can directly recruit CHMP2A or CHMP2B (Morita et al., 2011). However, if CHMP3 is present, the higher affinity of CHMP4B for CHMP3 may indicate that CHMP3 is preferentially recruited by CHMP4B, and can then subsequently engage CHMP2A or CHMP2B.
0.2 μM (Figure S8C), which is in agreement with the requirement of ∼ 7 μM concentrations of CHMP2A and CHMP3 for polymer formation (Lata et al., 2008b). Finally, CHMP4B and CHMP2B interacted with a KD of 0.56 μM and CHMP2B-CHMP3 interacted with a KD of 1.4 μM (Figure S8D). These data confirm that CHMP4B can directly recruit CHMP2A or CHMP2B (Morita et al., 2011). However, if CHMP3 is present, the higher affinity of CHMP4B for CHMP3 may indicate that CHMP3 is preferentially recruited by CHMP4B, and can then subsequently engage CHMP2A or CHMP2B.
Discussion
We have developed a low-resolution structural model for ESCRT-III CHMP2A-CHMP3 polymers, which form pleomorphic tubular or conical structures that sometimes have closed, dome-like end caps (Bajorek et al., 2009; Fabrikant et al., 2009; Lata et al., 2008b). The variability of the tube diameters is restricted, with a full range of 44.5 to 54.5 nm, and the majority of tubes falling into the narrow range of 47 to 51 nm. In contrast, helical CHMP2B membrane tubes show a larger diversity, with diameters ranging from 100 to 400 nm (Bodon et al., 2011). Despite their variability, we were able to use 2D classification of cryo-EM images of straight tube segments to isolate homogenous classes of CHMP2A-CHMP3 tubes. The 2D classes showed two types of tube morphology, which we refer to as line pattern and column pattern, respectively. Both patterns produced reliable layer lines at ∼ 30Å in Fourier transformation analyses, indicating the helical pitch. The line pattern is best described by a 1-start helix model, while the column patterns fit multiple (n) start helix models, with pitches of n times 30 Å. Because the group representing n=6 (i.e., the 6-start helix model) was the most populated among the 2-D classes, we were able to reconstruct a model to a resolution of 22 Å. The model revealed an elongated asymmetric unit that fits the 70 Å long helical hairpin structure of CHMP3 diagonally, although some ambiguity remains about the actual content and length of the asymmetric unit at this resolution. The presence of a single layer line in Fourier transforms hampers the precise determination of the number of subunits per turn and the helical parameters found by the IHRSR, while representing local minima, may not be completely reliable (Egelman, 2010). The map differs from a previous reconstruction, which was obtained at a lower 30-Å-resolution using lower quality data collected from MBP-CHMP2AΔC-CHMP3 (Lata et al., 2008b). Although the data corresponding to the 1-start helix model did not converge to a reliable structural model, we hypothesize that the molecular interactions making up the polymer strand are the same. In fact, it is most likely that the 1-start helix is the physiologically relevant assembly. 2D classification of CHMP2B membrane tubes produced the line pattern morphology consistent with a 1-start helix model. Because the Fourier transform of the CHMP2B data revealed a layer line at 32 Å corresponding to its helical pitch (Bodon et al., 2011), we suggest that CHMP2A-CHMP3 and CHMP2B employ similar structural principles to assemble into tubes in vitro and in vivo. The same linear assembly mode is most likely used by CHMP4 polymers that produce ∼30 Å thick loose coils with no lateral interactions evident in vitro (Pires et al., 2009). Notably, the length of the repeating unit of the CHMP4B polymers was estimated at 40 Å (Pires et al., 2009), which is consistent with the length of the CHMP2A-CHMP3 asymmetric unit. Similar sized CHMP4 strands have been observed in vivo (Hanson et al., 2008), indirectly supporting the physiological relevance of the head-to-tail interaction of ESCRT-III monomers seen in our study. We demonstrate further that CHMP2A can also form linear strands or coils on its own in the absence of membrane, which have the same width as CHMP2A-CHMP3, CHMP2B (Bodon et al., 2011) and CHMP4 strands (Pires et al., 2009). These narrow coils appear to interact laterally and constrict to nearly closed structures as viewed end on. In contrast larger spiral filaments were observed at the midbody, which were hypothesized to be formed by CHMP2 (Guizetti et al., 2011). It is also notable that there are other cases in which ESCRT-III filaments can have a much thicker appearance by negative staining electron microscopy (Pires et al., 2009; Ghazi-Tabatabai et al., 2008) and image analysis of such yeast Vps24p (CHMP3) filaments produced a two-stranded helical structure (CHMP3) (Ghazi-Tabatabai et al., 2008). Such alternative assemblies might either solely reflect the general polymerization propensities of ESCRT-III proteins or could have roles during earlier events of mitosis such as centrosome and spindle maintenance (Morita et al., 2010) or abscission control (Carlton et al., 2012).
Although we were able to fit the crystal structure of the CHMP3 core (helices 1 to 4) into the asymmetric unit of our reconstruction, it is possible that the structure is not present in this conformation for several reasons. First, we showed that CHMP2A and CHMP3 became protease resistant upon polymer formation. In contrast, the docked crystal structure still exposes several potential accessible trypsin cleavage sites in the current model. Second, CHMP2A residues such as V59 and V62 implicated in tube formation and thus CHMP3 interaction (Bajorek et al., 2009) do not interact with neighboring molecules in the docked model. In addition, the cluster of basic residues on helix 1 implicated in membrane binding (Muziol et al., 2006) are exposed, but it is difficult to imagine how the surface of the pseudo-atomic model that produced a step-like or staggered appearance (data not shown) could mediate efficient membrane interaction. Thus both CHMP2A and CHMP3 may undergo conformational changes that render them protease resistant and increase their thermostability when they co-polymerize. Notably, the increased stability could provide free energy for membrane bending.
Protease K digestion of CHMP2A-CHMP3 tubes left CHMP2A (residues 9-163) unchanged and trimmed CHMP3 residues 8 to 160, indicating that helices 1 to 4 including the linker region to helix 5 form the core of the polymer. This result is consistent with data showing that CHMP3 residues 1 to 150 suffice to form tubes (Bajorek et al., 2009), but it is remarkable that the linker region connecting helices 4 and 5, a region which is disordered in the crystal structures of CHMP3 (Bajorek et al., 2009; Muziol et al., 2006), becomes resistant to protease K digestion within the CHMP2A-CHMP3 polymer. This observation provides further support for the idea that CHMP3 may undergo a conformational change upon polymerization.
The asymmetric unit exposes a rather flat surface on the tube exterior, which presumably corresponds to the membrane-binding surface. Contacts between filaments are made in the peripheral adjacent region, but the overall contact surface is limited. In contrast, interactions between asymmetric units or monomers along the filament are more extensive. This arrangement permits transitions to smaller tube diameters as observed in cone-shaped tubes and end-capped dome-like structures (Bajorek et al., 2009; Lata et al., 2008b) by successively reducing the number of monomers per turn while maintaining the minimal lateral inter-strand interactions and requiring relatively few adjustments in the relative monomer orientations. Based on the similarities between images obtained for several CHMPs as well as their structural homology (Martinelli et al., 2012; Bajorek et al., 2009; Xiao et al., 2009; Muziol et al., 2006), we conclude that all CHMP polymers acting directly on membranes likely have the same molecular architecture whether they form hetero-polymers such as CHMP2A-CHMP3 or CHMP2A, CHMP2B or CHMP4 homo-polymers.
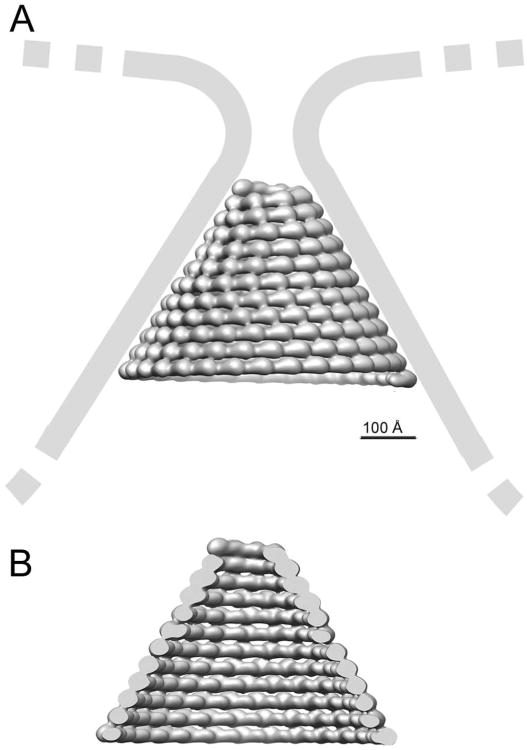
HIV-1 budding requires only one CHMP4 and one CHMP2 family member out of the 12 human ESCRT-III proteins for efficient budding (Morita et al., 2011), in contrast to cytokinesis (Morita et al., 2010). Importantly, CHMP3 depletion had a minor effect and reduced virus titer production only two-fold (Morita et al., 2011), despite the fact that C-terminally truncated CHMP3 and CHMP3-GFP exert potent dominant negative effects on HIV-1 budding (Muziol et al., 2006; Zamborlini et al., 2006; Strack et al., 2003). We extended the previous siRNA study to double knock-downs of CHMP2A, or CHMP2B and CHMP3, which confirmed that CHMP3 acts synergistically with CHMP2 proteins during HIV-1 budding. Notably, the synergy with CHMP2A is significantly higher than that with CHMP2B. Although a direct interaction between CHMP4B and CHMP2A is required for budding in the absence of CHMP3 (Morita et al., 2011), KD measurements revealed that CHMP3 has a ∼ 16-fold higher affinity for CHMP4B than does CHMP2A. In addition, CHMP2A interacts with similar micromolar KDs with both CHMP3 and CHMP4B. Thus our data indicate that when all the ESCRT-III subunits are present, CHMP3 preferentially interacts with CHMP4B, which then subsequently recruits CHMP2A. This sequence matches the order of yeast ESCRT-III recruitment (Teis et al., 2008). We further showed that CHMP2B also interacts directly with both CHMP4B and CHMP3, and that the dissociation constants are again similar to the CHMP2A-CHMP4B interaction. However, we suggest that CHMP3 may be less important for CHMP2B function than for CHMP2A function as supported by its lower synergistic effect in HIV-1 budding and by the higher affinity of CHMP4B for CHMP2B (vs. CHMP2A). This is consistent with the observation that CHMP2B membrane tube formation from the plasma membrane depends largely on CHMP4 (Bodon et al 2011). Although we showed here that CHMP2B can interact with CHMP3, CHMP3 is not important for CHMP2B membrane tube formation in vivo, and the two proteins did not form hetero-polymers in vitro (Bodon et al., 2011). These observations are in agreement with the idea that CHMP2A may function preferentially in combination with CHMP3, while CHMP2B function relies much less on CHMP3. Finally, although it is clear that the structural heterogeneity of the CHMP2A-CHMP3 polymers constituted the main limitation for cryo-EM image analysis, this heterogeneity may be important for ESCRT-III function. ESCRT-III polymer(s) presumably cannot be too static; because they may need to be remodeled by VPS4, and/or slide along membranes in order to remodel membranes in a process leading to fission (Elia et al., 2012). Furthermore the CHMP4 filaments appear to be ideally suited to form loose coils whose diameters can vary considerably and thus induce initial constriction that sets the stage for CHMP3-CHMP2A or CHMP3-CHMP2B recruitment. In contrast, more rigid structures may be required for the final constriction step catalyzed by dome-like end-caps (Fabrikant et al., 2009; Lata et al., 2008b). The tilted radial interactions between asymmetric units in our EM reconstruction are consistent with constricting a membrane bud to small diameter of 6 nm via the build-up of a dome-like polymer (Figure 8). Such a dome may be stabilized via lateral interactions of helical CHMP filaments observed in CHMP2A-CHMP3 and CHMP2B membrane tubes as well as potentially looser interfilament interactions as present in CHMP2A polymers that suffice to spiral the filaments into a constriction dome that can catalyze membrane fission (Fabrikant et al., 2009).
Figure 8. Model of a CHMP2A-CHMP3 dome-like polymer constricting a membrane bud.
(A) The arrangement is based on the EM reconstruction and indicates that the individual asymetric units may induce constriction via variations in their lateral interactions. Constriction is limited by the external length of the asymetric unit. (B) View inside the polymer that shows the long axis of the asymmetric unit perpendicular to the cone axis.
Experimental Procedures
Protein expression and purification
Expression and purification of full-length CHMP3, CHMP3ΔC (residues 9-183) (Muziol et al., 2006), MBP-CHMP2AΔC (residues 9-161) (Lata et al., 2008b) and MBP-CHMP4BΔC-Alix (Pires et al., 2009) were performed as described. A final purification step included gel filtration on a superdex 75 column (GE Healthcare) in a buffer containing 20 mM Hepes pH 7.6, 150 mM NaCl. For MBP-CHMP2AΔC-CHMP3 tube assembly, 10 to 15 μM of monomeric MBP-CHMP2AΔC were mixed with 20 to 30 μM of monomeric CHMP3 as described (Lata et al., 2008b). MBP-CHMP2AΔC-CHMP3 tubes were separated from monomeric protein by ultracentrifugation or by sucrose gradient centrifugation as described (Lata et al., 2008b). For cryo-EM, MBP was removed from the tubes by incubation with TEV protease at a molar ratio of 1: 5 (TEV:MBP-CHMP2AΔC). Full length CHMP2B was cloned into the expression vector pETM13 and expressed with a C-terminal His6 tag. Protein expression was induced in BL21 codon plus cells (Invitrogen), which produced a small amount of CHMP2B in the soluble fraction and the majority in the insoluble fraction. Cell pellets were sonicated in a buffer containing 50 mM Tris pH 7.4, 1 M NaCl, 1 M KCl, and soluble CHMP2B was purified by Ni2+ chromatography and eluted in a buffer containing 50 mM Tris pH 7.4, 300 mM NaCl and 300 mM imidazole. Final purification of the monomeric subunits was achieved by gel filtration on a superdex 75 column (GE Healthcare) in a 20 mM Hepes pH 7.4, 200 mM NaCl buffer.
Biosensor binding experiments
Biosensor binding experiments were performed with SPR detection on a Biacore × instrument (GE Healthcare). Flow rates were 5 μl/min in a running buffer containing 20 mM Hepes pH 7.6, 150 mM NaCl, 50 μM EDTA, 0.005% P20. MBP-CHMP2AΔC and MBP-CHMP4BΔC-Alix were biotinylated with biotin sulfo-LC-NHS (Thermo Scientific) for 1h at 4°C at a molar ratio of 1:5 (labeling only 1/5th of the total available lysine residues). Free biotin was removed by extensive dialysis. A CM5 chip (GE Healthcare) was coated with streptavidin (Sigma) (0.1μg/ml) to ∼6000-8000 response units in both flow cells, followed by ethanolamine injection. One of the flow cells containing the streptavidin-derivatized chip received approximately 200 RU of either biotinylated MBP-CHMP2AΔC or 800-1000 RU of biotinylated MBP-CHMP4BΔC-Alix. His-CHMP3 and MBP-CHMP2AΔC analytes were dialyzed against running buffer. 70 or 90 μl of CHMP solution dilutions (430 nM to 43 μM) were injected at the concentrations indicated (Figure S8. Global fitting of the curves using the BiaEvaluation software package (BIAcore Inc) and a 1:1 Langmuir model did not produce satisfactory fits, so we therefore averaged the equilibrium plateau responses of the binding curves (658 to 830 seconds for (CHMP2A-CHMP3); 1057 to 1197 seconds (CHMP4B-CHMP2A) and 1030 to 1170 seconds (CHMP4B-CHMP3)). The resulting RU values were used for non-linear least squares analyses to determine equilibrium binding constants. Chips were regenerated by 60 second washes in 20 mM Hepes pH 7.6, 1 m NaCl at a flow rate of 50μl/min.
Interactions between CHMP2B-His6 with MBP-CHMP4BΔC-Alix and CHMP3ΔC were performed in a buffer containing 10 mM Hepes pH 7.4, 150 mM NaCl, 3.4 mM EDTA, 0.005 % P20. MBP-CHMP4BΔC-Alix and CHMP2B-His6 were immobilized at levels of 500 and 600 RU in a sodium acetate buffer, pH 4.5, on an activated CM-5 sensor chip according to the manufacturer's instructions. Specific binding to the target protein was corrected for nonspecific binding to the deactivated control channel. The flow rate was 10 μl/min and dissociation was recorded over 10 minutes. The sensor chip was regenerated with 2 M NaCl for 60 seconds at a flow rate of 50 μl/min. Binding kinetics were evaluated using the BiaEvaluation software package (BIAcore, Inc.) using a Langmuir model 1:1 with no mass transfer.
HIV-1 production from cells depleted of CHMP2 and CHMP3 proteins
siRNA depletion experiments followed the protocol given in Morita et al. 2011 (Morita et al., 2011), and the following siRNAs were used: CHMP2-395 (AGGCAGAGAUCAUGGAU AUtt), CHMP2B-45 (GGAACAGAAUCGAGAGUUAtt), and CHMP3-474 (GGAAGAAGCA GAAAUGGAAtt), where “tt” denotes two overhanging deoxyribothymidines. Each siRNA was used at 10nM final concentrations in 2ml cultures (i.e., 20 nM total siRNA in the co-transfection reactions).
Cryo-EM image acquisition
3.5 μl of each sample was applied on the carbon side of a quantifoil grid, previously glow discharged for 1 min. The sample was then blotted and plunge-frozen in liquid ethane using a Vitrobot Mark II (FEI). Grids were stored in a liquid nitrogen dewar until they were loaded either in a Philips CM200 with LaB6 filament operating at 200 kV or in a FEI Polara with a FEG operating at 300 kV. Images were acquired under low dose conditions at magnifications of 39.5k for data recorded on CM200 (dataset 1), and 39 k (dataset 2) or 59 k (dataset 3) for data recorded on the FEI Polara. Cryo-EM micrographs were recorded on KODAK SO-163 Film.
Image preprocessing
Negatives were checked for drift and good spreading and tubes numbers. Films were digitized on a Zeiss scanner at either 14 μm for dataset 1 or 7 μm for datasets 1 and 2, giving pixel sizes of 3.54, 1.79 and 1.19 Å/pixel respectively at the specimen level. Scanned images were screened by evaluating their power spectra for the absence of drift or excessive astigmatism. Segments of tubes were picked from micrographs in consecutive overlapping (96.5 % overlap) boxes of 200, 400 and 600 pixels with boxer from the EMAN package, giving 17942, 23265 and 9184 boxes for datasets 1, 2 and 3, respectively (Ludtke et al., 1999). CTF parameters were determined for each micrograph using CTFFIND3 (Mindell and Grigorieff, 2003) and extracted images were phase-flipped with bctf, bandpass filtered to 20 Å and normalized with bfilter from the BSOFT package (Heymann and Belnap, 2007). The images were made vertical by calculating the in-plane rotation angle using the coordinates of the extremities of the tubes calculated from the boxer parameter file. A soft-edge mask was applied prior to further processing.
2D image classification
The same method was applied for the 3 datasets, but datasets 2 and 3 were binned 2 and 3 times, respectively. The vertical images were initially classified in SPIDER (Shaikh et al., 2008) by correspondence analysis, hierarchical clustering by the ward criteria, and averaging to generate first crude classes. These classes were used as references for a multi-reference alignment (Adams et al., 2009; Effantin et al., 2009; Hierro et al., 2007). An integer value was derived from the alignment parameter file for the translation in X and applied to the raw images to improve their centering. The same procedure of classification, multi-reference alignment and application of X shifts was repeated a second time. The vertically centered images were then classified in IMAGIC (van Heel et al., 1996) by multivariate statistical analysis, classification, averaging and multi-reference alignment. In the final round, the datasets were classified into 150 classes.
Helical symmetry analysis
2D Fourier transforms were calculated with bfft from BSOFT. Positions (from the equator and the meridian) and phase differences (0 or 180°) of the strongest layer line were measured in bshow. The first maximum of the Bessel function (hence its order n using the phase difference) was estimated according to the formula Jmax = 2πrR with r: distance from the meridian to the first major layer line in Å-1 and R: real space radius in Å.
To estimate the pitch of the structure, we follow a previously described method (Desfosses et al., 2011). The selected 2D class average was first carefully centered, vertically aligned and a soft-edged mask was applied. The method probes iteratively which pair of helical parameter (axial rise per subunit/azimuthal angle or pitch/number of subunit per pitch) gives the best match with the class average. For pitch estimation, we fixed the number of subunits per turn to 100 and varied only the pitch. Then, the helically related copies of the class average were generated using the helical parameter pair to be tested, and the stack of images was used to calculate a 3D reconstruction. 2D projections of the 3D model were computed and compared to the original stack of images to calculate an overall normalized correlation coefficient. The same procedure was repeated for all the pitch value to be tested and the maximum correlation is taken as the most likely pitch for the selected class average.
3D reconstruction
Dataset 2, being the largest, was selected for further processing. The 485Å diameter group was the most populated among the dataset after 2D classification. This group was combined with the 480Å group to give a 4910 segment dataset that was used for 3D reconstruction. The best 3D model derived from the pitch search described above was used as an initial reference for a projection matching approach in SPIDER, implementing hsearch and himpose programs from the IHRSR approach (Egelman, 2010; Egelman, 2007). The 3D reconstruction was arbitrarily chosen as right handed. Great care was taken to check that segments belonging to the same tube had the same polarity (or else they were discarded). Raw vertical images were used and images having an in-plane angle larger than 5° were also removed. Consecutive segments from the same tube that matched the same reference projection were also discarded. The distribution of the number of particles per reference was plotted at each iteration and any over represented views were truncated. The Fourier Shell Correlation between two independent reconstructions was calculated at each iteration and the 3D reconstruction filtered to the value found at FSC=0.5 before the helical symmetry search and imposition. Despite changing the starting value for the number of subunits per turn, the IHRSR helical parameter search converged repeatedly to an axial rise z = 6.03 Å and an azimuthal angle φ = 11.9°. The final 3D reconstruction includes 2145 segments, (corresponding to 19274 unique asymmetric units) for a calculated resolution of 22 Å at FSC = 0.5. The same procedure was applied to other groups having different diameter and a 3D reconstruction was also obtained for the 460 Å diameter tube. Its helical parameters determined by IHRSR were z = 6.03 Å and φ = 14.3°. The final reconstructions include 1601 out of 2973 particles for a resolution of 24 Å.
Docking
The fitting of CHMP3 core domain (residues 1-140, PDB: 2GD5 was performed with CHIMERA (Pettersen et al., 2004) and the SITUS package (Wriggers et al., 1999). The CHMP3 monomer was cropped to residues 1 – 140 to remove the auto-inhibitory region which was shown to prevent polymer formation and first placed semi-automatically in the cryo-EM density map using the Fit in map tool of CHIMERA. This position was then refined in SITUS. Helically symmetric copies of the monomer were generated with the pdbsymm command and the position of the individual copies was refined with the collage program. The two commands were repeated 10 cycles to ensure that the position of the fitted molecule did not change from iteration to iteration. The same procedure was followed for both hands of the cryo-EM reconstruction.
Supplementary Material
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft SPP1175 (W.W.), the Agence Nationale de la Recherche (ANR-08-BLAN-0271-01; W.W. and G.S.), the Labex GRAL (G.S.; W.W.) and NIH grants ROI AI051174 and P50 6M 082545 (W.I.S). G.E. is supported by a post doctoral fellowship from the Agence Nationale de Recherche sur le Sida (ANRS). We thank D. Lutje-Hulsik for help with SPR measurements and A. Desfosses for help and suggestions on helical image analysis. We acknowledge the Partnership for Structural Biology (PSB; http://www.psb-grenoble.eu) for access to the electron microscopy and biophysics platforms. The Polara microscope is part of the IBS Structural Biology and Dynamics GIS-IBISA-labeled platform.
References
- Adams P, Kandiah E, Effantin G, Steven AC, Ehrenfeld E. Poliovirus 2C protein forms homo-oligomeric structures required for ATPase activity. J Biol Chem. 2009;284:22012–22021. doi: 10.1074/jbc.M109.031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Davies BA, Katzmann DJ. Regulation of Vps4 during MVB sorting and cytokinesis. Traffic. 2011;12:1298–1305. doi: 10.1111/j.1600-0854.2011.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. ESCRT-III: An endosome-associated heterooligomeric protein complex required for MVB sorting. Developmental Cell. 2002;3:271–282. doi: 10.1016/s1534-5807(02)00220-4. [DOI] [PubMed] [Google Scholar]
- Bajorek M, Schubert HL, McCullough J, Langelier C, Eckert DM, Stubblefield WM, et al. Structural basis for ESCRT-III protein autoinhibition. Nat Struct Mol Biol. 2009;16:754–762. doi: 10.1038/nsmb.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartel V, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011;13:469–474. doi: 10.1038/ncb2215. [DOI] [PubMed] [Google Scholar]
- Bodon G, Chassefeyre R, Pernet-Gallay K, Martinelli N, Effantin G, Lutje Hulsik D, et al. Charged Multivesicular Body Protein 2B (CHMP2B) of the Endosomal Sorting Complex Required for Transport-III (ESCRT-III) Polymerizes into Helical Structures Deforming the Plasma Membrane. Journal of Biological Chemistry. 2011;286:40276–40286. doi: 10.1074/jbc.M111.283671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura E, Rozycki B, Chung HS, Herrick DZ, Canagarajah B, Cafiso DS, et al. Solution structure of the ESCRT-I and -II supercomplex: implications for membrane budding and scission. Structure. 2012;20:874–886. doi: 10.1016/j.str.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Caballe A, Agromayor M, Kloc M, Martin-Serrano J. ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science. 2012;336:220–225. doi: 10.1126/science.1217180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desfosses A, Goret G, Farias Estrozi L, Ruigrok RW, Gutsche I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J Virol. 2011;85:1391–1395. doi: 10.1128/JVI.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia N, Fabrikant G, Kozlov MM, Lippincott-Schwartz J. Computational Model of Cytokinetic Abscission Driven by ESCRT-III Polymerization and Remodeling. Biophysical journal. 2012;102:2309–2320. doi: 10.1016/j.bpj.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrikant G, Lata S, Riches JD, Briggs JA, Weissenhorn W, Kozlov MM. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5:e1000575. doi: 10.1371/journal.pcbi.1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Obita T, Pobbati AV, Perisic O, Samson RY, Bell SD, Williams RL. Evolution and assembly of ESCRTs. Biochem Soc Trans. 2009;37:151–155. doi: 10.1042/BST0370151. [DOI] [PubMed] [Google Scholar]
- Ghazi-Tabatabai S, Saksena S, Short JM, Pobbati AV, Veprintsev DB, Crowther RA, et al. Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure. 2008;16:1345–1356. doi: 10.1016/j.str.2008.06.010. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Gerlich DW. Cytokinetic abscission in animal cells. Semin Cell Dev Biol. 2010;21:909–916. doi: 10.1016/j.semcdb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Gerlich DW. ESCRT-III polymers in membrane neck constriction. Trends Cell Biol. 2012;22:133–140. doi: 10.1016/j.tcb.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, et al. Cortical Constriction During Abscission Involves Helices of ESCRT-III-Dependent Filaments. Science. 2011;331(6024):1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Lin Y, Heuser JE. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol. 2008;180(2):389–402. doi: 10.1083/jcb.200707031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Hierro A, Rojas AL, Rojas R, Murthy N, Effantin G, Kajava AV, et al. Functional architecture of the retromer cargo-recognition complex. Nature. 2007;449:1063–1067. doi: 10.1038/nature06216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer C, Skalicky JJ, Morita E, De Domenico I, Ward DM, Kaplan J, Sundquist WI. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev Cell. 2008;15:62–73. doi: 10.1016/j.devcel.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Lata S, Roessle M, Solomons J, Jamin M, Gottlinger HG, Svergun DI, Weissenhorn W. Structural basis for autoinhibition of ESCRT-III CHMP3. J Mol Biol. 2008a;378:818–827. doi: 10.1016/j.jmb.2008.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S, Schoehn G, Jain A, Pires R, Piehler J, Gottlinger HG, Weissenhorn W. Helical structures of ESCRT-III are disassembled by VPS4. Science. 2008b;321:1354–1357. doi: 10.1126/science.1161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Kimpler LA, Naismith TV, Lauer JM, Hanson PI. Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem. 2005;280:12799–12809. doi: 10.1074/jbc.M413968200. [DOI] [PubMed] [Google Scholar]
- Lindas AC, Karlsson EA, Lindgren MT, Ettema TJ, Bernander R. A unique cell division machinery in the Archaea. Proc Natl Acad Sci U S A. 2008;105(48):18942–18946. doi: 10.1073/pnas.0809467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Martinelli N, Hartlieb B, Usami Y, Sabin C, Dordor A, Miguet N, et al. CC2D1A Is a Regulator of ESCRT-III CHMP4B. J Mol Biol. 2012;419:75–88. doi: 10.1016/j.jmb.2012.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100:12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald B, Martin-Serrano J. No strings attached: the ESCRT machinery in viral budding and cytokinesis. J Cell Sci. 2009;122:2167–2177. doi: 10.1242/jcs.028308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Morita E, Sundquist WI. Retrovirus budding. Annu Rev Cell Dev Biol. 2004;20:395–425. doi: 10.1146/annurev.cellbio.20.010403.102350. [DOI] [PubMed] [Google Scholar]
- Morita E, Colf LA, Karren MA, Sandrin V, Rodesch CK, Sundquist WI. Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Nat Acad Sci USA. 2010;107:12889–12894. doi: 10.1073/pnas.1005938107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III Protein Requirements for HIV-1 Budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muziol T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Gottlinger H, Weissenhorn W. Structural basis for budding by the ESCRT-III factor CHMP3. Dev Cell. 2006;10:821–830. doi: 10.1016/j.devcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Peel S, Macheboeuf P, Martinelli N, Weissenhorn W. Divergent pathways lead to ESCRT-III catalyzed membrane fission. Trends Biochem Sci. 2011;36:199–210. doi: 10.1016/j.tibs.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pires R, Hartlieb B, Signor L, Schoehn G, Lata S, Roessle M, et al. A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure. 2009;17:843–856. doi: 10.1016/j.str.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29:1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saksena S, Sun J, Chu T, Emr SD. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci. 2007;32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. Functional reconstitution of ESCRT-III assembly and disassembly. Cell. 2009;136:97–109. doi: 10.1016/j.cell.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol Biol Cell. 2010;21:1059–1071. doi: 10.1091/mbc.E09-07-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S, Kimpler LA, Hanson PI. Structure/Function Analysis of Four Core ESCRT-III Proteins Reveals Common Regulatory Role for Extreme C-Terminal Domain. Traffic. 2007;8:1068–1079. doi: 10.1111/j.1600-0854.2007.00584.x. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Popova E, Gottlinger H. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Sundquist WI, Krausslich HG. HIV-1 Assembly, Budding, and Maturation. Cold Spring Harb Perspect Med. 2012;2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Saksena S, Emr SD. Ordered Assembly of the ESCRT-III Complex on Endosomes Is Required to Sequester Cargo during MVB Formation. Dev Cell. 2008;15(4):578–589. doi: 10.1016/j.devcel.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, G GH. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans. 2009;37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- Wollert T, Hurley JH. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature. 2010;464(7290):864–869. doi: 10.1038/nature08849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Milligan RA, McCammon JA. Situs: A package for docking crystal structures into low-resolution maps from electron microscopy. J Struct Biol. 1999;125:185–195. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- Xiao J, Chen XW, Davies BA, Saltiel AR, Katzmann DJ, Xu Z. Structural basis of Ist1 function and Ist1-Did2 interaction in the multivesicular body pathway and cytokinesis. Mol Biol Cell. 2009;20:3514–3524. doi: 10.1091/mbc.E09-05-0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Gottlinger H. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci U S A. 2006;103:19140–19145. doi: 10.1073/pnas.0603788103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.