Abstract
Nucleotide excision repair (NER) is the main pathway used by mammals to remove bulky DNA lesions such as those formed by UV light, environmental mutagens, and some cancer chemotherapeutic adducts from DNA. Deficiencies in NER are associated with the extremely skin cancer-prone inherited disorder xeroderma pigmentosum. Although the core NER reaction and the factors that execute it have been known for some years, recent studies have led to a much more detailed understanding of the NER mechanism, how NER operates in the context of chromatin, and how it is connected to other cellular processes such as DNA damage signaling and transcription. This review emphasizes biochemical, structural, cell biological, and genetic studies since 2005 that have shed light on many aspects of the NER pathway.
Bulky DNA lesions (e.g., those formed by UV light) are removed by nucleotide excision repair (NER). Recent studies have led to a detailed understanding of the NER mechanism and its regulation in the context of the cell.
Nucleotide excision repair (NER) is the main pathway responsible for the removal of bulky DNA lesions induced by UV irradiation, environmental mutagens, and certain chemotherapeutic agents. The history of the discovery of NER, its association with genetic disorders, mechanistic features, and relationship with other cellular pathways has been extensively reviewed in 2005 in several articles in DNA Repair and Mutagenesis (Friedberg et al. 2005). Here, I will briefly reiterate how the field of NER developed over the past 50 years and then focus on how our knowledge has progressed since 2005.
A BRIEF HISTORY OF NER RESEARCH
We now know that a short damage-containing oligonucleotide is released from DNA and the resulting gap filled during repair synthesis during NER. These two steps were observed in the earliest studies of NER in bacteria and humans, respectively, in 1964 and marked the beginnings of NER research 50 years ago (Boyce and Howard-Flanders 1964; Pettijohn and Hanawalt 1964; Rasmussen and Painter 1964; Setlow and Carrier 1964). The gap-filling step in NER can be monitored by unscheduled DNA synthesis (UDS), and this assay allowed the connection between NER and the genetic disorder xeroderma pigmentusum (XP) to be made (Cleaver 1968). UDS is used to date in the clinical diagnosis of XP patients and was instrumental in the elucidation of the NER pathway. XP patients display an extreme sensitivity to sunlight and an over 2000-fold increased risk of skin cancer, as they are unable to repair lesions induced by solar UV irradiation in their skin (DiGiovanna and Kraemer 2012). At least two other disorders, Cockayne syndrome and trichothiodystrophy (TTD), are also associated with defects in NER genes; but these patients do not present with skin cancer predisposition, but rather a host of developmental and neurological abnormalities (Lehmann 2003). These symptoms are generally ascribed to a defect in transcription-coupled NER and a mild or partial defect in transcription.
Studies of XP patient cell lines then revealed that UDS varied significantly among cell lines. Using cell fusion techniques, it was established that seven complementation groups with NER defects exist, XPA through XPG, each representing a different gene defect (De Weerd-Kastelein et al. 1972). The availability of the XP cell lines from patients, along with UV-sensitive yeast strains (Prakash and Prakash 2000) and Chinese hamster ovary cell lines (Thompson 1998), made it possible to clone the NER genes over the years (Gillet and Schärer 2006).
The next phase of discovery was facilitated by the development of assays of studying NER in cell-free extracts by either monitoring incorporation of radioactive deoxynucleotide triphosphates (dNTPs) into damage-containing plasmids during repair synthesis (Wood et al. 1988) or the excision of the labeled damage-containing oligonucleotide from a larger DNA fragment (Huang et al. 1992). In the mid-1990s, the use of these assays culminated in the reconstitution of the NER reaction with purified factors (Aboussekhra et al. 1995; Mu et al. 1995; Araujo et al. 2000). These studies revealed that the process also required proteins involved in replication, such as the single-stranded DNA- (ssDNA) binding protein RPA, the clamp loader PCNA, and polymerase δ. Biochemically derived models of NER received strong support from subsequent cell biological studies using UV irradiation through filters to generate sites of localized damage in cell nuclei in combination with green fluorescent protein-labeled proteins or immunohistological techniques to monitor the dynamic behavior of NER proteins (Houtsmuller et al. 1999; Volker et al. 2001). These studies revealed that NER functions through the sequential and coordinated assembly of the factors involved at the sites of UV damage.
In the following sections, I will discuss our current understanding of the NER process in the order in which the steps occur, followed by the connection to chromatin structure and DNA damage signaling. The reader is referred to other reviews in this collection that cover NER in bacteria (Kisker et al. 2013) and transcription-coupled NER (Vermeulen and Fousteri 2013).
THE CORE NER REACTION
Different Modes of Damage Recognition in NER
NER can be initiated by two subpathways: global genome NER (GG-NER) or transcription-coupled NER (TC-NER; Gillet and Schärer 2006; Hanawalt and Spivak 2008). GG-NER can occur anywhere in the genome, whereas TC-NER is responsible for the accelerated repair of lesions in the transcribed strand of active genes. GG-NER is initiated by the GG-NER specific factor XPC-RAD23B, in some cases with the help of UV-DDB (UV-damaged DNA-binding protein). TC-NER is initiated by RNA polymerase stalled at a lesion with the help of TC-NER specific factors CSA, CSB, and XAB2. Both pathways require the core NER factors to complete the excision process. Very recently, an additional class of enzymes, the alkyltransferase-like (ATL) proteins, have been shown to channel bulky O6-alkylguanine lesions into the NER pathway, at least in some lower eukaryotes and bacteria (Tubbs et al. 2009; Latypov et al. 2012). ATLs bind to these bulky alkyl lesion and facilitate their removal from DNA in an NER-dependent fashion. There is no other described repair pathway for bulky O6-alkylguanine lesions in humans, so it will be interesting to uncover whether an analogous pathway exists in mammals.
Substrate Specificity of Nucleotide Excision Repair
A key characteristic of the NER pathway is that one set of enzymes can recognize an extraordinarily wide range of substrates, including UV-induced photoproducts (cyclopyrimidine dimers [CPDs], 6-4 photoproducts [6-4PPs]), adducts formed by environmental mutagens such as benzo[a]pyrene or various aromatic amines, certain oxidative endogenous lesions such as cyclopurines and adducts formed by cancer chemotherapeutic drugs such as cisplatin (Friedberg et al. 2005; Gillet and Schärer 2006). Although these lesions do not share chemical structures that would lend themselves to a common lesion-binding pocket, a collective feature of good NER substrates is that they thermodynamically destabilize the DNA duplex and are bulky. These properties were originally framed in the context of the bipartite recognition model that posits that good NER substrates are destabilizing and bulky (Hess et al. 1997). Subsequent studies correlating the structures of adducts such as those formed by benzo[a]pyrene or acetylaminofluorene with their repair efficiencies confirmed these observations and showed that factors such as disrupted base-pairing, bending, and flexibility can contribute to repair outcomes (Mocquet et al. 2007; Cai et al. 2010; Mu et al. 2012; Yeo et al. 2012). As discussed in the next section, we now understand these principles in terms of how damage recognition is achieved in NER.
XPC-RAD23B Recognizes Lesions that Thermodynamically Destabilize DNA Duplexes
The core NER dual incision reaction has been reconstituted in vitro with purified factors using XPC-RAD23B, TFIIH, XPA, RPA, XPG, and ERCC1-XPF (Aboussekhra et al. 1995; Mu et al. 1995; Araujo et al. 2000). Functional studies revealed that XPC-RAD23B is the initial damage recognition factor in this system, as the presence of XPC-RAD23B is required for assembly of the other core NER factors and progression through the NER pathway both in vitro and in vivo (Fig. 1) (Evans et al. 1997b; Sugasawa et al. 1998; Volker et al. 2001; Riedl et al. 2003; Tapias et al. 2004). XPC-RAD23B is faced with the task of recognizing a wide range of structurally diverse lesions. XPC recognizes thermodynamically destabilized duplex DNA, irrespective of whether it contains a lesion (Sugasawa et al. 2001).
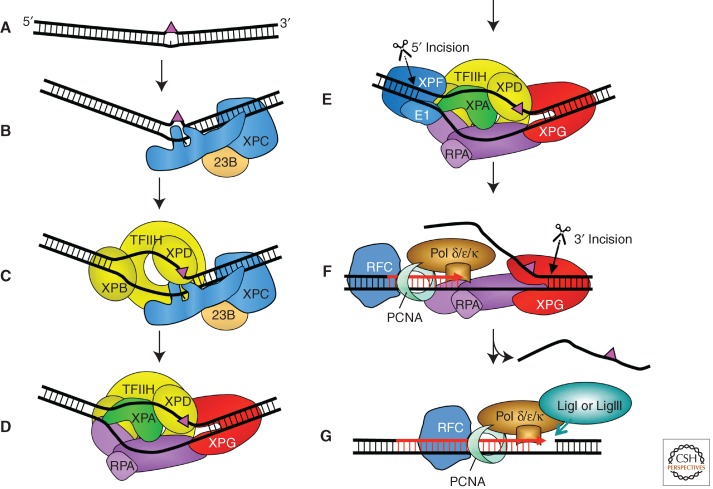
Figure 1.
Model for the core NER reaction. (A) Bulky DNA lesions that destabilize duplex DNA are induced by a number of damaging agents. (B) In global genome NER, strongly distorting lesions are directly recognized by XPC-RAD23B, which binds the nondamaged strand opposite the lesion. (C) TFIIH interacts with XPC-RAD23B and pries the DNA open with its XPB subunit allowing XPD to track along DNA until stalls at the damage and verifies the chemical modification (bulkiness) of the lesion. (D) Stalling of XPD at the lesion allows for the formation of the preincision complex by recruitment of XPA, RPA, and XPG. The endonuclease XPG does not make an incision at this point. (E) Recruitment of ERCC1-XPF by interaction with XPA to the complex leads to incision 5′ to the lesion. (F) Initiation of repair synthesis by Pol δ and Pol κ or Pol ε and associated factors, followed by 3′ incision by XPG. (G) Completion of repair synthesis and sealing of the nick by DNA ligase IIIα/XRCC1 or DNA ligase I completes the process.
Structural studies of the yeast homolog of XPC, Rad4, have provided a high-resolution view of how this is accomplished. XPC/Rad4 has two main functional domains (Min and Pavletich 2007): a lesion and sequence-unspecific DNA-binding domain made up of a transglutamase homology domain and a β-hairpin domain (BHD1) (blue in Fig. 2A) anchors the protein on the DNA. A double β-hairpin domain (BHD2/3, pink in Fig. 2A) binds the undamaged strand of the DNA without making direct contact with the lesion, which is disordered in this structure. Instead, BHD2/3 encircles two nucleotides opposite the damage that display increased ssDNA character due to the thermodynamic destabilization caused by the lesion. This binding pocket is specific for nondamaged DNA and it would not be able to accommodate bulky DNA adducts. Therefore, XPC is necessarily targeted to the nondamaged DNA strand. This mode of DNA binding makes it possible for XPC to bind a wide variety of structurally diverse bulky lesions, in keeping the broad substrate specificity of NER (Huang et al. 1994; Gunz et al. 1996; Liu et al. 2011). Biochemical studies confirmed the preference of XPC-RAD23B for binding duplexes with ssDNA character and showed that duplexes containing bulky lesions on both strands are not processed by NER (Sugasawa et al. 2002; Buterin et al. 2005; Maillard et al. 2007).
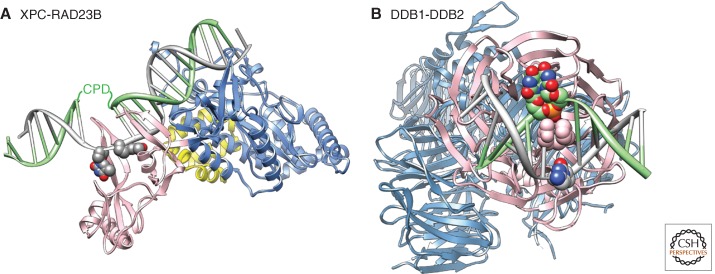
Figure 2.
Structural basis for damage recognition in NER. (A) Structure of RAD4(XPC)-RAD23 bound to a CPD lesion. The TDG/BHD1 domains (blue) of RAD4/XPC bind to undamaged DNA, the BHD2/BHD3 domains (pink) encircle two nucleotides in the nondamaged strand of DNA. DNA is represented in gray with the two thymidine residues opposite the lesion in atom color. The position of the CPD lesion that is disordered in this structure is indicated. A fragment of RAD23 is shown in yellow. Note that the CPD lesion is present in a two base pair mismatch in this structure. The damaged DNA strand is shown in green, the undamaged strand in gray. The figure was made using the Chimera extensible molecular modeling system located at UCSF (www.cgl.ucsf.edu/chimera), using the structure PDB 2QSG (Min and Pavletich 2007). (B) Structure of DDB1-DDB2 bound to a CPD lesion. The UV-DDB complex, made up of DDB1 (light blue) and DDB2 (pink), binds DNA (gray) containing a CPD (atom color, green) through the DDB2 subunit. The DNA-binding site in DDB2 is located on one site of the β-propeller; DDB1 binds DDB2 on the opposite side. A wedge made up of residues F371, Q372, and H373 in DDB2 (shown in pink van der Waals representation) inserts into the DNA helix at the lesion site from the minor groove. The two modified nucleotides of the 6-4PP insert into a shallow binding pocket in DDB2. The damaged DNA strand is shown in green, the undamaged strand in gray. The figure was made using the Chimera extensible molecular modeling system located at UCSF (www.cgl.ucsf.edu/chimera), using the structure PDB A408 (Fischer et al. 2011).
All the damage-binding specificity appears to reside in XPC, and it is thought that the main RAD23B (and to a lesser extent RAD23A, the other of two homologs of Saccharomyces cerevisiae Rad23) is to stabilize XPC. Although RAD23B is the main protein associated with XPC, RAD23B and RAD23A are partially redundant and the depletion of both proteins is required to render cells sensitive to UV irradiation (Ng et al. 2003). One of the main roles of RAD23A/B appears to be the stabilization of XPC, as cellular levels of XPC are reduced in the absence of RAD23A/B. RAD23B has a more specific role in promoting damage recognition as well, as the protein stimulates binding of XPC to lesions in vitro and in vivo (Sugasawa et al. 1996; Bergink et al. 2012). Interestingly, RAD23B dissociates from XPC following damage recognition, suggesting that the protein is not involved in preincision complex assembly. The role of RAD23B, therefore, appears to be to stabilize XPC and help deliver it to sites of UV damage (Bergink et al. 2012).
Although most studies have been performed with XPC-RAD23B, it is part of a trimeric complex with centrin-2, a calcium-binding protein of the calmodulin family (Nishi et al. 2005). The interaction with centrin-2 with XPC-RAD23B stimulates the NER activity, but the precise role of this protein in the process remains to be elucidated. Another open question in the study of XPC-RAD23B function is to what extent the human XPC-RAD23B differs from its yeast counterpart Rad4 (Bunick et al. 2006), whether the mode of lesion binding by XPC affects subsequent steps in NER (Mocquet et al. 2007), and how the protein finds DNA lesions in a cellular environment (Hoogstraten et al. 2008; Camenisch et al. 2009).
The UV-DDB Complex Facilitates Recognition of CPDs
Our knowledge of XPC-RAD23B does not explain how the most frequently formed UV-induced DNA adduct, the CPD is recognized in NER because XPC-RAD23B does not bind this adduct with any measurable specificity as this lesion does not dramatically destabilize DNA duplexes (Sugasawa et al. 2001; Reardon and Sancar 2003; Wittschieben et al. 2005). The answer to these issues is tightly connected to the UV-damage-binding proteins 1 and 2 (UV-DDB1/2). DDB2 has long been known to have an affinity for UV-damaged DNA and to be deficient in XP group E patients, the mildest XP complementation group (Tang and Chu 2002). DDB2 has, furthermore, been shown to be specifically required for the cellular repair of CPDs—in the absence of DDB2, XPC fails to be recruited to nuclear sites of CPDs, but not 6-4PPs (Tang et al. 2000; Wakasugi et al. 2001; Fitch et al. 2003).
Structural studies have revealed how UV-DDB2 interacts with 6-4PPs and CPDs (Fig. 2B) (Scrima et al. 2008; Fischer et al. 2011; Yeh et al. 2012). DDB2 has a shallow hydrophobic-binding pocket that is ideally suited to accommodate CPDs and also 6-4PPs through surface shape complementarity. The lesions are extruded from the duplex with the help of a wedge made up of three conserved residues (Phe371, Gln372, His373) that occupy the hole left by rotating the two nucleotides of the CPD/6-4-PP lesion out of the helix into the binding pocket of DDB2 (Schärer and Campbell 2009). This structure of the photolesions induces a compression of the adjacent phosphate groups, a feature readily accommodated and extended to a kink of ∼40% in the duplex (Scrima et al. 2008). This binding mode of DDB2 is highly complementary to the way lesions are bound by XPC (see above and Fig. 2A). Indeed, a direct role for UV-DDB in facilitating XPC binding to lesions has been shown (Sugasawa et al. 2005). UV-DDB is part of a large complex that includes the CUL4-ROC1 (also known as RBX1) ubiquitin ligase (Groisman et al. 2003). Following UV irradiation, this complex ubiquitinates DDB2, histones, and XPC (Sugasawa et al. 2005; Kapetanaki et al. 2006; Wang et al. 2006). Although DDB2 and histones appear to be degraded following ubiquitination, XPC is not, but displays enhanced DNA-binding properties (Sugasawa et al. 2005). These observations point to a key role of UV-DDB in facilitating NER in the context of chromatin, which could explain the modest effects of UV-DDB on the NER reaction on “naked” DNA substrates. Our current understanding of how NER occurs in the context of chromatin is discussed in more detail later in the review.
DNA Opening and Lesion Verification by TFIIH
The transcription and NER factor TFIIH is the next factor to join the NER complex and it is recruited by direct interaction with the XPC-RAD23B protein (Evans et al. 1997a; Yokoi et al. 2000; Araujo et al. 2001; Volker et al. 2001; Riedl et al. 2003; Friedberg et al. 2005). TFIIH consists of 10 subunits and can be divided up into the core (consisting of XPB, p52, p8, p62, p34, p44) and CAK (cyclin-activated kinase, consisting of CDK7, cyclin H, and MAT1) complexes and the XPD protein that bridges the two (Compe and Egly 2012). The CAK complex dissociates from TFIIH and is not required for NER (Araujo et al. 2000; Coin et al. 2008). Of particular importance for NER are the two helicase subunits, XPB and XPD, which are known to open the DNA around the lesion (Evans et al. 1997b; Tapias et al. 2004).
Structural studies of archaeal XPB and XPD homologs along with biochemical studies have made it possible to characterize the role of these two proteins in NER in more detail. The ATPase, but not the helicase activity of XPB, is required for NER and it is believed that the protein has a role in prying open the DNA to facilitate loading of TFIIH (Coin et al. 2007). Structures of the Archaeoglobus fulgidus XPB revealed unique domains in addition to the RecA-like domains commonly found in SF2 helicases (Fan et al. 2006). Two of these domains, termed RED and thumb domains (red and yellow, respectively, in Fig. 3A) have been shown to be important for NER and it has been suggested that they have a role in anchoring the protein to the DNA (Oksenych et al. 2009). XPB has to undergo a significant structural change to form the composite ATP-binding domain and this motion may be important for melting the DNA and anchoring TFIIH. The model for this conformation, displayed in Figure 3A, shows how the RED domain might insert into the DNA helix and the thumb domain anchors XPB in the major groove of DNA (Fan et al. 2006).
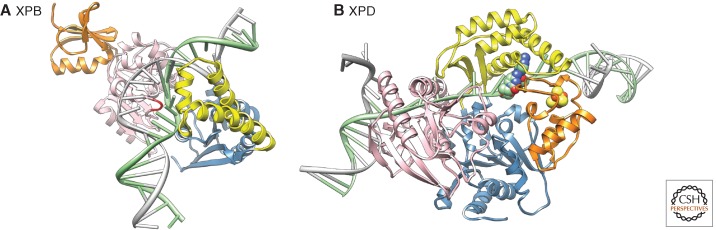
Figure 3.
Structural basis for helix opening and lesion verification in NER. (A) Model of an archaeal homolog of XPB bound to DNA. DNA-bound XPB undergoes a conformational change with respect to the apo protein, resulting in the insertion of the RED motif (red) into the duplex and binding of the DNA by the thumb domain (yellow). The two helicase domains are shown in pink (HD1) and blue (HD2), the damage-recognition domain (orange) is related to a similar domain found in MutS and aids initial DNA binding. (The figure was made using Chimera [www.cgl.ucsf.edu/chimera], based on data from Fan et al. 2006.) (B) Model of an archaeal XPD homolog tracking along DNA. The two helicase domains (HD1, blue; HD2, pink) bind the DNA at the ss/dsDNA (double-stranded DNA) junction, the FeS domain (orange, FeS cluster shown in yellow/red) and the arch domain encircle the ssDNA forming a tunnel through which the ssDNA translocate. Note that the tunnel is too narrow to permit bulky lesions to cross it, leading to stalling of the helicase and damage verification. Two native DNA bases in the tunnel are shown in green/atom color. (The figure was made using the Chimera extensible molecular modeling system located at UCSF [www.cgl.ucsf.edu/chimera], based on data from Fan et al. 2008.)
It is believed that this helix opening by XPB would then facilitate the engagement of XPD. The helicase and ATPase activity of XPD are required for NER, suggesting that this protein translocates along the DNA and opens the DNA (Winkler et al. 2000; Coin et al. 2007). XPD has long been a candidate as a damage-verification factor, as it had been shown that its helicase activity is blocked by the presence of DNA lesions (Naegeli et al. 1992). Structural studies of the archaeal XPD homologs revealed features of this class of proteins that support this idea (Fan et al. 2008; Liu et al. 2008; Wolski et al. 2008; Kuper et al. 2012; Pugh et al. 2012). In addition to the two RecA domains commonly found in SF2 helicases, XPD contains an iron–sulfur cluster and arch domain that form a tunnel through which the ssDNA is threaded during translocation (orange and yellow, respectively, in Fig. 3B). An attractive model for damage verification is that bulky lesions will not fit through this tunnel leading to a stalling of the XPD protein, whereas native DNA bases would readily be accommodated. It has indeed been shown that archaeal XPD stalls during translocation of a lesion-containing strand under certain conditions (Mathieu et al. 2010). Furthermore, mutations in a putative nucleotide-binding pocket at the entrance of the tunnel have been shown to impact damage verification raising the possibility of a more specific interaction between XPD and the lesion (Mathieu et al. 2013). Interestingly, it has been shown that in model systems XPC-RAD23B-mediated loading of TFIIH onto DNA at a distance from a lesion can initiate translocation in the 5′ to 3′ direction. This translocation ends at lesions, providing strong support that XPD is indeed responsible for damage verification (Sugasawa et al. 2009).
Although XPB and XPD are the catalytic engines that drive the role of TFIIH in NER, we are starting to learn about how other TFIIH subunits contribute to NER. p52 has been shown to associate and stimulate the activity of the XPB (Coin et al. 2007; Fregoso et al. 2007). p44, on the other hand, interacts tightly with and stimulates the XPD helicase.
Recent studies have revealed an essential and surprising role for p8, the smallest TFIIH subunit, in NER. p8 was originally isolated as the gene defective in the TTD-A complementation group of TTD (Giglia-Mari et al. 2004; Ranish et al. 2004). Because TTD is widely believed to be primarily a disease of (mild) transcription impairment, the main role of TTD was initially believed to be in the stabilization of the TFIIH complex (Vermeulen et al. 2000). Subsequent studies, however, revealed that p8/TTD-A is also an essential factor for NER, although it is dispensable for transcription in vitro. (Coin et al. 2006; Giglia-Mari et al. 2006). In the absence of p8, TFIIH-mediated duplex opening and recruitment of XPA is impaired. The study of Ttda−/− mice and cells provides a resolution of this apparent paradox. Compared to partially NER-deficient cells from TTD-A patients that express truncated forms of p8/TTD-A (Theil et al. 2011), Ttda−/− null cells are completely NER-deficient, revealing p8/TTD-A as an essential NER factor (Theil et al. 2013).
Completion of Preincision Complex Assembly
The engagement of XPD with the lesion enables the full assembly of the preincision complex. XPA, RPA, and XPG are next recruited to the site of the lesion independently of each other, and XPC-RAD23B departs from the complex at this point (Wakasugi and Sancar 1998; Volker et al. 2001; Rademakers et al. 2003; Riedl et al. 2003). XPA is a central component of the NER complex as it interacts with the TFIIH, RPA, XPC-RAD23B, DDB2, ERCC1-XPF, and PCNA proteins (Li et al. 1994; Park et al. 1995; Nocentini et al. 1997; You et al. 2003; Bunick et al. 2006; Wakasugi et al. 2009; Gilljam et al. 2012). Originally considered to be the primary damage recognition factor, it has been shown that XPA prefers to bind kinked, rather than lesion-containing DNA structures (Missura et al. 2001; Camenisch et al. 2006). This multitude of interactions makes XPA a central NER factor and its likely role is to make sure that all the NER factors are in the right place for the incision to occur. Model studies have suggested that XPA binds close to 5′ side of a bubble structure, in which it could be ideally positioned to interact with RPA and ERCC1-XPF to coordinate further steps in the NER reaction (Krasikova et al. 2010). The function of XPA furthermore seems to be regulated by a number of factors, including the histone deacetylase SIRT1, which deacetylates XPA at residues 63 and 67 (Fan and Luo 2010). In response to circadian rhythms, XPA is regulated at the transcriptional level by cryptochrome and at the posttranslational level by the HERC2 ubiquitin ligase (Kang et al. 2010, 2011).
XPA interacts tightly with the ssDNA-binding protein RPA in the NER complex and the two are believed to cooperate in their association with DNA (Li et al. 1995a; Matsuda et al. 1995; Patrick and Turchi 2002; Saijo et al. 2011). RPA is made up of three subunits (RPA70, RPA32, and RPA14). At least the two larger subunits interact with XPA through distinct regions and recent studies have shown that both of these interactions are needed for optimal NER activity (Saijo et al. 2011). The preferred binding site for RPA is ssDNA of approximately 30 nucleotides or about the size of the oligomer excised in NER (Fan and Pavletich 2012). RPA is believed to bind the nondamaged DNA strand, in which it helps position the two endonucleases ERCC1-XPF and XPG on their substrate, the damaged DNA strand (de Laat et al. 1998). RPA has an important role in coordinating excision and repair synthesis events (see next section) (Mocquet et al. 2008; Overmeer et al. 2011).
The structure-specific endonuclease XPG is recruited through interaction with TFIIH, the protein, in fact, seems to be constitutively associated with TFIIH at least for some of its roles in transcription (Araujo et al. 2001; Dunand-Sauthier et al. 2005; Zotter et al. 2006; Hohl et al. 2007; Ito et al. 2007). The first role of XPG in NER is a structural one and its catalytic activity is not required for completion of preincision complex assembly (Wakasugi et al. 1997; Constantinou et al. 1999). 3′ Incision by XPG is only triggered later in the NER reaction, following 5′ incision by ERCC1-XPF (see next section). Consistent with this observation, XPG has distinct requirements for DNA substrate binding and cleavage (Hohl et al. 2003). The protein binds many different substrates with ss/dsDNA junctions, including flaps and three-way junctions, but makes incisions only on those with an ssDNA stretch at the 3′ ssDNA overhang.
The complex consisting of TFIIH, XPA, RPA, and XPG is relatively stable (Wakasugi and Sancar 1998), and the dual excision reaction is only triggered once ERCC1-XPF joins the complex. ERCC1-XPF is recruited to NER complexes by interaction with the XPA protein (Li et al. 1994, 1995b). A short peptide of XPA containing a signature TGGGFI motif undergoes a disorder to order transition to bind in the central domain of ERCC1 and mutations in this region in XPA or the binding pocket in ERCC1 selectively abolish the NER reaction (Tripsianes et al. 2007; Tsodikov et al. 2007; Orelli et al. 2010).
Dual Incision and Repair Synthesis
Once the two endonucleases are in place, dual incision can be initiated. Although the order of dual incision has been unclear and it was thought that ERCC1-XPF or XPG might initiate the process, a more recent study has suggested that there is a defined order to the incisions (Fagbemi et al. 2011). Accordingly, the first incision is made by ERCC1-XPF, as it requires the presence, but not the catalytic activity of XPG. Indeed, repair synthesis can be initiated and proceed halfway through the gap in the absence of XPG incision (Staresincic et al. 2009). This mechanism is well-suited to ensure that the gap formed by dual incision does not persist. Incision by ERCC1-XPF generates a free 3′ hydroxyl group that the replication machinery can use to initiate repair synthesis. In contrast, incision by XPG would not produce an intermediate that can be used to initiate repair synthesis. Instead, a cut by XPG leaves a 5′ phosphate that is only productive at the ligation step. Biochemical and structural studies of XPG family of proteins revealed that they have to undergo a structural rearrangement following DNA binding to become catalytically active (Hohl et al. 2003; Orans et al. 2011; Tsutakawa et al. 2011; Grasby et al. 2012). It will be interesting whether regulatory mechanisms such as posttranslational modifications are in play that regulate the two incision reactions to ensure that no gaps or nicks are exposed.
Following the excision reaction, the lesion-containing oligonucleotide is released with TFIIH bound to it (Kemp et al. 2012) strengthening the model that XPD is the damage-verifying protein. TFIIH is then released from the oligo after binding ATP and the oligo is bound by RPA and eventually degraded. The fact that RPA may bind the product of the reaction suggests that this could have a role in damage signaling (see also section on The Role of NER in UV-Induced DNA Damage Signaling).
Based on in vitro studies using purified factors, it has been assumed that standard replication factors, DNA polymerases δ and ε, the sliding clamp PCNA, the pentameric clamp loader RFC, RPA, and DNA ligase are responsible for the repair synthesis and ligation steps to fill the gap (Shivji et al. 1995; Araujo et al. 2000). Studies in recent years have revealed that these steps too are more complex than originally assumed (Lehmann 2011). The first unexpected finding was the involvement of the error-prone translesion synthesis polymerase Pol κ in the repair synthesis step (Ogi and Lehmann 2006). Subsequent studies revealed that depletion of Pol δ and Pol κ, or both, lead to a 50% reduction of repair synthesis, suggesting that these two polymerases work together in one pathway and are responsible for about half of the repair synthesis (Ogi et al. 2010). The other 50% could be attributed to Pol ε, suggesting that there are two pathways for performing repair synthesis in human cells. Furthermore, to function in NER, these three polymerases require distinct interaction partners. The recruitment of Pol δ to NER complexes requires RFC and PCNA. Recruitment of Pol κ requires ubiquitinated PCNA, as is true for translesion synthesis, and XRCC1. Recruitment of Pol ε requires a modified form of the RFC containing Ctf18. Although evidence suggests that Pol ε may be important in dividing cells with high levels of dNTPs and open DNA structures (Ogi et al. 2010; Lehmann 2011), the roles of the two modes of repair synthesis remain subject of investigation.
The nature of the final step in the NER process, ligation of the nick left after repair synthesis, was recently also found to be dependent on the proliferative status of the cell. Initially believed to be mediated uniquely by the replicative DNA ligase I, it has been shown that this step also prominently involves DNA ligase IIIα and XRCC1 (Moser et al. 2007). Interestingly, and in line with a possible cell-cycle dependence of the repair synthesis step, DNA ligase IIIα and Pol δ are absolutely required for NER in quiescent cells and involved in replicating cells, whereas DNA ligase I and Pol ε are exclusively used in replicating cells.
NER IN THE CONTEXT OF CHROMATIN
Like all DNA transactions, NER acts on DNA in the context of chromatin, making it necessary for NER proteins to enlist the support of chromatin-modifying enzymes to gain access to lesions. In general, making DNA accessible in chromatin involves two components: the modification of histone tails to decrease the affinity of histones for the DNA and the action of ATP-dependent chromosome-remodeling enzymes to move the histones along DNA. It had been shown many years ago that chromatin undergoes rearrangement during NER, leading to the proposal of the “access–repair–restore” mechanism of NER in the context of chromatin (Smerdon and Lieberman 1978). In vitro studies furthermore show that, as expected, NER is slower in reconstituted nucleosomes and that repair rates can be enhanced in a relatively nonspecific fashion by chromatin-remodeling enzymes of the SNF2/SWI2 class (Hara et al. 2000; Ura et al. 2001; Hara and Sancar 2002). Studies in recent years have led to dramatic advances in our understanding of how NER is facilitated in the context of chromatin (see also Peterson and Almouzni 2013).
The UV-DDB Complex Coordinates NER in Chromatin
A number of observations suggested that UV-DDB (made up of the damage-binding protein DDB2 and the adaptor protein DDB1) has a key role in facilitating NER in the context of chromatin. DDB2 (deficient in the XP-E patient cell lines) is involved in the earliest step in NER and is required for targeting XPC-RAD23 to the site of UV lesions, in particular CPDs (Wakasugi et al. 2002; Fitch et al. 2003). UV-DDB forms part of a ubiquitin ligase complex with CUL4A and ROC1 (Groisman et al. 2003) that localizes to chromatin immediately after UV irradiation, in which it ubiquitinates histone H2A, H3, and H4 resulting in a destabilization of the nucleosome structure (Bergink et al. 2006; Kapetanaki et al. 2006; Wang et al. 2006; Guerrero-Santoro et al. 2008). This complex also ubiquitinates XPC and DDB2. Although ubiquitination leads to the degradation of DDB2, XPC is instead stabilized and its affinity for DNA is increased (Sugasawa et al. 2005; Nishi et al. 2009). Ubiquitination of XPC has also been suggested to ensure its localization to sites of damage in the accessible regions of the chromatin landscape where NER occurs (Fei et al. 2011).
Structural studies have revealed how the UV-DDB complex is anchored in chromatin (see Fig. 2B). This ubiquitin ligase complex does so by interaction of the DDB2 subunit with DNA lesions (Scrima et al. 2008; Fischer et al. 2011; Yeh et al. 2012). The ubiquitin ligase activity of the UV-DDB complex is autoinhibited by association of the COP9 signalosome (CSN) subunit. On binding to damaged DNA, CSN is released, activating the ubiquitin ligase activity and generating a defined ubiquitination zone on the chromatin determined by the flexible attachment of the CUL4/ROC1 factors to UV-DDB (Scrima et al. 2008; Fischer et al. 2011). One of the structures has shown that DDB2 binding to DNA can result in the formation of a dimer on the DNA via the amino termini of DDB2, thereby increasing its affinity for damage (Yeh et al. 2012). This dimerization may be important for proper positioning of the UV-DDB complex on chromatin.
More recently, a CUL4A/ROC1 ubiquitin ligase-independent function for DDB2 to mediate chromatin modification has been discovered (Luijsterburg et al. 2012; Robu et al. 2013). This activity is linked to poly(ADP-ribose) polymerase (PARP) activity and is dependent on ATP, but does not require XPC. DDB2 facilitates the PARP-mediated polyribosylation and recruitment of the chromatin-remodeling enzyme ALC1, a SNF2/SWI superfamily ATPase (Pines et al. 2012). DDB2 is poly(ADP)-ribosylated in this context and this modification prevents the ubiquitination and degradation of DDB2, presumably to give it time to facilitate chromatin decondensation (Pines et al. 2012; Robu et al. 2013). The stability of DDB2 is therefore controlled by competing posttranslational modifications that control its activity and residence time on UV-damaged chromatin. The contribution of PARP to NER may be even more complex as there is a direct interaction between PARylated PARP and XPA, and this has been suggested to facilitate NER (King et al. 2012).
Multiple Chromatin Remodeling Complexes Have a Role in NER
Considerable efforts have been deployed to determine how chromatin-remodeling enzymes contribute to NER (Lans et al. 2012). These studies revealed that there is not a single factor that mediates this role in NER because depletion of individual remodeling factors does not lead to UV sensitivity at levels comparable to deficiencies in the core NER factors. Instead, multiple remodeling factors have been associated with NER, and the role and relative contributions of the individual factors are just beginning to emerge.
The DDB2- and PARP-dependent recruitment ALC1 to sites of UV lesions was mentioned in the previous section. Depletion of ALC1 leads to a partial defect in UV adduct removal and UV sensitivity, showing that it is involved, but not absolutely required for the NER reaction (Pines et al. 2012). Additional SWI/SNF proteins, BRG1 and hSNF5 in humans and Snf6 in S. cerevisiae, have been implicated in NER (Gong et al. 2006; Ray et al. 2009; Zhao et al. 2009). These factors appear to be recruited to NER complexes in an XPC-dependent manner. They are, therefore, thought to act downstream of the initial DNA damage recognition step and are believed to facilitate the decompaction of chromatin to complete the NER reaction.
Depletion of INO80, another SWI/SNF ATPase, has also been shown to render cells sensitive to UV irradiation and to slow the repair of CPDs and 6-4PPs (Jiang et al. 2010). Recruitment of INO80 to NER lesions in human cells is not dependent on XPA or XPC, suggesting that it is an early factor. In line with this, INO80 interacts directly with DDB1, providing a possible mechanism for its recruitment to sites of NER (Jiang et al. 2010). As for other chromatin remodeling factors, the UV sensitivity conferred by its deficiency is relatively mild. Studies in S. cerevisiae suggest that INO80 interacts with Rad4-Rad23 in this organism and that it contributes to chromatin restoration and overall UV damage repair, acting perhaps at a different stage in the process than its human equivalent (Sarkar et al. 2010).
The repair of NER in the context of chromatin is therefore a complex process involving many components in addition to the central UV-DDB factor. It is likely that a number of parallel pathways are associated with making DNA lesions in chromatin accessible to NER lesions.
Restoration of Chromatin following NER
The completion of the NER reaction requires the full reassembly of the repaired DNA into chromatin. This processed involves the histone chaperone CAF-1 (chromatin assembly factor), which facilitates chromatin reassembly following the NER reaction (Gaillard et al. 1996, 1997). Although CAF-1 is not required for the excision of UV damage from DNA, its activity is not isolated from the earlier steps of the NER reactions. CAF-1 colocalizes with other NER factors at sites of UV damage and its recruitment to sites of NER is dependent on an interaction with PCNA, linking it with the repair synthesis step of NER (Green and Almouzni 2003). Interestingly, CAF-1 is even found at sites of NER in the absence of the 3′ incision by XPG, further linking CAF-1 to the core NER reaction (Staresincic et al. 2009). The activity of CAF-1 does not simply result in histone recycling (the reincorporation of the original histones at the site of damage), but also results in the incorporation of the histone variant H3.1, leaving a mark in an NER-dependent fashion (Polo et al. 2006).
THE ROLE OF NER IN UV-INDUCED DNA DAMAGE SIGNALING
Like most types of DNA damage, UV-induced lesions are a potent inducer of a DNA-damage-signaling response. This nature of this response differs depending of the phase of the cell cycle. Although it has been established that UV lesions trigger a cell cycle arrest during replication in the S phase, the discovery and characterization of UV-induced damaged response in G0/G1 cells is more recent (Novarina et al. 2011). Because only this latter response is connected to NER, it will be the exclusive subject of our discussion. The earliest indications of a connection of the DNA-damage response and NER came from studies in the yeast S. cerevisiae, which showed that a mutation in the XPA homolog Rad14, as well as homologs of other core NER factors abolished the UV-damage response in noncycling cells (Giannattasio et al. 2004). Subsequent investigations revealed that in human cells the activation of a UV-induced DNA-damage response outside of S phase also required functional NER (Marini et al. 2006; Marti et al. 2006). Interestingly, these studies revealed that this damage response led to the activation of many of the same markers more commonly associated with the double-strand break (DSB) damage response including phosphorylation of γ-H2AX, Chk1, and p53. Like in budding yeast, this response was absolutely dependent on core NER factors, suggesting that an incision reaction generates an intermediate that can activate signaling. The analogy to DSB repair signaling extends even further. In response to local UV irradiation in cell nuclei in G1 cells, the ATR kinase, the ubiquitin ligase RNF8, the adaptor protein MDC1, as well as 53BP1 and BRCA1 are localized at sites of UV damage in an NER-dependent manner (Fig. 4) (Marteijn et al. 2009). This response is increased in situations in which DNA repair synthesis is inhibited, either by addition of polymerase inhibitors, nucleoside analogs, or nucleotide depletion (Hanasoge and Ljungman 2007; Matsumoto et al. 2007; Marteijn et al. 2009; Overmeer et al. 2011), establishing a connection between incomplete repair synthesis and damage signaling. This response is also increased in cells expressing catalytically inactive XPG (Overmeer et al. 2011), showing that a single incision can be enough to trigger this signaling response.
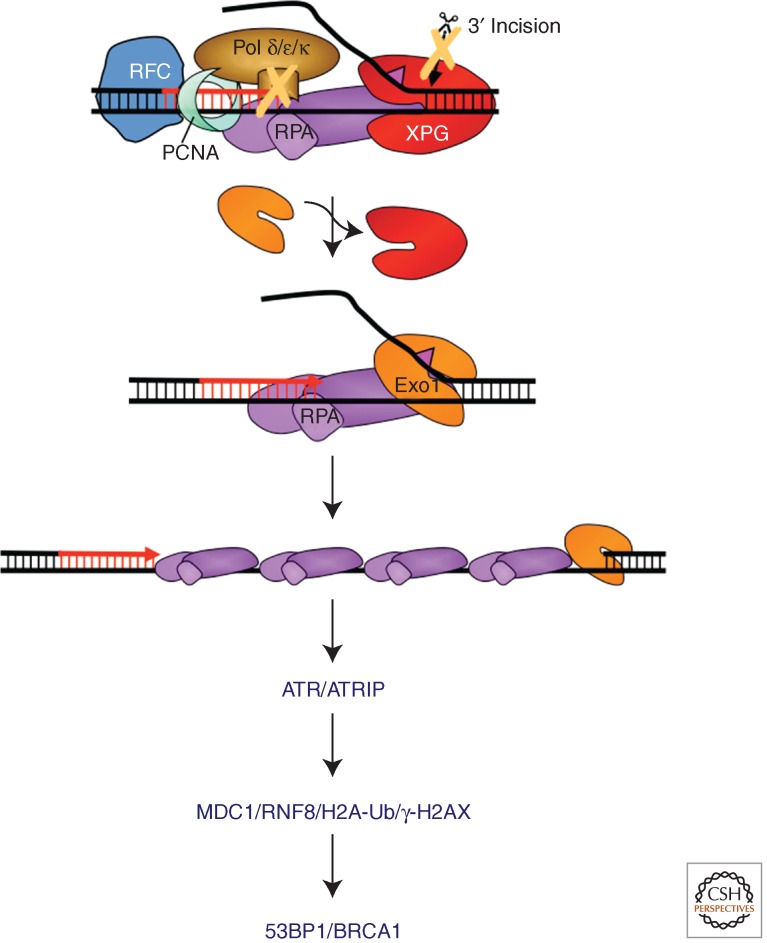
Figure 4.
Model for NER-mediated UV-induced DNA-damage signaling. Damage signaling can be triggered during the repair synthesis step of NER if (1) the damage load is too high, (2) repair synthesis is inhibited, and (3) 3′ incision does not occur. In these situations, XPG is replaced with EXO1, which processed the NER intermediate into an ssDNA gap of up to kb in length. RPA covers the ssDNA and binds ATRIP/ATR activating the ATR kinase and triggering a signal cascade that involves MDC1, RNF8, 53BP1, and BRCA1, and leads to the establishment of chromatin marks, including ubiquitination of histones and phosphorylation of γ-H2AX. This process does not share many common features with DSB-induced signaling.
These findings left one important question unanswered. It is well known that a principal signal for activation of the DNA-damage response is a long stretch of ssDNA covered with RPA. Such filaments bind the ATR-ATRIP complex activating the ATR kinase activity and triggering downstream signals (Zou and Elledge 2003; Novarina et al. 2011). Although an RPA-covered stretch of ssDNA is formed during NER following the incision reaction, this intermediate is short-lived and the ssDNA stretch not long enough to activate full ATR signaling. Recent work in S. cerevisiae and human cells has shown that long stretches of DNA (more than 1 kb) can be formed in an NER-dependent manner (Giannattasio et al. 2010; Sertic et al. 2011). This process involves the EXO1 nuclease, which has the ability to process products of incomplete NER repair synthesis into long stretches of ssDNA than are then covered by RPA and trigger ATR-mediated signaling. The localization to sites of UV damage and nuclease activity of EXO1 is also increased by inhibition of repair synthesis and occurs in the absence of the 3′ incision by XPG.
These studies provide the foundation for the following model that balances NER and DNA-damage signaling (Fig. 4). Under normal conditions (relatively low damage load, fully active NER factors, and conditions suitable for the completion of repair synthesis), NER can proceed normally as shown in Figure 1. If the damage load is too high, or incision by XPG or repair synthesis cannot go to completion, a subset of NER intermediates is processed by EXO1 to long stretches of ssDNA that are then covered by RPA and activates the signaling cascade. The last years have therefore shown that there is a universal signal to activate DNA-damage signaling in response to DSBs and UV damage, with a long stretch of ssDNA covered by RPA as the central signal.
CONCLUDING REMARKS
Although the basic NER reaction and roles of the core NER factors were well understood by the turn of the century, the past decade has led to a dramatic increase of our understanding of this process. Through structural, biochemical, and cell biological studies, we now understand in detail the molecular basis of the damage recognition and verification process as well as the dynamic assembly process underlying NER. We are beginning to understand the complex process of lesion recognition in the context of chromatin and the key role of UV-DDB in the process. A number of steps in NER that were previously assumed to be straightforward (dual incision, repair synthesis, ligation) were found to be much more complex and regulated, suggesting that every step in NER underlies the careful regulatory mechanism to coordinate this repair pathway with other aspects of cellular metabolism. The coupling of NER to the DNA damage response is one such example. The coupling of NER to transcription is another example and is covered by Vermeulen and Fousteri (2013).
ACKNOWLEDGMENTS
I thank Arthur J. Campbell for generating Figures 2 and 3, Laura Niedernhofer, Wim Vermeulen, and Hanspeter Naegeli for a critical reading of this manuscript, and Jill Fuss and John Tainer for providing coordinates of the model structures of XPB and XPD bound to DNA. Research on NER in the author’s laboratory is supported by National Institutes of Health grants GM080454 and CA092854.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aboussekhra A, Biggerstaff M, Shivji MK, Vilpo JA, Moncollin V, Podust VN, Protic M, Hubscher U, Egly JM, Wood RD 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80: 859–868 [DOI] [PubMed] [Google Scholar]
- Araujo SJ, Tirode F, Coin F, Pospiech H, Syvaoja JE, Stucki M, Hubscher U, Egly JM, Wood RD 2000. Nucleotide excision repair of DNA with recombinant human proteins: Definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev 14: 349–359 [PMC free article] [PubMed] [Google Scholar]
- Araujo SJ, Nigg EA, Wood RD 2001. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol Cell Biol 21: 2281–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. 2006. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev 20: 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Toussaint W, Luijsterburg MS, Dinant C, Alekseev S, Hoeijmakers JH, Dantuma NP, Houtsmuller AB, Vermeulen W 2012. Recognition of DNA damage by XPC coincides with disruption of the XPC-RAD23 complex. J Cell Biol 196: 681–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce RP, Howard-Flanders P 1964. Release of ultraviolet light-induced thymine dimers from DNA in E. coli K-12. Proc Natl Acad Sci 51: 293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunick CG, Miller MR, Fuller BE, Fanning E, Chazin WJ 2006. Biochemical and structural domain analysis of xeroderma pigmentosum complementation group C protein. Biochemistry 45: 14965–14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buterin T, Meyer C, Giese B, Naegeli H 2005. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol 12: 913–922 [DOI] [PubMed] [Google Scholar]
- Cai Y, Kropachev K, Xu R, Tang Y, Kolbanovskii M, Kolbanovskii A, Amin S, Patel DJ, Broyde S, Geacintov NE 2010. Distant neighbor base sequence context effects in human nucleotide excision repair of a benzo[a]pyrene-derived DNA lesion. J Mol Biol 399: 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camenisch U, Dip R, Schumacher SB, Schuler B, Naegeli H 2006. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nat Struct Mol Biol 13: 278–284 [DOI] [PubMed] [Google Scholar]
- Camenisch U, Trautlein D, Clement FC, Fei J, Leitenstorfer A, Ferrando-May E, Naegeli H 2009. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. EMBO J 28: 2387–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver JE 1968. Defective repair replication of DNA in xeroderma pigmentosum. Nature 218: 652–656 [DOI] [PubMed] [Google Scholar]
- Coin F, Proietti De Santis L, Nardo T, Zlobinskaya O, Stefanini M, Egly JM 2006. p8/TTD-A as a repair-specific TFIIH subunit. Mol Cell 21: 215–226 [DOI] [PubMed] [Google Scholar]
- Coin F, Oksenych V, Egly JM 2007. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol Cell 26: 245–256 [DOI] [PubMed] [Google Scholar]
- Coin F, Oksenych V, Mocquet V, Groh S, Blattner C, Egly JM 2008. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol Cell 31: 9–20 [DOI] [PubMed] [Google Scholar]
- Compe E, Egly JM 2012. TFIIH: When transcription met DNA repair. Nat Rev Mol Cell Biol 13: 343–354 [DOI] [PubMed] [Google Scholar]
- Constantinou A, Gunz D, Evans E, Lalle P, Bates PA, Wood RD, Clarkson SG 1999. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J Biol Chem 274: 5637–5648 [DOI] [PubMed] [Google Scholar]
- de Laat WL, Appeldoorn E, Sugasawa K, Weterings E, Jaspers NG, Hoeijmakers JH 1998. DNA-binding polarity of human replication protein A positions nucleases in nucleotide excision repair. Genes Dev 12: 2598–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weerd-Kastelein EA, Keijzer W, Bootsma D 1972. Genetic heterogeneity of xeroderma pigmentosum demonstrated by somatic cell hybridization. Nat New Biol 238: 80–83 [DOI] [PubMed] [Google Scholar]
- DiGiovanna JJ, Kraemer KH 2012. Shining a light on xeroderma pigmentosum. J Invest Dermatol 132: 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunand-Sauthier I, Hohl M, Thorel F, Jaquier-Gubler P, Clarkson SG, Schärer OD 2005. The spacer region of XPG mediates recruitment to nucleotide excision repair complexes and determines substrate specificity. J Biol Chem 280: 7030–7037 [DOI] [PubMed] [Google Scholar]
- Evans E, Fellows J, Coffer A, Wood RD 1997a. Open complex formation around a lesion during nucleotide excision repair provides a structure for cleavage by human XPG protein. Embo J 16: 625–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD 1997b. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. Embo J 16: 6559–6573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagbemi AF, Orelli B, Schärer OD 2011. Regulation of endonuclease activity in human nucleotide excision repair. DNA Repair (Amst) 10: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Luo J 2010. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell 39: 247–258 [DOI] [PubMed] [Google Scholar]
- Fan J, Pavletich NP 2012. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26: 2337–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA 2006. Conserved XPB core structure and motifs for DNA unwinding: Implications for pathway selection of transcription or excision repair. Mol Cell 22: 27–37 [DOI] [PubMed] [Google Scholar]
- Fan L, Fuss JO, Cheng QJ, Arvai AS, Hammel M, Roberts VA, Cooper PK, Tainer JA 2008. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell 133: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H 2011. Regulation of nucleotide excision repair by UV-DDB: Prioritization of damage recognition to internucleosomal DNA. PLoS Biol 9: e1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer ES, Scrima A, Bohm K, Matsumoto S, Lingaraju GM, Faty M, Yasuda T, Cavadini S, Wakasugi M, Hanaoka F, et al. 2011. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147: 1024–1039 [DOI] [PubMed] [Google Scholar]
- Fitch ME, Nakajima S, Yasui A, Ford JM 2003. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J Biol Chem 278: 46906–46910 [DOI] [PubMed] [Google Scholar]
- Fregoso M, Laine JP, Aguilar-Fuentes J, Mocquet V, Reynaud E, Coin F, Egly JM, Zurita M 2007. DNA repair and transcriptional deficiencies caused by mutations in the Drosophila p52 subunit of TFIIH generate developmental defects and chromosome fragility. Mol Cell Biol 27: 3640–3650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2005. DNA repair and mutagenesis. ASM Press, Washington, DC [Google Scholar]
- Gaillard PH, Martini EM, Kaufman PD, Stillman B, Moustacchi E, Almouzni G 1996. Chromatin assembly coupled to DNA repair: A new role for chromatin assembly factor I. Cell 86: 887–896 [DOI] [PubMed] [Google Scholar]
- Gaillard PH, Moggs JG, Roche DM, Quivy JP, Becker PB, Wood RD, Almouzni G 1997. Initiation and bidirectional propagation of chromatin assembly from a target site for nucleotide excision repair. EMBO J 16: 6281–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Lazzaro F, Longhese MP, Plevani P, Muzi-Falconi M 2004. Physical and functional interactions between nucleotide excision repair and DNA damage checkpoint. EMBO J 23: 429–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannattasio M, Follonier C, Tourriere H, Puddu F, Lazzaro F, Pasero P, Lopes M, Plevani P, Muzi-Falconi M 2010. Exo1 competes with repair synthesis, converts NER intermediates to long ssDNA gaps, and promotes checkpoint activation. Mol Cell 40: 50–62 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Coin F, Ranish JA, Hoogstraten D, Theil A, Wijgers N, Jaspers NG, Raams A, Argentini M, van der Spek PJ, et al. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat Genet 36: 714–719 [DOI] [PubMed] [Google Scholar]
- Giglia-Mari G, Miquel C, Theil AF, Mari PO, Hoogstraten D, Ng JM, Dinant C, Hoeijmakers JH, Vermeulen W 2006. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol 4: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet LC, Schärer OD 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev 106: 253–276 [DOI] [PubMed] [Google Scholar]
- Gilljam KM, Muller R, Liabakk NB, Otterlei M 2012. Nucleotide excision repair is associated with the replisome and its efficiency depends on a direct interaction between XPA and PCNA. PloS ONE 7: e49199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong F, Fahy D, Smerdon MJ 2006. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13: 902–907 [DOI] [PubMed] [Google Scholar]
- Grasby JA, Finger LD, Tsutakawa SE, Atack JM, Tainer JA 2012. Unpairing and gating: Sequence-independent substrate recognition by FEN superfamily nucleases. Trends Biochem Sci 37: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CM, Almouzni G 2003. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J 22: 5163–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, Kisselev AF, Tanaka K, Nakatani Y 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113: 357–367 [DOI] [PubMed] [Google Scholar]
- Guerrero-Santoro J, Kapetanaki MG, Hsieh CL, Gorbachinsky I, Levine AS, Rapic-Otrin V 2008. The cullin 4B-based UV-damaged DNA-binding protein ligase binds to UV-damaged chromatin and ubiquitinates histone H2A. Cancer Res 68: 5014–5022 [DOI] [PubMed] [Google Scholar]
- Gunz D, Hess MT, Naegeli H 1996. Recognition of DNA adducts by human nucleotide excision repair. Evidence for a thermodynamic probing mechanism. J Biol Chem 271: 25089–25098 [DOI] [PubMed] [Google Scholar]
- Hanasoge S, Ljungman M 2007. H2AX phosphorylation after UV irradiation is triggered by DNA repair intermediates and is mediated by the ATR kinase. Carcinogenesis 28: 2298–2304 [DOI] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G 2008. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol 9: 958–970 [DOI] [PubMed] [Google Scholar]
- Hara R, Sancar A 2002. The SWI/SNF chromatin-remodeling factor stimulates repair by human excision nuclease in the mononucleosome core particle. Mol Cell Biol 22: 6779–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R, Mo J, Sancar A 2000. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol Cell Biol 20: 9173–9181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess MT, Schwitter U, Petretta M, Giese B, Naegeli H 1997. Bipartite substrate discrimination by human nucleotide excision repair. Proc Natl Acad Sci 94: 6664–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl M, Thorel F, Clarkson SG, Schärer OD 2003. Structural determinants for substrate binding and catalysis by the structure-specific endonuclease XPG. J Biol Chem 278: 19500–19508 [DOI] [PubMed] [Google Scholar]
- Hohl M, Dunand-Sauthier I, Staresincic L, Jaquier-Gubler P, Thorel F, Modesti M, Clarkson SG, Schärer OD 2007. Domain swapping between FEN-1 and XPG defines regions in XPG that mediate nucleotide excision repair activity and substrate specificity. Nucleic Acids Res 35: 3053–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogstraten D, Bergink S, Ng JM, Verbiest VH, Luijsterburg MS, Geverts B, Raams A, Dinant C, Hoeijmakers JH, Vermeulen W, et al. 2008. Versatile DNA damage detection by the global genome nucleotide excision repair protein XPC. J Cell Sci 121: 2850–2859 [DOI] [PubMed] [Google Scholar]
- Houtsmuller AB, Rademakers S, Nigg AL, Hoogstraten D, Hoeijmakers JH, Vermeulen W 1999. Action of DNA repair endonuclease ERCC1/XPF in living cells. Science 284: 958–961 [DOI] [PubMed] [Google Scholar]
- Huang JC, Svoboda DL, Reardon JT, Sancar A 1992. Human nucleotide excision nuclease removes thymine dimers from DNA by incising the 22nd phosphodiester bond 5′ and the 6th phosphodiester bond 3′ to the photodimer. Proc Natl Acad Sci 89: 3664–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JC, Hsu DS, Kazantsev A, Sancar A 1994. Substrate spectrum of human excinuclease: Repair of abasic sites, methylated bases, mismatches, and bulky adducts. Proc Natl Acad Sci 91: 12213–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K 2007. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: Implications for Cockayne syndrome in XP-G/CS patients. Mol Cell 26: 231–243 [DOI] [PubMed] [Google Scholar]
- Jiang Y, Wang X, Bao S, Guo R, Johnson DG, Shen X, Li L 2010. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc Natl Acad Sci 107: 17274–17279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Lindsey-Boltz LA, Reardon JT, Sancar A 2010. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. Proc Natl Acad Sci 107: 4890–4895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang TH, Reardon JT, Sancar A 2011. Regulation of nucleotide excision repair activity by transcriptional and post-transcriptional control of the XPA protein. Nucleic Acids Res 39: 3176–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanaki MG, Guerrero-Santoro J, Bisi DC, Hsieh CL, Rapic-Otrin V, Levine AS 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci 103: 2588–2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp MG, Reardon JT, Lindsey-Boltz LA, Sancar A 2012. Mechanism of release and fate of excised oligonucleotides during nucleotide excision repair. J Biol Chem 287: 22889–22899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King BS, Cooper KL, Liu KJ, Hudson LG 2012. Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair. J Biol Chem 287: 39824–39833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kisker C, Kuper J, Van Houten B 2013. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol 5: a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasikova YS, Rechkunova NI, Maltseva EA, Petruseva IO, Lavrik OI 2010. Localization of xeroderma pigmentosum group A protein and replication protein A on damaged DNA in nucleotide excision repair. Nucleic Acids Res 38: 8083–8094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper J, Wolski SC, Michels G, Kisker C 2012. Functional and structural studies of the nucleotide excision repair helicase XPD suggest a polarity for DNA translocation. EMBO J 31: 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Vermeulen W 2012. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics Chromatin 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latypov VF, Tubbs JL, Watson AJ, Marriott AS, McGown G, Thorncroft M, Wilkinson OJ, Senthong P, Butt A, Arvai AS, et al. 2012. Atl1 regulates choice between global genome and transcription-coupled repair of O6-alkylguanines. Mol Cell 47: 50–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR 2003. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie 85: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Lehmann AR 2011. DNA polymerases and repair synthesis in NER in human cells. DNA Repair 10: 730–733 [DOI] [PubMed] [Google Scholar]
- Li L, Elledge SJ, Peterson CA, Bales ES, Legerski RJ 1994. Specific association between the human DNA repair proteins XPA and ERCC1. Proc Natl Acad Sci 91: 5012–5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Lu X, Peterson CA, Legerski RJ 1995a. An interaction between the DNA repair factor XPA and replication protein A appears essential for nucleotide excision repair. Mol Cell Biol 15: 5396–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peterson CA, Lu X, Legerski RJ 1995b. Mutations in XPA that prevent association with ERCC1 are defective in nucleotide excision repair. Mol Cell Biol 15: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rudolf J, Johnson KA, McMahon SA, Oke M, Carter L, McRobbie AM, Brown SE, Naismith JH, White MF 2008. Structure of the DNA repair helicase XPD. Cell 133: 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, et al. 2011. Probing for DNA damage with beta-hairpins: Similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 10: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijsterburg MS, Lindh M, Acs K, Vrouwe MG, Pines A, van Attikum H, Mullenders LH, Dantuma NP 2012. DDB2 promotes chromatin decondensation at UV-induced DNA damage. J Cell Biol 197: 267–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard O, Solyom S, Naegeli H 2007. An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS Biol 5: e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini F, Nardo T, Giannattasio M, Minuzzo M, Stefanini M, Plevani P, Muzi Falconi M 2006. DNA nucleotide excision repair-dependent signaling to checkpoint activation. Proc Natl Acad Sci 103: 17325–17330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marteijn JA, Bekker-Jensen S, Mailand N, Lans H, Schwertman P, Gourdin AM, Dantuma NP, Lukas J, Vermeulen W 2009. Nucleotide excision repair-induced H2A ubiquitination is dependent on MDC1 and RNF8 and reveals a universal DNA damage response. J Cell Biol 186: 835–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE 2006. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc Natl Acad Sci 103: 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Kaczmarek N, Naegeli H 2010. Strand- and site-specific DNA lesion demarcation by the xeroderma pigmentosum group D helicase. Proc Natl Acad Sci 107: 17545–17550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu N, Kaczmarek N, Ruthemann P, Luch A, Naegeli H 2013. DNA quality control by a lesion sensor pocket of the xeroderma pigmentosum group D helicase subunit of TFIIH. Curr Biol 23: 204–212 [DOI] [PubMed] [Google Scholar]
- Matsuda T, Saijo M, Kuraoka I, Kobayashi T, Nakatsu Y, Nagai A, Enjoji T, Masutani C, Sugasawa K, Hanaoka F, et al. 1995. DNA repair protein XPA binds replication protein A (RPA). J Biol Chem 270: 4152–4157 [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Yaginuma K, Igarashi A, Imura M, Hasegawa M, Iwabuchi K, Date T, Mori T, Ishizaki K, Yamashita K, et al. 2007. Perturbed gap-filling synthesis in nucleotide excision repair causes histone H2AX phosphorylation in human quiescent cells. J Cell Sci 120: 1104–1112 [DOI] [PubMed] [Google Scholar]
- Min JH, Pavletich NP 2007. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature 449: 570–575 [DOI] [PubMed] [Google Scholar]
- Missura M, Buterin T, Hindges R, Hubscher U, Kasparkova J, Brabec V, Naegeli H 2001. Double-check probing of DNA bending and unwinding by XPA-RPA: An architectural function in DNA repair. Embo J 20: 3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM 2007. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. Embo J 26: 2923–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM 2008. Sequential recruitment of the repair factors during NER: The role of XPG in initiating the resynthesis step. EMBO J 27: 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI 2007. Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III α in a cell-cycle-specific manner. Mol Cell 27: 311–323 [DOI] [PubMed] [Google Scholar]
- Mu D, Park CH, Matsunaga T, Hsu DS, Reardon JT, Sancar A 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J Biol Chem 270: 2415–2418 [DOI] [PubMed] [Google Scholar]
- Mu H, Kropachev K, Wang L, Zhang L, Kolbanovskiy A, Kolbanovskiy M, Geacintov NE, Broyde S 2012. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: Molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res 40: 9675–9690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naegeli H, Bardwell L, Friedberg EC 1992. The DNA helicase and adenosine triphosphatase activities of yeast Rad3 protein are inhibited by DNA damage. A potential mechanism for damage-specific recognition. J Biol Chem 267: 392–398 [PubMed] [Google Scholar]
- Ng JM, Vermeulen W, van der Horst GT, Bergink S, Sugasawa K, Vrieling H, Hoeijmakers JH 2003. A novel regulation mechanism of DNA repair by damage-induced and RAD23-dependent stabilization of xeroderma pigmentosum group C protein. Genes Dev 17: 1630–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R, Okuda Y, Watanabe E, Mori T, Iwai S, Masutani C, Sugasawa K, Hanaoka F 2005. Centrin 2 stimulates nucleotide excision repair by interacting with xeroderma pigmentosum group C protein. Mol Cell Biol 25: 5664–5674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi R, Alekseev S, Dinant C, Hoogstraten D, Houtsmuller AB, Hoeijmakers JH, Vermeulen W, Hanaoka F, Sugasawa K 2009. UV-DDB-dependent regulation of nucleotide excision repair kinetics in living cells. DNA Repair (Amst) 8: 767–776 [DOI] [PubMed] [Google Scholar]
- Nocentini S, Coin F, Saijo M, Tanaka K, Egly JM 1997. DNA damage recognition by XPA protein promotes efficient recruitment of transcription factor II H. J Biol Chem 272: 22991–22994 [DOI] [PubMed] [Google Scholar]
- Novarina D, Amara F, Lazzaro F, Plevani P, Muzi-Falconi M 2011. Mind the gap: Keeping UV lesions in check. DNA Repair (Amst) 10: 751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T, Lehmann AR 2006. The Y-family DNA polymerase kappa (pol κ) functions in mammalian nucleotide-excision repair. Nat Cell Biol 8: 640–642 [DOI] [PubMed] [Google Scholar]
- Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell 37: 714–727 [DOI] [PubMed] [Google Scholar]
- Oksenych V, de Jesus BB, Zhovmer A, Egly JM, Coin F 2009. Molecular insights into the recruitment of TFIIH to sites of DNA damage. EMBO J 28: 2971–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orans J, McSweeney EA, Iyer RR, Hast MA, Hellinga HW, Modrich P, Beese LS 2011. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell 145: 212–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orelli B, McClendon TB, Tsodikov OV, Ellenberger T, Niedernhofer LJ, Schärer OD 2010. The XPA-binding domain of ERCC1 is required for nucleotide excision repair but not other DNA repair pathways. J Biol Chem 285: 3705–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmeer RM, Moser J, Volker M, Kool H, Tomkinson AE, van Zeeland AA, Mullenders LH, Fousteri M 2011. Replication protein A safeguards genome integrity by controlling NER incision events. J Cell Biol 192: 401–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CH, Mu D, Reardon JT, Sancar A 1995. The general transcription-repair factor TFIIH is recruited to the excision repair complex by the XPA protein independent of the TFIIE transcription factor. J Biol Chem 270: 4896–4902 [DOI] [PubMed] [Google Scholar]
- Patrick SM, Turchi JJ 2002. Xeroderma pigmentosum complementation group A protein (XPA) modulates RPA-DNA interactions via enhanced complex stability and inhibition of strand separation activity. J Biol Chem 277: 16096–16101 [DOI] [PubMed] [Google Scholar]
- *.Peterson CL, Almouzni G 2013. Nucleosome dynamics as modular systems that integrate DNA damage and repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a012658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D, Hanawalt P 1964. Evidence for repair-replication of ultraviolet damaged DNA in bacteria. J Mol Biol 9: 395–410 [DOI] [PubMed] [Google Scholar]
- Pines A, Vrouwe MG, Marteijn JA, Typas D, Luijsterburg MS, Cansoy M, Hensbergen P, Deelder A, de Groot A, Matsumoto S, et al. 2012. PARP1 promotes nucleotide excision repair through DDB2 stabilization and recruitment of ALC1. J Cell Biol 199: 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G 2006. New histone incorporation marks sites of UV repair in human cells. Cell 127: 481–493 [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L 2000. Nucleotide excision repair in yeast. Mutat Res 451: 13–24 [DOI] [PubMed] [Google Scholar]
- Pugh RA, Wu CG, Spies M 2012. Regulation of translocation polarity by helicase domain 1 in SF2B helicases. EMBO J 31: 503–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademakers S, Volker M, Hoogstraten D, Nigg AL, Mone MJ, Van Zeeland AA, Hoeijmakers JH, Houtsmuller AB, Vermeulen W 2003. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol Cell Biol 23: 5755–5767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R 2004. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet 36: 707–713 [DOI] [PubMed] [Google Scholar]
- Rasmussen RE, Painter RB 1964. Evidence for repair of ultra-violet damaged deoxyribonucleic acid in cultured mammalian cells. Nature 203: 1360–1362 [DOI] [PubMed] [Google Scholar]
- Ray A, Mir SN, Wani G, Zhao Q, Battu A, Zhu Q, Wang QE, Wani AA 2009. Human SNF5/INI1, a component of the human SWI/SNF chromatin remodeling complex, promotes nucleotide excision repair by influencing ATM recruitment and downstream H2AX phosphorylation. Mol Cell Biol 29: 6206–6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon JT, Sancar A 2003. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev 17: 2539–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl T, Hanaoka F, Egly JM 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. Embo J 22: 5293–5303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu M, Shah RG, Petitclerc N, Brind’Amour J, Kandan-Kulangara F, Shah GM 2013. Role of poly(ADP-ribose) polymerase-1 in the removal of UV-induced DNA lesions by nucleotide excision repair. Proc Natl Acad Sci 110: 1658–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo M, Takedachi A, Tanaka K 2011. Nucleotide excision repair by mutant xeroderma pigmentosum group A (XPA) proteins with deficiency in interaction with RPA. J Biol Chem 286: 5476–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Kiely R, McHugh PJ 2010. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J Cell Biol 191: 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schärer OD, Campbell AJ 2009. Wedging out DNA damage. Nat Struct Mol Biol 16: 102–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH 2008. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell 135: 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sertic S, Pizzi S, Cloney R, Lehmann AR, Marini F, Plevani P, Muzi-Falconi M 2011. Human exonuclease 1 connects nucleotide excision repair (NER) processing with checkpoint activation in response to UV irradiation. Proc Natl Acad Sci 108: 13647–13652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow RB, Carrier WL 1964. The disappearance of thymine dimers from DNA: An error-correcting mechanism. Proc Natl Acad Sci 51: 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivji MK, Podust VN, Hubscher U, Wood RD 1995. Nucleotide excision repair DNA synthesis by DNA polymerase epsilon in the presence of PCNA, RFC, and RPA. Biochemistry 34: 5011–5017 [DOI] [PubMed] [Google Scholar]
- Smerdon MJ, Lieberman MW 1978. Nucleosome rearrangement in human chromatin during UV-induced DNA-reapir [sic] synthesis. Proc Natl Acad Sci 75: 4238–4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staresincic L, Fagbemi AF, Enzlin JH, Gourdin AM, Wijgers N, Dunand-Sauthier I, Giglia-Mari G, Clarkson SG, Vermeulen W, Schärer OD 2009. Coordination of dual incision and repair synthesis in human nucleotide excision repair. EMBO J 28: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Masutani C, Uchida A, Maekawa T, van der Spek PJ, Bootsma D, Hoeijmakers JH, Hanaoka F 1996. HHR23B, a human Rad23 homolog, stimulates XPC protein in nucleotide excision repair in vitro. Mol Cell Biol 16: 4852–4861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell 2: 223–232 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F 2001. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev 15: 507–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugasawa K, Shimizu Y, Iwai S, Hanaoka F 2002. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 1: 95–107 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Okuda Y, Saijo M, Nishi R, Matsuda N, Chu G, Mori T, Iwai S, Tanaka K, Tanaka K, et al. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121: 387–400 [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F 2009. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell 36: 642–653 [DOI] [PubMed] [Google Scholar]
- Tang J, Chu G 2002. Xeroderma pigmentosum complementation group E and UV-damaged DNA-binding protein. DNA Repair (Amst) 1: 601–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JY, Hwang BJ, Ford JM, Hanawalt PC, Chu G 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol Cell 5: 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapias A, Auriol J, Forget D, Enzlin JH, Schärer OD, Coin F, Coulombe B, Egly JM 2004. Ordered conformational changes in damaged DNA induced by nucleotide excision repair factors. J Biol Chem 279: 19074–19083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil AF, Nonnekens J, Wijgers N, Vermeulen W, Giglia-Mari G 2011. Slowly progressing nucleotide excision repair in trichothiodystrophy group A patient fibroblasts. Mol Cell Biol 31: 3630–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theil AF, Nonnekens J, Streuer B, Mari PO, de Wit J, Lemaitre C, Marteijn JA, Raams A, Maas A, Vermej M, et al. 2013. Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality. PLoS Genet 9: e1003431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH 1998. Chinese hamster cells meet DNA repair: An entirely acceptable affair. Bioessays 20: 589–597 [DOI] [PubMed] [Google Scholar]
- Tripsianes K, Folkers GE, Zheng C, Das D, Grinstead JS, Kaptein R, Boelens R 2007. Analysis of the XPA and ssDNA-binding surfaces on the central domain of human ERCC1 reveals evidence for subfunctionalization. Nucleic Acids Res 35: 5789–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsodikov OV, Ivanov D, Orelli B, Staresincic L, Shoshani I, Oberman R, Schärer OD, Wagner G, Ellenberger T 2007. Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA. EMBO J 26: 4768–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. 2011. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Latypov V, Kanugula S, Butt A, Melikishvili M, Kraehenbuehl R, Fleck O, Marriott A, Watson AJ, Verbeek B, et al. 2009. Flipping of alkylated DNA damage bridges base and nucleotide excision repair. Nature 459: 808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ura K, Araki M, Saeki H, Masutani C, Ito T, Iwai S, Mizukoshi T, Kaneda Y, Hanaoka F 2001. ATP-dependent chromatin remodeling facilitates nucleotide excision repair of UV-induced DNA lesions in synthetic dinucleosomes. EMBO J 20: 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Vermeulen W, Fousteri M 2013. Mammalian transcription-coupled excision repair. Cold Spring Harb Perspect Biol 5: a012625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W, Bergmann E, Auriol J, Rademakers S, Frit P, Appeldoorn E, Hoeijmakers JH, Egly JM 2000. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat Genet 26: 307–313 [DOI] [PubMed] [Google Scholar]
- Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell 8: 213–224 [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Sancar A 1998. Assembly, subunit composition, and footprint of human DNA repair excision nuclease. Proc Natl Acad Sci 95: 6669–6674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi M, Reardon JT, Sancar A 1997. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem 272: 16030–16034 [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Shimizu M, Morioka H, Linn S, Nikaido O, Matsunaga T 2001. Damaged DNA-binding protein DDB stimulates the excision of cyclobutane pyrimidine dimers in vitro in concert with XPA and replication protein A. J Biol Chem 276: 15434–15440 [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J Biol Chem 277: 1637–1640 [DOI] [PubMed] [Google Scholar]
- Wakasugi M, Kasashima H, Fukase Y, Imura M, Imai R, Yamada S, Cleaver JE, Matsunaga T 2009. Physical and functional interaction between DDB and XPA in nucleotide excision repair. Nucleic Acids Res 37: 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhai L, Xu J, Joo HY, Jackson S, Erdjument-Bromage H, Tempst P, Xiong Y, Zhang Y 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol Cell 22: 383–394 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wood RD, Timmers HT, Weeda G 2000. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem 275: 4258–4266 [DOI] [PubMed] [Google Scholar]
- Winkler GS, Araujo SJ, Fiedler U, Vermeulen W, Coin F, Egly JM, Hoeijmakers JH, Wittschieben BO, Iwai S, Wood RD 2005. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J Biol Chem 280: 39982–39989 [DOI] [PubMed] [Google Scholar]
- Wolski SC, Kuper J, Hanzelmann P, Truglio JJ, Croteau DL, Van Houten B, Kisker C 2008. Crystal structure of the FeS cluster-containing nucleotide excision repair helicase XPD. PLoS Biol 6: e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood RD, Robins P, Lindahl T 1988. Complementation of the xeroderma pigmentosum DNA repair defect in cell-free extracts. Cell 53: 97–106 [DOI] [PubMed] [Google Scholar]
- Yeh JI, Levine AS, Du S, Chinte U, Ghodke H, Wang H, Shi H, Hsieh CL, Conway JF, Van Houten B, et al. 2012. Damaged DNA induced UV-damaged DNA-binding protein (UV-DDB) dimerization and its roles in chromatinized DNA repair. Proc Natl Acad Sci 109: E2737–E2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo JE, Khoo A, Fagbemi AF, Scharer OD 2012. The efficiencies of damage recognition and excision correlate with duplex destabilization induced by acetylaminofluorene adducts in human nucleotide excision repair. Chem Res Toxicol 25: 2462–2468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F 2000. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J Biol Chem 275: 9870–9875 [DOI] [PubMed] [Google Scholar]
- You JS, Wang M, Lee SH 2003. Biochemical analysis of the damage recognition process in nucleotide excision repair. J Biol Chem 278: 7476–7485 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang QE, Ray A, Wani G, Han C, Milum K, Wani AA 2009. Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem 284: 30424–30432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zotter A, Luijsterburg MS, Warmerdam DO, Ibrahim S, Nigg A, van Cappellen WA, Hoeijmakers JH, van Driel R, Vermeulen W, Houtsmuller AB 2006. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced dna damage depends on functional TFIIH. Mol Cell Biol 26: 8868–8879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]