Abstract
The onset of genomic DNA synthesis requires precise interactions of specialized initiator proteins with DNA at sites where the replication machinery can be loaded. These sites, defined as replication origins, are found at a few unique locations in all of the prokaryotic chromosomes examined so far. However, replication origins are dispersed among tens of thousands of loci in metazoan chromosomes, thereby raising questions regarding the role of specific nucleotide sequences and chromatin environment in origin selection and the mechanisms used by initiators to recognize replication origins. Close examination of bacterial and archaeal replication origins reveals an array of DNA sequence motifs that position individual initiator protein molecules and promote initiator oligomerization on origin DNA. Conversely, the need for specific recognition sequences in eukaryotic replication origins is relaxed. In fact, the primary rule for origin selection appears to be flexibility, a feature that is modulated either by structural elements or by epigenetic mechanisms at least partly linked to the organization of the genome for gene expression.
Prokaryotic replication origins are specific DNA sequence motifs that position initiator proteins. In eukaryotes, the need for specific recognition sequences is relaxed.
Timely duplication of the genome is an essential step in the reproduction of any cell, and it is not surprising that chromosomal DNA synthesis is tightly regulated by mechanisms that determine precisely where and when new replication forks are assembled. The first model for a DNA synthesis regulatory circuit was described about 50 years ago (Jacob et al. 1963), based on the idea that an early, key step in building new replication forks was the binding of a chromosomally encoded initiator protein to specialized DNA regions, termed replication origins (Fig. 1). The number of replication origins in a genome is, for the most part, dependent on chromosome size. Bacterial and archaeal genomes, which usually consist of a small circular chromosome, frequently have a single replication origin (Barry and Bell 2006; Gao and Zhang 2007). In contrast, eukaryotic genomes contain significantly more origins, ranging from 400 in yeast to 30,000–50,000 in humans (Cvetic and Walter 2005; Méchali 2010), because timely duplication of their larger linear chromosomes requires establishment of replication forks at multiple locations. The interaction of origin DNA and initiator proteins (Fig. 1) ultimately results in the assembly of prereplicative complexes (pre-RCs), whose role is to load and activate the DNA helicases necessary to unwind DNA before replication (Remus and Diffley 2009; Kawakami and Katayama 2010). Following helicase-catalyzed DNA unwinding, replisomal proteins become associated with the single-stranded DNA, and new replication forks proceed bidirectionally along the genome until every region is duplicated (for review, see O’Donnell 2006; Masai et al. 2010).
Figure 1.
Revised versions of the replicon model for all domains of life. For cells of each domain type, trans-acting initiators recognize replication origins to assemble prereplicative complexes required to unwind the DNA and load DNA helicase. Eukaryotic initiators are preassembled into hexameric origin recognition complexes (ORCs) before interacting with DNA. In prokaryotes, single initiators (archaeal Orc1/Cdc6 or bacterial DnaA) bind to recognition sites and assemble into complexes on DNA. In all cases, the DNA helicases (MCMs or DnaB) are recruited to the origin and loaded onto single DNA strands. In bacteria, DNA-bending proteins, such as Fis or IHF, may modulate the assembly of pre-RC by bending the origin DNA. Two activities of DnaA are described in the figure. The larger version binds to recognition sites, and the smaller version represents DnaA required to assist DnaC in loading DnaB helicase on single-stranded DNA.
Initiator proteins from all forms of life share structural similarities, including membership in the AAA+ family of proteins (ATPases associated with various cellular activities) (Duderstadt and Berger 2008; Wigley 2009) that are activated by ATP binding and inactivated by ATP hydrolysis (Duderstadt and Berger 2008; Duncker et al. 2009; Kawakami and Katayama 2010). Despite these similarities, initiators assemble into prereplicative complexes in two fundamentally different ways (Fig. 2). In prokaryotes, initiator monomers interact with the origin at multiple repeated DNA sequence motifs, and the arrangement of these motifs (see below) can direct assembly of oligomers that mediate strand separation (Erzberger et al. 2006; Rozgaja et al. 2011). In eukaryotes, a hexameric origin recognition complex (ORC) binds to replication origins and then recruit additional factors (as Cdc6 and Cdt1) that will themselves recruit the hexameric MCM2-7 DNA helicase to form a prereplicative complex (for review, see Diffley 2011). This process occurs during mitosis and along G1 and is called “DNA replication licensing,” a crucial regulation of eukaryotic DNA replication (for review, see Blow and Gillespie 2008). Importantly, this complex is still inactive, and only a subset of these preassembled origins will be activated in S phase. This process is, therefore, fundamentally different from initiation of replication in bacteria. Moreover, because sequence specificity appears more relaxed in large eukaryotic genomes, prokaryotic mechanisms that regulate initiator–DNA site occupation must be replaced by alternative mechanisms, such as structural elements or the use of epigenetic factors.
Figure 2.
Functional elements in some well-studied prokaryotic replication origins. (A) Bacterial oriCs. The DNA elements described in the text are (arrows) DnaA recognition boxes or (boxes) DNA unwinding elements (DUEs). When recognition site affinities are known, colored arrows designate high- (Kd > 100 nm) and low- (Kd < 100 nm) affinity sites. (B) Archaeal oriCs. Arrows and boxes designate DNA elements as in A, but the initiator protein is Orc1/Cdc6 rather than DnaA. (Thick arrows) Long origin recognition boxes (ORBs); (thin arrows) shorter versions (miniORBs). Both ORBs and miniORBs are identified in Pyrococcus. DUEs are not yet well defined for Helicobacter or Sulfolobus genera and are not labeled in this figure.
Here, we describe replication origins on prokaryotic and eukaryotic genomes below, with a particular focus on the attributes responsible for orderly initiator interactions and origin selection specificity, as well as on the shift from origin sequence-dependent regulation to epigenetic regulation. You are also referred to other related articles in this collection and several recent reviews covering the topics of DNA replication initiation in more detail (Méchali 2010; Beattie and Bell 2011; Blow et al. 2011; Bryant and Aves 2011; Ding and MacAlpine 2011; Dorn and Cook 2011; Kaguni 2011; Leonard and Grimwade 2011; Sequeira-Mendes and Gomez 2012).
REPLICATION ORIGINS IN BACTERIA
Bacterial Origins Share Similar Nucleotide Sequence Motifs
The locations of replication origins are now predicted for more than 1300 bacterial genomes (Worning et al. 2006; Gao and Zhang 2007, 2008; see also http://tubic.tju.edu.cn/doric/). Putative oriCs are generally located at a unique site on the circular chromosome, often adjacent to the gene encoding the conserved bacterial initiator protein, DnaA (Mackiewicz et al. 2004; Gao and Zhang 2007). Among the small fraction of oriCs that are functionally characterized, there is considerable variability in both origin length (ranging from 100 to 1000 bp) and nucleotide sequence. Common features include an extremely A-T-rich DNA unwinding element (DUE) that facilitates DNA strand separation (Kowalski and Eddy 1989), and 9-bp motifs that are recognized by DnaA (DnaA boxes) (see Fig. 2A; Table 1) (for review, see Zakrzewska-Czerwinska et al. 2007). The number and arrangement of DnaA boxes vary among bacterial types, but DnaA boxes are often clustered within oriCs (e.g., see Fig. 2A) (Mackiewicz et al. 2004; Zawilak-Pawlik et al. 2005). The clustering of DnaA boxes shown among the examples in Figure 2A suggests that all bacterial origins play a role in directing the assembly of DnaA oligomers (see below), although the length and orientation of these oligomers vary.
Table 1.
Initiator recognition sequences in bacterial and archaeal oriCs
| Site | Affinity | |
|---|---|---|
| Bacteria representative DnaA boxes | ||
| Escherichia coli | ||
| TGTGGATAA | R1/R4 | High |
| TGTGTATAA | R2 | High |
| TGTCAGGAA | C1 | Low |
| TGTGAATGA | R5 | Low |
| TGGGATCAG | I2 | Low with DnaA-ATP preference |
| TTGGATCAA | I3 | Low with DnaA-ATP preference |
| GGTGATCCT | τ2 | Low with DnaA-ATP preference |
| AAAGAACAA | C3 | Low with DnaA-ATP preference |
| ACCGTATAA | I1 | Low with DnaA-ATP preference |
| GGTAGTTAT | C2 | Low with DnaA-ATP preference |
| Caulobacter crescentus | ||
| TGTGGATCA | G1 | Medium |
| TGTTAATCA | G2 | Medium |
| TCATCCCCA | W1 | Low |
| TCATCCCCA | W2 | Low |
| GCATCCCCC | W3 | Low |
| GAATCCCGA | W4 | Low |
| TCATCCCCA | W5 | Low |
| Mycobacterium tuberculosis | ||
| TGTGAGTGT | 3A | |
| TGTGCACAC | 3 | |
| TGTGCGCGG | 3B | |
| TGTGAACGG | 4 | |
| TGTGGGGTG | 6 | |
| TGTGGATGA | 8 | |
| TGTCCCCAA | 9 | |
| TGTGCAGTT | 10 | |
| Bacillus subtilis | ||
| oriII region | ||
| TGTGGATAA | ||
| TGTGAAAAA | ||
| TGTGGACAG | ||
| TGTGTATGA | ||
| TGTGAATAA | ||
| Helicobacter pylori | ||
| TGTCTATAA | H1 | |
| TGTGAATGA | H2/H3 | |
| TCATTCACC | H4 | |
| CGTGAATAA | H5 | |
| Representative archaeal miniORB (as 12-mer) and ORB sites (as 23-mer) | ||
| Pyrococcus furiosus | ||
| TTCCAATGGAGA | ||
| TTCCTCTGGAAA | ||
| TTCCAGTGGAAA | ||
| CTCCAGTGGAAATGAAACTCTGG(ORB2) | ||
| Aeropyrum pernix | ||
| CTCCACAGGAAA | ||
| CTCCAGAGGAAC | ||
| ATCCAGCGGAAA | ||
| CTCCACAGGAAACGGAGGGGTCG (ORB4) | ||
| Sulfolobus solfataricus | ||
| CTCCAGTGGAAA | ||
| TTCCAAACGAAA | ||
| TTCCAGAGGAAA | ||
| TTCCAGAGGAAATAGATGGGTCC (ORB3 in oriC1) | ||
| Methanothermobacter thermoautotrophicus | ||
| TTACAGTTGAAA | ||
| TTACACTTGAAA | ||
| TTACAGTTGAAC | ||
In Escherichia coli, the sequence 5′-TTATC/ACACA-3′ (boxes R1, R2, and R4) is occupied by the active initiator form, DnaA-ATP with high affinity (Kd = 4–100 nm) (Schaper and Messer 1995; Nievera et al. 2006). However, boxes deviating from this sequence at two or more positions coexist with high-affinity sites, and these nonconsensus boxes have a dramatically lower DnaA affinity (Kd > 200 nm) (Rozgaja et al. 2011). Many low-affinity DnaA boxes in E. coli also show a strong preference (fourfold) for DnaA-ATP compared with inactive DnaA-ADP (McGarry et al. 2004), suggesting a mechanism to couple pre-RC assembly to new DnaA-ATP synthesis. The DnaA recognition boxes presented in Table 1 suggest that the nucleotide sequence variations observed for E. coli are common features among bacterial oriC, and the affinity for DnaA at each site is likely to be important for directed pre-RC assembly and its coupling to cell growth (see below).
Replication origins of E. coli and other bacteria also carry recognition sites for proteins that modulate DnaA interactions within oriC (e.g., see Fig. 2A) (Hwang et al. 1992; Slater et al. 1995; Polaczek et al. 1997; Quon et al. 1998; Brassinga et al. 2002). These modulators ensure properly timed activation and inactivation of origin activity during the cell cycle via several modes of action, including bending origin DNA into configurations that promote or inhibit DnaA subassemblies, and by directly blocking DnaA access to low-affinity recognition sites (Marczynski and Shapiro 2002; Ryan et al. 2004; Nievera et al. 2006; Leonard and Grimwade 2011). In E. coli, the DNA-bending proteins Fis and IHF act as repressors and stimulators, respectively, of the initiation step, producing a switch in oriC DNA conformation as the origin loads with DnaA (Ryan et al. 2004). An interesting repercussion of these conformational changes is that high- and low-affinity DnaA recognition sites are moved into proximal positions to allow nucleation of new DnaA oligomers (Rozgaja et al. 2011). Although the DNA-bending proteins are not required for cell viability, their loss disturbs the timing of pre-RC assembly (Ryan et al. 2004).
Bacterial Origins Encode the Directions for Ordered pre-RC Assembly
In E. coli, high-affinity DnaA boxes in oriC are occupied throughout the majority of the cell cycle, whereas all low-affinity recognition sites become occupied only at the time of initiation of DNA synthesis (Nievera et al. 2006). Once all recognition sites become fully occupied, the resulting DnaA complex is sufficient both to unwind the DUE and assist in loading of DNA helicase (DnaB) onto the exposed single strands, completing bacterial pre-RC assembly (for review, see Kaguni 2011). The persistent occupation of DnaA at high-affinity boxes during the cell cycle serves as the bacterial equivalent of the eukaryotic ORC (origin recognition complex) (Miller et al. 2009), by both marking the origin throughout the cell cycle and by serving as a scaffold to recruit additional initiator molecules (Fig. 1).
The continuous occupation of high-affinity boxes raises questions regarding the mechanism responsible for ordered filling of low-affinity boxes during the cell cycle. A key component of this mechanism is encoded by oriC as a specific arrangement of low-affinity DnaA boxes (Fig. 2A) (Rozgaja et al. 2011; Taylor et al. 2011). DnaA occupying each high-affinity DnaA box is sufficient to promote DnaA interactions at proximal low-affinity boxes (Rozgaja et al. 2011). However, these initial steps in pre-RC assembly are limited by distance, and complete complexes of 10–20 DnaA molecules require additional DnaA–DNA interactions to span larger stretches of origin DNA (for review, see Leonard and Grimwade 2011). In E. coli, these DnaA interactions are guided along oriC DNA by cryptic low-affinity DnaA boxes separated by 2 nt and clustered into sets of two helically phased arrays facing in opposite directions (see Fig. 2A) (Rozgaja et al. 2011). Using this configuration, two DnaA oligomers (Duderstadt et al. 2010) are extended along the DNA gap regions from both R1 and R4 and ultimately anchored at R2 (Rozgaja et al. 2011).
This arrangement explains how protein modulators are able to regulate DnaA oligomer formation by altering the proximity of high- and low-affinity sites on bent DNA or by blocking the addition of monomers onto a growing oligomeric chain. Placing high-affinity DnaA boxes in or out of helical phase with low-affinity arrays, as is the case with E. coli, may also play a role in generating the torsional stress needed for initial unwinding of the DUE, because DNA twisting during oligomer assembly might be required to link a continuous DnaA oligomer to its high-affinity anchors at both ends of the origin. Further comparative analysis of low-affinity DnaA interactions in bacterial origins will be important to determine how different bacterial types use origin nucleotide sequences to direct pre-RC assembly.
REPLICATION ORIGINS IN ARCHAEA
Studies of the archaeal DNA replication machinery reveal a mixture of the attributes found in bacteria and eukarya (for review, see Grabowski and Kelman 2003; Barry and Bell 2006). Although most small circular archaeal genomes carry one copy of oriC, several genera carry multiple oriC copies (Kelman and Kelman 2004; Robinson et al. 2004; Robinson and Bell 2007), which may respond to distinct initiator complexes (SD Bell, pers. comm.). The archaeal initiator Cdc6/Orc1 (Matsunaga et al. 2010) shares extensive sequence homology with eukaryotic ORC proteins (for review, see Wigley 2009), but also shares attributes of DnaA, including regulation by bound ATP (SD Bell, pers. comm.) and interacting with oriC as a monomer rather than preassembling into a hexameric complex (for review, see Lundgren and Bernander 2005).
Cdc6/Orc1 contacts nucleotide sequence motifs termed “origin recognition boxes” (Taagepera et al. 1993) that are clustered within archaeal oriCs (Fig. 2B). Although number, orientation, and spacing of ORBs vary among archaeal genera, two predominant ORB motifs exist (Table 1). A long ORB motif (22–35 bp) with dyad symmetry is mapped at two or more locations in origins from Sulfolobus, Pyrococcus, and Aeropyrum (Grainge et al. 2006; Dueber et al. 2007; Gaudier et al. 2007). The orientation of each bound Cdc6/Orc1 is determined by the ORB sequence (Dueber et al. 2007; Gaudier et al. 2007). A shorter (12–13 bp) version, termed miniORB, coexists with ORBs in Pyrococcus (Matsunaga et al. 2010) and Sulfolobus (Dueber et al. 2007), or in the absence ORBs in Methanothermobacter oriCs (Capaldi and Berger 2004). MiniORBs are often found as multiple (seven to 15) direct repeats (Capaldi and Berger 2004; Matsunaga et al. 2010), analogous to some bacterial origins. Cdc6/Orc1 binding to proximal miniORBs is also cooperative (Capaldi and Berger 2004; Grainge et al. 2006), suggesting the assembly of oligomeric structures analogous to those formed by DnaA.
Although Cdc6/Orc1 higher-order complexes are reported to alter DNA topology and lead to origin unwinding (Matsunaga et al. 2010), Cdc6/Orc1 contact with ORBs differs from DnaA-oriC interactions in several ways. Although DnaA and Cdc6/Orc1 both contact double-stranded DNA through the carboxy-terminal domain, Cdc6/Orc1 make additional contacts with ORBS through the AAA+ domain of the protein. These contacts appear to rely less on specific base sequence contacts and more on DNA deformability than is the case for DnaA (Gaudier et al. 2007; Dueber et al. 2011). Each Cdc6/Orc1 interaction also distorts the DNA helix to a greater degree than is seen during bacterial pre-RC assembly (Gaudier et al. 2007; Dueber et al. 2011). It remains unclear whether the distinction between ORBs and miniORBS is due to the inclusion of sequences recognized or distorted by the AAA+ domain.
DNA REPLICATION ORIGINS IN EUKARYOTES
In Saccharomyces cerevisiae, DNA replication origins were first defined by their ability to confer autonomous replication to plasmids (Stinchcomb et al. 1979; Bell and Stillman 1992; Theis and Newlon 1994). These elements were called autonomous replication sequences (ARSs) and contain the specific consensus element autonomous consensus sequences (ACSs) of 11 bp further extended to 17 bp (for review, see Stinchcomb et al. 1979; Theis and Newlon 1997; Dhar et al. 2012). ACS is the binding site for ORC, the main factor that subsequently serves as a landing platform for the assembly of the other pre-RC proteins. In metazoans, despite the identification of several proteins involved in initiation of DNA replication (most, but not all, are conserved from yeast to humans), it is still not known how they target DNA replication origins. Studies are hampered by the absence of a single, really efficient genetic assay to isolate replication origins because DNA transferred into metazoan cells replicates very poorly. Thus, different biochemical approaches have been developed over the years, including two-dimensional (2D) gel electrophoresis of replication intermediates (Brewer and Fangman 1987) and quantitative PCR analysis of nascent strands (Giacca et al. 1994), a method greatly improved by the exploitation of the RNA-primed nature of nascent strands at origins (Gerbi and Bielinsky 1997; Bielinsky and Gerbi 1998). An interesting, but less used, bubble-trap method is based on the structure of replication origins (Mesner et al. 2011). Genome-wide sequencing of RNA-primed nascent DNAs presently appears as the more precise method.
DNA REPLICATION ORIGINS IN YEAST
Sequence Specificity
Specific consensus sequences for origins are identified only in S. cerevisiae. Although this 11-bp sequence [5/(A/T)TTTA(T/C)(A/G)TTT(A/T)-3] defines all the 12,000 ACSs present in this genome, only a few of them (∼400) are used in normal conditions (Linskens and Huberman 1988). Other elements close to the ACS motif contribute to its activity and provide a modular structure to origins (Fig. 3) (Marahrens and Stillman 1992). One example, the B domain, might serve as a DNA unwinding element (Huang and Kowalski 1993) and can be subdivided into three subdomains. B1 is adjacent to the ACS and is important for ORC binding (Rao and Stillman 1995; Rowley et al. 1995). It also contains an A/TTA/T motif that is found in 228 ARSs conserved in six Saccharomyces species and could be involved in ORC binding activity (Chang et al. 2008). The B2 element is suggested to be the binding site for the MCM2-7 helicase (Zou and Stillman 2000; Wilmes and Bell 2002). It may also contain a degenerate ANA/TA/TAAAT common sequence that contributes to the efficiency of origin recognition (Chang et al. 2011). B3 binds to Abf1 (ARS binding factor 1), a DNA-binding protein that plays a role in transcription and can influence chromatin structure and nucleosome assembly (Miyake et al. 2002; Ganapathi et al. 2011).
Figure 3.
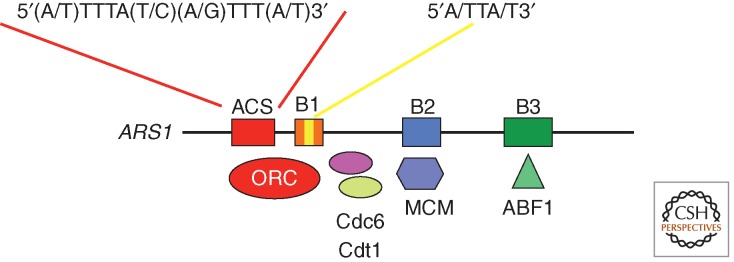
S. cerevisiae replication origins. Elements found at S. cerevisiae origins are represented, including the ACS and B elements. The main proteins that bind to these elements and that constitute the pre-RC are indicated.
In yeast strains other than S. cerevisiae, a clear consensus sequence is not found, and origin locations are not conserved in budding yeast species (Liachko et al. 2010; Xu et al. 2012). A 9-bp putative motif, similar to the ACS in S. cerevisiae, has been identified in Lachancea kluyveri, but it is not sufficient to confer ARS activity and must be embedded in a much larger DNA sequence (Liachko et al. 2011). The preference for poly(A) tracks was confirmed in Schizosaccharomyces pombe and Schizosaccharomyces octosporus, and AT-rich sequences were also found in Lachancea kluyveri (Liachko et al. 2011). In S. pombe, large DNA replication origins were found composed of several short AT-rich modules capable of initiating replication from ectopic chromosome positions (Cotobal et al. 2010). However, different motifs can be used in different unicellular organisms. Surprisingly, the A-T content is a strong negative predictor of origin function in Schizosaccharomyces japonicus (Xu et al. 2012) in which origins have a high G-C content. Because poly(G) motifs, like AT-rich elements, exclude nucleosomes (Tsankov et al. 2011; Fenouil et al. 2012), they are expected to have a similar role in the chromatin environment (see below).
Altogether, these results suggest more plasticity for origin recognition in unicellular eukaryotes than previously thought based only on the analysis of the S. cerevisiae genome. However, plasticity does not exclude site specificity. In S. pombe, there is a clear association of DNA replication origins with ORC-binding sites and nucleosome-free regions (Hayashi et al. 2007; Xu et al. 2012).
Chromatin Environment
In addition to sequence specificity, a second important feature of S. cerevisiae origins is nucleosome exclusion. The patterns of nucleosome occupancy around the ACS are similar to open transcription promoters, and genome-wide analyses of nucleosome positioning support the conclusions from studies on single origins (Thoma et al. 1984; Berbenetz et al. 2010; Eaton et al. 2010). The ACS is necessary to confer a nucleosome-free region. ORC binding is then required to induce a periodic positioning of nucleosomes adjacent to the ACS and might act as a barrier element to position and phase nucleosomes at precise intervals on both sides of the origin. Interestingly, nucleosome positioning around the ACS characterizes origins that will be active. The nucleosome-free feature of this region might be due at least in part to the A-T richness of this domain (Kaplan et al. 2009; Zhang et al. 2009). The size of the nucleosome-free region (∼130 bp) provides room for the assembly of the other pre-RC proteins and does not seem to vary during the cell cycle. However, if the ORC is depleted, the size of the nucleosome-free region decreases, and the positioning of adjacent nucleosomes is affected.
Although nucleosomes are depleted at the ACS, interaction of the ORC complex with adjacent chromatin regions and histone modifications are likely to contribute to the general mechanism of origin recognition in yeast. In agreement, if nucleosomes are moved away from the ORC-binding site, origin function is reduced (Lipford and Bell 2001), emphasizing the importance of precise nucleosome positioning for replication initiation. The exact set of histone modifications involved in DNA replication origin recognition or firing remains largely unknown in yeast as well as in multicellular organisms. This is partly because not all potential origins are active in each individual cell, making it difficult to identify the modifications specifically associated with origin recognition or firing. Nevertheless, using a mini-chromosome that contains an efficient yeast origin, increased histone H3 and H4 acetylation was correlated with origin activity (Unnikrishnan et al. 2010).
DNA REPLICATION ORIGINS IN MULTICELLULAR ORGANISMS
Organization of Metazoan Origins
In metazoans, because of the larger size of their genomes, thousands of replication origins are activated at each cell cycle (30,000–50,000 in human or mouse cells). Until very recently, only a few origins were clearly identified, but improved genome-wide analyses permit a better understanding of their nature (Cadoret et al. 2008; Sequeira-Mendes et al. 2009; Cayrou et al. 2011; Costas et al. 2011; Mesner et al. 2011). Several features are now emerging. First, there are many more origins than needed at each cell cycle. Only one out of five potential replication origins within a replicon is used in any cell during a given cell cycle, and, therefore, DNA replication origin activation appears to be a relatively inefficient process (Cayrou et al. 2011). This feature is not unique to metazoans because it is shared by yeast origins, including S. cerevisiae, where origins are defined by specific motifs (Friedman et al. 1997; Heichinger et al. 2006). In theory, this abundance of potential replication origins in each replication unit or replicon might favor the activation of at least one origin per replicon (Karschau et al. 2012). This low usage of replication origins results from a flexible choice of replication origins in each cell (Fig. 4). Thus, within a given cell population, each cell has the potential to activate a large number of origins (100,000 in mouse fibroblasts) and might use a different cohort of origins in an apparent stochastic manner. This situation results in initiation zones with an average of five potential origins that constitute the first order of organization of a replication unit: the flexible replicon (Cayrou et al. 2011). The second hierarchical level of organization is the cluster of replicons. DNA replication origins are activated in clusters where several replication origins are often activated simultaneously, as first observed by autoradiography and more recently by DNA combing methods (Lebofsky et al. 2006; Cayrou et al. 2011). In origin clusters, early- and late-firing origins are often separated by long, origin-poor regions, also called “temporal transition regions” (TTRs). TTRs were first detected in the mouse Ig heavy-chain locus (Norio et al. 2005) and then in genome-wide analysis of DNA replication origins (Cadoret et al. 2008; Cayrou et al. 2011). However, TTR replication might differ in different cell types because replication origins in TRR might be silenced in some cells, but gradually activated in others during the S-phase progression (Guilbaud et al. 2011).
Figure 4.
From replication origins to replicon clusters. Replicons are organized as functional domains that contain several potential DNA replication origins (on average five), already licensed. A single origin is activated per replicon during the S phase. The origin choice within each replicon can occur stochastically or can be influenced by specific cell fates or transcriptional activity. Replicon clusters include several consecutive replicons that are activated simultaneously (Berezney et al. 2000). Replicons could be organized in chromatin loops where activation of one origin silences the other origins within the same replicon.
The large number and the flexible activation of DNA replication origins have several advantages. Replication origins in excess (spare origins) can be used in case of poor growing conditions or when the replication fork experiences difficulties in passing through higher-order DNA or chromatin structures. Spare origins can also be activated in specific physiological conditions, such as during early embryonic development. Indeed, during early Drosophila or Xenopus development, the S phase lasts <15 min, and replication origins are activated every 10-15 kb (Blumenthal et al. 1974; Hyrien et al. 1995). It would be interesting to reinvestigate the site specificity of these origins in early embryos to determine whether all potential origins are used in every cell, as opposed to completely random initiation. The large number of flexible origins also include dormant origins that normally are not activated unless the cell is under replication stress (such as DNA damage and in response to checkpoint signals) (Branzei and Foiani 2005; Woodward et al. 2006; Blow et al. 2011). Finally, the large number of potential origins also includes those that allow the replication program to adapt to the transcription program. The number of identified DNA replication origins that are cell or developmental stage specific and that are normally inactive in other cell types is relatively limited at present (Hyrien et al. 1995; Norio et al. 2005), but they are likely to increase in the next years.
Active transcription may have two potentially opposite consequences on the choice of DNA replication origins. Indeed, it can provide a nucleosome-free area at the promoter region to which the replication complex might bind opportunely. On the other hand, active transcription may also not be favorable to the assembly of a replication complex (for review, see Méchali 2010; Sequeira-Mendes and Gomez 2012). This cross talk between genome organization for DNA replication and gene expression is likely to be crucial for the maintenance of cell identity during development and differentiation.
Sequence Specificity in Metazoans
The question of sequence specificity remains unresolved in metazoan cells. The ORC is the first replication factor known to bind to replication origins, but it does not recognize a specific sequence, although in vitro it shows a preference for supercoiled DNA and poly(dA)–poly(dT) tracts (Vashee et al. 2003; Remus et al. 2004). A recent genome-wide distribution of ORCs in Drosophila could identify motifs that have a predictive value for origins (MacAlpine et al. 2010). However, ORC also binds to sites other than replication origins, such as centrosomes, centromeres, or heterochromatin (Prasanth et al. 2004; Hemerly et al. 2009). Sequence specificity for replication origins is more directly addressed by recent genome-wide identification of active sites of initiation of DNA replication. They confirmed a preference for CpG islands (Delgado et al. 1998; Cadoret et al. 2008; Sequeira-Mendes et al. 2009; Cayrou et al. 2011). Association with GC-rich regions was also observed in plant replication origins (Costas et al. 2011). In addition, in mouse and human cells, an origin G-rich repeat element (OGRE) is present in 80%–90% of DNA replication origins and can form G quadruplexes (Cayrou et al. 2011, 2012) in which guanines can form unusual four-stranded DNA structures through hydrogen bonding. This finding suggests that metazoan origins may be rather different from yeast origins, which are mainly A-T rich. At metazoan origins, a DNA structure formed by a specific positioning of bases, similar to the case for G quadruplexes, might be more important than strict sequence specificity for initiation of DNA replication. It now remains to show experimentally that these conserved elements are components of the metazoan replication origins.
Finally, a completely different approach to map DNA replication origins, based on in silico analysis of the nucleotide composition skew, showed an abrupt change in the skew profile at known replication origins, possibly because of different mutation rates at the leading or lagging strand of DNA synthesis (Chen et al. 2011). Based on this feature, very large interacting chromatin domains with efficient origins at the borders could be defined.
Transcription, Chromatin, and Nuclear Organization in the Selection of DNA Replication Origins in Metazoans
A permissive chromatin environment is a major characteristic of active transcription promoters and has been considered an important feature of replication origins for many years (for review, see Sequeira-Mendes and Gomez 2011). This may explain the general correlation between transcription activity and enrichment in DNA replication origins found by several of the recent genome-wide analyses of DNA replication. Nucleosome-free or unstable chromatin regions of active promoter regions are frequently associated with origins because AT-rich or G-rich regions are not favorable to nucleosome formation. These regions are also marked by specific histone variants, such as H2AZ or H3.3 (Jin et al. 2009; MacAlpine et al. 2010; Stroud et al. 2012), and by the association of chromatin remodelers that contribute to the recruitment of the pre-RC complex, such as the histone acetylase HBO1 (Miotto and Struhl 2010), the histone methyl transferase PR-Set7 (Tardat et al. 2010), or the chromatin-remodeling complex SNF2H (Sugimoto et al. 2011).
However, a nucleosome-free region does not exclude the possibility that interactions of pre-RC proteins with adjacent nucleosomes might be important, at least in specific chromatin environments. For instance, ORC interacts with heterochromatin and HP1 in Drosophila (Pak et al. 1997) in agreement with the finding that on Drosophila chromosome 4, which is organized mainly into heterochromatin, origins are associated with HP1 sites (Cayrou et al. 2011). Similarly, ORC interactions (regulated by the BAH domain of the ORC1 subunit) with chromatin and with adjacent nucleosomes were also described in S. cerevisiae and in human cells (Noguchi et al. 2006; Muller et al. 2010). The BAH domain in human Orc1 facilitates the ability of Orc1 to activate replication origins in vivo (Noguchi et al. 2006) and links H4K20me2 to DNA replication licensing and to Meier–Gorlin syndrome, a genetic problem leading to primordial dwarfism (Kuo et al. 2012). However, the role of this domain in ORC association with chromatin varies at different origins, emphasizing the non-unique nature of replication origins.
Histone modifications are essential for modulating gene expression and might also play an important role in the choice of DNA replication origins and in the regulation of their activation. Histone lysine acetylation has often been associated with the regulation of replication timing, but links with DNA replication origins activation have also been reported (Aggarwal and Calvi 2004; Liu et al. 2012). Histone lysine methylations have been recently reviewed (Dorn and Cook 2011), and H4K20 methylation appears to be involved in the control of DNA replication origins. H4K20me1 is high during pre-RC formation, from mitosis to G1, but low in S phase (Rice et al. 2002), and is found at some human DNA replication origins (Tardat et al. 2010). In addition, tethering PR-Set7, the methylase responsible for H4K20 methylation, to a specific genomic locus promotes pre-RC assembly. Another methylation mark found at replication origins is H3K4 di- and trimethylation (Costas et al. 2011; Eaton et al. 2011).
At a higher level of chromosomal organization, the timing of replication domains strongly correlates with long-range chromatin interactions, as measured by using the Hi-C technique (Ryba et al. 2010), supporting the hypothesis that replication domains localized near each other replicate at similar times. These domains may correspond to clusters of replication origins forming the DNA replication foci or factories that are observed in all nuclei (Fig. 4) (for review, see Jackson and Pombo 1998; Berezney et al. 2000; Maya-Mendoza et al. 2010). How DNA replication origins are organized in such structures remains hypothetical. Each replicon, which is constituted by several potential origins, might form a chromatin loop (Cayrou et al. 2011), and several clustered chromatin loops could constitute replication domains or replication foci. The chromatin loop hypothesis is supported by the correlation between the size of the loop and the size of replicons in different species (Buongiorno-Nardelli et al. 1982) and under different growing conditions (Lemaitre et al. 2005; Courbet et al. 2008). Cohesin, a protein involved in sister-chromatid cohesion, might be involved in the stabilization of these loops and is enriched at some replication origins (Guillou et al. 2010), although it appears to be loaded via pre-RC assembly-independent mechanisms. A concordance between ORC-binding sites and cohesin sites confirmed this relationship between replication origins, cohesin, and chromatin loops (MacAlpine et al. 2010).
CONCLUDING REMARKS
The differences in the amount of information encoded by prokaryotic and eukaryotic replication origins and the contribution of chromatin structure have obvious impacts on both the nucleotide stringency and length of required chromosomal DNA. In the case of bacteria, much of the encoded information has emerged only recently, and as the nucleotide sequences of origins are examined in different species, additional instructions for assembling ordered DnaA prereplicative complexes and even components of the cell cycle timing mechanism will be revealed. It is likely that metazoan replication origins are characterized by a combination of different elements, including some DNA motifs or structures, and that different classes of origins coexist in the metazoan genomes. This modular nature of replication origins might provide the flexibility required to organize chromosomes for their replication, depending on the cell fate, as well as to provide the ability to complete DNA replication of the whole genome in a wide range of physiological conditions. Further analysis of archaeal origins is needed to understand the important bridge from origins with high information content to more streamlined versions, ultimately leading to the minimal sequence specificity found among the higher eukaryotes. The study of eukaryotic genomes must continue in order to determine the requirements of “origin-ness,” and the bulk of the effort will surely be focused on unraveling the epigenetic regulators that allow ORC to gain DNA access. It is clear that a great deal of the story remains to be told.
ACKNOWLEDGMENTS
We are grateful to many colleagues for their helpful discussions and insights. Research in our laboratories is supported by National Institutes of Health grant GM054042 (to A.C.L.) and the European Research Council (FP7/2007-2013 grant agreement no. 233339). This work is also supported by the ANR, the ARC, and the Ligue Nationale Contre le Cancer (LNCC).
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Barry ER, Bell SD 2006. DNA replication in the archaea. Microbiol Mol Biol Rev 70: 876–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beattie TR, Bell SD 2011. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 15: 614–619 [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 [DOI] [PubMed] [Google Scholar]
- Berbenetz NM, Nislow C, Brown GW 2010. Diversity of eukaryotic DNA replication origins revealed by genome-wide analysis of chromatin structure. PLoS Genet 6: e1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Bielinsky AK, Gerbi SA 1998. Discrete start sites for DNA synthesis in the yeast ARS1 origin. Science 279: 95–98 [DOI] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ 2008. Replication licensing and cancer—A fatal entanglement? Nat Rev Cancer 8: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal AB, Kriegstein HJ, Hogness DS 1974. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol 38: 205–223 [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M 2005. The DNA damage response during DNA replication. Curr Opin Cell Biol 17: 568–575 [DOI] [PubMed] [Google Scholar]
- Brassinga AK, Siam R, McSween W, Winkler H, Wood D, Marczynski GT 2002. Conserved response regulator CtrA and IHF binding sites in the α-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J Bacteriol 184: 5789–5799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51: 463–471 [DOI] [PubMed] [Google Scholar]
- Bryant JA, Aves SJ 2011. Initiation of DNA replication: Functional and evolutionary aspects. Ann Bot 107: 1119–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buongiorno-Nardelli M, Micheli G, Carri MT, Marilley M 1982. A relationship between replicon size and supercoiled loop domains in the eukaryotic genome. Nature 298: 100–102 [DOI] [PubMed] [Google Scholar]
- Cadoret JC, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, Prioleau MN 2008. Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc Natl Acad Sci 105: 15837–15842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi SA, Berger JM 2004. Biochemical characterization of Cdc6/Orc1 binding to the replication origin of the euryarchaeon Methanothermobacter thermoautotrophicus. Nucleic Acids Res 32: 4821–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. 2011. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21: 1438–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Coulombe P, Puy A, Rialle S, Kaplan N, Segal E, Mechali M 2012. New insights into replication origin characteristics in metazoans. Cell Cycle 11: 658–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Theis JF, Miller J, Nieduszynski CA, Newlon CS, Weinreich M 2008. Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site. Mol Cell Biol 28: 5071–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, May CD, Hoggard T, Miller J, Fox CA, Weinreich M 2011. High-resolution analysis of four efficient yeast replication origins reveals new insights into the ORC and putative MCM binding elements. Nucleic Acids Res 39: 6523–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Duquenne L, Audit B, Guilbaud G, Rappailles A, Baker A, Huvet M, d’Aubenton-Carafa Y, Hyrien O, Arneodo A, et al. 2011. Replication-associated mutational asymmetry in the human genome. Mol Biol Evol 28: 2327–2337 [DOI] [PubMed] [Google Scholar]
- Costas C, de la Paz Sanchez M, Stroud H, Yu Y, Oliveros JC, Feng S, Benguria A, Lopez-Vidriero I, Zhang X, Solano R, et al. 2011. Genome-wide mapping of Arabidopsis thaliana origins of DNA replication and their associated epigenetic marks. Nat Struct Mol Biol 18: 395–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotobal C, Segurado M, Antequera F 2010. Structural diversity and dynamics of genomic replication origins in Schizosaccharomyces pombe. EMBO J 29: 934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courbet S, Gay S, Arnoult N, Wronka G, Anglana M, Brison O, Debatisse M 2008. Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature 455: 557–560 [DOI] [PubMed] [Google Scholar]
- Cvetic C, Walter JC 2005. Eukaryotic origins of DNA replication: Could you please be more specific? Semin Cell Dev Biol 16: 343–353 [DOI] [PubMed] [Google Scholar]
- Dhar MK, Sehgal S, Kaul S 2012. Structure, replication efficiency and fragility of yeast ARS elements. Res Microbiol 163: 243–253 [DOI] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 17: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF 2011. Quality control in the initiation of eukaryotic DNA replication. Philos Trans R Soc Lond B Biol Sci 366: 3545–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, MacAlpine DM 2011. Defining the replication program through the chromatin landscape. Crit Rev Biochem Mol Biol 46: 165–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn ES, Cook JG 2011. Nucleosomes in the neighborhood: New roles for chromatin modifications in replication origin control. Epigenetics 6: 552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duderstadt KE, Berger JM 2008. AAA+ ATPases in the initiation of DNA replication. Crit Rev Biochem Mol Biol 43: 163–187 [DOI] [PubMed] [Google Scholar]
- Duderstadt KE, Mott ML, Crisona NJ, Chuang K, Yang H, Berger JM 2010. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J Biol Chem 285: 28229–28239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM 2007. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science 317: 1210–1213 [DOI] [PubMed] [Google Scholar]
- Dueber EC, Costa A, Corn JE, Bell SD, Berger JM 2011. Molecular determinants of origin discrimination by Orc1 initiators in archaea. Nucleic Acids Res 39: 3621–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncker BP, Chesnokov IN, McConkey BJ 2009. The origin recognition complex protein family. Genome Biol 10: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM 2011. Chromatin signatures of the Drosophila replication program. Genome Res 21: 164–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM 2006. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol 13: 676–683 [DOI] [PubMed] [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza JC, Innocenti C, Ferrier P, Spicuglia S, Gut M, Andrau JC 2012. CpG islands and GC contents dictate nucleosome depletion in a transcription independent manner at mammalian promoters. Genome Res 10.1101/gr.138776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman KL, Brewer BJ, Fangman WL 1997. Replication profile of Saccharomyces cerevisiae chromosome VI. Genes Cells 2: 667–678 [DOI] [PubMed] [Google Scholar]
- Ganapathi M, Palumbo MJ, Ansari SA, He Q, Tsui K, Nislow C, Morse RH 2011. Extensive role of the general regulatory factors, Abf1 and Rap1, in determining genome-wide chromatin structure in budding yeast. Nucleic Acids Res 39: 2032–2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Zhang CT 2007. DoriC: A database of oriC regions in bacterial genomes. Bioinformatics 23: 1866–1867 [DOI] [PubMed] [Google Scholar]
- Gao F, Zhang CT 2008. Ori-Finder: A web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics 9: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier M, Schuwirth BS, Westcott SL, Wigley DB 2007. Structural basis of DNA replication origin recognition by an ORC protein. Science 317: 1213–1216 [DOI] [PubMed] [Google Scholar]
- Gerbi SA, Bielinsky AK 1997. Replication initiation point mapping. Methods 13: 271–280 [DOI] [PubMed] [Google Scholar]
- Giacca M, Zentilin L, Norio P, Diviacco S, Dimitrova D, Contreas G, Biamonti G, Perini G, Weighardt F, Riva S, et al. 1994. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci 91: 7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski B, Kelman Z 2003. Archeal DNA replication: Eukaryal proteins in a bacterial context. Annu Rev Microbiol 57: 487–516 [DOI] [PubMed] [Google Scholar]
- Grainge I, Gaudier M, Schuwirth BS, Westcott SL, Sandall J, Atanassova N, Wigley DB 2006. Biochemical analysis of a DNA replication origin in the archaeon Aeropyrum pernix. J Mol Biol 363: 355–369 [DOI] [PubMed] [Google Scholar]
- Guilbaud G, Rappailles A, Baker A, Chen CL, Arneodo A, Goldar A, d’Aubenton-Carafa Y, Thermes C, Audit B, Hyrien O 2011. Evidence for sequential and increasing activation of replication origins along replication timing gradients in the human genome. PLoS Comput Biol 7: e1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, Mendez J 2010. Cohesin organizes chromatin loops at DNA replication factories. Genes Dev 24: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Katou Y, Itoh T, Tazumi A, Yamada Y, Takahashi T, Nakagawa T, Shirahige K, Masukata H 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J 26: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bahler J, Nurse P 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly AS, Prasanth SG, Siddiqui K, Stillman B 2009. Orc1 controls centriole and centrosome copy number in human cells. Science 323: 789–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang RY, Kowalski D 1993. A DNA unwinding element and an ARS consensus comprise a replication origin within a yeast chromosome. EMBO J 12: 4521–4531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang DS, Thony B, Kornberg A 1992. IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J Biol Chem 267: 2209–2213 [PubMed] [Google Scholar]
- Hyrien O, Maric C, Mechali M 1995. Transition in specification of embryonic metazoan DNA replication origins. Science 270: 994–997 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Brenner J, Cuzin F 1963. On the regulation of DNA replication in bacteria. Cold Spring Harbor Symp Quant Biol 28: 329–348 [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G 2009. H3.3/H2A.Z double variant-containing nucleosomes mark “nucleosome-free regions” of active promoters and other regulatory regions. Nat Genet 41: 941–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni JM 2011. Replication initiation at the Escherichia coli chromosomal origin. Curr Opin Chem Biol 15: 606–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. 2009. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458: 362–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karschau J, Blow JJ, de Moura AP 2012. Optimal placement of origins for DNA replication. Phys Rev Lett 108: 058101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami H, Katayama T 2010. DnaA, ORC, and Cdc6: Similarity beyond the domains of life and diversity. Biochem Cell Biol 88: 49–62 [DOI] [PubMed] [Google Scholar]
- Kelman LM, Kelman Z 2004. Multiple origins of replication in archaea. Trends Microbiol 12: 399–401 [DOI] [PubMed] [Google Scholar]
- Kowalski D, Eddy MJ 1989. The DNA unwinding element: A novel, cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J 8: 4335–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo AJ, Song J, Cheung P, Ishibe-Murakami S, Yamazoe S, Chen JK, Patel DJ, Gozani O 2012. The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome. Nature 484: 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebofsky R, Heilig R, Sonnleitner M, Weissenbach J, Bensimon A 2006. DNA replication origin interference increases the spacing between initiation events in human cells. Mol Biol Cell 17: 5337–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M 2005. Mitotic remodeling of the replicon and chromosome structure. Cell 123: 1–15 [DOI] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE 2011. Regulation of DnaA assembly and activity: Taking directions from the genome. Annu Rev Microbiol 65: 19–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Bhaskar A, Lee C, Chung SC, Tye BK, Keich U 2010. A comprehensive genome-wide map of autonomously replicating sequences in a naive genome. PLoS Genet 6: e1000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liachko I, Tanaka E, Cox K, Chung SC, Yang L, Seher A, Hallas L, Cha E, Kang G, Pace H, et al. 2011. Novel features of ARS selection in budding yeast Lachancea kluyveri. BMC Genomics 12: 633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linskens M, Huberman JA 1988. Organization of replication in ribosomal DNA in Saccharomyces cerevisiae. Mol Cell Biol 8: 4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Bell SP 2001. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell 7: 21–30 [DOI] [PubMed] [Google Scholar]
- Liu J, McConnell K, Dixon M, Calvi BR 2012. Analysis of model replication origins in Drosophila reveals new aspects of the chromatin landscape and its relationship to origin activity and the prereplicative complex. Mol Biol Cell 23: 200–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren M, Bernander R 2005. Archaeal cell cycle progress. Curr Opin Microbiol 8: 662–668 [DOI] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM 2010. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz P, Zakrzewska-Czerwinska J, Zawilak A, Dudek MR, Cebrat S 2004. Where does bacterial replication start? Rules for predicting the oriC region. Nucleic Acids Res 32: 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B 1992. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255: 817–823 [DOI] [PubMed] [Google Scholar]
- Marczynski GT, Shapiro L 2002. Control of chromosome replication in Caulobacter crescentus. Annu Rev Microbiol 56: 625–656 [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M 2010. Eukaryotic chromosome DNA replication: Where, when, and how? Annu Rev Biochem 79: 89–130 [DOI] [PubMed] [Google Scholar]
- Matsunaga F, Takemura K, Akita M, Adachi A, Yamagami T, Ishino Y 2010. Localized melting of duplex DNA by Cdc6/Orc1 at the DNA replication origin in the hyperthermophilic archaeon Pyrococcus furiosus. Extremophiles 14: 21–31 [DOI] [PubMed] [Google Scholar]
- Maya-Mendoza A, Olivares-Chauvet P, Shaw A, Jackson DA 2010. S phase progression in human cells is dictated by the genetic continuity of DNA foci. PLoS Genet 6: e1000900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry KC, Ryan VT, Grimwade JE, Leonard AC 2004. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci 101: 2811–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méchali M 2010. Eukaryotic DNA replication origins: Many choices for appropriate answers. Nature Rev Mol Cell Biol 11: 728–738 [DOI] [PubMed] [Google Scholar]
- Mesner LD, Valsakumar V, Karnani N, Dutta A, Hamlin JL, Bekiranov S 2011. Bubble-chip analysis of human origin distributions demonstrates on a genomic scale significant clustering into zones and significant association with transcription. Genome Res 21: 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DT, Grimwade JE, Betteridge T, Rozgaja T, Torgue JJ, Leonard AC 2009. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc Natl Acad Sci 106: 18479–18484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Struhl K 2010. HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell 37: 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake T, Loch CM, Li R 2002. Identification of a multifunctional domain in autonomously replicating sequence-binding factor 1 required for transcriptional activation, DNA replication, and gene silencing. Mol Cell Biol 22: 505–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Park S, Shor E, Huebert DJ, Warren CL, Ansari AZ, Weinreich M, Eaton ML, MacAlpine DM, Fox CA 2010. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev 24: 1418–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievera C, Torgue JJ, Grimwade JE, Leonard AC 2006. SeqA blocking of DnaA–oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell 24: 581–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi K, Vassilev A, Ghosh S, Yates JL, DePamphilis ML 2006. The BAH domain facilitates the ability of human Orc1 protein to activate replication origins in vivo. EMBO J 25: 5372–5382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL 2005. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell 20: 575–587 [DOI] [PubMed] [Google Scholar]
- O’Donnell M 2006. Replisome architecture and dynamics in Escherichia coli. J Biol Chem 281: 10653–10656 [DOI] [PubMed] [Google Scholar]
- Pak DT, Pflumm M, Chesnokov I, Huang DW, Kellum R, Marr J, Romanowski P, Botchan MR 1997. Association of the origin recognition complex with heterochromatin and HP1 in higher eukaryotes. Cell 91: 311–323 [DOI] [PubMed] [Google Scholar]
- Polaczek P, Kwan K, Liberies DA, Campbell JL 1997. Role of architectural elements in combinatorial regulation of initiation of DNA replication in Escherichia coli. Mol Microbiol 26: 261–275 [DOI] [PubMed] [Google Scholar]
- Prasanth SG, Prasanth KV, Siddiqui K, Spector DL, Stillman B 2004. Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J 23: 2651–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quon KC, Yang B, Domian IJ, Shapiro L, Marczynski GT 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci 95: 120–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Stillman B 1995. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci 92: 2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Diffley JF 2009. Eukaryotic DNA replication control: Lock and load, then fire. Curr Opin Cell Biol 21: 771–777 [DOI] [PubMed] [Google Scholar]
- Remus D, Beall EL, Botchan MR 2004. DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC–DNA binding. EMBO J 23: 897–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD 2002. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev 16: 2225–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD 2007. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci 104: 5806–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Dionne I, Lundgren M, Marsh VL, Bernander R, Bell SD 2004. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell 116: 25–38 [DOI] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF 1995. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J 14: 2631–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozgaja TA, Grimwade JE, Iqbal M, Czerwonka C, Vora M, Leonard AC 2011. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol Microbiol 82: 475–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan VT, Grimwade JE, Camara JE, Crooke E, Leonard AC 2004. Escherichia coli prereplication complex assembly is regulated by dynamic interplay among Fis, IHF and DnaA. Mol Microbiol 51: 1347–1359 [DOI] [PubMed] [Google Scholar]
- Ryba T, Hiratani I, Lu J, Itoh M, Kulik M, Zhang J, Schulz TC, Robins AJ, Dalton S, Gilbert DM 2010. Evolutionarily conserved replication timing profiles predict long-range chromatin interactions and distinguish closely related cell types. Genome Res 20: 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaper S, Messer W 1995. Interaction of the initiator protein DnaA of Escherichia coli with its DNA target. J Biol Chem 270: 17622–17626 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Gomez M 2011. On the opportunistic nature of transcription and replication initiation in the metazoan genome. Bioessays 34: 119–125 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Gomez M 2012. On the opportunistic nature of transcription and replication initiation in the metazoan genome. Bioessays 34: 119–125 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82: 927–936 [DOI] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282: 39–43 [DOI] [PubMed] [Google Scholar]
- Stroud H, Otero S, Desvoyes B, Ramirez-Parra E, Jacobsen SE, Gutierrez C 2012. Genome-wide analysis of histone H3.1 and H3.3 variants in Arabidopsis thaliana. Proc Natl Acad Sci 109: 5370–5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N, Yugawa T, Iizuka M, Kiyono T, Fujita M 2011. Chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with Cdc10 protein-dependent transcript 1 (Cdt1) and promotes pre-replication complex formation. J Biol Chem 286: 39200–39210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taagepera S, Rao PN, Drake FH, Gorbsky GJ 1993. DNA topoisomerase IIa is the major chromosomal protein recognized by the mitotic phosphoprotein antibody MPM-2. Proc Natl Acad Sci 90: 8407–8411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E 2010. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol 12: 1086–1093 [DOI] [PubMed] [Google Scholar]
- Taylor JA, Ouimet MC, Wargachuk R, Marczynski GT 2011. The Caulobacter crescentus chromosome replication origin evolved two classes of weak DnaA binding sites. Mol Microbiol 82: 312–326 [DOI] [PubMed] [Google Scholar]
- Theis JF, Newlon CS 1994. Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol Cell Biol 14: 7652–7659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis JF, Newlon CS 1997. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc Natl Acad Sci 94: 10786–10791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F, Bergman LW, Simpson RT 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol 177: 715–733 [DOI] [PubMed] [Google Scholar]
- Tsankov A, Yanagisawa Y, Rhind N, Regev A, Rando OJ 2011. Evolutionary divergence of intrinsic and trans-regulated nucleosome positioning sequences reveals plastic rules for chromatin organization. Genome Res 21: 1851–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17: 430–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, Walter JC 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev 17: 1894–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley DB 2009. ORC proteins: Marking the start. Curr Opin Struct Biol 19: 72–78 [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Bell SP 2002. The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proc Natl Acad Sci 99: 101–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ 2006. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biol 173: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worning P, Jensen LJ, Hallin PF, Staerfeldt HH, Ussery DW 2006. Origin of replication in circular prokaryotic chromosomes. Environ Microbiol 8: 353–361 [DOI] [PubMed] [Google Scholar]
- Xu J, Yanagisawa Y, Tsankov AM, Hart C, Aoki K, Kommajosyula N, Steinmann KE, Bochicchio J, Russ C, Regev A, et al. 2012. Genome-wide identification and characterization of replication origins by deep sequencing. Genome Biol 13: R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawilak-Pawlik A, Kois A, Majka J, Jakimowicz D, Smulczyk-Krawczyszyn A, Messer W, Zakrzewska-Czerwinska J 2005. Architecture of bacterial replication initiation complexes: Orisomes from four unrelated bacteria. Biochem J 389: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewka-Czerwinska J, Jalimowicz D, Zawilak-Pawlik A, Messer W 2007. Regulation of the initiation of chromosomal replication in bacteria. FEMS Microbiol Rev 31: 378–387 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K 2009. Intrinsic histone–DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol 16: 847–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B 2000. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p–Dbf4p kinase. Mol Cell Biol 20: 3086–3096 [DOI] [PMC free article] [PubMed] [Google Scholar]





