Abstract
Polyamines are ubiquitous components of all living cells, and their depletion usually causes cytostasis, a strategy employed for treatment of West African trypanosomiasis. To evaluate polyamine depletion as an anti-malarial strategy, cytostasis caused by the co-inhibition of S-adenosylmethionine decarboxylase/ornithine decarboxylase in Plasmodium falciparum was studied with a comprehensive transcriptome, proteome, and metabolome investigation. Highly synchronized cultures were sampled just before and during cytostasis, and a novel zero time point definition was used to enable interpretation of results in lieu of the developmentally regulated control of gene expression in P. falciparum. Transcriptome analysis revealed the occurrence of a generalized transcriptional arrest just prior to the growth arrest due to polyamine depletion. However, the abundance of 538 transcripts was differentially affected and included three perturbation-specific compensatory transcriptional responses as follows: the increased abundance of the transcripts for lysine decarboxylase and ornithine aminotransferase and the decreased abundance of that for S-adenosylmethionine synthetase. Moreover, the latter two compensatory mechanisms were confirmed on both protein and metabolite levels confirming their biological relevance. In contrast with previous reports, the results provide evidence that P. falciparum responds to alleviate the detrimental effects of polyamine depletion via regulation of its transcriptome and subsequently the proteome and metabolome.
Polyamines such as putrescine, spermidine, and spermine are small organic compounds containing two or more amino groups. At physiological pH, these polycations interact electrostatically with numerous anionic macromolecules, thereby stabilizing DNA, RNA, nucleoside triphosphates (e.g. ATP), phospholipids, and proteins (1, 2). These interactions with polyamines can alter DNA conformation, regulate replication and transcription, strengthen membranes, regulate ion channels, and protect DNA and phospholipids from oxidative stress (1–5). Yet polyamines are also implicated in apoptosis (5). Polyamine depletion generally causes cytostasis or growth arrest, which implies that these molecules are involved in cell cycle progression and regulation, and it is speculated that polyamines regulate cyclin degradation (1, 6, 7). Therefore, polyamines are essential for cellular growth, differentiation, and macromolecular synthesis and are ubiquitous components of all living cells, except two orders of Archaea (1). Polyamine metabolism is particularly important in rapidly proliferating cells and has been exploited in the treatment of cancer (1) and parasitic diseases (8). Polyamine metabolism of the malaria parasite Plasmodium falciparum is also a potential target for therapeutic intervention (9, 10).
Polyamine and methionine metabolism are closely connected. This is particularly evident in Plasmodium where the two rate-limiting enzymes of polyamine biosynthesis, ornithine decarboxylase (ODC)2 and S-adenosylmethionine decarboxylase (AdoMetDC), form a single bifunctional protein (PfAdoMetDC/ODC, PF10_0322 (11)). ODC inhibition decreases putrescine levels in P. falciparum, whereas AdoMetDC inhibition decreases the levels of both spermidine and spermine (12, 13). The unique bifunctional nature of this complex could lead to the selective treatment of malaria. Inhibition of either ODC or AdoMetDC of P. falciparum in vitro causes cytostatic arrest in the trophozoite stage of the intraerythrocytic developmental cycle (IDC), but it does not cure Plasmodium berghei-infected mice in vivo (12, 14, 15). This is possibly because of exogenous polyamine salvage from the host (16). However, when ODC, AdoMetDC, and exogenous polyamine import are inhibited by the combination of bis(benzyl)polyamine analogues and dl-α-difluoromethylornithine (DFMO), 100% of P. berghei-infected mice were cured (17). A polyamine transporter has not yet been identified in the P. falciparum genome, but a drug combination selectively inhibiting both polyamine biosynthesis and transport may provide a promising anti-malarial strategy (16).
Transcriptional profiling can be used to assess the response of cells or organisms to environmental stress and to identify feedback mechanisms, alternative pathways, and metabolic buffering systems activated to cope with a perturbation (18, 19). This approach was applied with great success in the case of Mycobacterium tuberculosis, revealing transcriptional signatures specific to the mode-of-action for several antimycobacterial drugs (20). However, the correlation of perturbation-specific events in the transcriptome of the malaria parasite has been limited to a few studies (21–25), and support for a more prominent role of post-transcriptional control in plasmodial gene regulation is mounting (26–29).
Reported here is a comprehensive functional genomics investigation of P. falciparum during cytostasis after the co-inhibition of both catalytic sites of PfAdoMetDC/ODC with selective enzyme-activated, irreversible inhibitors (30). Cautious experimental design enabled transcriptional profiling and revealed very prominent polyamine metabolism-specific compensatory responses induced in the parasite to circumvent the perturbation. Convincingly, these were subsequently confirmed by proteomic and metabolomic analyses. This study therefore indicates that perturbation-specific events can be observed in the P. falciparum transcriptome and clearly links this to regulation in the parasite proteome and metabolome.
EXPERIMENTAL PROCEDURES
Parasite Cultures—Parasite culturing and sampling for the transcriptomics, proteomics, and metabolomics occurred independently, but perturbation conditions were replicated in terms of the parasite population, treatment, and sampling times. 3D7 P. falciparum parasites were cultured and synchronized (for three generations) according to established methods (31, 32). Parasites were treated in the late schizont stage (42 h post-invasion (hpi)) with 5 mm DFMO (IC50 = 1 mm) and 5 μm 5′-{[(Z)-4-amino-2-butenyl]methylamino}-5′-deoxyadenosine (MDL73811, IC50 = 1 μm) to ensure complete parasite arrest to prevent parasites escaping cytostasis and causing asynchrony (14). At these dosages the combined drug effect was additive and cytostatic (12, 14), but complete enzyme inhibition was confirmed with radiolabeled substrate assays (see below). DFMO was kindly provided by P. Woster (Wayne State University, MI) and MDL73811 by Sanofi-Aventis. A preceding small scale morphological study was performed to determine the exact sampling times, just before and during growth arrest. For the actual experiments treatment was performed in duplicate (i.e. two biological replicates, A and B) alongside untreated controls. Drug treated (T) and untreated (UT) cultures, at about 8–10% parasitemia and 2–3% hematocrit, were harvested at three time points within the trophozoite stage (t1 = 19 hpi, t2 = 27 hpi, and t3 = 34 hpi), based on the morphology of untreated parasites after microscopic inspection of Giemsa-stained thin smears. Culture medium (with and without drug) was replaced halfway through the time course but before the first sampling, to prevent metabolic stress of the parasites.
Radiolabeled Substrate Decarboxylase Assays—To ensure complete enzyme inhibition and presumably arrest of the cultures at the specified dosages, AdoMetDC and ODC activity of DFMO/MDL73811-treated and untreated cultures sampled at t1 = 19 hpi and t3 = 34 hpi were assessed as described (33), with uninfected erythrocytes as negative controls.
RNA Isolation and Microarray—Total RNA was isolated from frozen phosphate-buffered saline-washed pellets from 15-ml culture samples (two biological replicates) with the Qia-gen RNeasy kit, using a modified protocol including TRI Reagent® (Sigma) in the lysis step. Contaminating genomic DNA was removed with DNase I (Qiagen). A reference RNA pool was prepared from all three treated and untreated time points, including both sets of biological replicates. First strand cDNA synthesis was initiated from 12 μg of total RNA with 775 pmol of random primer 9 (New England Biolabs) and 250 pmol of oligo(dT)25 by incubation at 70 °C for 10 min followed by cooling on ice for 10 min. Reverse transcription and amino-allyl incorporation were performed at 42 °C as described (34), but the reaction was performed overnight using 480 units of SuperScript III (Invitrogen). Contaminating RNA was removed by hydrolysis with 0.5 m EDTA and 1 m NaOH at 65 °C for 15 min, and the reactions were purified with the Wizard SV Gel and PCR Clean-Up system (Promega). The cDNA obtained was divided into aliquots of 2 μg each, and where possible two technical array replicates were performed. The 2 μg of cDNA was coupled to either Cy3 (reference pool) or Cy5 (samples) at pH 9.0 (Amersham Biosciences). Free dye was removed with DNA Clean and Concentrator-5 columns (Zymo Research). DeRisi style 70-mer oligonucleotide arrays (34) were spotted in-house at the Lewis-Sigler Institute Microarray Facility (Princeton University). Post-processing, overnight hybridization at 65 °C, and washing were performed as described (34). Twenty arrays were scanned with an Axon GenePix 4000A scanner, and the images were analyzed with GenePix Pro 6.0 software (Axon Instruments).
Microarray Data Analysis—The GenePix default flagging parameters were applied in combination with visual inspection to assess spot quality, and flagged values received a zero weight. Array data were stored in the Princeton University Microarray data base and the NCBI Gene Expression Omnibus (35), accession number GSE13578. Exploratory data analysis was performed using CLUSTER and TREEVIEW software (36). Data were normalized, log transformed, and mean centered in Prin-ceton University Microarray data base and ordered according to the phase of expression (37, 38). Pearson correlation coefficients (r) were calculated in Excel. For differential abundance analysis, data quality and normalization methods were evaluated using data diagnostic tools from the MARRAY software package (39) in R on GenePix data. Background subtraction (offset = 50) and robust spline normalization were applied within each array, followed by Gquantile normalization between arrays because of the reference array design of the experiment. Differential abundance analysis was performed with linear models for microarray data (LIMMA) analysis within R (40, 41). With a common reference design LIMMA is similar to ordinary analysis of variance or multiple regression except that a linear model is fitted to the data for every oligonucleotide. Significance was calculated with moderated t statistics using a simple Bayesian model to make the analyses robust even for a small number of arrays (41). Differential abundance was calculated compared with UTt1, defined as relative time 0 (t0). Genes in at least one treated time point with transcript abundance greater than 1.7-fold (log2 ratio ≥0.75 or ≤ −0.75) in either direction compared with relative t0, and p values (adjusted for multiple comparison false discovery rate) of less than 0.05, were regarded as differentially affected. Data within these limits of transcripts represented by multiple oligonucleotides were averaged. The differentially affected transcripts were classified into functional groups using gene ontology terms obtained from DAVID (42) and PlasmoDB 5.3 (43). Furthermore, the transcript data were compared with the PlasmoDB 5.3 general feature format file to search for clusters of adjacently located genes with differentially affected transcripts, where a cluster was regarded as q4 or more genes within a window of six adjacent genes.
Microarray Validation with Real Time PCR—The differential abundance analysis was validated using a LightCycler 1.5 and FastStart DNA MasterPLUS SYBR Green I kit (Roche Applied Science). Six transcripts of interest were amplified, three transcripts with increased (PFL1885c, PFD0285c, and PFF0435w) and three with decreased (PF08_0131, PFD0830w, and PFI1090w) abundance. Starting levels across different samples were equalized relative to a putative cyclophilin (PFE0505w), which remained unchanged in the array data as well as in the IDC transcriptome (38).
Protein Extraction and Two-dimensional Gel Electrophoresis Separation—Parasites for two-dimensional gel electrophoresis were released with 0.05% saponin from 60-ml culture samples (two biological replicates) and washed with phosphate-buffered saline to reduce contamination with erythrocyte proteins. Two-dimensional gel electrophoresis was performed as described (44) with a few modifications. Two biological replicate extracts were combined to obtain enough protein for three to four replicate gels. The protein concentration was determined with a two-dimensional Quant kit (Amersham Biosciences), and 400 μg of total protein was applied to 18-cm Immobilon DryStrip gels (pH 3–10, GE Healthcare). The first dimension separation was performed with a Ettan IPGphor II isoelectric focusing system with active rehydration at 30 V for 10 h followed by a gradual step-and-hold increase to 8000 V for a total of 24,000 V h. The run was terminated after 35,000 V h. The second dimension separation was performed on 10% vertical SDS-polyacrylamide gels with a Hoefer SE 600 vertical system at 80 mA and 20 °C. The gels were fixed with 40% ethanol, 10% acetic acid overnight and stained with in Flamingo fluorescent stain (Bio-Rad). Twenty one gels were scanned with a Pharos FX Plus molecular imager at high and medium intensity (photomultiplier tube voltage), and the best three of four were selected for spot matching and differential spot analysis.
Proteomics Data Analysis—Data analysis was performed with PDQuest 8.0 Advanced software (Bio-Rad). Roller-ball background subtraction, loess normalization, and automated spot detection and matching were performed. The automated spot matching was carefully checked by visual inspection and comparison for maximum consensus within each replicate group. A master image was generated, including all replicate groups to be compared. Differential protein abundance was calculated in comparison with UTt1 (relative t0). Proteins in at least one treated time point with an abundance greater than 2-fold in either direction compared with relative t0, and p values of less than 0.05 (Student’s t test), were regarded as differentially affected. Correlation coefficients (R) of the regression line between replicate groups were calculated in PDQuest.
Spot Excision and MALDI-MS/MS—Spots of interest were excised, purified, trypsin-digested and prepared for MALDI-MS/MS as described (44), followed by peptide analysis with a QStar Elite instrument (Applied Biosystems) with a MALDI source. The instrument was calibrated with a commercially available peptide calibration standard ranging from ∼1000 to 3200 Da (Bruker Daltonics). Peptide mass finger prints and MS/MS fragments were compared with a nonredundant protein data base (Swiss-Prot/TrEMBL) using MASCOT with oxidation (methionine) and carbamidomethylation set as protein modifications, only one missed trypsin cleavage accepted and a mass tolerance of 50 ppm. The probability-based MOWSE score was used to estimate the significance of the identification with p < 0.05.
Metabolite Extraction and LC-MS/MS—From 20-ml culture samples (two biological replicates), 10 ml was used for general metabolite analysis, and 10 ml was derivatized for polyamine analysis. Cultures were pelleted and serially extracted immediately thereafter, first with 4 volumes of 100% methanol at −75 °C for 15 min and then twice more with 1 volume 80:20 methanol/water at 4 °C. For the second and third extractions, the cell/methanol mixture was sonicated for 15 min on ice in a water bath sonicator. The supernatants from each extraction were pooled and centrifuged free of cell debris and protein. All samples were analyzed within 24 h of their generation and were analyzed for 167 metabolites as described (45, 46), as well as putrescine, spermidine, spermine, cadaverine, and decarboxylated AdoMet (dcAdoMet). The dcAdoMet standard used for calibration was a kind gift by K. Samejima (Josai University, Japan).
For polyamine analysis, 10 μl of triethylamine was added to 100 μl of cell extract and mixed. A few crystals of solid succinic anhydride were then added, and the mixture was vortexed vigorously. The reaction was allowed to proceed for 1 h at room temperature, with vortexing every 10 min. After this incubation the samples were centrifuged to pellet any remaining solid succinic anhydride, and the liquid portion of the sample was subjected to LC-MS/MS analysis using parameters previously determined with pure stock solutions of putrescine, cadaverine, spermidine, and spermine.
LC-MS/MS was performed using an LC-10A high pressure liquid chromatography system (Shimadzu) and a Luna aminopropyl column (250 × 2 mm with a 5-μm particle size) from Phenomenex coupled to the mass spectrometer. The LC parameters were as follows: autosampler temperature, 4 °C; injection volume, 20 μl; column temperature, 15 °C; and flow rate, 150 μl/min. The LC solvents were solvent A (20 mm ammonium acetate + 20 mm ammonium hydroxide in 95:5 water/acetonitrile, pH 9.45) and solvent B, acetonitrile. The gradients are as follows: positive mode, t =0, 85% B; t =15 min, 0% B; t = 28 min, 0% B; t = 30 min, 85% B; t = 40 min, 85% B; and negative mode, t = 0, 85% B; t = 15 min, 0% B; t = 38 min, 0% B; t = 40 min, 85% B; t = 50 min, 85% B.
Mass spectrometric analyses were performed on a Finnigan TSQ Quantum Ultra triple-quadrupole mass spectrometer (Thermo Electron Corp.), equipped with an ESI source. ESI spray voltage was 3200 V in positive mode and 3000 V in negative mode. Nitrogen was used as sheath gas at 30 p.s.i. and as the auxiliary gas at 10 p.s.i., and argon as the collision gas at 1.5 millitorr, with the capillary temperature 325 °C. Scan time for each single reaction monitoring event transition was 0.1 s with a scan width of 1 m/z. The LC runs were divided into time segments, with the single reaction monitoring scans within each time segment limited to those compounds eluting during that time interval. For compounds eluting at the boundaries between time segments, the single reaction monitoring scan corresponding to the compound is conducted in both time segments. The instrument control, data acquisition, and data analysis were performed by the Xcalibur software (Thermo Electron Corp., version 1.4 SR1), which also controlled the chromatography system.
Metabolomics Data Analysis—Raw data peak quantitation was performed by Xcalibur software with 103 as the quantitation limit. Biological replicate data were averaged, background subtracted, and normalized to relative t0 in Excel. Metabolites with a fold change of 2 were regarded as changed.
RESULTS
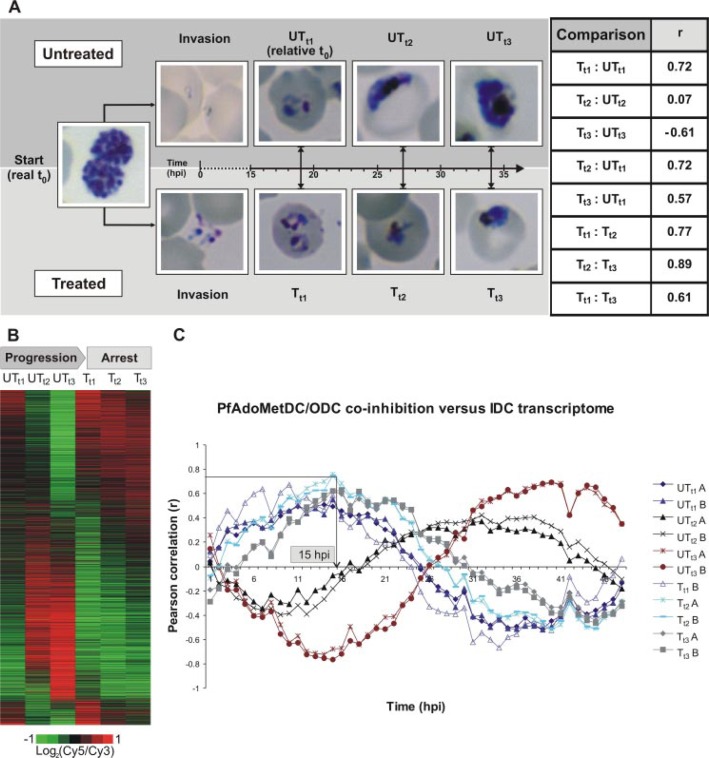
The global response of P. falciparum during cytostasis resulting from PfAdoMetDC/ODC co-inhibition was examined. Both catalytic sites of the bifunctional enzyme were simultaneously inhibited with MDL73811 (AbeAdo) and DFMO, respectively. The functional genomics investigation was preceded by enzyme activity and morphological studies of drug-treated (T) versus untreated (UT) parasites to ensure complete growth arrest at the treatment dosage and to determine sampling times. Complete enzyme inhibition of both AdoMetDC and ODC was confirmed because no decarboxylase activity was observed in the treated samples (Tt1 to Tt3) compared with increasing enzyme activity in UTt1 to UTt3 (results not shown). Growth arrest was observed morphologically from the trophozoite stage with no visible effect during the ring stage, corresponding with previous reports (12, 14). Parasites were subsequently sampled in the early (t1 = 19 hpi), mid (t2 = 27 hpi), and mature (t3 = 34 hpi) trophozoite stages such that the normal expression period of PfAdoMetDC/ODC (12–40 hpi) was spanned (Fig. 1A).
FIGURE 1.
Transcriptional arrest prior to cytostasis. A, Giemsa-stained thin smears of treated (T) and untreated (UT) P. falciparum cultures. DFMO/MDL73811 treatment was initiated in the schizont stage at about 42 hpi (real t0) and sampled at t1 = 19 hpi, t2 = 27 hpi, and t3 = 34 hpi. Pearson correlation coefficients (r) of transcriptome data are tabled. The close correlations between UTt1 and Tt1 to Tt3 compared with the low correlation with the matched untreated controls at t2 and t3 is the result of transcriptional arrest at t1 (technical replicates correlated at 0.93 and biological replicates at 0.88 on average). However, growth arrest was morphologically observed only from t2 (Tt2 to Tt3). The transcriptional arrest negated direct comparison of parallel treated and untreated time points at t2 and t3, and UTt1 was defined as a relative t0 for quantitative differential abundance analysis. B, phaseogram depicting the transcriptional profiles over the three time points (t1 to t3) by ordering 3206 oligonucleotides according to the phase of expression. Transcriptional arrest is visible in Tt1 to Tt3. C, Pearson correlation between the PfAdoMetDC/ODC co-inhibition data and the 1-h time points of the 3D7 IDC transcriptome. All three treated samples have a correlation profile similar to UTt1 (relative t0), which corroborates the transcriptional arrest and also indicates the approximate time thereof as ∼15–16 hpi. Data of the respective biological replicates (A and B) are shown separately (Tt1 had only one biological replicate because of technical difficulty).
Transcriptome Analysis Reveals Transcriptional Arrest—Transcriptome analyses of treated and untreated parasites harvested at the abovementioned time points were performed with oligonucleotide-based DNA microarray containing 8088 70-mer oligonucleotides and using a reference design (34). Fast Fourier analysis has been applied previously to calculate the apparent phase and frequency of gene expression in the IDC transcriptome (37, 38). Subsequently, by ordering the PfAdoMetDC/ODC co-inhibition transcriptome data according to the phase of expression, as determined for the 3D7 strain (38), cytostasis was revealed as a generalized transcriptional arrest across Tt1 to Tt3 with normal IDC progression visible in UTt1 to UTt3 (Fig. 1B). In general, expression of genes that were already transcribed before the effect of the treatment (IDC peak expression before or around t1) was unchanged, whereas genes that were not yet transcribed (IDC peak expression in second half of 48-h cycle, e.g. t2 and t3) showed no or low transcript abundance.
Pearson correlation (r) of the data with each of the 1-h time points of the 3D7 IDC transcriptome (38) indicated the highest correlation of UTt1 (19 hpi) at 14–15 hpi of the IDC data with the three treated samples following closely thereafter at 15–16 hpi (Fig. 1C). This correlation with the IDC aligned the two data sets in terms of expression times and indicated the approximate time of transcriptional arrest. Thus, all three treated samples were arrested at around t1 (Fig. 1, B and C), and correlation between the treated data was relatively close (Tt1 versus Tt2, r = 0.77; Tt2 versus Tt3, r = 0.89; Tt1 versus Tt3, r = 0.61 (Fig. 1A)). Pearson correlation was also calculated between the treated and untreated data to estimate the impact of the drug treatment on the parasite transcriptome. UTt1 versus Tt1 correlated at 0.72, but due to transcriptional arrest of treated and normal progression of untreated parasites, UTt2 versus Tt2 was uncor-related (r = 0.07) and UTt3 versus Tt3 was anti-correlated (r = −0.61, Fig. 1A) after 40 h of treatment. Conversely, in the absence of transcriptional arrest array data of doxycycline-treated versus untreated P. falciparum was still highly correlated (r = 0.8) after 55 h (47). As expected, growth arrest occurred after transcriptional arrest and was morphologically visible only from Tt2 with parasites looking distressed compared with their untreated counterparts (Fig. 1A).
Differential Transcript Abundance Despite Transcriptional Arrest—To identify the transcripts with differential abundance within the transcriptome as a result of the perturbation, a quantitative approach was followed with LIMMA analysis within R. The global transcriptional arrest caused by PfAdoMetDC/ODC co-inhibition negated the direct comparison of treated and untreated data at t2 and t3, as this standard approach would have detected mainly growth/stage differences. Therefore, in differential transcript abundance analysis, all treated time points were compared with UTt1, regarded as a relative t0, mediated by the reference design of the microarray. Comparison to the real t0 would also primarily have identified stage differences because drug treatment was performed in the late schizont stage, but cytostasis and sampling occurred in the subsequent trophozoite stage. Over the time course, the abundance of 538 transcripts from 5332 unique genes represented on the array were significantly affected compared with the relative t0 (supplemental Table S1). Of these, 171 transcripts were increased (up to 3.2-fold), and 377 were decreased (down to 5-fold), with the transcripts of 10 apparently unrelated genes falling in both categories by displaying an increase in t1 as well as a decrease in t2 and/or t3. A selected subset of the differentially affected transcripts is presented (Table 1), including eight transcripts from polyamine and methionine metabolism and three methyltransferases.
TABLE 1.
Biological functions of a subset of transcripts with differential abundance due to PfAdoMetDC/ODC co-inhibition
| PlasmoDB ID | Annotation | Fold changea | IDC time of peak expression |
|---|---|---|---|
| hpi | |||
| Polyamine and methionine metabolism | |||
| PF10_0322 | PfAdoMetDC/ODC | −1.9 | 24 |
| PFD0285c | LDC | 2.8 | 25 |
| PFF0435w | OAT | 1.9 | 18 |
| PFI1090w | AdoMet synthetase | −2.4 | 27 |
| PFE1050w | Adenosylhomocysteinase | −1.9 | 33 |
| PF10_0289 | Adenosine deaminase, putative | −2.6 | 27 |
| PFE0660c | Uridine phosphorylase, putative | −3.2 | 27 |
| PF10_0340 | Methionine-tRNA ligase | −1.7 | 33 |
|
| |||
| Methyltransferases | |||
| MAL13P1.214 | Phosphoethanolamine N-methyltransferase, putative | −2.8 | 40 |
| PF14_0309 | Protein-l-isoaspartate O-methyltransferase, putative | −1.9 | 41 |
| PF14_0526 | Generic methyltransferase | −2.9 | 37 |
|
| |||
| Potential polyamine associated effects | |||
| PF14_0316 | DNA topoisomerase II | −1.7 | 50 |
| PFL1885c | Calcium/calmodulin-dependent protein kinase 2, putative | 2.4 | 50 |
| PF07_0065 | Zinc transporter, putative | −2.4 | 40 |
|
| |||
| Oxidative stress defense | |||
| PF08_0131 | 1-Cys-peroxidoxin | −3.2 | 34 |
| PF14_0192 | Glutathione reductase | −1.7 | 34 |
| PF14_0187 | Glutathione S-transferase | −1.8 | |
|
| |||
| Energy metabolism | |||
| Oxidative phosphorylation | |||
| CoI | Cytochrome oxidase I, putative | −2.0 | |
| CoxI | Cytochrome oxidase I, putative | −1.7 | 53 |
| CoxIII_2 | Mitochondrial encoded cytochrome oxidase subunit 3 | −2.0 | |
| PF11_0412 | Vacuolar ATP synthase subunit F, putative | −1.8 | 34 |
| vMAL7P1.75 | Mitochondrial ATP synthase F1, ∊ subunit, putative | −1.8 | |
| PFE0970w | Cytochrome c oxidase assembly, putative | −1.7 | 24 |
| PF13_0121 | Dihydrolipoamide succinyltransferase | −1.7 | 27 |
| Glycolysis | |||
| PF10_0155 | Enolase | −2.0 | 18 |
| PF13_0141 | l-Lactate dehydrogenase | −1.8 | 18 |
| PF14_0378 | Triose-phosphate isomerase | −1.8 | 18 |
| PF14_0598 | Glyceraldehyde-3-phosphate dehydrogenase | −2.1 | 27 |
| PFF1300w | Pyruvate kinase | −1.9 | 28 |
|
| |||
| DNA replication | |||
| PF11_0117 | Replication factor C subunit 5, putative | −2.1 | 34 |
| PF11_0087 | Rad51 homologue, putative | −2.0 | 35 |
| PF13_0095 | DNA replication licensing factor mcm4-related | −2.2 | 42 |
| PF13_0291 | Replication licensing factor, putative | −1.8 | 34 |
| PF14_0081 | DNA repair helicase, putative | −1.7 | |
| PF14_0112 | POM1, putative | −2.0 | 37 |
| PF14_0254 | DNA mismatch repair protein Msh2p, putative | −1.8 | 32 |
| PF14_0314 | Chromatin assembly factor 1 p55 subunit, putative | 2.4 | |
| PF14_0601 | Replication factor C3 | −2.0 | 34 |
| PFB0180w | 5′–3′-Exonuclease, putative | −1.9 | 26 |
| PFB0895c | Replication factor C subunit 1, putative | −2.0 | 33 |
| PFD0470c | Replication factor A protein, putative | −2.5 | 34 |
| PFD0685c | Chromosome-associated protein, putative | −2.0 | 40 |
| PFD0830w | Dihydrofolate reductase-thymidylate synthase | −2.0 | 33 |
| PFD0950w | Ran binding protein 1 | 2.0 | |
| PFE0675c | DNA photolyase | −2.2 | 36 |
| PFF1470c | DNA polymerase ∊, catalytic subunit A, putative | −1.7 | 36 |
| PFI0235w | Replication factor A-related protein, putative | −1.8 | 33 |
| PFI0530c | DNA primase, large subunit, putative | −1.8 | 35 |
|
| |||
| Transcription factors | |||
| PF11_0241 | Hypothetical protein with Myb-like domains | 1.8 | |
| PFL0465c | C2H2-type zinc finger transcription factor, krox1 | 2.0 | |
| PFE1245w | CCCH-type zinc finger protein | 1.7 | 26 |
| PFD0560w | Hypothetical protein with a TATA box-like domain | 1.7 | 32 |
| PFE0415w | Transcription factor IIB, putative | −1.8 | |
|
| |||
| Translation | |||
| MAL13P1.327 | Ribosomal protein S17 homologue, putative | 1.7 | 22 |
| PF07_0080 | 40 S ribosomal protein S10, putative | 1.9 | 16 |
| PF10_0038 | Ribosomal protein S20e, putative | 2.3 | 15 |
| PF11_0454 | Ribosomal protein, 40 S subunit, putative | 2.0 | |
| PF13_0014 | 40 S ribosomal protein S7 homologue, putative | 1.8 | 15 |
| PF13_0171 | 60 S ribosomal protein L23, putative | 2.0 | 13 |
| PF13_0228 | 40 S ribosomal subunit protein S6 | 1.8 | 13 |
| PF14_0205 | Ribosomal protein S25 | 2.4 | 23 |
| PF14_0231 | Ribosomal protein L7a, putative | 1.8 | 21 |
| PF14_0579 | Ribosomal protein L27, putative | 2.1 | 21 |
| PF14_0709 | Ribosomal protein L20, putative | −2.6 | 32 |
| PFB0455w | Ribosomal L37ae protein, putative | 1.7 | 16 |
| PFC0535w | 60 S ribosomal protein L26, putative | 2.0 | |
| PFC1020c | 40 S ribosomal protein S3A, putative | 1.7 | 15 |
| PFE0185c | 60 S ribosomal subunit protein L31, putative | 1.8 | 14 |
| PFI1585c | 30 S ribosomal protein S6-like protein, putative | 1.8 | |
|
| |||
| Cell cycle mediators | |||
| PF13_0328 | Proliferating cell nuclear antigen | −3.3 | 40 |
| PF14_0604 | Hypothetical protein with cyclin homology | −1.7 | 2 |
| PFL1330c | Hypothetical protein with cyclin homology | 1.8 | 37 |
Average fold change was calculated at the time point of maximum change.
The 538 transcripts were classified into 14 functional groups (supplemental Fig. S1) using gene ontology terms obtained from DAVID and PlasmoDB. The transcripts with the most increased abundance were related to RNA metabolism (9%), translation (10%), and host/parasite interaction (11%), whereas those with decreased abundance mostly represented DNA (7%) and primary metabolism (8%, including carbohydrate, lipid, and energy metabolism). The increase of transcripts associated with host/parasite interaction (including surface antigens) is regarded as a general stress response (22, 48). Transcripts related to mitochondrial and plastid metabolism, including organellar translation, were generally decreased, whereas those related to ribosomal translation were increased. As expected with growth arrest, cell cycle regulators were affected, which included three cyclin-associated transcripts (Table 1). The limited annotation status (only ∼40%) of the P. falciparum genome (49) was also reflected in the dataset with 51% of the transcripts encoding hypothetical proteins with unknown biological function.
The accuracy of the dataset was validated with real time PCR, which confirmed the differential abundance of three increased and three decreased transcripts, including lysine decarboxylase (LDC, PFD0285c), OAT (ornithine aminotransferase, PFF0435w), and S-adenosylmethionine synthetase (AdoMet synthetase, PFI1090w) (supplemental Table S2). Relatively low abundance transcripts within the dataset were validated by the inclusion of LDC and dihydrofolate reductase/thymidylate synthase (DHFR/TS, PFD0830w) in the real time PCR strategy.
Perturbation-specific Transcriptional Responses—The majority (70%) of the differentially affected transcripts were decreased and similarly so for polyamine and methionine metabolism, with the abundance of only two transcripts, LDC and OAT, being increased by 2.8- and ∼2-fold, respectively. The transcript level for PfAdoMetDC/ODC, the protein that was targeted by DFMO and MDL73811, was decreased by ∼2-fold. Thus, despite the transcripts for LDC and PfAdoMetDC/ODC being expressed at approximately the same time in the IDC (25 and 24 hpi, respectively), the transcript for LDC was increased and that of PfAdoMetDC/ODC was decreased, which illustrates the differential effects of the co-inhibition on the abundance of specific transcripts. Based on reports in other systems, the increase of the transcripts for LDC (50) and OAT (51) and the decrease of that for AdoMet synthetase (52) were regarded as compensatory to alleviate the effects of the perturbation.
Interestingly, several of the decreased abundance transcripts translate to proteins that are known to require polyamines for optimal functioning, protection, or gene expression in other organisms (Table 1). These, for example, include the transcript for DNA topoisomerase II (PF14_0316) (53), three oxidative stress defense transcripts (54), and transcripts involved with zinc transport and energy metabolism (55), which were all decreased.
The possible enrichment of the differentially affected transcript data for transcripts of proteins functionally connected to polyamine and methionine metabolism were investigated by comparison with the in silico predicted interactome of PfAdoMetDC/ODC (56). These networks were constructed using among others the IDC transcriptome (37, 56). Sixty percent (12:20) of the top 20 (highest probability) scored binding partners of PfAdoMetDC/ODC was found within the dataset of 538 (supplemental Table S3). In contrast, the transcripts of only 10% (2:20) of the top 20 binding partners of another unrelated bifunctional protein (dihydropteroate synthase/dihydroxym-ethylpterin pyrophosphokinase (PF08_0095)) were present among the 538, thereby excluding random overlap (supplemental Table S3). Although the interactome is a theoretical interaction prediction that requires experimental verification, these analyses may suggest enrichment of the differentially affected transcript data for transcripts of proteins that interact with PfAdoMetDC/ODC and/or are functionally related to polyamine and methionine metabolism.
During data analysis, it was noticed that several of the differentially affected transcripts were encoded by genes that were physically located adjacently or in close proximity at the chromosomal level. The dataset of 538 was therefore evaluated for clusters of adjacently located genes, where a cluster was defined as four or more genes within a window of six adjacent genes with transcripts within the dataset. Seven such adjacent gene clusters were found among the decreased abundance transcripts and none among the increased abundance transcripts. The seven clusters were distributed over chromosomes 7, 10, and 11, respectively (supplemental Table S4). Particularly significant was a cluster of 11 genes on chromosome 10 (PF10_0014 to PF10_0025) that lie back-to-back on the same strand, with only one gene (PF10_0018) missing in the entire stretch of ∼41,000 bases. PF10_0018 produces a low abundance transcript, which was most likely affected in the same way but was not detected. Decreased transcription of co-localized genes may be due to a common transcription factor that became nonfunctional in the absence of polyamines. However, the genes within these clusters are not all co-expressed according to the IDC transcriptome and nuclear expression of contiguous genes are rarely co-regulated in P. falciparum (37).
Perturbation-specific Compensatory Mechanisms Confirmed in the Proteome—To further probe the effects of the perturbation, PfAdoMetDC/ODC co-inhibition was repeated, and the effects were assessed on the parasite proteome with two-dimensional gel electrophoresis. As in the transcriptome, cytostasis resulted in a high correlation between the gels of the relative t0 (UTt1) and the three treated time points (UTt1 versus Tt1, r = 0.93; UTt1 versus Tt2, r = 0.88; UTt1 versus Tt3, r = 0.88) and a lower correlation with UTt2 and UTt3 (UTt1 versus UTt2, r =0.78; UTt1 versus UTt3, r = 0.70). The effects of cytostasis on the proteome were more subtle than in the transcriptome, but the perturbation caused an overall decrease in the number of proteins detected over the time course (UTt1 = 483 spots; Tt1 = 461; Tt2 = 409; Tt3 = 416).
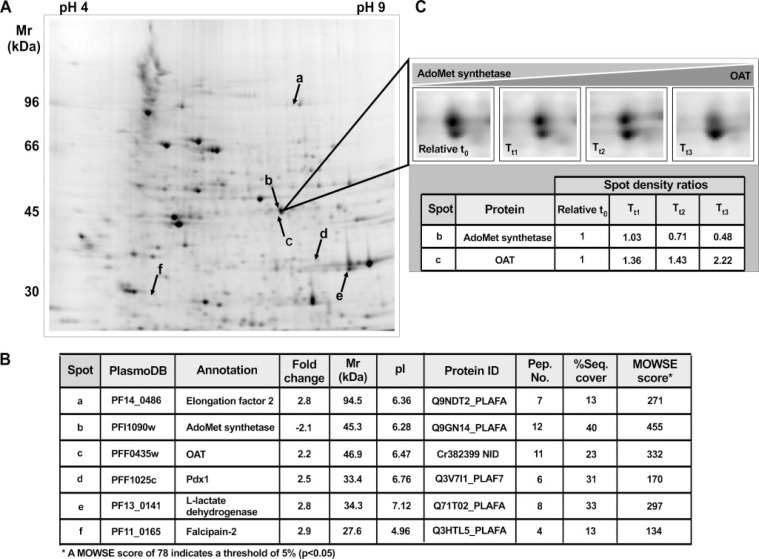
As with the transcriptome analysis, the effects of PfAdoMetDC/ODC co-inhibition on the proteome were quantified with differential protein abundance analysis compared with the defined relative t0. Forty one spots with significant differential abundance were excised (supplemental Table S5), of which the majority had low molecular weight and low intensity. Similar to another two-dimensional gel electrophoresis plasmodial study (57), around 30% of the spots could be identified with MALDI-MS/MS. The identification scores and characteristics of a subset of the differentially affected proteins are provided (Fig. 2B). These include proteins involved with poly-amine and methionine metabolism, namely AdoMet synthetase, OAT, and pyridoxal 5′-phosphate (PLP) synthase (Pdx1, PFF1025c). The latter protein was increased 2.5-fold and synthesizes PLP, which is an important co-factor for both PfAdoMetDC/ODC and LDC (58). The differential protein abundance of OAT and AdoMet synthetase (Fig. 2C) correlated with their transcript abundance after PfAdoMetDC/ODC co-inhibition (Table 1), which confirmed these compensatory mechanisms in the proteome and furthermore suggested transcriptional control of these proteins. For other proteins (e.g. falcipain-2 (PF11_0165) and Pdx1), incremental changes on the transcript level (therefore not present in supplemental Table S1) resulted in significant changes on the protein level (Fig. 2B). Furthermore, the transcriptional response was sometimes delayed in the proteome, e.g. the lactate dehydrogenase (PF13_0141) transcript was decreased 1.8-fold in the treated parasites at t1, whereas the protein was increased 2.8-fold. However, during the time course the lactate dehydrogenase protein gradually decreased to the same level as that of the relative t0 (i.e. unchanged). The same effect was observed for elongation factor 2 (PF14_0486). This delay in translation (translational gap) was also reported, specifically for these two proteins, in a comparative study of the Plasmodium transcriptome and proteome (59), and implicates post-transcriptional regulation of expression for these specific proteins (59).
FIGURE 2.
Two-dimensional gel electrophoresis illustrated with a typical gel image (UTt1_tech_repl_1) indicating the positions of a subset of proteins (A), the protein identities and characteristics (B), and an enlarged view of AdoMet synthetase and OAT (just below) over the time course, including the respective spot densities (C). Spot density ratios were calculated compared with UTt1 (relative t0) with p < 0.05. Protein ID = the MASCOT search identifier; Pep. No. = the number of peptides identified in the mass spectrum.
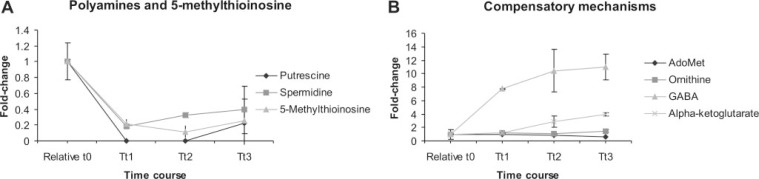
Perturbation-specific Compensatory Mechanisms Confirmed in the Metabolome—The effects of PfAdoMetDC/ODC co-inhibition were also evaluated in the parasite metabolome by assaying 172 metabolites over the time course with LC-MS/MS as described (45, 46). Reliable data were obtained for 92 metab-olites (supplemental Table S6) with the balance excluded because of levels below the detection or set quantitation limit. Differential metabolite abundance was again quantified compared with the relative t0, and 24 metabolites were changed at least 2-fold (supplemental Table S6), although many of these were similarly affected in the untreated controls. Compellingly, the perturbation-specific effects of PfAdoMetDC/ODC co-inhibition were specifically observed in the parasite metabolome by the significant decrease of the polyamines (putrescine and spermidine, Fig. 3A) in the treated parasites compared with increasing levels in the untreated controls (supplemental Table S6). Moreover, downstream metabolites, including 5-methyl-thioinosine (Fig. 3A), were also decreased, corroborating the complete metabolic halt of polyamine metabolism after the co-inhibition.
FIGURE 3.
Selected metabolite profiles during cytostasis induced by PfAdoMetDC/ODC co-inhibition. A, putrescine, spermidine, and 5-methylthioinosine levels decreased as expected, but cadaverine and spermine were too low for reliable detection. B, AdoMet and ornithine levels did not accumulate despite PfAdoMetDC/ODC co-inhibition, but ornithine enters glutamate metabolism via OAT, and the glutamate metabolites, α-ketoglutarate and GABA, increased.
The proposed compensatory responses of LDC, OAT, and AdoMet synthetase were also investigated in the metabolome. The increased abundance in LDC transcripts could indicate the functional production of cadaverine from lysine by this protein in P. falciparum, but cadaverine could not be detected. A reduced production of AdoMet synthetase was observed after complete inhibition of PfAdoMetDC/ODC, and metabolome analysis revealed no change in the levels of AdoMet (Fig. 3B). In other organisms, inhibition of AdoMetDC caused an increase of the substrate AdoMet (60); however, in Plasmodium AdoMet homeostasis is apparently maintained by decreasing the synthesis thereof through a reduced production of AdoMet synthetase as reflected in the decreased transcript and protein abundance. Moreover, ornithine levels remained unchanged (Fig. 3B) and appeared to be regulated through a compensatory increase in OAT as also detected in both the transcriptome and the proteome. The observed increase of the glutamate metab-olites, γ-aminobutyrate (GABA) and α-ketoglutarate, may additionally be due to increased ornithine degradation via OAT (Fig. 3B). However, both these metabolites were also significantly increased in the untreated parasites, and GABA was recently reported to be one of the most abundant metabolites in isolated trophozoites (61). Several other relevant metabolites, namely spermine, dcAdoMet, and PLP, were not detected or could not be reliably quantified.
DISCUSSION
Cytostatic drugs, including DFMO, have been therapeutically used in the treatment of protozoan diseases such as West African sleeping sickness (8). Global expression profiling after treatment with cytostatic drugs has been performed in cancer (62, 63) but not in multistage organisms such as P. falciparum. Although the cytostatic effects of DFMO and MDL73811 are well established, the exact mechanism by which the induced polyamine depletion results in growth inhibition is not clear (64). In this investigation, the transcriptional arrest preceding and resulting in cytostatic growth arrest due to polyamine depletion was demonstrated for the first time to our knowledge in any organism. P. falciparum is a multistage organism, and the transcriptional arrest of highly synchronized treated parasites compared with normal transcriptional progression of untreated parasites was clearly visible when the transcript data were ordered according to peak expression times within the IDC. Pearson correlation calculations indicated the approximate time of transcriptional arrest to occur at about 15–16 hpi, thus late ring/early trophozoite stage, which correlates to the estimated time of PfAdoMetDC/ODC expression, indicating the perturbation-specific effect only after production of the targeted protein. The exact mechanism by which polyamine depletion results in transcriptional arrest requires further elucidation, but their importance in macromolecular synthesis (including RNA and proteins, e.g. transcription factors) (33), optimal ribosome functioning (65), and the association of the main fraction of polyamines with RNA (2) is well known.
The transcriptional arrest of treated and normal progression of untreated parasites negated direct comparison at t2 and t3, as these would have mainly encompassed growth/stage differences. Therefore, in differential abundance analysis, all treated time points were compared with UTt1, regarded as a relative t0 and reference point for quantitative analysis. This critical principle was applied through the whole functional genomics investigation. Despite the generalized transcriptional arrest, the abundance of 538 transcripts was significantly changed with fold changes ranging between 3.2-fold up and 5-fold down. This is in agreement with other transcriptome reports of perturbed Plasmodium where relatively small amplitude transcriptional responses were detected, especially in the increased abundance datasets (22, 48).
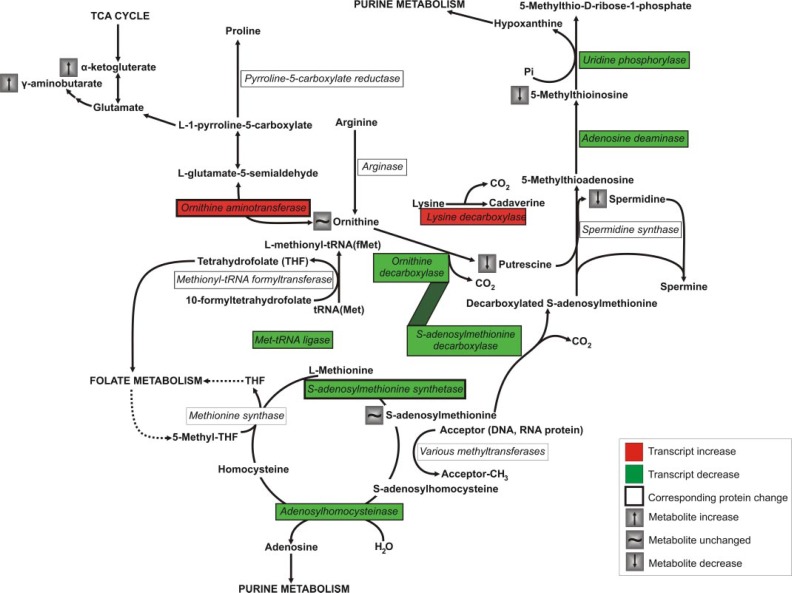
The most dramatic perturbation-specific transcriptional responses were the significant increase of the transcripts for LDC and OAT and the decrease of the transcript for AdoMet synthetase (Fig. 4). Lysine decarboxylation produces cadaver-ine, a diamine, and structural analogue of putrescine. LDC activity and cadaverine accumulation have been reported to alleviate ethylene inhibition of arginine decarboxylase and AdoMetDC in pea seedlings (50) and 0.4 mm cadaverine partially reversed DFMO-induced growth arrest in Plasmodium (14). LDC induction as compensatory mechanism for polyamine depletion in Plasmodium remains to be confirmed because the protein could not be detected by two-dimensional gel electrophoresis or cadaverine with LC-MS/MS. However, the possible inhibition of the induced LDC via DFMO, as in Selenomonas ruminantum (66), cannot be excluded. The transcripts for LDC and PfAdoMetDC/ODC are expressed at approximately the same time in P. falciparum (25 and 24 hpi, respectively), which is expected should LDC serve as a compensatory mechanism for polyamine/diamine biosynthesis. The increased transcript abundance of LDC may indicate a potential resistance mechanism should PfAdoMetDC/ODC be clinically targeted in the future.
FIGURE 4.
Polyamine and methionine metabolism (adapted from Malaria Parasite Metabolic Pathways). Plasmodial spermine synthesis is currently believed to be catalyzed by spermidine synthase as indicated (75). Enzymes of which the transcript abundance was significantly increased are indicated in red and those significantly decreased are indicated in green, whereas proteins with confirmed corresponding abundance are framed with a thick border. Metabolites that were unchanged or that were increased or decreased at least 2-fold are indicated.
The increased levels of OAT transcripts and protein observed in this study and the maintenance of ornithine concentrations in the metabolome provide evidence for compensatory effects of OAT in PfAdoMetDC/ODC co-inhibited P. falciparum. OAT catalyzes both the synthesis of ornithine from glutamate-5-semialdehyde when levels are low as well as its degradation to proline and glutamate when present in excess (67) to prevent toxic ornithine accumulation (51).
DFMO/MDL73811 treatment of trypanosomes caused a 40-fold accumulation of AdoMet (68), which resulted in speculation on hypermethylation of nucleic acids and/or proteins being the main anti-trypanosomal mechanism of MDL73811 (60). However, this study shows that AdoMet concentrations were maintained in P. falciparum. Several mechanisms could restore such metabolic homeostasis during a perturbation such as regulation of enzyme activity or protein production. This study revealed a decrease of both the transcript and protein of AdoMet synthetase that may act as a compensatory strategy induced to maintain AdoMet levels. The exact mechanism behind this regulation needs to be elucidated, but in MDL73811-treated mammalian cells the AdoMet concentration was effectively regulated through substrate feedback inhibition of AdoMet synthetase activity (52, 60). In contrast, the trypanosomal enzyme is apparently poorly regulated resulting in the substantial accumulation of AdoMet after AdoMetDC inhibition (60). Plasmodial AdoMet synthetase activity may therefore also be regulated by AdoMet similar to mammalian systems, but this needs to be confirmed. Moreover, the proposed difference in regulation of AdoMet synthetase between Trypanosoma and Plasmodia, resulting in hypermethylation with polyamine depletion in the first case and possibly only polyamine depletion in the second case, may be the reason for the success of MDL73811 in Trypanosoma brucei rhodesiense-infected mice (69) and failure of MDL73811 in P. berghei-infected mice (12).
The coordinated response on the transcript, protein, and metabolite levels as demonstrated for AdoMet synthetase and OAT is evidence that the parasite is able to respond in a programmed manner on the transcriptional level to perturbation of polyamine and methionine metabolism. This is corroborated by other transcriptome studies of polyamine-depleted P. falciparum. Increased transcript levels of OAT was also reported after treatment with DFMO alone (70), and inhibition of spermidine synthase (the enzyme downstream to PfAdoMetDC/ODC) also resulted in increased transcript levels of LDC.3 In contrast, the transcripts for neither OAT, LDC, nor AdoMet synthetase were differentially affected after exposure of P. falciparum to a variety of other perturbations including a series of anti-malarial drugs and environmental stressors (23, 25, 48).4 It therefore appears as if these transcriptional responses are specific to the perturbation of polyamine and methionine metabolism in P. falciparum.
A number of transcriptome studies of environmentally perturbed Plasmodium detected compensatory transcriptional responses (21–23), but few of these were confirmed on the proteome/metabolome level, and others failed to detect such programmed responses (48, 71, 72). An important difference of this study is that highly synchronized cultures were used compared with the asynchronous cultures used in most perturbations of Plasmodium reported up to now. The transcriptional arrest and general cytostasis demonstrated would have been masked if asynchronous cultures had been used, and defining a reference point for quantitative analysis (relative t0) would have been difficult. The use of synchronized cultures thus enabled comparison with the IDC transcriptome, and transcripts from treated samples with profiles that deviated from their IDC profiles further substantiated the findings of the differential abundance analysis.
The limited evidence of compensatory feedback and small amplitude of transcriptional responses upon perturbation of P. falciparum compared with other organisms, e.g. M. tuberculosis (20), have been attributed to the dominant role of post-transcriptional mechanisms of gene regulation (27, 71, 73). However, recently a cascade of AP2 transcription factors were proposed to control transcription during the IDC (74). In this study, the transcript and protein levels correlated well for some genes (e.g. OAT and AdoMet synthetase), indicating a predominant transcriptional level of regulation. However, others (e.g. lactate dehydrogenase, elongation factor 2, falcipain, and Pdx1) seem to have a delay between transcript and protein production, which may implicate involvement of post-transcriptional regulatory mechanisms. Additionally, in this study compensatory responses specific to polyamine and methionine metabolism were detected in the transcriptome, which provide convincing evidence of regulation occurring at the transcriptional level in P. falciparum after exogenous perturbations. Comprehensively, this was also confirmed in the proteome and metabolome (Fig. 4).
In conclusion, evidence was presented here of the response of a multistage organism, P. falciparum, to cytostasis upon polyamine depletion. There was a clear correlation between the perturbation and the transcriptional response observed in the parasite, which indicates the presence of transcriptional regulatory mechanisms in P. falciparum. This was translated in certain cases to the parasite proteome and metabolome. Together, the results imply that compensatory mechanisms are induced in P. falciparum after polyamine depletion, which corroborates the biological importance of this pathway to the malaria parasite.
Acknowledgments
We thank J. de Ridder, L. Dzikiti, and C. Stutzer (University of Pretoria), B. Crampton and S. Stoychev (CSIR), J. D. Rabinowitz and the Princeton University Microarray data base curators (Princeton University) for their assistance. The Princeton University Microarray Data Base was supported in part by National Institutes of Health Grant P50 GM071508 (NIGMS).
Footnotes
This work was supported by National Research Foundation Grant FA2004051300055, Thuthuka TTK2006061500031, and Prestigious Bursary (to A. C. v. B.), the South African Malaria Initiative, and the University of Pretoria. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: ODC, ornithine decarboxylase; AdoMetDC, S-adenosylmethionine decarboxylase; IDC, intraerythrocytic developmental cycle; DFMO, dl-α-difluoromethylornithine; T, drug treated; UT, untreated; hpi, hours post-invasion; LIMMA, linear models for microarray data; dcAdoMet, decarboxylated S-adenosylmethionine; LDC, lysine decarbox-ylase; OAT, ornithine aminotransferase; AdoMet synthetase, S-adenosyl-methionine synthetase; PLP, pyridoxal 5′-phosphate; Pdx1, PLP synthase; GABA, γ-aminobutyrate; LDC, lysine decarboxylase; LC-MS/MS, liquid chromatography- tandem mass spectrometry; MALDI, matrix-assisted laser desorption ionization time-of-feight.
J. Becker, personal communication.
M. Lliná s, unpublished data.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1 and Tables S1–S6.
Supplemental material: http://www.jbc.org/content/suppl/2008/12/11/M807085200.DC1.html
REFERENCES
- 1.Wallace H, Fraser A, Huges A. Biochem J. 2003;376:1–14. doi: 10.1042/BJ20031327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Igarashi K, Kashiwagi K. Biochem Biophys Res Commun. 2000;271:559–564. doi: 10.1006/bbrc.2000.2601. [DOI] [PubMed] [Google Scholar]
- 3.Tadolini B. Biochem J. 1988;249:33–36. doi: 10.1042/bj2490033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muscari C, Guarnieri C, Giaccari A, Caldarera C. Mol Cell Biochem. 1995;144:125–129. doi: 10.1007/BF00944391. [DOI] [PubMed] [Google Scholar]
- 5.Seiler N, Raul F. J Cell Mol Med. 2005;9:623–642. doi: 10.1111/j.1582-4934.2005.tb00493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heby O. Differentiation. 1981;19:1–20. doi: 10.1111/j.1432-0436.1981.tb01123.x. [DOI] [PubMed] [Google Scholar]
- 7.Thomas T, Thomas TJ. Cell Mol Life Sci. 2001;58:244–258. doi: 10.1007/PL00000852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro T, Goldberg D. In: Goodman and Gilman’s The Pharmacological Basis of Therapeutics. 11th Ed. Brunton L, Lazo J, Parker K, editors. McGraw-Hill Inc; New York: 2006. pp. 1053–1055. [Google Scholar]
- 9.Yeh I, Altman R. Mini Rev Med Chem. 2006;6:177–202. doi: 10.2174/138955706775475957. [DOI] [PubMed] [Google Scholar]
- 10.Müller I, Das Gupta R, Luërsen K, Wrenger C, Walter R. Mol Biochem Parasitol. 2008;160:1–7. doi: 10.1016/j.molbiopara.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 11.Müller S, Da’dara A, Lüersen K, Wrenger C, Gupta RD, Madhubala R, Walter R. J Biol Chem. 2000;275:8097–8102. doi: 10.1074/jbc.275.11.8097. [DOI] [PubMed] [Google Scholar]
- 12.Wright P, Byers T, Cross-Doersen D, McCann P, Bitonti A. Biochem Pharmacol. 1991;41:1713–1718. doi: 10.1016/0006-2952(91)90174-4. [DOI] [PubMed] [Google Scholar]
- 13.Das Gupta R, Krause-Ihle T, Bergmann B, Müller I, Khomutov A, Müller S, Walter R, Lüersen K. Antimicrob Agents Chemother. 2005;49:2857–2864. doi: 10.1128/AAC.49.7.2857-2864.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Assaraf Y, Golenser J, Spira D, Messer G, Bachrach U. Parasitol Res. 1987;73:313–318. doi: 10.1007/BF00531084. [DOI] [PubMed] [Google Scholar]
- 15.Bitonti A, McCann P, Sjoerdsma A. Exp Parasitol. 1987;64:237–243. doi: 10.1016/0014-4894(87)90148-2. [DOI] [PubMed] [Google Scholar]
- 16.Reguera R, Tekwani B, Balaña-Fouce R. Comp Biochem Physiol C Toxicol Pharmacol. 2005;140:151–164. doi: 10.1016/j.cca.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Bitonti A, Dumont J, Bush T, Edwards M, Stemerick D, McCann P, Sjoerdsma A. Proc Natl Acad Sci U S A. 1989;86:651–655. doi: 10.1073/pnas.86.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boshoff H, Manjunatha U. Microbes Infect. 2006;8:1654–1661. doi: 10.1016/j.micinf.2005.11.018. [DOI] [PubMed] [Google Scholar]
- 19.Birkholtz L-M, van Brummelen A, Clark K, Niemand J, Maréchal E, Llinas M, Louw A. Acta Trop. 2007;105:113–123. doi: 10.1016/j.actatropica.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 20.Boshoff H, Myers T, Copp B, McNeil M, Wilson M, Clifton B. J Biol Chem. 2004;279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 21.Gunasekera A, Patankar S, Schug J, Eisen G, Wirth D. Mol Microbiol. 2003;50:1229–1239. doi: 10.1046/j.1365-2958.2003.03787.x. [DOI] [PubMed] [Google Scholar]
- 22.Fang J, Zhou H, Rathore D, Sullivan M, Su X-Z, McCutchan T. Mol Biochem Parasitol. 2003;133:125–129. doi: 10.1016/j.molbiopara.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Oakley M, Kumar S, Anantharaman V, Zheng H, Mahajan B, Haynes J, Moch J, Fairhurst R, McCutchan T, Aravind L. Infect Immun. 2007;75:2012–2025. doi: 10.1128/IAI.01236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cui L, Miao J, Furuya T, Fan Q, Li X, Rathod P, Su X-Z, Cui L. Eukaryot. Cell. 2008;7:1200–1210. doi: 10.1128/EC.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Natalang O, Bischoff E, Deplaine G, Proux C, Dillies M-A, Sismeiro O, Guigon C, Bonnefoy S, Patarapotikul J, Mercereau-Puijalon O, Coppee J-Y, David P. BMC Genomics. 2008;9:388. doi: 10.1186/1471-2164-9-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coulson M, Hall N, Ouzounis C. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deitsch K, Duraisingh R, Dzikowski A, Gunaseker A, Khan S, Le Roch K, Llinás M, Mair G, McGovern V, Roos D, Shock J, Sims J, Wiegland R, Winzeler E. Am J Trop Med Hyg. 2007;77:201–208. [PubMed] [Google Scholar]
- 28.Mair G, Braks J, Garver L, Dimopoulos G, Hall N, Wiegant J, Dirks R, Khan S, Janse C, Waters A. Science. 2006;313:667–669. doi: 10.1126/science.1125129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shock J, Fischer K, DeRisi J. Genome Biol. 2007;8:R134. doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seiler N. Curr Drug Targets. 2003;4:537–564. doi: 10.2174/1389450033490885. [DOI] [PubMed] [Google Scholar]
- 31.Trager W, Jensen J. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- 32.Lambros C, Vanderberg J. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- 33.Assaraf Y, Golenser J, Spira D, Bachrach U. Biochem J. 1984;222:815–819. doi: 10.1042/bj2220815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozdech Z, Zhu J, Joachimiak M, Cohen F, Pulliam B, DeRisi J. Genome Biol. 2003;4:R9. doi: 10.1186/gb-2003-4-2-r9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R, Domrachev M, Lash A. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eisen M, Spellman P, Brown P, Botstein D. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bozdech Z, Llinás M, Pulliam B, Wong E, Zhu J, DeRisi J. PLoS Biology. 2003;1:E5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Llinás M, Bozdech Z, Wong E, Adai A, DeRisi J. Nucleic Acids Res. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang Y, Paquet A. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. 1st Ed. Gentleman R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Springer-Verlag Inc; New York: 2005. pp. 49–69. [Google Scholar]
- 40.Smyth G. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 41.Smyth G. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Springer-Verlag Inc; New York: 2005. pp. 397–420. [Google Scholar]
- 42.Dennis GJ, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. Genome Biol. 2003;4(5):P3. [PubMed] [Google Scholar]
- 43.Stoeckert C, Fischer S, Kissinger J, Heiges M, Aurrecoechea C, Gajria B, Roos D. Trends Parasitol. 2006;22:543–546. doi: 10.1016/j.pt.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Nirmalan N, Sims P, Hyde J. Mol Microbiol. 2004;52:1187–1199. doi: 10.1111/j.1365-2958.2004.04049.x. [DOI] [PubMed] [Google Scholar]
- 45.Bajad S, Kimball E, Yuan J, Peterson C, Rabinowitz J. J. Chromatogr. A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 46.Lu W, Kimball E, Rabinowitz J. J Am Soc Mass Spectrom. 2006;17:37–50. doi: 10.1016/j.jasms.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Dahl E, Shock J, Shenai B, Gut J, Derisi J, Rosenthal P. Antimicrob Agents Chemother. 2006;50:3124–3131. doi: 10.1128/AAC.00394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gunasekera A, Myrick A, Le Roch K, Winzeler E, Wirth D. Exp Parasitol. 2007;117:87–92. doi: 10.1016/j.exppara.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 49.Gardner M, Hall N, Fung E, White O, Berriman M, Hyman R, Carlton J, Pain A, Nelson K, Bowman S, Paulsen I, James K, Eisen J, Rutherford K, Salzberg S, Craig A, Kyes S, Chan M-S, Nene V, Shallom S, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather M, Vaidya A, Martin D, Fairlamb A, Fraunholz M, Roos D, Ralph S, McFadden G, Cummings L, Subramanian G, Mungall C, Venter J, Carucci D, Hoffman S, Newbold C, Davis R, Fraser C, Barrell B. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Icekson I, Bakhanashvili M, Apelbaum A. Plant Physiol. 1986;82:607–609. doi: 10.1104/pp.82.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ueda M, Masu Y, Ando A, Maeda H, Del Monte M, Uyama M, Ito S. Investig Ophthalmol Vis Sci. 1998;39:820–827. [PubMed] [Google Scholar]
- 52.Oden K, Clarke S. Biochemistry. 1983;22:2978–2986. doi: 10.1021/bi00281a030. [DOI] [PubMed] [Google Scholar]
- 53.Alm K, Berntsson A, Oredsson S. J Cell Biochem. 1999;75:46–55. doi: 10.1002/(sici)1097-4644(19991001)75:1<46::aid-jcb5>3.3.co;2-e. [DOI] [PubMed] [Google Scholar]
- 54.Tkachenko A, Nesterova L. Biochemistry (Mosc) 2002;68:850–856. doi: 10.1023/a:1025790729797. [DOI] [PubMed] [Google Scholar]
- 55.Yoshida M, Kashiwagi K, Shigemasa A, Taniguchi S, Yamamoto K, Makinoshima H, Ishihama A, Igarashi K. J Biol Chem. 2004;279:46008–46013. doi: 10.1074/jbc.M404393200. [DOI] [PubMed] [Google Scholar]
- 56.Date S, Stoeckert C. Genome Res. 2006;16:542–549. doi: 10.1101/gr.4573206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Makanga M, Bray P, Horrocks P, Ward S. Proteomics. 2005;5:1849–1858. doi: 10.1002/pmic.200401076. [DOI] [PubMed] [Google Scholar]
- 58.Sandmeier E, Hale T, Christen P. Eur J Biochem. 1994;221:997–1002. doi: 10.1111/j.1432-1033.1994.tb18816.x. [DOI] [PubMed] [Google Scholar]
- 59.Le Roch K, Johnson J, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan S, Williamson K, Holder A, Carucci D, Yates J, III, Winzeler E. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Byers T, Bush T, McCann P, Bitonti A. Biochem J. 1991;274:527–533. doi: 10.1042/bj2740527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Teng R, Junankar P, Bubb W, Rae C, Mercier P, Kirk K. NMR Biomed. Nov 19, 2008. [DOI] [PubMed]
- 62.Efferth T, Gillet J, Sauerbrey A, Zintl F, Bertholet V, De Longueville F, Remacle J, Steinbach D. Mol Cancer Ther. 2006;5:1986–1994. doi: 10.1158/1535-7163.MCT-06-0086. [DOI] [PubMed] [Google Scholar]
- 63.Johnsson A, Vallon-Christensson J, Strand C, Litman T, Eriksen J. Anticancer Res. 2005;25:2661–2668. [PubMed] [Google Scholar]
- 64.Nemoto T, Kamel S, Seyama Y, Kubota S. Biochem Bio-phys Res Commun. 2001;280:848–854. doi: 10.1006/bbrc.2000.4227. [DOI] [PubMed] [Google Scholar]
- 65.Tabor C, Tabor H. Microbiol Rev. 1985;49:81–99. doi: 10.1128/mr.49.1.81-99.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takatsuka Y, Yamaguchi Y, Ono M, Kamio Y. J Bacteriol. 2000;182:6732–6741. doi: 10.1128/jb.182.23.6732-6741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu G, Morris SJ. Biochem J. 1998;336:1–17. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bacchi C, Nathan H, Yarlett N, Goldberg B, McCann P, Bitonti A, Sjoerdsma A. Antimicrob Agents Chemother. 1992;36:2736–2740. doi: 10.1128/aac.36.12.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bitonti A, Byers T, Bush T, Casara P, Bacchi C, Clarkson A, Mc-Cann P, Sjoerdsma A. Antimicrob Agents Chemother. 1990;34:1485–1490. doi: 10.1128/aac.34.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clark K, Dhoogra M, Louw A, Birkholtz L-M. Biol Chem. 2008;389:111–125. doi: 10.1515/BC.2008.014. [DOI] [PubMed] [Google Scholar]
- 71.Le Roch K, Winzeler E. In: Molecular Approaches to Malaria. Sherman I, editor. American Society for Microbiology; Washington, D.C: 2005. pp. 68–84. [Google Scholar]
- 72.Nirmalan N, Wang P, Sims P, Hyde J. Mol Microbiol. 2002;46:179–190. doi: 10.1046/j.1365-2958.2002.03148.x. [DOI] [PubMed] [Google Scholar]
- 73.Young J, Winzeler E. Pharmacogenomics. 2005;6:1–26. doi: 10.1517/14622416.6.1.17. [DOI] [PubMed] [Google Scholar]
- 74.De Silva E, Gehrke A, Olszewski K, León I, Chahal J, Bulyk M, Llinás M. Proc Natl Acad Sci U S A. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haider N, Eschbach M-L, De Souza Dias S, Gilberger T-W, Walter R, Luërsen K. Mol Biochem Parasitol. 2005;142:224–236. doi: 10.1016/j.molbiopara.2005.04.004. [DOI] [PubMed] [Google Scholar]