Multilocus variable number of tandem repeats analysis genotyping of Clostridium difficile isolates from screening tests and patients with symptomatic C. difficile infection reveals that asymptomatic carriers and environmental transmission from both carriers and patients with C. difficile infection contribute to new hospital-acquired cases.
Keywords: Clostridium difficile, screening, transmission, genotyping, MLVA
Abstract
Background. Previous studies have suggested that asymptomatic carriers of toxigenic Clostridium difficile are a source of hospital-associated (HA) infections. Multilocus variable number of tandem repeats analysis (MLVA) is a highly discriminatory molecular subtyping tool that helps to determine possible transmission sources.
Methods. Clostridium difficile isolates were recovered from perirectal swabs collected for vancomycin-resistant Enterococcus (VRE) surveillance as well as from clinical C. difficile toxin–positive stool samples from July to November 2009 at the University of Pittsburgh Medical Center Presbyterian (UPMC). MLVA was performed to determine the genetic relationships between isolates from asymptomatic carriers and patients with HA C. difficile infection (HA-CDI). Asymptomatic carriage and HA-CDI isolates were considered to be associated if the carriage isolate was collected before the HA-CDI isolate and if the MLVA genotypes had a summed tandem-repeat difference of ≤2.
Results. Of 3006 patients screened, 314 (10.4%) were positive for toxigenic C. difficile, of whom 226 (7.5%) were detected only by VRE surveillance cultures. Of 56 incident cases of CDI classified as HA at UPMC during the study with available isolates, 17 (30%) cases were associated with CDI patients, whereas 16 (29%) cases were associated with carriers. Transmission events from prior bed occupants with CDI (n = 2) or carriers (n = 2) were identified in 4 of 56 cases.
Conclusions. In our hospital with an established infection control program designed to contain transmission from symptomatic CDI patients, asymptomatic carriers appear to have played an important role in transmission. Identification and isolation of carriers may be necessary to further reduce transmission of C. difficile in such settings.
(See the Editorial Commentary by McDonald on pages 1103–5.)
Previous studies have established that asymptomatic carriers of Clostridium difficile outnumber symptomatic patients in medical wards [1–4]. Shim et al found that carriers, defined as a patient with at least 2 culture-positive specimens 7 days apart, were at a decreased risk of symptomatic C. difficile infection (CDI) [5]. In one study, C. difficile from carriers appeared subsequently in hospital-associated (HA) CDIs attributable to the same strain in 16 of 19 (84%) cases [1].
Active surveillance testing to identify methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) carriers coupled with isolation precautions has been effective in reducing infections with these organisms [6, 7]. A similar approach for prevention of CDI has not been practical until the recent development of molecular tests that allow detection of the C. difficile carrier state with a reasonable turnaround time [8].
Although previous studies of CDI transmission based on restriction endonuclease analysis (REA) as the primary molecular epidemiologic approach have shown that asymptomatic carriers have some role in CDI transmission, [1, 9] current infection control guidelines focus on isolation measures for symptomatic CDI patients. Multilocus variable number of tandem repeats analysis (MLVA) for C. difficile typing has more discriminatory power than REA and so better facilitates tracking of C. difficile transmission within hospitals [10]. We performed MLVA genotyping of C. difficile isolates from colonized and infected patients to assess potential of each to HA-CDI and to determine the potential role of active surveillance for C. difficile carriage in a hospital with an established infection control program that has been associated with successful control of HA-CDI [11].
METHODS
Patient Population and Setting
This study was conducted at the University of Pittsburgh Medical Center Presbyterian–Shadyside (UPMC), a 762-bed (156 intensive care unit [ICU] beds) teaching hospital affiliated with the University of Pittsburgh. Patients undergoing VRE surveillance testing over a 117-day period in July 2009 through November 2009 were also screened for C. difficile. VRE surveillance cultures were performed at admission for patients admitted from other healthcare facilities; VRE surveillance was also performed at weekly intervals for all ICU patients and for all other inpatients with length of stay >20 days (>5 days if receiving antibiotics) or who tested positive for MRSA, C. difficile, or multidrug-resistant organisms [8]. Patients identified as VRE positive were placed in contact isolation for current and subsequent healthcare and were not rescreened.
Clostridium difficile reduction interventions did not change throughout the study, including contact isolation for the duration of hospitalization for patients identified as colonized or infected by C. difficile stool toxin testing [11]. Since 2006, the HA-CDI rate at UPMC has never exceeded our target rate of 8 per 10 000 patient-days. The average HA-CDI rate per year (2006–2012) was 5.6 cases per 10 000 patient-days. In addition to previously reported measures, 5000 ppm sodium hypochlorite was routinely used to disinfect all patient care areas. This study was approved by the University of Pittsburgh Institutional Review Board.
Clostridium difficile Diagnosis and Screening
Laboratory diagnosis of CDI was done using cell culture cytotoxicity assays (stool toxins) ordered by physicians (Diagnostic Hybrids, Athens, Ohio) of formed and unformed stool specimens. Both perirectal swabs collected for VRE surveillance (screening tests) and positive stool toxins were cultured for C. difficile using 48-hour broth enrichment and anaerobic culture [8]. Positive stool toxin specimens were cultured for C. difficile ±30 days relative to the 117-day study period. Toxigenic C. difficile was confirmed by tcdC genotyping for both stool toxins and screening tests [12]. The field sensitivity of screening tests for detection of C. difficile was calculated as the number of screening tests positive for toxigenic C. difficile divided by the total number of screening tests collected patients with positive stool toxins from −1 to 0 days prior to collection the stool toxin specimen. The results of C. difficile screening were not released to clinicians.
Molecular Typing of C. difficile
Clostridium difficile isolates were typed using MLVA and tcdC genotyping [10, 12, 13]. The tcdC genotypes in this study were assigned according to the PubMLST database (http://pubmlst.org/cdifficile) and were used to infer ribotype as described by Dingle et al [14]. Allele designations for tcdC conform to those previously described with the exception of genotypes 0, A1, and B, which have been renumbered as 19, 20, and 47 respectively [13, 15]. Minimum-spanning-tree analysis of all isolates recovered in the study was performed using BioNumerics software version 6.6 (Applied Maths, Austin, Texas), using a Manhattan coefficient to calculate the summed tandem-repeat difference (STRD) between isolates [10]. Complexes containing ≥2 isolates whose MLVA genotypes generated an STRD of ≤2 were defined as highly related isolates.
Epidemiologic Definitions and Analysis
Patients with positive stool toxins collected at UPMC during the study period plus a lag period of 10 days (to account for the potential incubation period of CDI) were epidemiologically classified by UPMC infection control according to modified criteria for CDI surveillance [16]. Hospital-associated CDI was defined by toxin positive stool and CDI symptom onset ≥3 days after admission to UPMC or present on readmission with documented UPMC exposure (without exposures to other healthcare facilities) in the preceding 12 weeks. CDIs occurring within 6 months of any previous CDI were classified as recurrent CDI. Patients with positive stool toxins but without signs of CDI on chart review were classified as carriers by infection control personnel.
Patients with C. difficile–positive screening tests underwent electronic record review of stool toxins sent during the study period ±12 weeks, and patients without any stool toxins sent were defined as carriers and were categorized into 3 groups: (1) persistent carriers: patients with at least 2 screening tests positive for toxigenic C. difficile collected ≥7 days apart and no stool toxins ordered by clinicians; (2) transient carriers: patients with only 1 screening test positive for toxigenic C. difficile (or >1 test collected within a 7-day period), and negative screening tests collected before and afterward; and (3) indeterminate carriers: patients with single screening cultures (or >1 culture collected within a 7-day period) positive for toxigenic C. difficile. Patients who had ≥1 negative stool toxin sent during the study ±12 weeks were defined as discrepant carriers. Chart review to exclude a history of CDI diagnosed at non-UPMC facilities was performed for carriers linked to incident HA-CDI by MLVA.
TheraDoc 4.3.0 software (Hospira, Salt Lake City, Utah) was used to determine patient locations for all patients with isolates highly related by MLVA to HA-CDI. These data were used to categorize each HA-CDI case to the most likely transmission route: (1) ward transmission: patients shared a common ward occupancy with a symptomatic CDI patient or carrier within 30 days; (2) non–ward transmission: patient hospitalized concurrently with carrier or CDI patient within 30 days without wards in common; (3) environmental: patient developed CDI after placement in a room previously occupied by a carrier or CDI patient; or (4) indeterminate: patients with no time–place epidemiologic links to patients with highly related isolates. For each patient, the collection date of the presumed source isolate had to precede that of the patient's C. difficile symptom onset date by at least 1 calendar day.
Environmental Cultures
To determine whether carriers had room contamination with C. difficile, environmental cultures were performed for all patients meeting the following criteria: (1) screening test positive for toxigenic C. difficile; (2) not in contact isolation precautions; and (3) inpatient at the time of culture confirmation. Five sites in each room were cultured with selective broth amplification using sterile 2 × 2-inch gauze pads premoistened with phosphate-buffered saline [17]. For ICUs, the sites were the patient call bell, bed rails, bedside table, toilet seat, and computer keyboard. For non-ICU rooms, the sites were the same except the patient bedside telephone receiver was substituted for the computer keyboard.
RESULTS
Incident HA-CDI
During the screening period, 158 patients had positive stool toxin assays for C. difficile ordered by clinicians, of whom 34 (22%) were classified as carriers. Of the remaining 124 symptomatic patients, 53 (43%), were classified as HA-CDI at UPMC, 13 (10%) as community-acquired, and 57 (46%) as acquired at other facilities or relapses.
Of 61 cases of HA-CDI during the screening period plus the 10-day lag period, isolates were obtained from 56 (92%) cases. Of these, 17 cases were unrelated by MLVA to any isolate recovered during the study and 5 were chronologically first within a complex of highly related isolates; these latter were recovered as late as study day 74. Detailed molecular combined with time–place epidemiologic analysis was thus performed for 33 of 61 (54%) HA-CDI cases (Table 1).
Table 1.
Epidemiologic Analysis of 33 Patients With Clostridium difficile Infections Classified as Hospital-Acquired at the University of Pittsburgh Medical Center, July–November 2009
| Study Day | MLVA Complex | tcdC Genotype | Same Unit | Same Bed | Concurrent Hospital Stay | Dates of Exposure to Sourcea | Classification of Presumed Sourceb | No. of Days Between Source and Patient Culture | Classification of Most Likely Transmission Category | STRD (Source to Case) |
|---|---|---|---|---|---|---|---|---|---|---|
| 9 | F | 1 | No | No | Yes | −6 to 0 | 2 | −6 | Non–ward CDI | 3 |
| 10 | K | 1 | Yes | No | Yes | −2 to −1 | 6 | −28 | Ward CDI | 0 |
| 12 | F | 1 | Yes | Yes | Yes | −7 to −3 | 2 | −9 | Environmental | 1 |
| 16 | A | 3 | Yes | No | Yes | −5 to 0 | 3 | −8 | Ward CDI | 0 |
| 19 | M | 1 | No | No | Yes | −7 to 0 | 6 | −9 | Non–ward CDI | 5 |
| 27 | C | 1 | Yes | No | Yes | −6 to 0 | 1 | −1 | Ward carrier | 0 |
| 33 | F | 1 | No | No | Yes | −11 to −5 | 4 | −11 | Non–ward carrier | 0 |
| 37c | M | 1 | Yes | No | Yes | −6 to −6 | 6 | −12 | Ward CDI | 5 |
| 44 | S | 1 | Yes | Yes | Yes | −24 to −22 | 2 | −7 | Environmental | 0 |
| 45 | M | 1 | Yes | No | Yes | −7 to −4 | 2 | −8 | Ward CDI | 0 |
| 47 | W | 5 | No | No | Yes | −12 to −11 | 5d | −11 | Non–ward carriera | 0 |
| 48c | M | 1 | Yes | No | Yes | −13 to −1 | 1 | −4 | Ward carrier | 3 |
| 62 | C | 1 | No | No | Yes | −17 to 0 | 6 | −1 | Non–ward CDI | 2 |
| 68 | Q | 20 | No | No | Yes | −24 to −21 | 1 | −25 | Non–ward carriere | 3 |
| 71 | V | 19 | No | No | Yes | −34 to −14 | 7 | −14 | Non–ward carrier | 0 |
| 71c | U | 42 | No | No | No | Unknown | 1 | −34 | Indeterminatee | 1 |
| 75 | N | 1 | Yes | Yes | No | −6 to −6 | 1 | −18 | Environmental | 0 |
| 90 | C | 1 | No | No | Yes | −16 to 0 | 7 | −10 | Non–ward carrier | 1 |
| 93c | B | 3 | No | No | Yes | −25 to −8 | 1 | −15 | Non–ward carriere | 2 |
| 94c | J | 1 | No | No | Yes | −50 to −39 | 4 | −43 | Non–ward carriere | 1 |
| 94 | J | 1 | No | No | Yes | −55 to −40 | 4 | −44 | Non–ward carrier | 1 |
| 94 | M | 1 | No | No | Yes | −12 to −12 | 4 | −16 | Non–ward carriere | 4 |
| 101 | M | 1 | Yes | No | Yes | −6 to −1 | 2 | −6 | Ward CDI | 0 |
| 101 | H | 1 | No | No | No | Unknown | 1 | −35 | Indeterminate | 3 |
| 114 | J | 1 | No | No | Yes | −4 to 0 | 4 | −4 | Non–ward carrier | 0 |
| 117 | J | 1 | No | No | Yes | −2 to 0 | 2 | −3 | Non–ward CDI | 0 |
| 119 | G | 1 | Yes | No | Yes | −3 to 0 | 6 | −8 | Ward CDI | 0 |
| 120 | H | 1 | Yes | No | Yes | −6 to −6 | 2 | −14 | Ward CDI | 2 |
| 120 | J | 1 | No | No | Yes | −21 to 0 | 2 | −7 | Non–ward CDI | 0 |
| 120 | J | 1 | No | No | Yes | −21 to 0 | 2 | −7 | Non–ward CDI | 0 |
| 125 | J | 1 | No | No | Yes | −10 to 0 | 2 | −5 | Non–ward CDI | 0 |
| 126 | D | 1 | Yes | Yes | Yes | −12 to −12 | 4 | −15 | Environmental | 3 |
| 127 | H | 1 | No | No | Yes | −12 to −4 | 2 | −7 | Non–ward CDI | 1 |
Patients for whom an isolate was unavailable for genotyping (n = 6) and with Clostridium difficile having unique MLVA genotypes (n = 17) or isolates that were chronologically first within a complex of highly related isolates (n = 5) are not depicted. MLVA cluster designations conform to those in Figure 2 and Supplementary Table 1, whereas source classification and presumed transmission category conform to definitions outlined in the Methods section.
Abbreviations: CDI, Clostridium difficile infection; MLVA, multilocus variable number of tandem repeats analysis; STRD, summed tandem-repeat difference.
a Relative to patient's toxin testing date.
b 1 = discrepant carrier; 2 = C. difficile infection acquired at University of Pittsburgh Medical Center (UPMC); 3 = hospital-acquired colonization with C. difficile detected by toxin test ordered by clinician; 4 = indeterminate carrier; 5 = other CDI (infection not acquired at UPMC)a; 6 = recurrent; 7 = persistent carrier.
c Indicates CDI that was attributable to UPMC after discharge (community-onset, healthcare facility–associated).
d Source patient's first positive toxin test occurred on study day 50 after having positive screening cultures on days 15, 22, 36, and 49.
e Source patient was positive for other organism requiring isolation prior to screening positive for C. difficile.
Carriers identified by screening tests only were identified as the sources for a total of 16 of 56 (29%) HA-CDI cases in the study period; of these, 9 were classified as non–ward transmissions, 2 as ward transmissions, 2 as environmental transmissions, and 2 as indeterminate. The presumed source for 1 HA-CDI case classified as a non–ward carrier transmission was a patient who was later diagnosed with CDI; this source patient's screening tests were positive 35 days prior to the collection of the first stool toxin assay, and the patient was not in contact isolation at the time of the presumed transmission event. Four of the carriers identified as source patients were already in contact isolation for infections with other organisms (Table 1).
One carrier detected by positive stool toxin testing was implicated as the source of a ward transmission (Table 1). No other HA-CDI cases could be traced to a source that was determined to be colonized by stool toxin testing.
Transmission events from CDI patients comprised 17 of 56 (30%) cases; of these, 9 were classified as non–ward transmissions, 7 as ward CDI transmissions, and 2 as environmental transmissions (Table 1). The ward transmission events occurred in 7 different units.
Screening Cultures
During the study period, 422 of 4979 (8.5%) screening tests performed on 3006 patients were positive for toxigenic C. difficile. Of 3006 patients screened, 314 (10.4%) and 506 (16.8%) were positive for toxigenic C. difficile and VRE on at least 1 specimen, respectively. Of the 3006 patients, 1957 (65.1%), 598 (19.9%), 229 (7.6%), 104 (3.5%), 53 (1.8%), and 65 (2.2%) patients had a total of 1, 2, 3, 4, 5, and >5 screening tests performed, respectively. Of 314 patients positive for toxigenic C. difficile, 120 (38.2%) patients were also positive for VRE at least once, whereas 194 patients had no positive VRE tests. Positive screening for VRE during the study was associated with increased risk of being positive for toxigenic C. difficile on at least 1 specimen (odds ratio [OR], 3.7; 95% confidence interval [CI], 2.9–4.8). Of the 120 patients positive for both C. difficile and VRE, C. difficile was identified first in 17 of 120 (14%) patients, on the same day in 95 of 120 patients (80%), and afterward in 6 of 120 (6%) patients.
During the 117-day screening period, 417 C. difficile isolates were obtained from the positive stool toxins and/or screening cultures of 384 patients. Of these patients, 226 of 384 (58.9%) were detected using screening cultures only. Of these patients, 131 of 226 (58.0%), 58 of 226 (25.7%), 27 of 226 (11.9%), and 10 of 226 (4.4%) were classified as indeterminate-length, discrepant, persistent, and transient carriers, respectively. During the screening period, 3006 of 12 054 (24.9%) admissions were tested for VRE.
Environmental Cultures
Five of 6 patients who met the criteria for environmental culturing had recovery of C. difficile from at least 1 site. The patient with no recovery of C. difficile was classified as a transient carrier and had been moved to a different room by the date of room sampling. All 5 patients with room sites positive for C. difficile had isolates highly related by MLVA to isolates from 1 or more screening cultures (Table 2). Additionally, 3 environmental isolates from 2 patients were unrelated (STRD >10) to any of their screening culture isolates. The most frequently contaminated site was the patient call bell.
Table 2.
Clostridium difficile Recovered From Environmental Cultures of Asymptomatic Patients
| Patient | Source/Site Cultured | Interval, d | tcdC Genotype | MLVA Locus (CDR) |
MLVA Cluster | Epidemiologic Classification | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 5 | 9 | 48 | 49 | 60 | STRD | ||||||
| 1 | Screening culture | 0 | 9 | 27 | 6 | 12 | 5 | 18 | 2 | NA | u | Persistent carrier |
| 1 | Call bell #1 | 9 | 9 | 41 | 6 | 12 | 5 | 12 | 3 | 21 | w | Environmental |
| 1 | Call bell #2 | 9 | 9 | 27 | 6 | 12 | 5 | 18 | 2 | 0 | u | Environmental |
| 1 | Computer keyboard #2 | 9 | 9 | 42 | 6 | 12 | 5 | 12 | 3 | 22 | w | Environmental |
| 1 | Screening culture | 19 | 9 | 28 | 6 | 12 | 5 | 18 | 2 | 1 | u | Persistent carrier |
| 2 | Screening culture | 0 | 9 | 49 | 6 | 17 | 5 | 15 | 3 | NA | x | Discrepant |
| 2 | Screening culture | 15 | 9 | 48 | 6 | 17 | 5 | 15 | 3 | 1 | x | Discrepant |
| 2 | Screening culture | 22 | 9 | 48 | 6 | 17 | 5 | 15 | 3 | 1 | x | Discrepant |
| 2 | Bedside table | 29 | 9 | 49 | 6 | 17 | 5 | 15 | 3 | 0 | x | Environmental |
| 2 | Toilet seat | 29 | 9 | 40 | 6 | 8 | 4 | 13 | 11 | 29 | None | Environmental |
| 3 | Screening culture | 0 | 1 | 22 | 3 | 8 | 9 | 12 | 9 | NA | C | Persistent carrier |
| 3 | Screening culture | 12 | 1 | 24 | 3 | 8 | 9 | 13 | 9 | 3 | C | Persistent carrier |
| 3 | Call bell | 30 | 1 | 24 | 3 | 8 | 9 | 13 | 9 | 3 | C | Environmental |
| 4 | Screening culture | 0 | 5 | 37 | 6 | 6 | 4 | 10 | 18 | NA | None | Persistent carrier |
| 4 | Call bell | 16 | 19 | 16 | 6 | 9 | 2 | 16 | 13 | 37 | V | Environmental |
| 4 | Screening culture | 19 | 19 | 16 | 6 | 9 | 2 | 16 | 13 | 0 | V | Persistent carrier |
| 5 | Screening culture | 0 | 1 | 21 | 3 | 16 | 9 | 12 | 4 | NA | N | Discrepant |
| 5 | Screening culture | 17 | 1 | 20 | 3 | 16 | 9 | 13 | 4 | 2 | N | Discrepant |
| 5 | Screening culture | 17 | 1 | 20 | 3 | 16 | 9 | 12 | 4 | 1 | N | Discrepant |
| 5 | Bed rail | 21 | 1 | 20 | 3 | 16 | 9 | 12 | 4 | 1 | N | Environmental |
| 5 | Bedside table | 21 | 1 | 21 | 3 | 16 | 9 | 12 | 4 | 0 | N | Environmental |
| 5 | Telephone | 21 | 1 | 20 | 3 | 16 | 9 | 12 | 4 | 1 | N | Environmental |
| 5 | Toilet seat | 21 | 1 | 20 | 3 | 16 | 9 | 12 | 4 | 1 | N | Environmental |
| 5 | Screening culture | 24 | 1 | 20 | 3 | 16 | 9 | 12 | 4 | 1 | N | Discrepant |
Patients were either still in the room or sampled before terminal disinfection. All Clostridium difficile isolates from patients' environment and screening cultures are included. Five sites (toilet seat, call bell, telephone, bedrail, and bedside table [computer keyboard instead of telephone for intensive care unit beds]) were sampled per patient. Patient 1 had 2 intensive care unit rooms sampled, as the patient had just been transferred <60 minutes before sampling. Interval is the number of days between the patient's first screening culture and any subsequent culture. The MLVA complexes conform to Figure 2 and Supplementary Table 1. The STRD listed for each isolate is the STRD between the patient's first screening culture and the listed isolate. Epidemiological classifications conform to those outlined for C. difficile carriers in the Methods section.
Abbreviations: MLVA, multilocus variable number of tandem repeats analysis; NA, not applicable; STRD, summed tandem-repeat distance.
Molecular Epidemiology
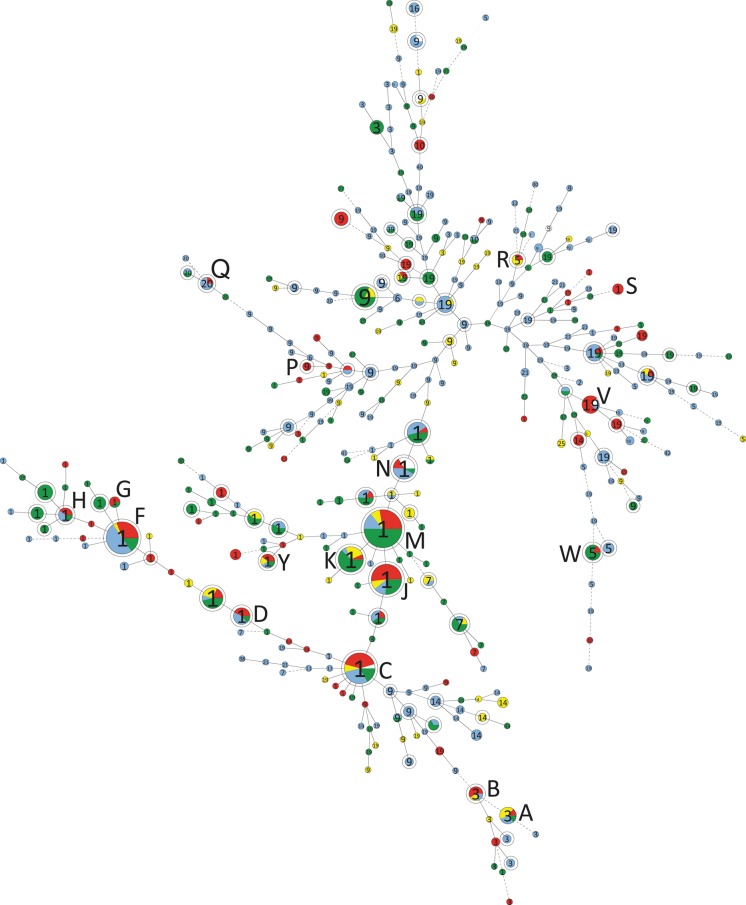
MLVA genotyping of all 739 toxigenic C. difficile isolates in this study yielded 524 unique genotypes from 479 patients and 13 environmental samples (Figure 1). Two hundred thirty of 739 isolates formed 78 complexes of highly related isolates (STRD ≤2). In 5 of 18 complexes, genotypes contributed by carriers were essential to formation of the complex (Figure 2).
Figure 1.
Minimum spanning tree depicting the relationships between 739 isolates in 524 genotypes by multilocus variable number of tandem repeats analysis (MLVA) recovered at the University of Pittsburgh Medical Center (UPMC) July–November 2009. Duplicate isolates from the same patient are included. Single circles represent 1 MLVA genotype; double circles represent complexes of highly related isolates by MLVA (summed tandem-repeat distances of ≤2), each labeled by capital letter designations conforming to those in Table 1. The numbers within each complex/genotype represent the tcdC genotype. Circle/complex size is proportional to the number of isolates included. Complexes labeled by capital letters contain cases of Clostridium difficile infection (CDI) acquired at UPMC according to epidemiologic criteria occurring within the screening period +10 days; complexes with hospital-associated (HA) UPMC case isolates only from the 30 days before and after this period remain unlabeled. Complexes and genotypes are color-coded according to epidemiologic classifications: red = HA-CDI at UPMC; yellow = colonization detected by toxin testing; blue = C. difficile colonization detected on screening tests; white = environmental isolates; green = all other C. difficile infections detected on toxin testing (recurrent CDI, community-acquired CDI, and HA-CDI not acquired at UPMC).
Figure 2.
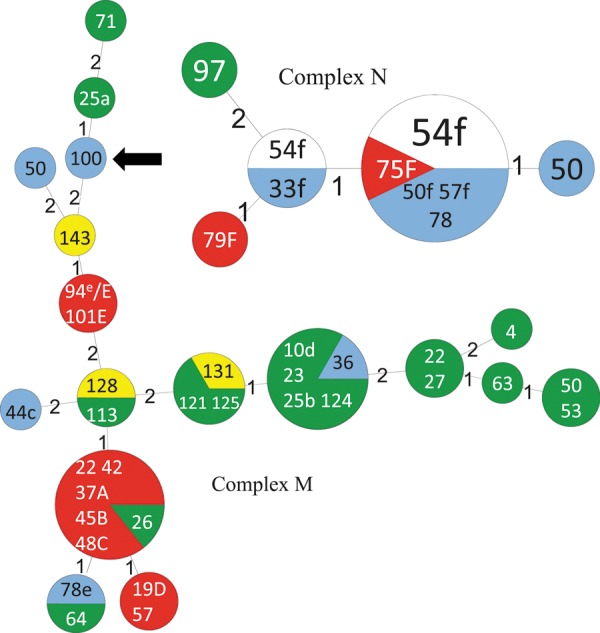
Minimum spanning tree depicting the detailed relationships between multilocus variable number of tandem repeats analysis (MLVA) genotypes in complexes M and N (Figure 1). Each circle represents 1 MLVA genotype scaled in size to the number of isolates comprising it. Numbers between genotypes indicate the summed tandem-repeat difference between them. The color coding of MLVA genotypes conforms to that in Figure 1. Numbers within genotypes represent the study day(s) on which each isolate within the genotype was collected. Isolates with capital letters (A–F) indicate isolates from patients with HA C. difficile infection (HA-CDI) listed in Table 1; the source isolates for these cases based on epidemiological analysis are labeled by corresponding lower-case letters (a–f). One isolate labeled e/E is both a hospital-associated CDI case isolate as well as a source for a subsequent case. The MLVA genotype indicated by the arrow shows an example of a C. difficile genotype recovered by screening tests only that was essential to the formation of a complex.
The 17 isolates from HA-CDI cases that were unrelated to other isolates comprised tcdC genotypes 1 (n = 6), 19 (n = 5), 14 (n = 2), 10 (n = 2), 9 (n = 1), and 3 (n = 1). Epidemic strain isolates (tcdC genotype 1, inferred ribotype 027) comprised 302 of 739 (40.9%) isolates in this study; these were discriminated by MLVA into 170 genotypes, 26 of which formed complexes (Table 1; Figure 2). Epidemic strain isolates were found in 62 of 225 (28%) carriers and in 105 of 194 (54%) of patients with CDI (OR, 3.1; 95% CI, 2.1–4.7; P < .001). No other significant differences were seen in strain prevalence by any other tcdC genotype over different epidemiological classes. Detailed typing data for all patient isolates in this study are found in Supplementary Table 1.
Sensitivity of Screening for C. difficile and Patterns of Carriage
The field sensitivity of screening cultures for C. difficile was 21 of 24 (88%) and 12 of 13 (92%) collected at intervals of −1 and 0 days from the date of the patient's first toxin-positive stool, respectively. One hundred fourteen screening cultures were performed on patients ≥8 days before their first toxin-positive stool; of these, 100 (87.7%) were negative for C. difficile, 4 (3.5%) were positive for unrelated C. difficile isolates, and 10 (8.8%) were positive for highly related isolates compared to isolates from the patient's first positive stool toxin. The highly related isolates came from 7 patients at intervals of −35 to −14 days (3 patients) and −8 to −13 days (4 patients).
Of 35 patients who were screened >14 days after the collection date of their first positive stool toxin assays, 15 patients (43%) remained positive for C. difficile, and 8 of 15 patients collected at this interval remained positive for C. difficile that was highly related to the isolate from the patient's first toxin test over intervals as long as 139 days (median, 33 days).
DISCUSSION
This study supports the hypothesis that carriers of C. difficile contribute to transmission within hospitals. Approximately a quarter of isolates from HA-CDI cases were highly related to isolates from asymptomatic patients identified from screening tests. This finding suggests that screening and isolating patients with carriage could potentially lead to further reductions in CDI incidence. Some CDI patients had isolates that could be traced by MLVA to carriers or other CDI patients in isolation at the time of screening. These patients may have contaminated the environment prior to isolation initiation or may represent noncompliance with contact precautions. However, it is conceivable that earlier isolation initiation for some of these sources of new cases through screening for C. difficile carriage might have prevented spread. These cases would have then required hand hygiene with soap and water in addition to existing contact isolation precautions.
This study supports our practice of extending C. difficile isolation precautions for the duration of hospitalization [11]. As >50% of CDI patients screened at >14 days after their first positive toxin were negative by perirectal swab culture, an active surveillance screening program for C. difficile may have a role in determining the timing of contact precaution discontinuation.
This study also supports the hypothesis that environmental transmission events from prior room occupants with CDI or carriage can occur. Room surfaces were positive with MLVA genotypes closely matching that of the carrier in 5 of 6 patients tested in this study. Despite the use of universal sodium hypochlorite cleaning, environmental transmission may have accounted for 4 of 61 incident HA-CDI cases. This finding is in line with the study of Shaughnessy et al, who reported that occupancy of a hospital room previously containing a CDI patient is an independent risk factor for CDI [18]. In a recent study, housekeeping staff were ineffective in disinfecting rooms contaminated with C. difficile in 7 of 9 instances [19]. Given the indefinite viability of C. difficile spores, remote room contamination from carriers may account for the large number of HA-CDI cases that were unrelated to any isolate in this short study. These data suggest that further comparative effectiveness research in hospital disinfection is needed.
This study suggests that the incubation period for CDI may be >1 week, as 7 symptomatic patients tested 8–28 days prior to their first stool toxin were positive for highly related C. difficile. In contrast to the meta-analysis of Shim et al [5], where only 2 patients with C. difficile carriage developed CDI in an analysis of 810 patients, 12 patients meeting their definition of colonization developed symptomatic disease in our study. These data suggest that colonization may long precede other factors that precipitate CDI such as antimicrobials.
Because our screening cultures used an existing surveillance program for a hospital-acquired organism, we did not screen the entire population at risk for C. difficile carriage; prior ward-based studies have shown that patients without recognized C. difficile risk factors comprise 30%–50% of the population of C. difficile carriers [1, 20]. In addition, many patients in this study who were at high risk for C. difficile carriage were not screened because they acquired VRE. We were unable to track common epidemiologic exposures beyond common inpatient wards such as exposure to diagnostic tests or personnel such as physicians. Such exposures may account for patients identified as non-ward or indeterminate transmissions in this study. Future intervention studies of active surveillance testing for C. difficile may need to include all inpatients to avoid these limitations.
Our study confirms that of Walker et al, who showed that ward-based contact with CDI patients can only account for approximately 25% of CDI transmission events using a less discriminatory typing method than MLVA [21]. In this study, we showed that a substantial balance of transmissions arises from asymptomatic carriers; whether expanding screening to all inpatients would identify more ward transmission events or, conversely, show more non–ward transmissions is a matter of conjecture, but the large number of HA-CDI cases with non–ward transmissions suggests that C. difficile spores may be widely transmitted in time and space within hospitals because of their near-indefinite viability and low infective dose [22, 23].
In summary, there could be added value in performing surveillance testing for C. difficile carriage. A controlled trial is required to evaluate the utility of screening for C. difficile carriage with incident HA-CDI as the main study endpoint.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Jamie L. Grey for her maintenance of the infection control database used in this study.
Financial support. This work was supported by the Pennsylvania Department of Health (number 4100047864, all authors) and by a research career award from the National Institute of Allergy and Infectious Diseases (K24 AI52788 to L. H. H.).
Potential conflicts of interests. L. H. H, J. W. M., and K. A. S. receive research funding from ViroPharma and Merck. C. A. M. serves on the speakers' bureau for the Robert Michael Educational Institute and Postgraduate Institute for Medicine. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Clabots CR, Johnson S, Olson MM, Peterson LR, Gerding DN. Acquisition of Clostridium difficile by hospitalized patients: evidence for colonized new admissions as a source of infection. J Infect Dis. 1992;166:561–7. doi: 10.1093/infdis/166.3.561. [DOI] [PubMed] [Google Scholar]
- 2.Johnson S, Clabots CR, Linn FV, Olson MM, Peterson LR, Gerding DN. Nosocomial Clostridium difficile colonisation and disease. Lancet. 1990;336:97–100. doi: 10.1016/0140-6736(90)91605-a. [DOI] [PubMed] [Google Scholar]
- 3.McFarland LV, Mulligan ME, Kwok RY, Stamm WE. Nosocomial acquisition of Clostridium difficile infection. N Engl J Med. 1989;320:204–10. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 4.Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis. 1994;18:181–7. doi: 10.1093/clinids/18.2.181. [DOI] [PubMed] [Google Scholar]
- 5.Shim JK, Johnson S, Samore MH, Bliss DZ, Gerding DN. Primary symptomless colonisation by Clostridium difficile and decreased risk of subsequent diarrhoea. Lancet. 1998;351:633–6. doi: 10.1016/S0140-6736(97)08062-8. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqui AH, Harris AD, Hebden J, Wilson PD, Morris JG, Jr, Roghmann MC. The effect of active surveillance for vancomycin-resistant enterococci in high-risk units on vancomycin-resistant enterococci incidence hospital-wide. Am J Infect Control. 2002;30:40–3. doi: 10.1067/mic.2002.118616. [DOI] [PubMed] [Google Scholar]
- 7.Robicsek A, Beaumont JL, Paule SM, et al. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann Intern Med. 2008;148:409–18. doi: 10.7326/0003-4819-148-6-200803180-00003. [DOI] [PubMed] [Google Scholar]
- 8.Curry SR, Schlackman JL, Hamilton TM, et al. Perirectal swab surveillance for Clostridium difficile by use of selective broth preamplification and real-time PCR detection of tcdB. J Clin Microbiol. 2011;49:3788–93. doi: 10.1128/JCM.00679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mekonen ET, Gerding DN, Sambol SP, et al. Predominance of a single restriction endonuclease analysis group with intrahospital subgroup diversity among Clostridium difficile isolates at two Chicago hospitals. Infect Control Hosp Epidemiol. 2002;23:648–52. doi: 10.1086/501988. [DOI] [PubMed] [Google Scholar]
- 10.Marsh JW, O'Leary MM, Shutt KA, et al. Multilocus variable-number tandem-repeat analysis for investigation of Clostridium difficile transmission in hospitals. J Clin Microbiol. 2006;44:2558–66. doi: 10.1128/JCM.02364-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muto CA, Blank MK, Marsh JW, et al. Control of an outbreak of infection with the hypervirulent Clostridium difficile BI strain in a university hospital using a comprehensive “bundle” approach. Clin Infect Dis. 2007;45:1266–73. doi: 10.1086/522654. [DOI] [PubMed] [Google Scholar]
- 12.Marsh JW, O'Leary MM, Shutt KA, et al. Multilocus variable-number tandem-repeat analysis and multilocus sequence typing reveal genetic relationships among Clostridium difficile isolates genotyped by restriction endonuclease analysis. J Clin Microbiol. 2010;48:412–8. doi: 10.1128/JCM.01315-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. tcdC genotypes associated with severe TcdC truncation in an epidemic clone and other strains of Clostridium difficile. J Clin Microbiol. 2007;45:215–21. doi: 10.1128/JCM.01599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dingle KE, Griffiths D, Didelot X, et al. Clinical Clostridium difficile: clonality and pathogenicity locus diversity. PLoS One. 2011;6:e19993. doi: 10.1371/journal.pone.0019993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spigaglia P, Mastrantonio P. Molecular analysis of the pathogenicity locus and polymorphism in the putative negative regulator of toxin production (TcdC) among Clostridium difficile clinical isolates. J Clin Microbiol. 2002;40:3470–5. doi: 10.1128/JCM.40.9.3470-3475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald LC, Coignard B, Dubberke E, et al. Recommendations for surveillance of Clostridium difficile–associated disease. Infect Control Hosp Epidemiol. 2007;28:140–5. doi: 10.1086/511798. [DOI] [PubMed] [Google Scholar]
- 17.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis. 2007;45:992–8. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 18.Shaughnessy MK, Micielli RL, DePestel DD, et al. Evaluation of hospital room assignment and acquisition of Clostridium difficile infection. Infect Control Hosp Epidemiol. 2011;32:201–6. doi: 10.1086/658669. [DOI] [PubMed] [Google Scholar]
- 19.Eckstein BC, Adams DA, Eckstein EC, et al. Reduction of Clostridium difficile and vancomycin-resistant Enterococcus contamination of environmental surfaces after an intervention to improve cleaning methods. BMC Infect Dis. 2007;7:61. doi: 10.1186/1471-2334-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loo VG, Bourgault AM, Poirier L, et al. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med. 2011;365:1693–703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 21.Walker AS, Eyre DW, Wyllie DH, et al. Characterisation of Clostridium difficile hospital ward-based transmission using extensive epidemiological data and molecular typing. PLoS Med. 2012;9:e1001172. doi: 10.1371/journal.pmed.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawley TD, Clare S, Deakin LJ, et al. Use of purified Clostridium difficile spores to facilitate evaluation of health care disinfection regimens. Appl Environ Microbiol. 2010;76:6895–900. doi: 10.1128/AEM.00718-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez J, Springthorpe VS, Sattar SA. Clospore: a liquid medium for producing high titers of semi-purified spores of Clostridium difficile. J AOAC Int. 2011;94:618–26. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.