Abstract
Some molecular analyses require microgram quantities of DNA, yet many epidemiologic studies preserve only the buffy coat. In Frederick, Maryland, in 2010, we estimated DNA yields from 5 mL of whole blood and from equivalent amounts of all-cell-pellet (ACP) fraction, buffy coat, and residual blood cells from fresh blood (n = 10 volunteers) and from both fresh and frozen blood (n = 10). We extracted DNA with the QIAamp DNA Blood Midi Kit (Qiagen Sciences, Germantown, Maryland) for silica spin column capture and measured double-stranded DNA. Yields from frozen blood fractions were not statistically significantly different from those obtained from fresh fractions. ACP fractions yielded 80.6% (95% confidence interval: 66, 97) of the yield of frozen whole blood and 99.3% (95% confidence interval: 86, 100) of the yield of fresh blood. Frozen buffy coat and residual blood cells each yielded only half as much DNA as frozen ACP, and the yields were more variable. Assuming that DNA yield and quality from frozen ACP are stable, we recommend freezing plasma and ACP. Not only does ACP yield twice as much DNA as buffy coat but it is easier to process, and its yield is less variable from person to person. Long-term stability studies are needed. If one wishes to separate buffy coat before freezing, one should also save the residual blood cell fraction, which contains just as much DNA.
Keywords: all-cell-pellet fraction, buffy coat, DNA extraction yield, residual blood cells, whole blood
New technologies for whole-genome scans, DNA sequencing, and studies of DNA methylation can require microgram quantities of high-quality DNA. Studies of DNA adducts have required tens of micrograms (1). In the past, many epidemiologic researchers stored buffy coat as the DNA source, and this has been recommended (2), but we noticed in preliminary unpublished data that fresh buffy coat contained only about 39% of the DNA available from an equivalent quantity of fresh whole blood. We hypothesized that one could substantially increase the amount of DNA available for epidemiologic studies by storing an “all-cell-pellet” (ACP) fraction composed of all cells spun down by centrifugation, rather than buffy coat. This idea is supported by a recent study (3) showing that the “residual cell pack” from fresh whole blood yielded substantial quantities of DNA. Therefore, we sought to determine how much DNA could be extracted from equivalent amounts of fresh whole blood, ACP, buffy coat, and residual blood cells. Because samples would need to be frozen and stored for most epidemiologic investigations, we also estimated the amount of DNA extractable from these 4 sources after freezing matched aliquots obtained from the same people and the same blood draw as that from which the fresh samples were taken. We chose the QIAamp DNA Blood Midi Kit (Qiagen Sciences, Germantown, Maryland) DNA extraction procedure because it works well for both fresh and frozen samples.
MATERIALS AND METHODS
Blood donors
Twenty healthy volunteers (7 male and 13 female) were recruited in 2010 from the Research Donor Program administered by the Frederick National Laboratory's Occupational Health Services in Frederick, Maryland, under an approved protocol. The donors ranged in age from 31 years to 65 years. Donors were identified only by code, and the specimens were further deidentified using laboratory accession numbers.
Sample preparation
From each donor, whole blood was collected into three 10-mL acid-citrate-dextrose–containing BD Vacutainer ACD Solution A Blood Collection Tubes (Becton Dickinson & Company, Franklin Lakes, New Jersey). Two experiments were performed, each requiring whole blood from 10 donors. The first set of 10 donors had DNA extracted only from fresh blood samples; the second set of 10 donors had DNA extracted from both fresh and frozen samples. Once blood was collected, each vacutainer was inverted 8 times before it was transferred to the processing laboratory. The tubes were inverted an additional 5 times once they arrived in the processing laboratory to ensure complete mixing before processing. The total time of processing was less than 2 hours from collection time for fresh samples. All samples were placed on wet ice during processing and fresh blood laboratory transfer steps. The 3 acid-citrate-dextrose tubes from each donor were randomly assigned to one of 3 processing groups: 1) whole blood, 2) ACP, and 3) buffy coat/residual blood cells. The volume of blood in each vacutainer was measured and recorded before any processing was performed.
From the vacutainer labeled “whole blood,” approximately eight 1-mL aliquots and one 0.5-mL aliquot were made. A white blood cell count was obtained from the 0.5-mL aliquot of whole blood with the Sysmex XT-2000i Automated Hematology Analyzer (Sysmex, Richmond, Virginia) at the AIDS Monitoring Laboratory of SAIC-Frederick, Frederick National Laboratory for Cancer Research (SAIC-Frederick, Inc., Frederick, Maryland). Four 1-mL aliquots of whole blood had their DNA extracted immediately as a “fresh” sample, and four 1-mL aliquots were frozen and stored at −80°C, and later had their DNA extracted.
The vacutainer identified as “ACP” was centrifuged at 1,200 × g for 15 minutes at 10°C. Plasma was removed, except for approximately 0.5 mL, which remained on top of the pelleted cellular material. The residual plasma and cellular material were mixed well and referred to as “ACP,” from which 1-mL aliquots were made. A 0.5-mL aliquot was used to determine the ACP white cell count. One 1-mL aliquot of ACP had its DNA extracted immediately as a “fresh” sample. The remaining aliquots of ACP were frozen at −80°C for future DNA extraction.
The vacutainer labeled “buffy coat/residual blood cells” was centrifuged at 1,200 × g for 15 minutes at 10°C. The plasma was removed, except for approximately 0.5 mL, which remained on top of the pelleted cellular material. The residual plasma and the uppermost 1.4 mL of cellular material was collected, mixed, and aliquoted equally into 2 Nunc CryoTube Vials (Nunc A/S, Roskilde, Denmark) (buffy coat). One cryovial immediately had 0.2 mL removed for a white cell count, and DNA was extracted from the remainder of the sample. This aliquot was the “fresh” buffy coat sample, and the other cryovial was frozen at −80°C until its DNA was extracted. The residual cellular material (residual blood cells) was mixed well, and then 1-mL aliquots were made. One aliquot was identified as the “fresh” residual blood cell sample, which had 0.2 mL removed for a white cell count, with the remaining sample undergoing immediate DNA extraction. The other cryovials were frozen at −80°C for future DNA extraction.
All fresh cryovials were transferred to the extraction laboratory on wet ice for immediate DNA extraction. All frozen cryovials were stored at −80°C for a minimum of 24 hours before DNA extraction. Frozen samples were transferred to the extraction laboratory on dry ice, where they were thawed for use. Frozen vials were placed in a 37°C water bath for 30 minutes and blotted, and DNA was immediately extracted.
DNA extraction and quantification
DNA was extracted using the QIAamp DNA Blood Midi Kits (for 0.3–2 mL of whole or processed blood). The QIAamp DNA Blood Midi Kits are based on silica spin column capture. DNA was quantified with the Quant-iT PicoGreen dsDNA Reagent and Kits (Invitrogen, Eugene, Oregon), which measures double-stranded DNA (dsDNA) by fluorescein excitation and emission at 480 nm and 520 nm, respectively. The coefficient of variation is less than or equal to 3% for dsDNA concentrations in the range of 25 pg/mL–1,000 ng/mL.
Statistical methods
To compare yields from the various blood fractions, we calculated the amount of DNA in micrograms for a given fraction that would be derived from 5 mL of fresh whole blood. These calculations required measuring the volumes of material in each of the 3 tubes mentioned above. Detailed definitions and formulas are given in the Appendix, which also includes a table of variable definitions. Most calculations were performed in SAS 9.2 (SAS Institute Inc., Cary, North Carolina) (4). To obtain the most precise information on DNA yields from fractions of fresh blood, we combined the data from the 10 volunteers whose samples were used for fresh and frozen analyses with data from 10 additional volunteers whose blood was used only for analyses of fresh blood fractions.
Because 10 volunteers provided samples that were used for both fresh and frozen analyses, resulting correlations needed to be taken into account in Table 1. Blood that was used for both fresh and frozen samples induced a correlation between the fresh and frozen yields, but blood samples taken from other persons that were used only for fresh blood analyses produced data independent of the yields from frozen samples. This data structure was taken into account for computing the standard error of the difference between the mean of all 20 fresh samples with the mean of 10 frozen samples. Let the first 10 components of a dsDNA measurement vector correspond to fresh samples, the next 10 components to the corresponding paired frozen samples, and the last 10 to the unpaired fresh samples. Define a vector λ with value 1/20 for the first 10 rows and the last 10 rows and −1/10 for the middle 10 rows. Then the inner product of λ with the vector of 30 dsDNA measurements estimates the difference between mean fresh yields and mean frozen yields. Let Σ be a 30 × 30 covariance matrix with 3 variances corresponding to the 3 groups of measurements on the diagonal, correlation ρ between paired fresh and frozen samples, and correlation 0 elsewhere. Then the estimated variance of the difference in means is 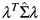 , where
, where 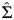 is the estimated covariance of the dsDNA measurement vector. The correlation ρ was estimated from the paired fresh and frozen measurements. The 3 variances for the 3 groups of 10 measurements were estimated via their respective sample variances, each with 9 degrees of freedom.
is the estimated covariance of the dsDNA measurement vector. The correlation ρ was estimated from the paired fresh and frozen measurements. The 3 variances for the 3 groups of 10 measurements were estimated via their respective sample variances, each with 9 degrees of freedom.
Table 1.
Mean Double-Stranded DNA Yields (μg) Equivalent to Those Obtained From 5-mL Whole-Blood Samples, Frederick, Maryland, 2010
| Fraction | Sample Source |
Fresh Blood Mean Minus Frozen Blood Meana |
Fresh Yield as % of Fresh Whole-Blood Yield (n = 20) |
Frozen Yield as % of Frozen Whole-Blood Yield (n = 10) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fresh Blood Only (n = 10) | Fresh Blood From Fresh and Frozen Samples (n = 10) | All Fresh Blood (n = 20) | Frozen Blood (n = 10) | 95% CI | P Value | % | 95% CI | % | 95% CIb | |
| Whole blood | 89.9 (7.96)c | 84.2 (11.5) | 87.0 (6.85) | 89.3 (8.86) | −18.7, 14.3 | 0.79 | 100 | 100 | ||
| All-cell-pellet | 76.0 (5.51) | 96.8 (12.4) | 86.4 (7.01) | 71.9 (8.24) | −6.6, 35.6 | 0.18 | 99.3 | 86, 100 | 80.6 | 66, 97 |
| Buffy coat | 40.4 (3.75) | 37.5 (5.34) | 38.9 (3.19) | 38.0 (7.91) | −12.9, 14.7 | 0.90 | 44.7 | 37, 54 | 42.6 | 26, 57 |
| Residual blood cells | 27.7 (3.38) | 40.1 (5.41) | 33.9 (3.42) | 34.6 (5.72) | −10.4, 11.0 | 0.90 | 39.0 | 30, 50 | 38.8 | 24, 56 |
Abbreviation: CI, confidence interval.
a Blood that was used for both fresh and frozen samples induced a correlation in the fresh and frozen yields, but blood samples from other persons that were used only for fresh blood analyses produced data that were independent of the yields from frozen samples. This data structure was taken into account for computing the standard error of the difference between the mean from all 20 fresh samples and the mean from 10 frozen samples, as described in “Statistical methods.”
b Test-based confidence intervals take correlations into account by computing the variance of 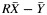 , where
, where 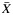 is the average yield from whole blood,
is the average yield from whole blood, 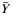 is the average yield from the blood fraction, and R is the ratio of the yields. Dividing
is the average yield from the blood fraction, and R is the ratio of the yields. Dividing 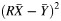 by the variance of
by the variance of 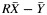 , we have a quantity that follows an F distribution with 1 and n – 1 degrees of freedom. By setting this quantity equal to the critical value of the F distribution, we solved for upper and lower confidence limits for R.
, we have a quantity that follows an F distribution with 1 and n – 1 degrees of freedom. By setting this quantity equal to the critical value of the F distribution, we solved for upper and lower confidence limits for R.
c Numbers in parentheses, standard error.
DNA quality
DNA samples from each of the above components were stored at 4°C for 2.3–2.7 years and reanalyzed using the Quantifiler Human DNA Quantification Kit (Life Technologies, Grand Island, New York) and NanoDrop 2000 (Thermo Scientific, Wilmington, Delaware) assays. The Quantifiler assay measures amplifiable single- and double-stranded DNA. The NanoDrop assay measures optical density to estimate DNA concentration, and the absorbance ratio (260 nm/280 nm) is a measure of DNA purity. The genomic DNA was also electrophoresed in 1.2% agarose gels to determine average DNA molecular weight and to detect DNA degradation and fragmentation.
RESULTS
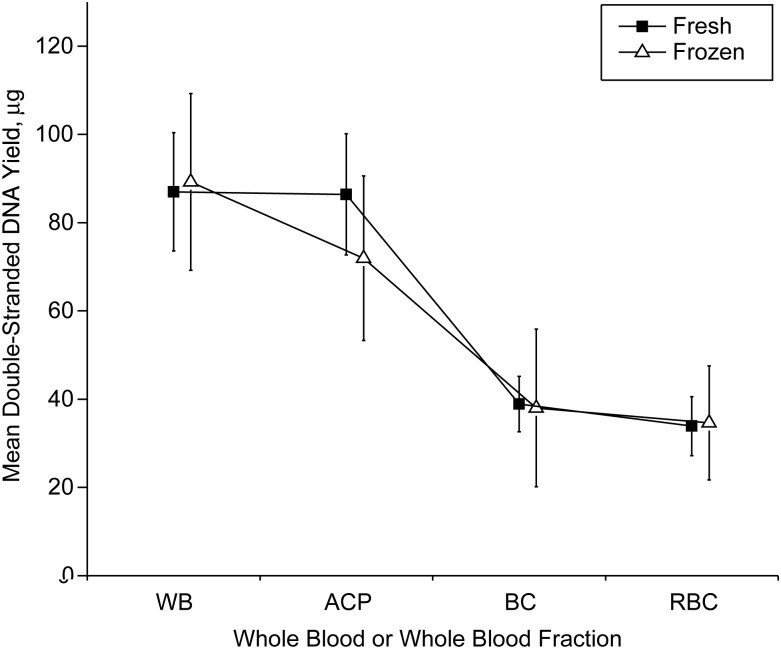
Average absolute dsDNA yields equivalent to 5 mL of whole blood are shown in Table 1, together with standard errors of the mean. The percentage yields from various fractions in comparison with whole blood are also shown. The average dsDNA yield from fresh whole blood (5 mL) was 87.0 μg, and it was 89.3 μg from frozen whole blood. There were no statistically significant differences between yields from fresh and frozen samples for whole blood, ACP, buffy coat, or residual blood cells (Table 1 and Figure 1). Thus, the freezing process did not lead to a loss of extractable dsDNA.
Figure 1.
Mean double-stranded DNA yield (μg) from components derived from 5 mL of fresh whole blood, Frederick, Maryland, 2010. Data are for whole blood (WB), all-cell-pellet (ACP), buffy coat (BC), and residual blood cells (RBC). The 2 loci correspond to the frozen samples from 10 volunteers (open triangles) and fresh samples from all 20 volunteers (solid squares). Bars, 95% confidence interval.
The fresh ACP fraction contained 99.3% (95% confidence interval (CI): 86, 100) of the dsDNA extractable from an equivalent amount (5 mL) of whole fresh blood, and frozen ACP contained 80.6% (95% CI: 66, 97) of the dsDNA in frozen whole blood (Table 1). Buffy coat yielded 44.7% (95% CI: 37, 54) of the dsDNA extractable from whole blood in fresh samples and 42.6% (95% CI: 26, 57) in frozen samples. The residual blood cell fraction yielded 39.0% (95% CI: 30, 50) of the dsDNA extractable from whole blood in fresh samples and 38.8% (95% CI: 24, 56) in frozen samples. Thus, whether analyzing fresh blood or frozen samples, buffy coat and the residual blood cell material each contained only about half as much extractable dsDNA as ACP, which contained most of the dsDNA extractable from whole blood.
These patterns are evident in Figure 1, which plots yields separately for frozen samples (open triangles) and all fresh samples (solid squares). Except for ACP, the yields from frozen samples are close to the yields from fresh samples for each blood fraction. The yield from fresh ACP is close to that from fresh whole blood, but the yield from frozen ACP is a little smaller than that from frozen whole blood. The yields from buffy coat and residual blood cells are each only about half those from ACP, in both fresh and frozen samples.
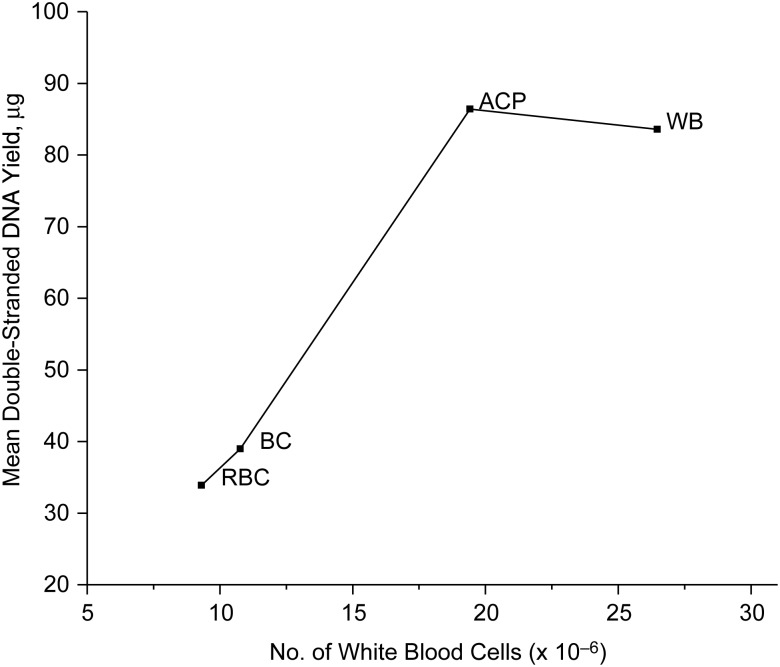
Mean dsDNA yield equivalent to 5 mL of fresh whole blood increased with the mean white cell count that corresponded to the same volume of fresh whole blood (Figure 2). Thus, the fact that ACP contains approximately twice as much dsDNA as buffy coat reflects the larger number of white cells in ACP. We could not present similar data for frozen samples, because counts of intact white cells were only available for fresh samples.
Figure 2.
Mean double-stranded DNA yield (μg) from components derived from 5 mL of fresh whole blood versus corresponding mean white blood cells (× 10−6) derived from 5 mL of fresh whole blood, Frederick, Maryland, 2010. The graph shows averages from the 20 fresh samples in Table 1, separately for whole blood (WB), all-cell-pellet (ACP), buffy coat (BC), and residual blood cells (RBC), each derived from the same volume (5 mL) of fresh whole blood. White blood cell counts equivalent to 5 mL of whole blood were derived in a manner similar to DNA yields (see Appendix).
From the data in Table 1, one can calculate the coefficient of variation of yield (standard deviation divided by the mean) for the frozen samples, which are most relevant for clinical and population studies. Because we have only 1 estimate of dsDNA yield for each subject and component, we cannot compute a within-subject coefficient of variation. Instead, the coefficients of variation of 0.31, 0.36, 0.66, and 0.52, respectively, for frozen whole blood, ACP, buffy coat, and residual blood cells reflect between-subject variability, compared with average yield across all subjects. Thus, the amount of dsDNA extractable from frozen buffy coat from various subjects was much more variable, compared with the mean yield, than was the case for ACP or whole blood. Relative yields from frozen residual blood cells were also more variable. The coefficients of variation from the 20 fresh samples were 0.35, 0.36, 0.37, and 0.45 for whole blood, ACP, buffy coat, and residual blood cells, respectively. Thus, the higher relative variability in yield for buffy coat and residual blood cells may be related to freezing and thawing after separating the buffy coat from the residual blood cells.
From the mean values and standard errors shown in Table 1, one can estimate the proportion of samples that yield insufficient dsDNA for a series of experiments. Suppose that one is planning a series of experiments that requires at least 20 μg of dsDNA for each subject with a frozen sample derived from 5 mL of whole blood. Assuming a normal distribution of yields, we calculated that the probability that a buffy coat sample will fail to reach or exceed this threshold is Φ {(20 − 38.0)/(7.91 ×101/2)} = 0.24, whereas, if ACP is used, the probability is only Φ {(20 − 79.1)/(8.24 × 101/2)} = 0.01. Here Φ is the standard normal distribution. Thus, approximately one in 4 buffy coat samples will yield insufficient dsDNA, as compared with only 1 in 100 ACP samples. These calculations depend not only on the mean yield of each component but also on the standard deviation measuring person-to-person variability.
We have data on the quality of these DNA samples after 2.3–2.7 years of storage at 4°C. All 30 absorbance ratios for DNA from whole blood and from ACP, whether fresh or frozen, fell within the desirable range of 1.7–2.0. One of 20 fresh buffy coat samples had an absorbance ratio greater than 2.0, and 1 had an absorbance ratio less than 1.7. Three of 10 frozen buffy coat samples had ratios less than 1.7. Two of 20 fresh residual blood cell samples had ratios greater than 2.0, and 1 had a ratio less than 1.7. Two of 9 frozen residual blood cell samples had ratios less than 1.7. Mean DNA sizes across all samples were 45,619 base pairs (standard deviation (SD), 4,313) for whole blood, 45,727 base pairs (SD, 4,127) for ACP, 43,158 base pairs (SD, 8,016) for buffy coat, and 41,360 base pairs (SD, 8,958) for residual blood cells. The Quantifiler assay confirmed that all samples were amplifiable by polymerase chain reaction. Electrophoresis gels revealed clear banding at the expected high-molecular-weight range, with minimal smearing or extra bands, indicating that the DNA samples were not degraded or fragmented, even after 2 years in storage.
DISCUSSION
Whole-blood fractions are the preferred DNA source for most epidemiologic studies, because they provide comparatively large quantities of high-quality DNA and other valuable biological material. Microgram quantities of DNA are needed for whole-genome scans, DNA sequencing, DNA methylation, and DNA adduct detection. Although DNA requirements for some assays have decreased as technology has improved, future assays for other analytes may require large amounts of DNA. Moreover, ample amounts of DNA are useful for performing several different assays on the same subject, repeating failed assays, assessing assay reliability, and performing separate confirmatory studies.
Frozen buffy coat has often been stored as a source of DNA and was recommended in earlier guidelines (2). We found that frozen buffy coat yields only about half the DNA that can be extracted from frozen ACP, and ACP is easier to obtain. The ACP fraction has about twice as many white blood cells as buffy coat, which can explain the difference in DNA yield (Figure 2). Our quality assessments after 2 years’ storage at 4°C indicated that ACP samples are at least as good as buffy coat, and perhaps better, as judged by absorbance ratios. Thus, we would recommend freezing ACP, which contained 80% of the DNA extractable from an equivalent quantity of whole frozen blood in our experiments. Alternatively, if one desires to separate buffy coat before freezing, one should take care to save the residual blood cells as well, because our data indicate that residual blood cells contain about half the extractable DNA found in ACP. Indeed, because standard methods not involving gradient centrifugation were used to isolate the buffy coat, the buffy coat contained both white and red cells and was not qualitatively different from the residual cell pellet, which also contained white and red cells.
Another advantage of ACP, compared with buffy coat and residual blood cells, is that the yields from frozen ACP are less variable, as measured by the coefficient of variation, than yields from buffy coat or from residual blood cells. Using mean dsDNA yields and person-to-person standard deviations in dsDNA yields, we found that not only does frozen ACP yield twice as much DNA as buffy coat, but the chance that a given individual will have an insufficient yield is also smaller for ACP. For example, if 20 μg of dsDNA are required for a series of measurements, we estimated that 1 in 4 frozen buffy coat samples from 5 mL of whole blood would be insufficient, compared with 1 in 100 ACP samples.
We are not aware of other reports describing dsDNA yields from whole blood, ACP, buffy coat, and residual blood cell fractions, either for fresh specimens or for frozen specimens. In a recent paper, Rosinger et al. (3) reported an average yield of single- and double-stranded DNA of 144 μg from the “residual cell pack” from 5 mL of fresh whole anticoagulated blood. A study of 120 samples of buffy coat (5), each collected from 8.5 mL of anticoagulated blood, yielded an average of 79.4 μg (135 × 5/8.5 = 79.4) of single- and double-stranded DNA per 5 mL of blood, or 55% of the yield for the “residual cell pack” in the study by Rosinger et al. (3). This percentage is consistent with our findings.
We found an average of 87.0 μg of dsDNA per 5 mL of fresh whole blood. This is lower than the average yield measured by absorbance spectrometry, which also detects single-stranded DNA, nucleotides, and even some signals from protein and RNA. Rosinger et al.'s report (3) of 144 μg per 5 mL of fresh whole blood seems to be based on normalizing absorbance spectrometry and PicoGreen fluorescence on the assumption that the DNA detection ratio comparing absorbance with fluorescence is 2:1. If this ratio holds, then 2 times our yield of 87.0 μg of dsDNA, namely 174 μg, is consistent with the value of 144 μg given by Rosinger et al. (3). The actual ratio depends on how the DNA was extracted and stored. An advantage of using the Quant-iT PicoGreen dsDNA Reagent and Kits to determine dsDNA is that dsDNA is the substrate for widely used polymerase chain reaction–based assays.
Our findings have important implications for the handling of anticoagulated whole blood for epidemiologic studies. If plasma will also be needed, one can separate the plasma from anticoagulated whole blood and either extract DNA from the ACP immediately or freeze the ACP at −80°C for future case-cohort or nested case-control studies, for example. This approach is less labor-intensive, and it doubles yield and reduces person-to-person variability in relative yield, compared with freezing only the buffy coat. If there is a special need to have buffy coat, one should also freeze the residual blood cells. If plasma is not needed, one could obtain maximal DNA by extracting it from the fresh blood directly or freezing the blood for future extraction.
Hemoglobin is needed for studies of hemoglobin adducts and glycans. For such studies, one must freeze intact red cells or extract hemoglobin from fresh blood before freezing it. Frozen ACP and residual blood cells contain red blood cells and are potential sources of hemoglobin for such studies, but further research is needed to confirm their utility for this purpose. Erythrocyte membrane lipids can be measured from fresh blood or from frozen red cells. Such assays might work on red cell membranes isolated from ACP or from residual blood cells, but further research is needed to show this. Red blood cell folate is estimated by subtracting serum or plasma folate concentrations from whole-blood folate concentrations, and it would be difficult to measure in fresh or frozen ACP.
Some researchers extract DNA from clotted blood. Although the DNA yield may be near 40 μg from a clot from a 10-mL tube of fresh blood, yields decline to less than 1 μg after 1 month of storage at 4°C (6). The amount and quality of the DNA may also depend on how the clot was disrupted and digested, which can introduce variability in yield.
Although we have studied the effects of freezing on yields of dsDNA from whole blood, ACP, buffy coat, and residual blood cells and although we have data on DNA quality after 2 years’ storage at 4°C, we do not have data on the long-term effects of freezing. Recent data indicate that yields from buffy coat remain fairly constant over a 9-year period (5). Further long-term studies are needed to confirm the quality and stability of DNA yields from ACP over time. Another potential limitation of our study is that we have used only 1 method to extract DNA, namely silica spin column capture with the Qiagen QIAamp DNA Blood Midi Kit. This method was chosen because it works well in fresh and frozen samples. This method yields a lower distribution of DNA lengths in unfrozen samples than phenol-chloroform extraction, but these distributions have similar modes after 18 freeze-thaw cycles (7).
Our data show that twice as much dsDNA can be obtained from frozen ACP as from frozen buffy coat or from residual blood cells, that ACP captures most of the dsDNA available from an equivalent quantity of frozen whole blood, and that compared with buffy coat or residual blood cells, the yields from ACP have a smaller coefficient of variation from person to person. Moreover, the processing of ACP is less labor-intensive and time-consuming. Higher yields can also reduce the amount of blood that needs to be drawn, which might improve participation rates in epidemiologic studies and clinical studies. Thus, epidemiologists and clinical scientists should consider storing ACP, or, if they decide to separate buffy coat initially, they should consider freezing the residual blood cell material as well, because it contains as much DNA as the buffy coat.
ACKNOWLEDGMENTS
Author affiliations: Division of Cancer Epidemiology and Genetics, National Cancer Institute, Rockville, Maryland (Mitchell H. Gail, Neil E. Caporaso, Karen Pitt, Regina G. Ziegler); Laboratory of Biospecimen and Biorepository Research, SAIC-Frederick Inc., Frederick National Laboratory for Cancer Research, Frederick, Maryland (Mark Cosentino, Norma A. Diaz-Mayoral); Information Management Services, Rockville, Maryland (David Pee); Promega, Global Genomics, Madison, Wisconsin (Tim Sheehy); and Division of Genetics and Epidemiology, The Institute of Cancer Research, Sutton, United Kingdom (Montserrat Garcia-Closas).
This project was funded in whole or in part by the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E and by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
We thank Dr. Bharathi Anekella of SeraCare Life Sciences (Gaithersburg, Maryland) for work on preliminary studies that led to this investigation; David Check of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, for graphics and data analysis; Marianne K. Henderson of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, for administrative support; and Dr. Meredith Yaeger of the Cancer Genomics Research Laboratory, Frederick National Laboratory for Cancer Research, for helpful comments on DNA quantitation.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Conflict of interest: none declared.
APPENDIX
Determination of DNA Yields Equivalent to 5 mL of Fresh Whole Blood
Using measured volumes in each of the 3 tubes labeled whole blood, all-cell-pellet (ACP), and buffy coat/residual blood cells, we calculated the DNA yield in micrograms that could be obtained for each blood fraction starting from 5 mL of fresh whole blood. Symbols for these volumes and corresponding DNA yields are shown in Appendix Table 1.
First we discuss the samples from which both fresh and frozen aliquots were analyzed. The DNA (in μg) extracted from the Vfrwb mL of fresh whole blood was Yfrwb, and the DNA extracted from the Vfzwb mL of frozen whole blood was Yfzwb. The concentration of DNA in μg/mL was Cfrwb = Yfrwb/Vfrwb for fresh whole-blood samples and Cfzwb = Yfzwb/Vfzwb for frozen whole-blood samples. Thus, the equivalent yield from 5 mL of whole blood is Xfrwb = 5 × Cfrwb for fresh whole blood and Xfzwb = 5 × Cfzwb for frozen whole blood.
To estimate the DNA yield from fresh ACP equivalent to a starting volume of 5 mL of whole blood, we use the formula Xfrapc = (5 × Cfrapc){(Vwbcacp + Vfracp + Vfzapc)/Vtube2}, where Cfrapc = Yfrapc/Vfrapc. Here the factor {(Vwbcacp + Vfracp + Vfzapc)/Vtube2} is needed because the volume Vtube2 is reduced to a volume Vwbcacp + Vfracp + Vfzapc in the process of preparing ACP material. Likewise, the yield for frozen ACP is given by Xfzapc = (5 × Cfzapc){(Vwbcacp + Vfracp + Vfzapc)/Vtube2}, where Cfzapc = Yfzapc/Vfzapc.
To estimate the DNA yield from fresh buffy coat equivalent to a starting volume of 5 mL of whole blood, we use the formula Xfrbc = (5 × Cfrbc){(Vwbcbc + Vfrbc + Vfzbc)/Vtube3}, where Cfrbc = Yfrbc/Vfrbc. Here the factor {(Vwbcbc + Vfrbcp + Vfzbc)/Vtube3 is needed because the volume Vtube3 is reduced to a volume (Vwbcbc + Vfrbc + Vfzbc) in the process of preparing buffy coat. Likewise, the yield for frozen buffy coat is given by Xfzbc = (5 × Cfzbc){(Vwbcbc + Vfrbc + Vfzbc)/Vtube3}, where Cfzbc = Yfzbc/Vfzbc. DNA yields from fresh residual blood cells equivalent to 5 mL of fresh whole blood are Xfrrbc = (5 × Cfrrbc){(Vwbcrbc + Vfrrbc + Vfzrbc)/Vtube3}, and those from frozen residual blood cells are Xfzrbc = (5 × Cfzrbc){(Vwbcrbc + Vfrrbc + Vfzrbc)/Vtube3}, where Cfrrbc = Cfzrbc/Vfrrbc and Cfzrbc = Yfzrbc/Vfzrbc.
For studies of the samples used for fresh DNA extraction only, the analogous formulas are Xfrwb = 5 × Cfrwb; Xfrapc = (5 × Cfrapc)(Vfrapc/Vtube2); Xfrbc = (5 × Cfrbc)(Vfrbc/Vtube3); and Xfrrbc = (5 × Cfrrbc)(Vfrrbc/Vtube3).
Appendix Table 1.
Symbols for Computation of DNA Yields Equivalent to 5 mL of Whole Blood
| Source | Volume Symbol, mL | Approximate Volume, mL | DNA Yield Symbol, μg |
|---|---|---|---|
| Samples used for fresh and frozen DNA extraction | |||
| Whole blood (tube has fresh whole blood volume = Vtube1) | |||
| White cell (fresh) | Vwbcwb | 1 | Ywbcwb |
| Fresh DNA extraction | Vfrwb | 4 | Yfrwb |
| Frozen DNA extraction | Vfzwb | 4 | Yfzwb |
| All-cell-pellet (tube has fresh whole blood volume = Vtube2) | |||
| White cell (fresh) | Vwbcacp | 1 | Ywbcacp |
| Fresh DNA extraction | Vfracp | 2 | Yfracp |
| Frozen DNA extraction | Vfzacp | 2 | Yfzacp |
| Buffy coat/residual blood cells (tube has fresh whole blood volume = Vtube3) | |||
| For buffy coat white cell (fresh) | Vwbcbc | 0.5 | Ywbcbc |
| For buffy coat fresh DNA extraction | Vfrbc | 1 | Yfrbc |
| For buffy coat frozen DNA extraction | Vfzbc | 1 | Yfzbc |
| Residual blood cells white cell (fresh) | Vwbcrbc | 0.5 | Ywbcrbc |
| Residual blood cells fresh DNA extraction | Vfrrbc | 1 | Yfrrbc |
| Residual blood cells frozen DNA extraction | Vfzrbc | 1 | Yfzrbc |
| Samples used for fresh DNA extraction only | |||
| Whole blood | Vfrwb | 8.5 | Yfrwb |
| All-cell-pellet | Vfracp | 5 | Yfracp |
| Buffy coat | Vfrbc | 1.5 | Yfrbc |
| Residual blood cells | Vfrrbc | 1.5 | Yfrrbc |
REFERENCES
- 1.Reddy MV, Randerath K. Nuclease-P1-mediated enhancement of sensitivity of P-32 postlabeling test for structurally diverse DNA adducts. Carcinogenesis. 1986;7(9):1543–1551. doi: 10.1093/carcin/7.9.1543. [DOI] [PubMed] [Google Scholar]
- 2.Austin MA, Ordovas JM, Eckfeldt JH, et al. Guidelines of the National Heart, Lung, and Blood Institute Working Group on Blood Drawing, Processing, and Storage for Genetic Studies. Am J Epidemiol. 1996;144(5):437–441. doi: 10.1093/oxfordjournals.aje.a008948. [DOI] [PubMed] [Google Scholar]
- 3.Rosinger S, Nutland S, Mickelson E, et al. Collection and processing of whole blood for transformation of peripheral blood mononuclear cells and extraction of DNA: the Type 1 Diabetes Genetics Consortium. Clin Trials. 2010;7(1 suppl):S65–S74. doi: 10.1177/1740774510373493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.SAS Institute Inc. SAS 9.2 Software. Cary, NC: SAS Institute Inc; 2010. [Google Scholar]
- 5.Mychaleckyj JC, Farber EA, Chmielewski J, et al. Buffy coat specimens remain viable as a DNA source for highly multiplexed genome-wide genetic tests after long term storage. J Transl Med. 2011;9:91–98. doi: 10.1186/1479-5876-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SSF, Kuei JJ, Prasad N, et al. A simple method for DNA isolation from clotted blood extricated rapidly from serum separator tubes. Clin Chem. 2007;53(3):522–524. doi: 10.1373/clinchem.2006.078212. [DOI] [PubMed] [Google Scholar]
- 7.Shao W, Khin S, Kopp WC. Characterization of effect of repeated freeze and thaw cycles on stability of genomic DNA using pulsed field gel electrophoresis. Biopreserv Biobank. 2012;10(1):4–11. doi: 10.1089/bio.2011.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]