Abstract
The mineralocorticoid receptor (MR) is a ligand-induced transcription factor belonging to the steroid receptor family and involved in water-electrolyte homeostasis, blood pressure regulation, inflammation and fibrosis in the renocardiovascular system. The MR shares a common hormone-response-element with the glucocorticoid receptor but nevertheless elicits MR-specific effects including enhanced epidermal growth factor receptor (EGFR) expression via unknown mechanisms. The EGFR is a receptor tyrosine kinase that leads to activation of MAP kinases, but that can also function as a signal transducer for other signaling pathways. In the present study, we mechanistically investigate the interaction between a newly discovered MR- but not glucocorticoid receptor- responsive-element (=MRE1) of the EGFR promoter, specificity protein 1 (SP1) and MR to gain general insights into MR-specificity. Biological relevance of the interaction for EGFR expression and consequently for different signaling pathways in general is demonstrated in human, rat and murine vascular smooth muscle cells and cells of EGFR knockout mice. A genome-wide promoter search for identical binding regions followed by quantitative PCR validation suggests that the identified MR-SP1–MRE1 interaction might be applicable to other genes. Overall, a novel principle of MR-specific gene expression is explored that applies to the pathophysiologically relevant expression of the EGFR and potentially also to other genes.
INTRODUCTION
The mineralocorticoid receptor (MR) is a ligand-bound transcription factor that shares its classical hormone-response-element, the glucocorticoid-response-element (GRE), with the glucocorticoid receptor (GR) but elicits different effects involved in stress and immune response and metabolism. The GR closely resembles the MR in structure but not in function, and the mechanisms for MR specificity over GR remain ambiguous. Of the MR-mediated actions, its pathophysiological effects on the cardiovascular system and the kidney are of special interest. In these tissues, inappropriate activation of the MR leads to inflammation, hypertrophy and tissue remodeling, and in several clinical trials, MR antagonists reduced mortality and morbidity of patients suffering for instance from congestive heart failure or myocardial infarction most likely due to a reduction in vascular remodeling (1,2). Interestingly, some of the mechanistically unexplained pathological MR effects in the cardiovascular system and the kidney (2–8) are mimicked by the epidermal growth factor receptor (EGFR), raising the possibility of them being mediated by the cross-talk between the two signaling pathways.
The EGFR (ERBB1) is the most prominent member of the membrane tyrosine kinase family including ERBB2, ERBB3 and ERBB4. Depending on the genetic background, EGFR knockout mice die at peri-implantation, midgestation or within the first 3 weeks, showing the importance of the EGFR for embryonic development and cell differentiation (9). Furthermore, a crucial role in cell proliferation, migration and pathological tissue remodeling has been demonstrated (10). For the vasculature, we could recently show its importance for physiological tone and vessel reactivity in vivo (11). The underlying signaling network of the EGFR is intricate and includes ligand-dependent transmembrane signal transduction and transactivation of the EGFR through other signaling pathways (12). For transmembrane signaling, the EGFR forms homo- or heterodimers with its family members on binding of one of its various ligands (e.g. EGF, heparin-binding EGF, tumour necrosis factor a, amphiregulin, betacellulin, epiregulin). This leads to activation of important cytosolic downstream targets such as mitogen-activated protein kinases, phospholipase Cδ, PI3 kinase or cSrc (13). Alternatively, the EGFR can be transactivated by cross-talk with other signaling pathways, for example with G-protein-coupled receptors or with steroid receptors like the MR (14,15). Overall, the EGFR functions as an important relay station for a wide variety of different signaling molecules, highlighting the potential impact of changes in its expression.
So far, both MR transactivation of the EGFR and also modulation of genomic MR activity by downstream kinases of the EGFR have been described (15–17). As an additional mechanism, we recently reported an MR-dependent increase in EGFR expression that was mediated by binding of MR to the EGFR promoter. Reporter gene assays with deletion constructs of the EGFR promoter revealed an MR- but not GR-responsive element (= MRE1) stretching from −316 to 163 bp, thus being a putative MR-specific element. The region contained no typical GRE (18). In the current article, we use the MR-MRE1 model to investigate the mechanisms underlying the MR–DNA interaction to characterize specificity-conferring molecular parameters in more detail. We determined the responsive DNA regions and identified specificity protein 1 (SP1) as the necessary cofactor for this MR–DNA interaction. Furthermore, a bioinformatical genome-wide screen for similar promoter elements revealed additional MR-target genes that were validated by quantitative PCR (qPCR), suggesting that the mechanism is a more general regulatory mechanism for conferring MR-specificity. The importance of the EGFR for other signaling pathways involving cell proliferation and tissue remodeling is highlighted.
MATERIALS AND METHODS
Cell culture
Human embryonic kidney cells (HEK-293), Human kidney 2 cells (HK2) and A7r5 cells were acquired from American Type Culture Collection (Rockville, MD) and cultivated in Dulbecco’s modified Eagle’s medium/Ham’s F-12 medium supplemented with 10% fetal calf serum (FCS) and Dulbecco’s modified Eagle’s medium supplemented with 1.5 g/l bicarbonate and 4.5 g/l glucose and 10% FCS, respectively. HEK cells do not possess detectable levels of endogenous MR but GR. OK cells, a proximal tubule cell line from opossum, obtained from Dr Biber, Dept of Physiology, University of Zurich were grown in MEM medium (pH 7.4), supplemented with Earl’s salts, non-essential amino acids and 10% (v/v) FCS. Human aortic smooth muscle (HAoSMC) primary cells were purchased from Promocell, Heidelberg, Germany and were cultivated in Smooth Muscle Cell Growth Medium 2 (Promocell, Heidelberg, Germany) supplemented with 5% FCS. Cells were maintained at 37°C in a humidified 5% CO2 atmosphere. Transient transfections were performed with Polyfect Reagent (Qiagen, Hilden, Germany) for HEK and HK2 cells and Effectene (Qiagen, Hilden, Germany) for OK cells, according to the manufacturer’s instructions in medium without supplements. Before experiments, cells were made quiescent by incubating them for at least 24 h in medium without serum and steroids. Primary culture of murine vascular smooth muscle cells (VSMC) were obtained from mice with SM22-dependent deletion of EGFR and wild-type littermates and cultured as described previously (11).
Plasmids
pEGFP-hMR is a kind gift of N. Farman (Paris). The truncated versions of pEGFP-hMR have been described previously (18). To exclude that the EGFP-tag of the human mineralocorticoid receptor (hMR) changes the characteristics of the receptor, we compared its GRE activation and nuclear translocation properties with that of untagged hMR but could not find significant differences (15).
For in vitro synthesis of hMR and Human glucocorticoid receptor (hGR), pcDNA3.1His (Invitrogen, Darmstadt, Germany) was used as backbone. Briefly, hMR from pEGFP-hMR was ligated into pcDNA3.1HisC after restriction with BamHI/BglII and ApaI. Human glucocorticoid receptor was excised from pcDNA1-hGR (gift from M. Govindan) with EcoRI and inserted into pcDNA3.1HisB. pcDNAHisLacz was purchased from Invitrogen.
As a reporter gene, we used secretory alkaline phosphatase (SEAP) encoded on a pSEAP-basic vector obtained from Clontech (Mountain View, CA). The fragments of the EGFR promoter tested in the reporter gene assay were cloned into this vector by PCR cloning with the help of a T4 DNA Ligase (Invitrogen, Karlsruhe, Germany). Promoter fragments with appropriate restriction sites were created according to their length either by PCR or oligonucleotide hybridization (Supplement 1). All cloned promoter fragments were sequenced (Eurofins MWG Operon, Ebersberg, Germany) to confirm sequence identity.
Reporter gene assay
Reporter gene assays with secretory alkaline phosphatase as reporter were performed as described previously (15). SEAP activity was measured by fluorescence measurements with the AttoPhos System (Promega, Mannheim, Germany). Beta-galactosidase encoded by pcDNA3.1Hislacz (Invitrogen, Darmstadt, Germany) was cotransfected as an internal control. Beta-galactosidase activity was measured by a colorimetric assay with ortho-nitrophenyl-β-galactoside as substrate and photometric measurement at 405 nm. For the reporter gene assay with the classical GRE, we applied the pGRE-SEAP plasmid from Clontech consisting of three glucocorticoid response elements. Inhibition experiments with the SP1 inhibitor WP631 dimethanesulfonate (Santa Cruz Biotechnology, Heidelberg, Germany) and the aldosterone antagonist eplerenone were performed with 5 μM unless indicated otherwise.
siRNA experiments
Highly specific SP1 and SP3 ON-TARGETplus SMARTpool siRNA and as negative control ON-TARGETplus Non-targeting Pool from Thermo Scientific Dharmacon (Epsom, UK) was used for knockdown of human SP1 and SP3. HEK cells were transiently transfected with DharmaFECT (Thermo Scientific Dharmacon, Epsom, UK) in six-well plates according to the manufacturer’s instructions. Cells were collected after 48 h, 72 h and 96 h for total RNA isolation and real-time PCR analysis. Total proteins for western blot analysis were isolated after 72 h, 96 h and 120 h. HEK cells for SEAP assays were harvested in 24-well plates and incubated with siRNA for 96 h after transient transfection with DharmaFECT.
Western blot
Cells were washed, harvested and lysed in Cell Signaling Technology (CST) buffer [20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM Na3VO4]. Subsequently, cell lysates were matched for protein content, separated by 10% SDS–PAGE and transferred to nitrocellulose membrane. Membranes were incubated with anti-SP1 (Abcam, Cambridge, UK), anti-SP3 (Millipore, USA), anti-EGFR, anti-pp42/44, β-Actin (both from Cell Signaling Technology, NEB Frankfurt, Germany) or HSP90α/β (Santa Cruz Biotechnology Inc., USA).
The bound primary antibody was visualized using horseradish peroxidase-conjugated secondary IgG and the Immun-Star WesternC Chemiluminescent Kit (Biorad, Munich, Germany). The signal was detected with the chemiluminescence detection system ChemiDoc XRS (Biorad, Munich, Germany) in the linear detection range. Densitometry analysis was performed with Quantity One (Biorad, Munich, Germany).
Enzyme-linked immunosorbent assay-based transcription factor DNA-binding assay
In vitro hMR, hGR and LacZ (negative control) were synthesized using the TNT® T7 Quick Coupled Transcription/Translation system from Promega (Mannheim, Germany) and using pcDNA3.1-HisC-hMR, pcDNA3.1-His-hGR or pcDNA3.1His-Lacz. Nuclear extracts were isolated from HEK cells with the Nuclear Extraction Kit (Active Motif, Rixensart, Belgium) and desalted using centrifugal filter units (Millipore, Billerica, USA). Biotinylated probes were constructed by hybridization or PCR with bio-dUTP (Roche, Mannheim, Germany) and MRE1-SEAP reporter plasmids as template (Primer see Supplement 1). These probes were immobilized in streptavidin-coated wells. Wells without probes were used as blanks. Either in vitro synthesized hMR, hGR with or without human recombinant SP1 (Promega, Mannheim, Germany) or nuclear extracts were added to the wells and incubated for 1 h at room temperature with an equal volume of blocking buffer [5% bovine serum albumin (BSA) in PBS/Tween 0.05%]. After three washes, the transcription factor bound to the biotinylated probe was incubated with anti-XPRESS antibody (1:2000, Invitrogen, Darmstadt, Germany), anti-GR (1:500 Active Motif, Rixenart, Belgium) or anti-rMR1-18 1D5 (1:200), a kind gift from C. Gomez-Sanchez (19). After three further washes, a horseradish peroxidase-coupled secondary antibody was added and detected by colorimetric assay (0.5 mg/ml o-phenylenediamine, 11.8 mg/ml Na2HPO4 • 2H2O plus 7.3 mg/ml citric acid and 0.015% H2O2) at 490 nm in a multiwell reader.
Electromobility shift assay
Electromobility shift assays (EMSAs) were performed with biotinylated probes obtained by PCR with biotinylated dUTPs or oligo hybridization (primers see Supplement 1) and the LightShift Chemiluminescence EMSA Kit (Thermo Fisher Scientific, Bonn, Germany). Binding reactions were carried out for 30 min at room temperature in the presence of 10 mM Tris, 50 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol and 1 μg/μl poly (dI-dC). Biotinylated probes were used in a concentration of 5 fmol per reaction and incubated with 300 ng of recombinant human SP1 (rhSP1) (Promega, Mannheim, Germany), 100 ng of rhSP3 (Abnova, Taiwan) or BSA as negative control. Nuclear extracts were isolated from HEK cells with the Nuclear Extraction Kit (Active Motif, Rixensart, Belgium) and desalted using centrifugal filter units (Millipore, Billerica, USA). Electrophoresis of the DNA–protein complexes was carried out with a 10% non-denaturing polyacrylamide gel at 140 V for 1:15 h. Afterwards, the samples were transferred to a nylon-membrane at 250 mA for 45 min and cross-linked on the membrane at 120 mJ/cm2 for 1 min. Detection was done with the Chemiluminescent Nucleic Acid Detection Module (Thermo Fisher Scientific, Bonn, Germany) according to the manufacturer’s instructions with extended washing steps.
Computation of optimal semi-global alignments
We computed optimal semi-global alignments of MRE1.3 or infixes thereof with promoter regions of the human genome by dynamic programming using a match score of −1, a mismatch score of +2 and an affine gap cost function with a gap opening cost of +2 and a gap elongation cost of +1 (www.jstacs.de).
Quantification of mRNA by real-time RT-PCR
Total RNA from transiently transfected hMR-HK2 cells was isolated using InviTrap® Spin Tissue RNA Mini Kit (Invitek, Berlin, Germany) following the manufacturer’s protocol. To quantify mRNA levels, 1 µg of RNA was converted to cDNA, and equal amounts of cDNA were applied to RT-PCR reaction. Used primers are listed in Supplement 1.
Statistics
The data are presented as mean values ± SEM. Significance of difference was tested by Student’s t-test with P < 0.05 considered statistically significant. N represents the number of individual experiments and n the number of wells or culture dishes investigated per experiment.
RESULTS
Importance of EGFR for additional signaling pathways
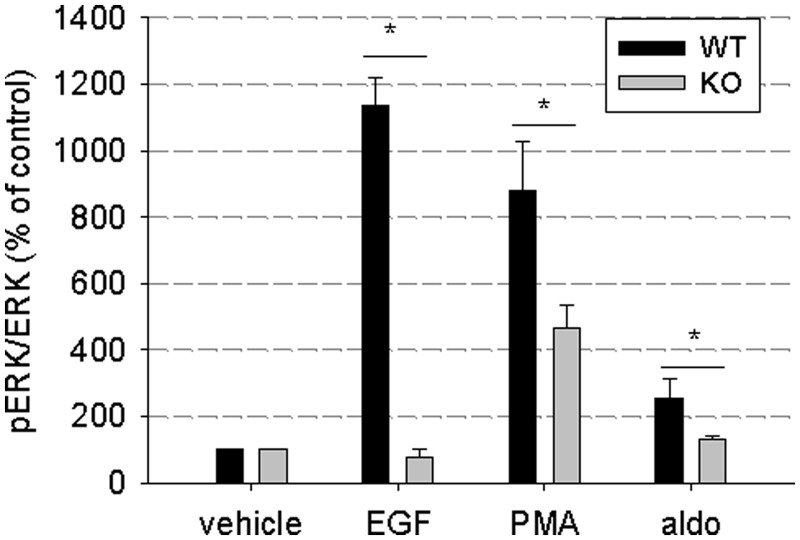
To emphasize the importance of EGFR for different signaling pathways, stimulation of extracellular signal-regulated kinase (ERK) phosphorylation via different pathways was compared in VSMC of EGFR-knockout mice or wild-type littermates. In VSMC of wild-type mice, ERK phosphorylation could be induced by the EGFR ligand EGF, by protein kinase C (PKC) activator phorbol-12-myristate-13-acetate or by the MR ligand aldosterone. In VSMC of EGFR-knockout mice, activation of ERK phosphorylation was impaired after stimulation of EGFR, PKC or MR signaling, confirming the widespread importance of EGFR expression for various signaling pathways (Figure 1).
Figure 1.
Importance of EGFR as a mediator for other signaling pathways. VSMC of EGFR knockout or wild-type mice were incubated for 5 min with the EGFR ligand EGF (10 µg/l), the PKC activator phorbol-12-myristate-13-acetate (1 µM) or the MR ligand aldosterone (10 nM aldosterone). ERK phosphorylation was assessed by western blot (n = 4–5, data represented as mean ± SEM).
MR interaction site on the EGFR promoter
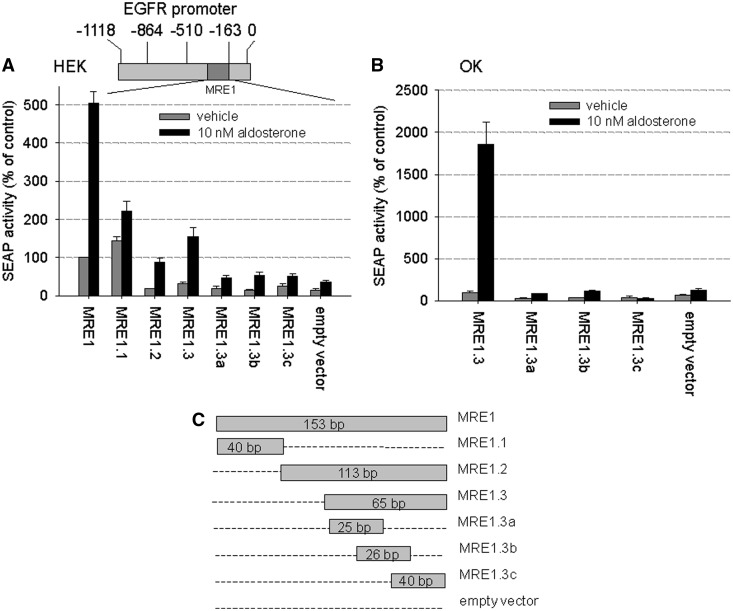
To determine the binding region necessary for the interaction between EGFR promoter and activated MR, we cloned parts of the EGFR promoter fragment MRE1 (Figure 2A, top) into a reporter gene vector (empty vector = SEAPbasic) as indicated in Figure 2C. We then performed reporter gene assays in transiently hMR-transfected HEK cells that were incubated with vehicle or 10 nM aldosterone (Figure 2A). The fold induction achieved by aldosterone was largest for the fragments MRE1.2 and MRE1.3, suggesting that MRE1 contains an inhibitory element. As MRE1.2 comprises MRE1.3, the 65 bp long MRE1.3 fragment seems to be sufficient to confer MR-responsiveness to MRE1. Neither MRE1.3a nor MRE1.3b nor MRE1.3c showed aldosterone/MR-responsiveness comparable with that of the complete MRE1.3, suggesting that smaller fragments are not sufficient to mediate MR-induced EGFR expression. Key experiments were repeated in OK cells in which the stimulation of MRE1.3 compared with MRE1.3a, b and c or empty vector is more pronounced (Figure 2B).
Figure 2.
MR interaction site on EGFR promoter (A) HEK cells top: Scheme of the EGFR promoter showing localization of MRE1, bottom: HEK cells transiently transfected with MRE1 reporter plasmid and hMR were incubated with 10 nM aldosterone or vehicle for 48 h, and then reporter gene activities were measured and depicted as percentage of control (MRE1 with vehicle) (N = 3–18, n = 9–45, data represented as mean ± SEM), (B) OK cells transiently transfected with MRE1 reporter plasmid and hMR were incubated with 10 nM aldosterone or vehicle for 48 h, and then reporter gene activities were measured and depicted as percentage of control (MRE1.3 with vehicle) (N = 4–16, n = 15–47, data represented as mean ± SEM), (C) Scheme of the different MRE1 promoter deletion constructs tested in the reporter gene assay.
MR specificity and dose dependency of MRE1.3
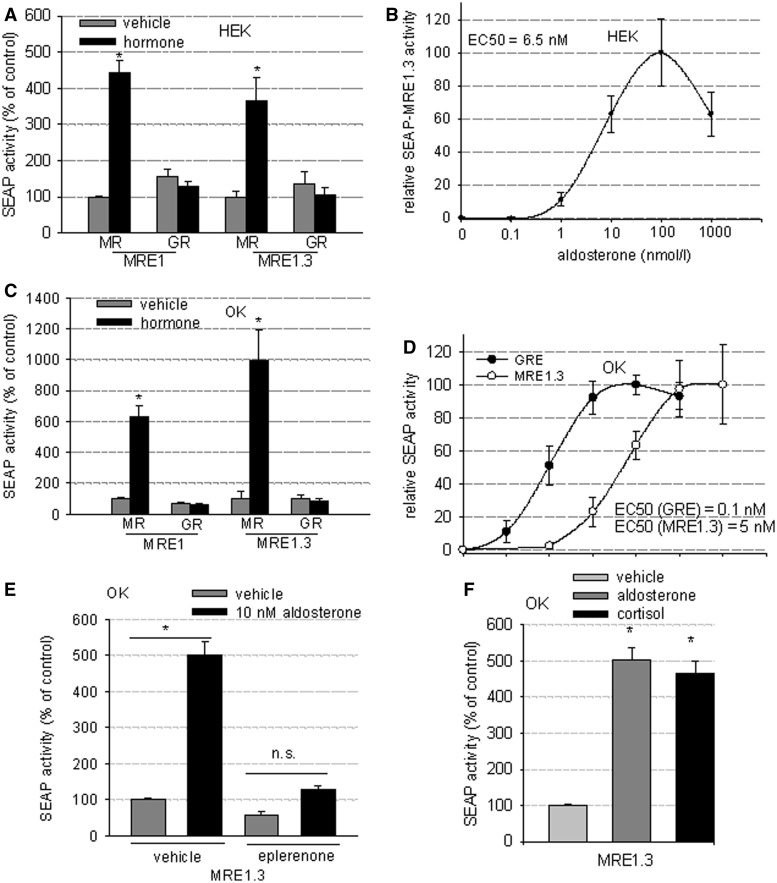
To verify MR specificity, we tested MRE1 and MRE1.3 in a reporter gene assay in HEK cells transiently transfected with either hMR or hGR expression plasmid. In HEK cells overexpressing hGR and stimulated with 100 nM dexamethasone, no increase in reporter gene activity was detected compared with unstimulated cells. In contrast, HEK cells overexpressing hMR and stimulated with 10 nM aldosterone showed a dose-dependent increase in reporter activity as depicted in Figure 3A and B (EC50 = 6.5 nM). To rule out cell-specific effects, experiments were repeated in OK cells (Figure 3C and D), which rendered similar qualitative results and quantitatively an even greater stimulation of MRE1.3 by aldosterone/hMR. In OK cells transiently transfected with hGR, incubation with 100 nM dexamethasone did not increase reporter gene activity of MRE1 or MRE1.3, whereas OK cells transfected with hMR and stimulated with 10 nM aldosterone showed a dose-dependent induction of reporter gene activity (EC50 = 5 nM). Dose-response curves with classical GRE-SEAP reporter plasmid showed an over 10-fold lower EC50 in OK cells (EC50 = 0.1 nM) (Figure 3D) and also HEK cells as published previously (20). For further evidence that the EGFR promoter is activated specifically by MR, the MR antagonist eplerenone (5 µM) was applied. In the presence of eplerenone, 10 nM aldosterone was no longer able to stimulate MRE1.3 (Figure 3E). Because the glucocorticoid cortisol can also function as an MR ligand and is able to induce transactivation of a classical GRE, the effect of cortisol on OK cells transfected with MR and the MRE1.3 reporter plasmid (Figure 3F) was also tested. Cortisol could stimulate MRE1.3 reporter gene activity to a similar extent as aldosterone.
Figure 3.
MR specificity and dose dependency of MRE1.3 in HEK and OK cells. (A) MR-specificity of MRE1.3 was tested in a reporter gene assay in HEK cells transfected either with MR or GR plus MRE1.3 reporter plasmid and then stimulated with 10 nM aldosterone or 100 nM dexamethasone accordingly (N = 6–8, n = 14–24, data represented as mean ± SEM, *P ≤ 0.05). (B) A dose-response-curve for rising aldosterone concentrations was constructed for MR transactivation activity at MRE1.3 in hMR-HEK cells and EC50 was calculated (N = 3–8, n = 9–27, data represented as mean ± SEM). (C) Experiment (A) was repeated with OK cells to exclude cell-specific effects (N = 4, n = 8–12, data represented as mean ± SEM, *P ≤ 0.05). (D) Experiment (B) was also repeated with OK cells transfected with MR and MRE1.3 or GRE reporter plasmid (N = 3–6, n = 9–18 data represented as mean ± SEM), (E) Influence of eplerenone (antagonist of aldosterone) on MR/aldosterone induced response of MRE1.3 was tested in a reporter gene assay in transiently transfected hMR OK cells after incubation with either vehicle or 10 nM aldosterone after 48 h and depicted as percentage of control (MRE1.3 with vehicle) (N = 3, n = 9, data represented as mean ± SEM), (F) Activation of MRE1.3 by cortisol and aldosterone was analyzed in a reporter gene assay in OK cells transiently transfected with hMR and MRE1.3 and incubation with vehicle, 10 nM aldosterone or 1 µM cortisol for 48 h (N = 3, n = 9, data represented as mean ± SEM).
MR binding to MRE1
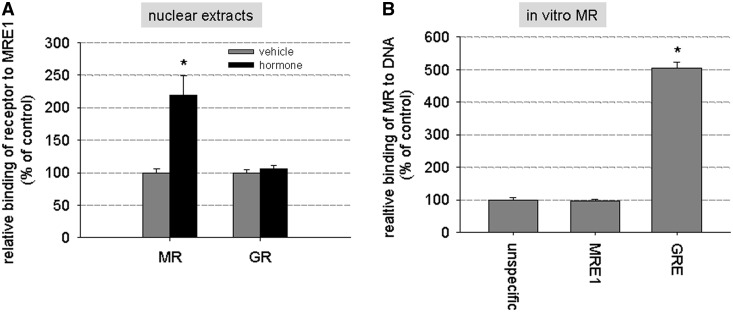
Binding of hMR to MRE1 was tested in a transcription factor ELISA with biotinylated MRE1 probe. hMR binding to MRE1 was significantly increased when using nuclear extracts from hMR-overexpressing HEK cells incubated with 10 nM aldosterone (Figure 4A). No increase in binding of GR to MRE1 could be detected in nuclear extracts of GR-overexpressing HEK cells incubated with 1 µM hydrocortisone compared with unstimulated cells. However, binding of in vitro synthesized hMR to MRE1 did not exceed unspecific DNA binding (Figure 4B) as was tested with biotinylated eNOS exon DNA of approximately the same length as MRE. As positive control, a probe with three classical GREs was applied (Figure 4B).
Figure 4.
In vitro binding of MR to MRE1 (A) In a transcription factor-binding ELISA, nuclear extracts of hMR- and hGR-HEK cells that had been previously stimulated with vehicle, 10 nM aldosterone or 1 µM hydrocortisone, respectively, were tested for binding of hMR or hGR to biotinylated MRE1. (N = 1–3, n = 4–11, data represented as mean ± SEM, *P ≤ 0.05). (B) Binding of in vitro synthesized MR to MRE1 was tested and compared with unspecific binding of MR to DNA (unspecific = binding of MR to DNA probes of exon of eNOS) and binding of MR to a probe containing three classical GREs, which was used as positive control for this experiment (N = 2–3, n = 4–6, data represented as mean ± SEM, *P ≤ 0.05).
Binding of cofactor to MRE1.3
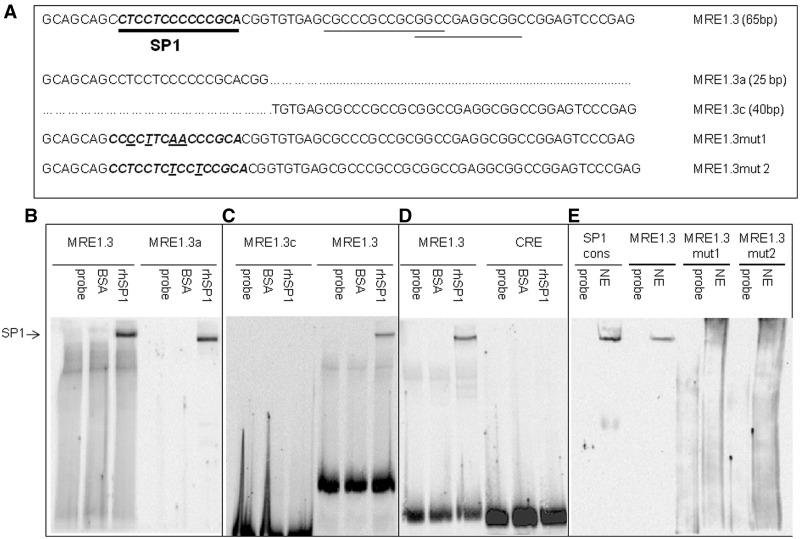
Both the length of MRE1.3 as the smallest element with full MR-responsiveness and the discrepancy in the binding of in vitro hMR and hMR from nuclear extracts to MRE1 suggest the involvement of one or more cofactors in the binding of MR to MRE1.3. To identify such potential cofactors, we performed an in silico search for transcription factor-binding sites in the sequence of MRE1.3 (AliBaba 2.1, http://www.gene-regulation.com/pub/programs.html). This search identified three possible binding sites of SP1 in the MRE1.3 sequence indicated by underlining in Figure 5A. To validate these sites, we performed electromobility shift experiments with rhSP1 and biotinylated MRE1.3, MRE1.3a, MRE1.3c or CRE as a negative control. Incubation of MRE1.3 or MRE1.3a with rhSP1 resulted in a retention band not detectable when testing unspecific protein binding with BSA (Figure 5B). No binding of rhSP1 could be detected to MRE1.3c, indicating that SP1 only binds to the first of the three predicted SP1-binding sites (Figure 5C). To exclude unspecific binding of rhSP1 to DNA, we repeated the experiments with a biotinylated cAMP response element (CRE) probe of comparable length that did not yield a retention band (Figure 5D). To provide evidence for direct binding of SP1 to MRE1.3 in cells, EMSAs for MRE1.3 were repeated with nuclear extracts of transiently MR-transfected HEK cells. Sp1 consensus binding sequence was used as a positive control. Binding of biotinylated MRE1.3 probe was detected as a specific retention band comparable with our results using recombinant Sp1 and nuclear extracts with Sp1 consensus sequence. Experiments with nuclear extracts and MRE1.3 probe with mutated SP1-binding sites (see Figure 5A) did not result in a retention band, suggesting specific binding of SP1 to MRE1.3a (Figure 5E).
Figure 5.
Binding of SP1 to MRE1.3, MRE1.3a and MRE1.3c in EMSAs. (A) Sequences of MRE1.3 with in silico predicted SP1-binding sites, different MRE1.3 deletion constructs and constructs with mutated SP1-binding site are depicted. (B) EMSAs for detection of SP1 binding were performed with biotinylated MRE1.3 (lanes 1–3) or MRE1.3a (lanes 4–6) probe. Probes were either incubated with buffer (probe), 300 ng BSA as control for unspecific protein interactions (BSA) or with 300 ng of rhSP1. (C) To identify the relevant SP1-binding site for the interaction, we compared biotinylated MRE1.3c (lanes 1–3) with biotinylated MRE1.3 (lanes 4–6). Probes were either incubated with only buffer (probe), 300 ng of BSA as control for unspecific protein interactions (BSA) or with 300 ng of rhSP1. (D) To exclude unspecific SP1-DNA interactions, EMSAs with biotinylated MRE1.3 (lanes 1–3) were compared with EMSAs with biotinylated CRE (cAMP response element) (lanes 4–6) probe. Probes were either incubated with only buffer (probe), 300 ng of BSA as control for unspecific protein interactions (BSA) or with 300 ng of rhSP1 (N = 6). (E) To verify in vivo binding of SP1 to MRE1.3, we analyzed binding of protein from nuclear extracts to MRE1.3 and MRE1.3 with mutations in the SP1-binding sequence, MRE1.3mut1 and MRE1.3mut2. Biotinylated SP1 consensus sequence was used as a positive control. Probes were either incubated with only buffer (probe) or nuclear extracts (NE) (N = 4).
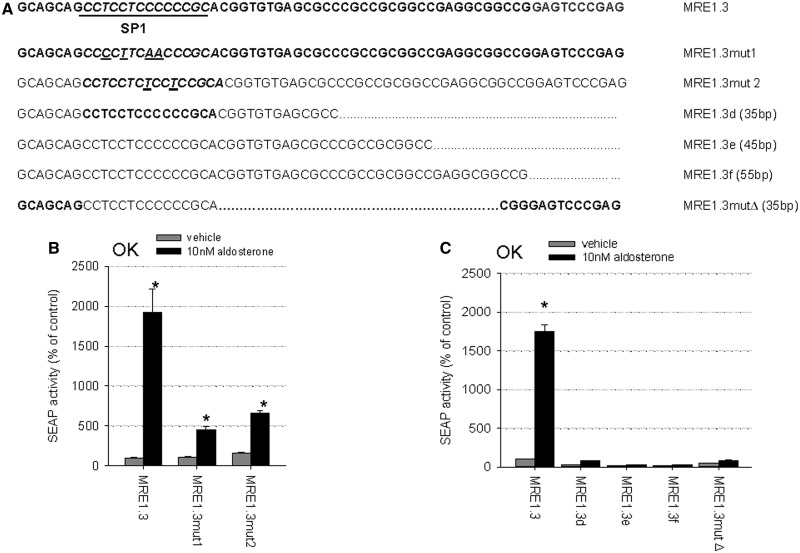
To confirm the importance of the SP1-binding site located in MRE1.3a for MR-induced EGFR promoter activation, reporter gene assays with the MRE1.3 point mutations depicted in Figure 6A were performed. The reporter gene activities after stimulation with activated MR were markedly reduced compared with the intact MRE1.3 sequence (Figure 6B). Despite the importance of the 5′-located SP1 site, it was not possible to delete parts of the 3′-end of MRE1.3 without losing MR responsiveness (Figure 6C). Furthermore, deleting the 30 base pairs located between the SP1-binding site and the 3′-end led to a decline in MR transactivation so that also the middle part seems to be of functional importance.
Figure 6.
Importance of full-length MRE1.3 and the SP1-binding site (A) Sequences of MRE1.3, different MRE1.3 deletion constructs and constructs with mutated SP1-binding site are depicted. (B) OK cells transiently transfected with hMR and reporter plasmids were incubated with 10 nM aldosterone or vehicle for 48 h and then reporter gene activities were measured and depicted as percentage of control (MRE1.3 with vehicle). MRE1.3mut1 and MRE1.3mut2, both mutated in the SP1-binding site, were compared with MRE1.3 (N = 3–6, n = 9–18, data represented as mean ± SEM). (C) Transiently hMR- and reporter plasmid transfected OK cells were incubated with 10 nM aldosterone or vehicle for 48 h. Reporter gene activities of four different deletion constructs (MRE1.3b, e and f and MRE1.3mut Δ) were measured and compared with full-length MRE1.3 transactivation (N = 3–6, n = 9–18, data represented as mean ± SEM).
SP1 as cofactor for aldosterone-induced MRE1.3 promoter activity
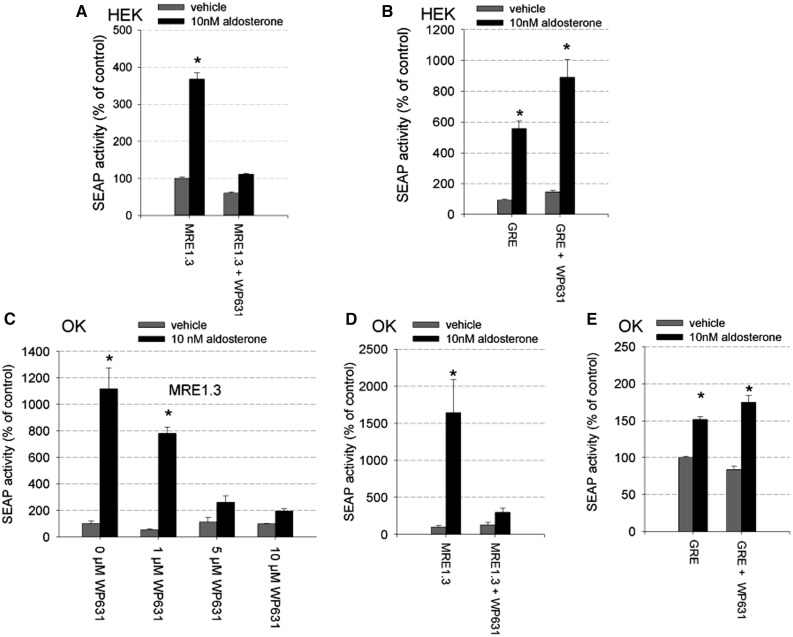
To investigate the role of SP1 for hMR responsiveness of MRE1.3 promoter activity further, we repeated the reporter gene assays in HEK cells with transiently transfected hMR, but this time added the SP1 inhibitor WP631 to the incubation solution. Compared with hMR-overexpressing cells incubated with aldosterone alone, cells incubated additionally with WP631 no longer showed a significant reporter gene induction (Figure 7A). No general inhibition of transcription caused by WP631 was detectable because aldosterone-induced GRE reporter gene activity was not affected by the presence of WP631 (Figure 7B). Again cell-type-specificity was ruled out by repeating the experiments in OK cells. OK cells showed a dose-dependent reduction in MRE1.3 promoter activity by rising concentrations of WP631 (Figure 7C). With 5 µM WP631, aldosterone-hMR-induced activation of the reporter gene activity could be inhibited (Figure 7D), whereas GRE reporter gene activiation by activated hMR was not affected (Figure 7E).
Figure 7.
Effect of SP1 inhibition by WP631 on MR-induced MRE1.3 reporter gene activity. (A) HEK cells transfected with hMR and MRE1.3 reporter plasmid were incubated for 48 h with vehicle or 10 nM aldosterone in the presence and absence of 5 µM WP631, a potent inhibitor of SP1-dependent transcription (N = 3, n = 6–9, data represented as mean ± SEM, *P ≤ 0.05). (B) To exclude unspecific effects on transcription, reporter gene activity was also measured in HEK cells transfected with hMR and classical GRE reporter plasmid after incubating the cells with vehicle or 10 nM aldosterone in the presence and absence of 5 µM WP631 (N = 3, n = 9, data represented as mean ± SEM, *P ≤ 0.05). (C) The effect of rising concentrations of WP631 on aldosterone-induced MRE1.3 promoter activity was tested in OK cells transfected with hMR (N = 1–11, n = 3–33, data represented as mean ± SEM, *P ≤ 0.05), (D) OK cells transfected with hMR and MRE1.3 reporter plasmid were incubated for 48 h with vehicle or 10 nM aldosterone in the presence and absence of 5 µM WP631 (N = 3–4, n = 9–12, data represented as mean ± SEM, *P ≤ 0.05), (E) To exclude unspecific effects on transcription, reporter gene activity was also measured in OK cells transfected with hMR and classical GRE reporter plasmid after incubating the cells with vehicle or 10 nM aldosterone in the presence and absence of 5 µM WP631 (N = 10, n = 30, data represented as mean ± SEM, *P ≤ 0.05).
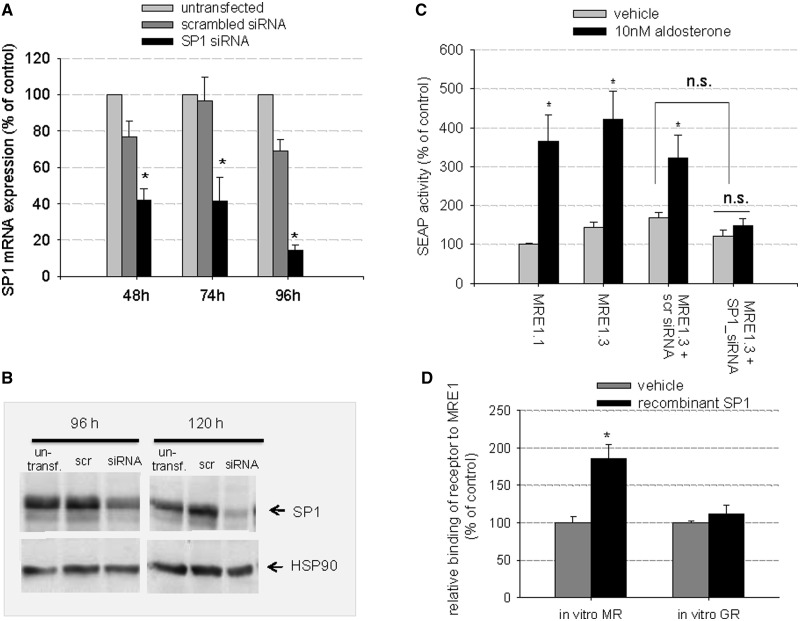
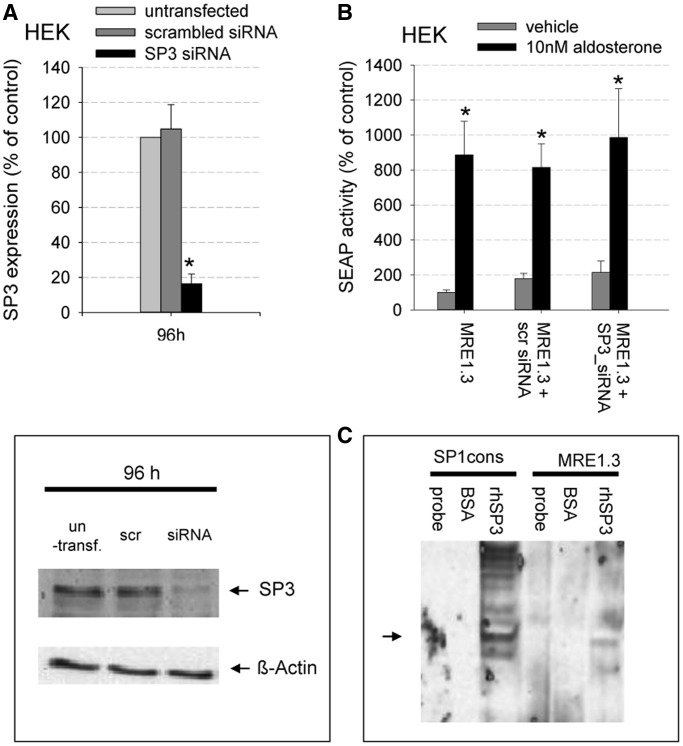
Next, we repeated the MRE1.3 reporter gene assays in the presence of SP1-specific siRNA or scrambled siRNA. Scrambled siRNA did not lead to significant reduction in SP1 mRNA or protein expression within 96 h (mRNA) or 120 h (protein) after transfection (Figure 8A and B). SP1-specific siRNA, on the other hand, reduced SP1 levels increasingly with time with a maximal reduction of mRNA levels to 20.9 ± 4% 96 h after transfection and a decrease in SP1 protein levels to 34.5 ± 6% 120 h after transfection with siRNA. Consequently, we cotransfected our hMR-HEK cells with SP1-siRNA and then incubated the cells with siRNA or scrambled siRNA before the reporter gene assay. Aldosterone-mediated induction of MRE1.3 reporter gene activity was inhibited by SP1-specific siRNA but not by scrambled siRNA (Figure 8C). Of note, basal SEAP activity was not significantly reduced after either treatment with SP1 inhibitor or specific SP1-siRNA, suggesting that SP1 is not essential for basal promoter activity but for MR-induced gene regulation.
Figure 8.
Importance of SP1 for MR-induced MRE1.3 reporter activity and binding of MR to MRE1: (A–C) Effect of SP1 inhibition by SP1-siRNA on MR-induced MRE1.3 reporter gene activity (A) HEK cells were transiently transfected with hMR and with either scramble-siRNA or SP1-specific siRNA. SP1- mRNA levels were then quantified by qPCR 48, 72, and 96 h after transfection and compared to untransfected cells (N = 3, n = 6, data represented as mean ± SEM, *P ≤ 0.05), (B) A representative western blot of SP1 protein levels 96 and 120 h after transfection of HEK cells is shown (N = 3, n = 6). (C) HEK cells transfected with hMR and reporter plasmid were stimulated with vehicle or 10 nM aldosterone. Aldosterone-induced MRE1.3 reporter activity was additionally assessed in the presence of scramble-siRNA and specific SP1-siRNA (N = 3, n = 9, data represented as mean ± SEM, *P ≤ 0.05). (D) Importance of SP1 for MR binding to MRE1 (D) Binding of in vitro synthesized hMR to MRE1 was quantified in the presence and absence of 100 ng of rhSP1 in a transcription factor-binding assay. Binding of in vitro synthesized hGR to MRE1 was also detected in the presence and absence of rhSP1 (N = 2–5, n = 4–12, data represented as mean ± SEM, *P ≤ 0.05).
SP1 as cofactor for MR binding to MRE1
To test the effect of SP1 on MR binding to MRE1, we again used our transcription factor-binding ELISA with biotinylated MRE1 probe. When adding rhSP1, binding of in vitro hMR to MRE1 reached levels comparable with those obtained previously with nuclear extracts. Binding of in vitro GR to MRE1.3 was not affected by this maneuver (Figure 8D).
SP3 as cofactor for MR binding to MRE1
To analyze a potential involvement of SP3 to our proposed mechanism, we knocked down SP3 using SP3-specific siRNA and repeated MRE1.3 reporter gene assays (Figure 9A–C). Protein expression level of SP3 was reduced by SP3-specific siRNA to a level of 16.4 ± 5% 96 h after transfection. As a negative control, we used scrambled siRNA, which did not lead to a significant reduction in SP3 protein level after 96 h. Human recombinant SP3 showed weak binding to MRE1.3 in an EMSA, but aldosterone-mediated induction of MRE1.3 reporter gene activity was not inhibited by SP3-specific siRNA (Figure 9B and C). This suggests that SP3 is not involved in MR-induced EGFR promoter activation.
Figure 9.
Relevance of SP3 for MR-induced MRE1.3 reporter activity (A) top: Transiently transfected hMR-HEK cells were incubated with either SP3-specific siRNA or scrambled siRNA for 96 h. SP3 protein levels were quantified by western blot and compared with untransfected cells (N = 3, n = 6, data represented as mean ± SEM, *P ≤ 0.05), bottom: representative western blot. (B) HEK cells transfected with hMR and reporter plasmid were stimulated with vehicle or 10 nM aldosterone. Aldosterone-induced MRE1.3 reporter activity was additionally analyzed in the presence of scrambled-siRNA and specific SP3-siRNA (N = 3, n = 6–48), data represented as mean ± SEM, *P ≤ 0.05). (C) Binding of human recombinant SP3 to MRE1.3 and SP1 consensus sequence was analyzed. Probes were either incubated with only buffer (probe), 100 ng of BSA as control for unspecific protein interactions or 100 ng rhSP3 (N = 5).
MR domains involved in MRE binding
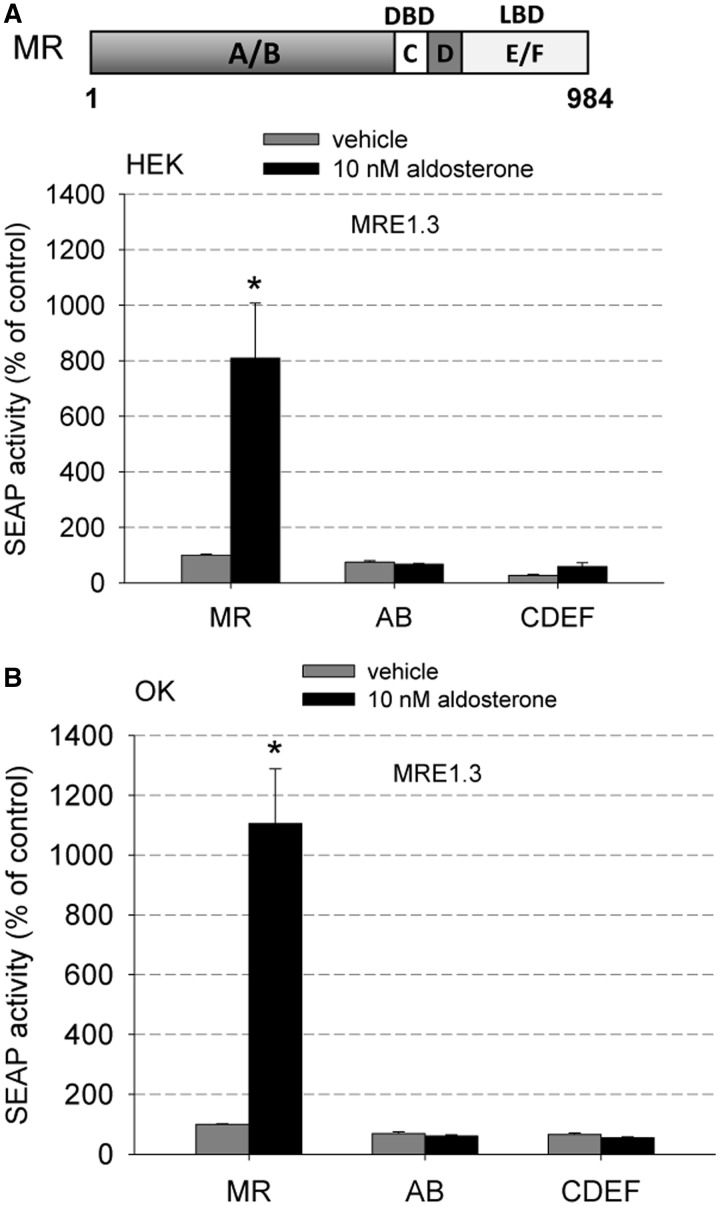
To identify hMR domains participating in the interaction with MRE1.3, we tested deletion constructs of the hMR in our MRE1.3 reporter gene assay. Deletion constructs either possessed the domain A/B (=AB), which is a cofactor binding site or the domains C, D, E and F (=CDEF), which include the DNA binding, ligand binding and dimerization domain. None of the deletion constructs by themselves could MR-dependently activate MRE1.3 either in HEK cells (Figure 10A) or OK cells (Figure 10B). In previous investigations, we already characterized the expression level and function of the applied deletion constructs (20). Interestingly, CDEF-MR lacking the AB domain is able to translocate into the nucleus and transactivate a classical GRE in a reporter gene assay while it is not able to activate MRE1.3. Thus, the DNA-binding domain and the N-terminal domain seem to be important for transactivating MRE1.3. Because the N-terminus is also the most variable amongst the steroid receptors, its requirement may be responsible for the MR specificity of the interaction between MR and EGFR promoter. This suggests that the activation function 1 (AF-1) located in the N-terminus of the MR and not the activation function 2 (AF-2) of the E/F domain may be responsible for MR transactivation at MRE1.3 elements. The AB domain by itself is not activated by aldosterone, is located ubiquitously and cannot bind to DNA.
Figure 10.
MR domains involved in the interaction with MRE1.3 (A) Deletion constructs of hMR consisting of the domains A and B (AB) or the domains C, D, E, F (CDEF) were tested for their ability to increase MRE1.3 promoter activity in HEK cells (N = 3, n = 9, data represented as mean ± SEM, *P ≤ 0.05). (B) The MRE1.3 promoter activity of the MR deletion constructs used in (A) was also tested in OK cells to exclude cell-specific effects (N = 4–8, n = 12–24, data represented as mean ± SEM, *P ≤ 0.05).
Inhibition of SP1 prevents aldosterone-induced upregulation of EGFR
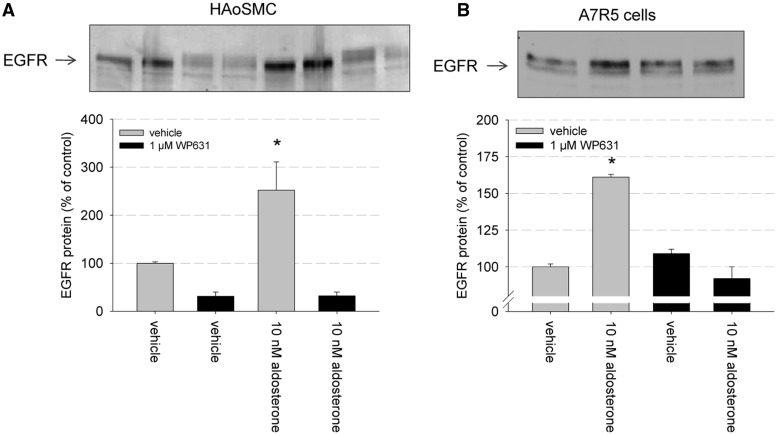
To explore the biological relevance of SP1 for MR-mediated genomic signaling, we chose two additional types of VSMCs and tested the effect of chemical SP1 inhibition on EGFR protein expression (Figure 11). We chose HAoSMC and A7r5 cells from rat aorta to support a more comprehensive nature of our findings. In HAoSMC, 10 nM aldosterone induced an upregulation of EGFR protein expression as detected by western blot. The SP1 inhibitor WP631 (1 µM) reduced basal EGFR protein expression per se but also inhibited aldosterone-induced EGFR expression (Figure 11A). In A7r5 cells, EGFR protein expression was also stimulated by 10 nM aldosterone (Figure 11B). WP631 (1 µM) had no effect on basal EGFR expression, and simultaneous incubation with 10 nM aldosterone and 1 µM WP631 inhibited the stimulatory effect of aldosterone on EGFR expression.
Figure 11.
Relevance of SP1 for aldosterone-induced EGFR expression in primary culture. (A) HAoSMC were incubated for 24 h with either vehicle, 10 nM aldosterone, 1 µM WP631 or 10 nM aldosterone and 1 µM WP631 and EGFR protein expression was quantified by western blot and densitometric analysis (n = 5, data represented as mean ± SEM, *P ≤ 0.05). (B) Primary rat aortic smooth muscle cells (A7r5) were incubated for 24 h with either vehicle, 10 nM aldosterone, 1 µM WP631 or 10 nM aldosterone and 1 µM WP631 and EGFR protein expression was quantified by western blot and densitometric analysis (n = 3, data represented as mean ± SEM, *P ≤ 0.05).
Genome-wide search for promoter regions similar to MRE1.3
As mentioned previously, no classical GRE could be detected within the MR-specific MRE1.3, suggesting the presence of a binding site different from classical GRE. Therefore, we wanted to investigate whether our results are applicable in a broader context. With the goal of finding promoter regions in the human genome that are similar to MRE1.3 or infixes thereof, we computed optimal semi-global alignments between the three fragments
MRE1.3 = GCAGCAGCCTCCTCCCCCCGCACGGTGTGAGCGCCCGACGCGGCCGAGGCGGCCGGAGTCCCGAG
MRE1.3′ = CCTCCTCCCCCCGCACGGTGTGAGCGCCCG
MRE1.3′′ = CCTCCTCCCCCCGCAC
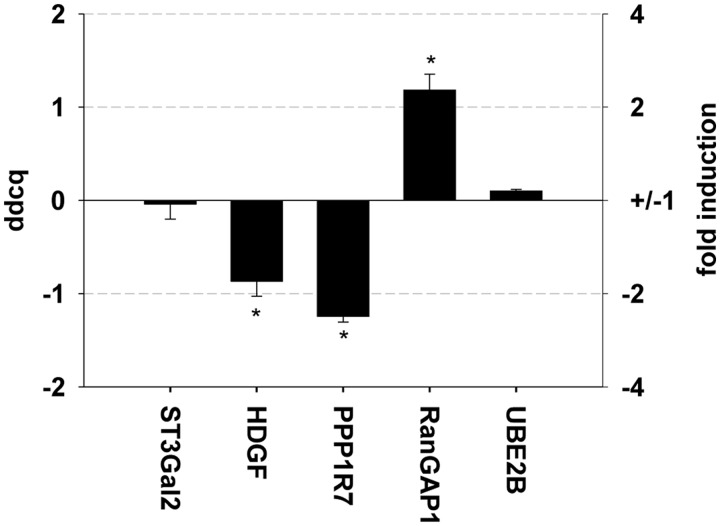
and all human promoters (see ‘Materials and Methods’ section). MRE1.3′′ was chosen because it comprises the EMSA-verified SP1-binding site that is important for the interaction with MR. Furthermore, the terminal 40 bp of MRE1.3 by themselves (=MRE1.3c) did not show any reporter gene activity, supporting that the initial 25 bp in the beginning are of importance. Because the initial 25 bp alone also did not possess any MR-mediated reporter gene activity, the slightly larger infix MRE1.3′ was additionally used for alignment analysis. As there is no unique definition and no precise annotation of human promoters, we operationally defined a promoter to be the sequence starting 250 bp upstream of the annotated transcription start site and ending at the 3′end of the 5′UTR, i.e. ending 1 bp upstream of the translation start site. For each optimal semi-global alignment, we computed the ratio of the alignment score and the alignment length, and we used that ratio as a measure of the degree of similarity. We found that the promoter sequence with the highest degree of similarity to MRE1.3, MRE1.3′ and MRE1.3′′ has a ratio of −0.15, −0.44 and −0.81, respectively, with the highest possible ratio being −1. Consequently, for all three sequences, no perfect match could be found, but several targets containing sequences similar to MRE1.3′′ in their promoter regions are listed in Supplement 2 (ratio 0.817). The MR dependency of gene expression was explored in HK2 cells by qPCR for five promoters with ratios of 0.81 (Figure 12). For three genes a significant regulation by active MR could be verified by qPCR, suggesting that the mechanism underlying binding of MR to the EGFR promoter uses a mechanism with a more general applicability.
Figure 12.
Influence of MR/aldosterone on regulation of promoter regions similar to MRE1.3. An immortalized proximal tubule epithelial cell line from normal adult human kidney (HK2) was transfected with hMR and incubated either with 10 nM aldosterone or vehicle for 24 h. mRNA expression levels of different genes containing promoter regions similar to MRE1.3 and judged to be of potential pathophysiological relevance by literature were detected and depicted as fold induction and ddcq. (N = 3–5, n = 3–15, data represented as mean ± SEM, *P ≤ 0.05).
DISCUSSION
The EGFR is a receptor highly relevant for cell growth, proliferation, migration and vasoconstriction and has been associated with tumor progression and tissue remodeling processes. Activation of the EGFR can be achieved by extracellular ligands but also by interaction with other signaling pathways through transactivation. Transactivation resulting in tissue remodeling processes has been studied in detail for G-protein-coupled receptors like the angiotensin-II-type-1-receptor and the endothelin-1-receptor (21–24). Recently, transactivation of the EGFR has been also demonstrated for the MR and has been linked to vascular dysfunction (11,25,26). As an additional mechanism of interaction, an increase in EGFR expression mediated specifically by activated MR and not by the closely related GR has been proposed.
Because MR and GR classically share a common hormone response element, regulation of EGFR by MR has to be mediated by an additional mechanism that may also help explain other MR-specific effects. In general, identifying pathophysiologically relevant target genes of the MR has proven difficult. Genes identified so far often play a role in sodium handling (like Na+-K+-ATPase alpha-1 subunit, channel inducing factor, epithelial sodium channel 1 alpha, serum glucocorticoid regulated kinase 1, glucocorticoid-induced leucine zipper, ubiquitin-specific protease 2-45, 14-3-3 protein isoform beta) but also include circadian clock protein period 1, endothelin-1, striatin, plasminogen activator inhibitor-1, n-myc downstream regulated gene 2, kras2, connector enhancer of kinase suppressor of RAS3 and the EGFR (18,27–31). Nevertheless, these genes have not been verified to be strictly MR- and not GR-responsive except for the EGFR, and so far, no specifically MR-responsive element has been identified.
Indications that the EGFR might be a pathophysiological relevant MR-regulated gene come from reports showing that mineralocorticoids enhance the arterial contraction in response to EGF in rats and that in vitro incubation of aortas with 10 nM aldosterone increases EGFR mRNA by >50% (32). These observations were extended by Dorrance et al. (33) who found that the infarct size after middle artery occlusion could be reduced in SHRSP (spontaneously hypertensive rats stroke prone) by pretreatment with the MR antagonist spironolactone supposedly owing to less vascular remodeling. Concomitantly, spironolactone reduced the EGFR-mRNA expression in the aorta. In Wistar rats, aldosterone led to cardiovascular remodeling and perivascular fibrosis of the coronary arteries while simultaneously increasing EGFR mRNA and protein expression in the left ventricle (34). Both effects could be attenuated by spironolactone. Furthermore, aldosterone and ageing also increase EGFR protein expression in rat VSMC, inducing a proinflammatory phenotype (16). Interestingly, only organs prone to pathophysiological aldosterone effects like heart, vasculature and kidney reacted to aldosterone with enhanced EGFR expression (18).
In the present article, we investigated the mechanism of the interaction between activated MR and the EGFR promoter in more detail and explored the relevance of the identified DNA-binding site for additional genes. The importance of the EGFR for various signaling pathways in VSMC was demonstrated by comparing ERK phosphorylation in VSMC of EGFR-knockout mice to wild-type mice. Experiments were routinely performed in MR-transfected HEK cells, and results were confirmed in OK cells to exclude cell-specific effects. To ensure the relevance of our mechanistic findings for differentiated vascular cells, which mediate pathophysiological MR effects, the influence of aldosterone/MR and the importance of SP1 for this interaction were also tested in differentiated rat aortic smooth muscle cells (A7r5) and HAoSMC.
With reporter gene assays, we could identify a 65 bp hormone-responsive and MR-specific element on the EGFR promoter (=MRE1.3). MRE1.3 interacts dose dependently with aldosterone-activated MR with an EC50-value in the low nanomolar range. Normal aldosterone levels in human plasma vary from ∼0.1 to 1 nM, and intracellular values are not known; therefore, the concentrations necessary to activate the EGFR promoter seem to be in the upper range or slightly above normal aldosterone concentrations. This supports the hypothesis that aldosterone-induced EGFR expression mediates pathophysiological effects of the MR like organ dysfunction, remodeling or inflammation as has been suggested by investigations in animal models (25,32–34). Furthermore, the EC50 of aldosterone at a classical GRE is ∼10-fold lower for both HEK and OK cells, suggesting that GRE activation might be already important under physiological conditions while MRE1.3 activation occurs in pathological situations. The glucocorticoid cortisol, known to activate classical GREs, was also able to enhance MRE1.3 promoter activity. Dexamethasone/GR had no effect on the MRE1.3 promoter activity, highlighting the MR specificity of MRE1.3. The dependence of MRE1.3 activation on MR is further shown by the inhibition achieved by the selective MR antagonist eplerenone.
The minimal DNA sequence necessary for the interaction of MR and EGFR promoter is much longer than a classical GRE, which consists of 15 bp arranged in two 6 bp long inverted repeats with three base pairs in between. This suggests that additional cofactors may be important for MR-induced EGFR promoter activation. Our transcription factor-binding assays support this hypothesis because in vitro MR does not bind specifically to MRE1 while MR from nuclear extracts of cells transfected with MR and stimulated with aldosterone specifically binds to MRE1. By bioinformatical analysis, SP1 was identified as a possible transcription factor binding to MRE1.3, and this hypothesis was validated by EMSA. SP1 is a ubiquitously expressed transcription factor in mammalian cells and is the original member of the SP1-like/KLF family of zinc-finger transcription factors. Overall, SP1 is involved in many different cellular processes, such as cell growth, apoptosis, differentiation and immune responses. By increasing the stringency of the search conditions and by reducing the length of the promoter fragment, the 5′-SP1 site was identified as relevant. Support for in vivo binding of SP1 to MRE1.3 is further provided by a technical note recently published by Schuch et al. (35) in which binding of SP1 to an element on the EGFR promoter that corresponds to MRE1.3a is demonstrated by chromatin immunoprecipitation. Binding of MR to the EGFR promoter was previous shown by us by chromatin immunoprecipitation in HEK cells and could now be confirmed by transcription factor-binding ELISA with nuclear HEK cell extracts (18). Interestingly, binding of in vitro synthesized MR to MRE1 was dependent on the presence of rhSP1, suggesting the formation of an SP1-MR-DNA-complex.
The importance of this SP1 site for MRE1.3 activation was further demonstrated by showing that respective mutations impair reporter gene activity. Using the chemical SP1 inhibitor WP631 and specific SP1-siRNA, the importance of SP1 for aldosterone-induced EGFR expression was shown in both HEK and OK cells. Because classical GRE transactivation activity was not affected by WP631 an unspecific effect of SP1 on transcription or translation in general is also not supported by our data. Additionally, the use of SP1-specific siRNA did not lead to a significant decrease in basal transcription activity, making it unlikely that SP1 is essential for basal promoter activity but rather implying that SP1 elicits an effect on MR action per se. An interaction between SP1 and other steroid receptors has been suggested for GR, progesterone receptor and androgen receptor (36–38) but the exact nature of the interaction between these other steroid receptors and SP1 is not clear. Furthermore, the MR chaperone protein HSP90 is also important for stabilizing SP1 (39). For the estrogen receptor, studies have identified both a mechanism in which estrogen receptor and SP1 both bind to DNA and a second mechanism in which SP1 alone binds to DNA and the estrogen receptor then interacts with SP1 (40–42).
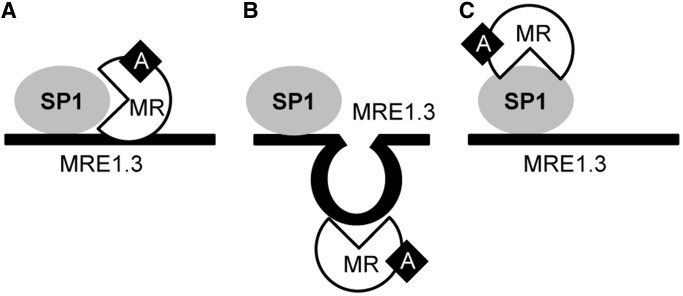
For MR and SP1, there are also several possibilities for the mechanism of interaction. MR may bind to the DNA adjacent to SP1 with or without additional interaction with SP1 (Figure 13A) or SP1 may modulate the 3D DNA structure to enable MR binding (Figure 13B). Alternatively, MR could bind to SP1 and thereby influence the transcription of genes (Figure 13C). It is unclear whether SP1 and MR act as monomers or multimers, and it is also not clear if additional coregulatory factors are involved. For example, even a competition of MR with an inhibitory factor is a possible explanation for a stimulatory effect of activated MR. Because the first 25 bp of MRE1.3 were sufficient for binding of SP1 to DNA but not sufficient for complete transactivation, simple binding of MR to SP1 in a piggyback fashion is unlikely. Instead the length of the DNA necessary for the full MR-SP1 interaction would suggest adjacent binding of SP1 and MR, possibly also involving additional cofactors. The closely related SP3 showed only weak binding of MRE1.3, and its knockdown did not influence EGFR promoter activation. Additional experiments will be necessary to elucidate the MR-SP1-MRE1.3 interaction further.
Figure 13.
Model of MR-SP1-MRE1.3 Interaction. (A) MR may bind to the DNA adjacent to SP1. (B) SP1 may modulate DNA structure to enable MR binding. (C) MR could bind to SP1.
Interestingly, the CDEF-MR mutant lacking the regulatory N-terminus and therefore the activator function 1 (AF-1) is not able to enhance MRE1.3 promoter activity. CDEF-MR has been extensively characterized previously (20) and possesses many similarities to complete MR. Both are located in the cytosol in their unliganded state and on binding of ligand shuttle into the nucleus where they lead to transactivation of classical GREs with comparable aldosterone-response curves. Furthermore, both contain the AF-2 domain. However, CDEF-MR not only is able to activate GREs but can also transactivate the EGFR and induce ERK phosphorylation similar to complete MR. Therefore, the lack of stimulation of MRE1.3 by CDEF-MR highlights that the underlying mechanism is different to classical transactivation and non-genomic signaling and likely involves AF-1 of the MR N-terminus and not AF-2 of the C-terminal E/F domain, which fits well to our observation of MR over GR specificity. Structurally, MR and GR both possess a cofactor binding domain A/B, a DNA-binding domain C, a hinge region D and a ligand binding and dimerization domain E/F. Amino acid comparison between GR and MR shows a 94% identity between the DNA-binding domain and a 57% identity of the ligand-binding and dimerization domain. In contrast, the N-terminus has <15% identity, which makes it an ideal candidate for mediating MR specific effects (43). Because it is not involved in DNA binding, an interaction of the N-terminus of the MR with SP1 while both are bound to DNA with their zinc finger domains seems an attractive hypothesis but needs to be investigated further. Preliminary coimmunoprecipitation experiments suggest an interaction between MR and SP1. However, these experiments are complex to interpret because under unstimulated conditions, both transcription factors are clients of HSP90 and therefore part of a common multiprotein complex. After stimulation, MR and SP1 rapidly bind to DNA, and co-immunoprecipitation would then require cross-linking and shearing of DNA, which would facilitate false-positive results in an overexpression system like ours with transcription factors binding in close vicinity.
To explore whether the MR-SP1-MRE1.3 interaction could also be of relevance for other genes, we performed an alignment between all human promoters with MRE1.3 or infixes thereof. Several promoters with a score of −0.81 could be identified for the 16 bp long region containing the SP1-binding site (see Supplement 2). Because this approach is not specific, we validated five of the predicted targets by qPCR and found the expression of three of them aldosterone/MR-dependently regulated, thus suggesting relevance of our screening approach. Candidate genes regulated by aldosterone include the hepatoma-derived growth factor (HDGF), protein phosphatase 1 regulatory subunit 7 (PPP1R7) and ran GTPase-activating protein 1 (RanGAP1). HDGF is the prototype of a family of proteins that are mitogenic for various cell lines including fibroblasts, smooth muscle cells and endothelial cells (44). Additionally, HDGF has been shown to modulate the expression of matrix metalloproteases and to regulate cardiovascular growth and differentiation (45,46). Protein Phosphatase 1 is known to participate in cell cycle regulation, and downregulation is associated with tumor progression (47,48). Ran GTPase activating protein 1 influences nucleocytoplasmic transport and is associated with microtubuli formation during mitosis (49,50). Furthermore, the expression of RanGAP1 has been shown to be enhanced in patients with cardiac ischemia and dilation (51). Overall, involvement of these genes in pathophysiological MR effects in the cardiovascular system is an attractive hypothesis but of course needs further validation.
To summarize, we characterized the mechanism by which the expression of the EGFR, which mediates signaling of a variety of signaling pathways, can be regulated by activated MR. We could show that a 65 bp fragment (=MRE1.3) of the EGFR promoter is required for MR transactivation, and that SP1 is a compulsory cofactor for the interaction. To investigate whether this interaction between MR, SP1 and DNA also occurs in the promoters of other genes, alignments between MRE1.3 and its infixes and all human promoters were performed, and several candidates of interest were determined and validated by qPCR. Thus, we identified and characterized the molecular nature of a new mechanism of MR-mediated gene expression for a pathophysiological relevant gene, the EGFR, and we could also show that this interaction may also be of relevance for additional genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [GE 905/13-1, GR 3415/1-1]; Ministry of Culture Saxony-Anhalt [XP3624HP/0606T]. Funding for open access charge: Medical Faculty, University Halle-Wittenberg.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Edgar Wingender for valuable discussion.
REFERENCES
- 1.Pitt B, Zannad F, Remme WJ, Cody R, Castaigne A, Perez A, Palensky J, Wittes J. The effect of spironolactone on morbidity and mortality in patients with severe heart failure. Randomized aldactone evaluation study investigators. N. Engl. J. Med. 1999;341:709–717. doi: 10.1056/NEJM199909023411001. [DOI] [PubMed] [Google Scholar]
- 2.Pitt B, Remme W, Zannad F, Neaton J, Martinez F, Roniker B, Bittman R, Hurley S, Kleiman J, Gatlin M. Eplerenone, a selective aldosterone blocker, in patients with left ventricular dysfunction after myocardial infarction. N. Engl. J. Med. 2003;348:1309–1321. doi: 10.1056/NEJMoa030207. [DOI] [PubMed] [Google Scholar]
- 3.Chrysostomou A, Pedagogos E, MacGregor L, Becker GJ. Double-blind, Placebo-controlled study on the effect of the aldosterone receptor antagonist spironolactone in patients who have persistent proteinuria and are on long-term angiotensin-converting enzyme inhibitor therapy, with or without an angiotensin II receptor blocker. Clin. J. Am. Soc. Nephrol. 2006;1:256–262. doi: 10.2215/CJN.01040905. [DOI] [PubMed] [Google Scholar]
- 4.Sartorio CL, Fraccarollo D, Galuppo P, Leutke M, Ertl G, Stefanon I, Bauersachs J. Mineralocorticoid receptor blockade improves vasomotor dysfunction and vascular oxidative stress early after myocardial infarction. Hypertension. 2007;50:919–925. doi: 10.1161/HYPERTENSIONAHA.107.093450. [DOI] [PubMed] [Google Scholar]
- 5.Blasi ER, Rocha R, Rudolph AE, Blomme EA, Polly ML, McMahon EG. Aldosterone/salt induces renal inflammation and fibrosis in hypertensive rats. Kidney Int. 2003;63:1791–1800. doi: 10.1046/j.1523-1755.2003.00929.x. [DOI] [PubMed] [Google Scholar]
- 6.Brilla CG, Weber KT. Mineralocorticoid excess, dietary sodium, and myocardial fibrosis. J. Lab. Clin. Med. 1992;120:893–901. [PubMed] [Google Scholar]
- 7.Brown NJ. Aldosterone and vascular inflammation. Hypertension. 2008;51:161–167. doi: 10.1161/HYPERTENSIONAHA.107.095489. [DOI] [PubMed] [Google Scholar]
- 8.Young M, Fullerton M, Dilley R, Funder J. Mineralocorticoids, hypertension, and cardiac fibrosis. J. Clin. Invest. 1994;93:2578–2583. doi: 10.1172/JCI117269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, et al. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- 10.Sibilia M, Kroismayr R, Lichtenberger BM, Natarajan A, Hecking M, Holcmann M. The epidermal growth factor receptor: from development to tumorigenesis. Differentiation. 2007;75:770–787. doi: 10.1111/j.1432-0436.2007.00238.x. [DOI] [PubMed] [Google Scholar]
- 11.Schreier B, Döhler M, Rabe S, Schneider B, Schwerdt G, Ruhs S, Sibilia M, Gotthardt M, Gekle M, Grossmann C. Consequences of epidermal growth factor receptor (ErbB1) loss for vascular smooth muscle cells from mice with targeted deletion of ErbB1. Arterioscler Thromb. Vasc. Biol. 2011;31:1643–1652. doi: 10.1161/ATVBAHA.111.223537. [DOI] [PubMed] [Google Scholar]
- 12.Daub H, Weiss FU, Wallasch C, Ullrich A. Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature. 1996;379:557–560. doi: 10.1038/379557a0. [DOI] [PubMed] [Google Scholar]
- 13.Holbro T, Hynes NE. ErbB receptors: directing key signaling networks throughout life. Annu. Rev. Pharmacol. Toxicol. 2004;44:195–217. doi: 10.1146/annurev.pharmtox.44.101802.121440. [DOI] [PubMed] [Google Scholar]
- 14.Prenzel N, Zwick E, Daub H, Leserer M, Abraham R, Wallasch C, Ullrich A. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402:884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 15.Grossmann C, Benesic A, Krug AW, Freudinger R, Mildenberger S, Gassner B, Gekle M. Human mineralocorticoid receptor expression renders cells responsive for nongenotropic aldosterone actions. Mol. Endocrinol. 2005;19:1697–1710. doi: 10.1210/me.2004-0469. [DOI] [PubMed] [Google Scholar]
- 16.Krug AW, Allenhöfer L, Monticone R, Spinetti G, Gekle M, Wang M, Lakatta EG. Elevated mineralocorticoid receptor activity in aged rat vascular smooth muscle cells promotes a proinflammatory phenotype via extracellular signal-Regulated Kinase 1/2 mitogen-activated protein kinase and epidermal growth factor receptor-dependent pathways. Hypertension. 2010;55:1476–1483. doi: 10.1161/HYPERTENSIONAHA.109.148783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazak I, Fiebeler A, Muller DN, Park JK, Shagdarsuren E, Lindschau C, Dechend R, Viedt C, Pilz B, Haller H, et al. Aldosterone potentiates angiotensin II-induced signaling in vascular smooth muscle cells. Circulation. 2004;109:2792–2800. doi: 10.1161/01.CIR.0000131860.80444.AB. [DOI] [PubMed] [Google Scholar]
- 18.Grossmann C, Krug AW, Freudinger R, Mildenberger S, Voelker K, Gekle M. Aldosterone-induced EGFR expression: interaction between the human mineralocorticoid receptor and the human EGFR promoter. Am. J. Physiol. Endocrinol. Metab. 2007;292:E1790–E1800. doi: 10.1152/ajpendo.00708.2006. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Sanchez CE, de Rodriguez AF, Romero DG, Estess J, Warden MP, Gomez-Sanchez MT, Gomez-Sanchez EP. Development of a panel of monoclonal antibodies against the mineralocorticoid receptor. Endocrinology. 2006;147:1343–1348. doi: 10.1210/en.2005-0860. [DOI] [PubMed] [Google Scholar]
- 20.Grossmann C, Freudinger R, Mildenberger S, Husse B, Gekle M. EF domains are sufficient for nongenomic mineralocorticoid receptor actions. J. Biol. Chem. 2008;283:7109–7116. doi: 10.1074/jbc.M708751200. [DOI] [PubMed] [Google Scholar]
- 21.Bokemeyer D, Schmitz U, Kramer HJ. Angiotensin II-induced growth of vascular smooth muscle cells requires an Src-dependent activation of the epidermal growth factor receptor. Kidney Int. 2000;58:549–558. doi: 10.1046/j.1523-1755.2000.t01-1-00201.x. [DOI] [PubMed] [Google Scholar]
- 22.Flamant M, Tharaux PL, Placier S, Henrion D, Coffman T, Chatziantoniou C, Dussaule JC. Epidermal growth factor receptor transactivation mediates the tonic and fibrogenic effects of endothelin in the aortic wall of transgenic mice. FASEB J. 2003;17:327–329. doi: 10.1096/fj.02-0115fje. [DOI] [PubMed] [Google Scholar]
- 23.Francois H, Placier S, Flamant M, Tharaux PL, Chansel D, Dussaule JC, Chatziantoniou C. Prevention of renal vascular and glomerular fibrosis by epidermal growth factor receptor inhibition. FASEB J. 2004;18:926–928. doi: 10.1096/fj.03-0702fje. [DOI] [PubMed] [Google Scholar]
- 24.Kagiyama S, Eguchi S, Frank GD, Inagami T, Zhang YC, Phillips MI. Angiotensin II-induced cardiac hypertrophy and hypertension are attenuated by epidermal growth factor receptor antisense. Circulation. 2002;106:909–912. doi: 10.1161/01.cir.0000030181.63741.56. [DOI] [PubMed] [Google Scholar]
- 25.Griol-Charhbili V, Fassot C, Messaoudi S, Perret C, Agrapart V, Jaisser F. Epidermal growth factor receptor mediates the vascular dysfunction but not the remodeling induced by aldosterone/salt. Hypertension. 2011;57:238–244. doi: 10.1161/HYPERTENSIONAHA.110.153619. [DOI] [PubMed] [Google Scholar]
- 26.Schreier B, Rabe S, Schneider B, Bretschneider M, Rupp S, Ruhs S, Neumann J, Rueckschloss U, Sibilia M, Gotthardt M, et al. Loss of epidermal growth factor receptor in vascular smooth muscle cells and cardiomyocytes causes arterial hypotension and cardiac hypertrophy. Hypertension. 2013;61:333–340. doi: 10.1161/HYPERTENSIONAHA.112.196543. [DOI] [PubMed] [Google Scholar]
- 27.Chen S-Y, Bhargava A, Mastroberardino L, Meijer OC, Wang J, Buse P, Firestone GL, Verrey F, Pearce D. Epithelial sodium channel regulated by aldosterone-induced protein sgk. Proc. Natl Acad. Sci. USA. 1999;96:2514–2519. doi: 10.1073/pnas.96.5.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gumz ML, Stow LR, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Weaver DR, Wingo CS. The circadian clock protein Period 1 regulates expression of the renal epithelial sodium channel in mice. J. Clin. Invest. 2009;119:2423–2434. doi: 10.1172/JCI36908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kolla V, Litwack G. Transcriptional regulation of the human NA/K ATPase via the human mineralocorticoid receptor. Mol. Cell. Biochem. 2000;204:35–40. doi: 10.1023/a:1007009700377. [DOI] [PubMed] [Google Scholar]
- 30.Stow LR, Gumz ML, Lynch IJ, Greenlee MM, Rudin A, Cain BD, Wingo CS. Aldosterone modulates steroid receptor binding to the endothelin-1 gene (edn1) J. Biol. Chem. 2009;284:30087–30096. doi: 10.1074/jbc.M109.030718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chun TY, Pratt JH. Aldosterone increases plasminogen activator inhibitor-1 synthesis in rat cardiomyocytes. Mol. Cell. Endocrinol. 2005;239:55–61. doi: 10.1016/j.mce.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 32.Florian JA, Dorrance A, Webb RC, Watts SW. Mineralocorticoids upregulate arterial contraction to epidermal growth factor. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R878–R886. doi: 10.1152/ajpregu.2001.281.3.R878. [DOI] [PubMed] [Google Scholar]
- 33.Dorrance AM, Osborn HL, Grekin R, Webb RC. Spironolactone reduces cerebral infarct size and EGF-receptor mRNA in stroke-prone rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2001;281:R944–R950. doi: 10.1152/ajpregu.2001.281.3.R944. [DOI] [PubMed] [Google Scholar]
- 34.Nakano S, Kobayashi N, Yoshida K, Ohno T, Matsuoka H. Cardioprotective mechanisms of spironolactone associated with the angiotensin-converting enzyme/epidermal growth factor receptor/extracellular signal-regulated kinases, NAD(P)H Oxidase/Lectin-like oxidized Low-density lipoprotein receptor-1, and Rho-kinase pathways in aldosterone/salt-induced hypertensive rats. Hypertens Res. 2005;28:925–936. doi: 10.1291/hypres.28.925. [DOI] [PubMed] [Google Scholar]
- 35.Schuch R, Agelopoulos K, Neumann A, Brandt B, Burger H, Korsching E. Site-specific chromatin immunoprecipitation: a selective method to individually analyze neighboring transcription factor binding sites in vivo. BMC Res. Notes. 2012;5:109. doi: 10.1186/1756-0500-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan H, Young C, Tian Y, Liu Z, Zhang M, Lou H. Suppression of the androgen receptor function by quercetin through protein-protein interactions of Sp1, c-Jun, and the androgen receptor in human prostate cancer cells. Mol. Cell. Biochem. 2010;339:253–262. doi: 10.1007/s11010-010-0388-7. [DOI] [PubMed] [Google Scholar]
- 37.Owen GI, Richer JK, Tung L, Takimoto G, Horwitz KB. Progesterone regulates transcription of the p21 WAF1 cyclindependent kinase inhibitor gene through Sp1 and CBP/p300. J. Biol. Chem. 1998;273:10696–10701. doi: 10.1074/jbc.273.17.10696. [DOI] [PubMed] [Google Scholar]
- 38.Chen K, Ou XM, Wu JB, Shih JC. Transcription factor E2F-associated phosphoprotein (EAPP), RAM2/CDCA7L/JPO2 (R1), and simian virus 40 promoter factor 1 (Sp1) cooperatively regulate glucocorticoid activation of monoamine oxidase B. Mol. Pharmacol. 2011;79:308–317. doi: 10.1124/mol.110.067439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang SA, Chuang JY, Yeh SH, Wang YT, Liu YW, Chang WC, Hung JJ. Heat shock protein 90 is important for sp1 stability during mitosis. J. Mol. Biol. 2009;387:1106–1119. doi: 10.1016/j.jmb.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 40.Petz LN, Nardulli AM. Sp1 binding sites and an estrogen response element half-site are involved in regulation of the human progesterone receptor A promoter. Mol. Endocrinol. 2000;14:972–985. doi: 10.1210/mend.14.7.0493. [DOI] [PubMed] [Google Scholar]
- 41.Li D, Mitchell D, Luo J, Yi Z, Cho SG, Guo J, Li X, Ning G, Wu X, Liu M. Estrogen regulates KiSS1 gene expression through estrogen receptor alpha and SP protein complexes. Endocrinology. 2007;148:4821–4828. doi: 10.1210/en.2007-0154. [DOI] [PubMed] [Google Scholar]
- 42.Porter W, Saville B, Hoivik D, Safe S. Functional synergy between the transcription factor Sp1 and the estrogen receptor. Mol. Endocrinol. 1997;11:1569–1580. doi: 10.1210/mend.11.11.9916. [DOI] [PubMed] [Google Scholar]
- 43.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang CH, Davamani F, Sue SC, Lee SC, Wu PL, Tang FM, Shih C, Huang TH, Wu WG. Cell surface heparan sulfates mediate internalization of the PWWP/HATH domain of HDGF via macropinocytosis to fine-tune cell signalling processes involved in fibroblast cell migration. Biochem. J. 2011;433:127–138. doi: 10.1042/BJ20100589. [DOI] [PubMed] [Google Scholar]
- 45.Everett AD. Identification, cloning, and developmental expression of hepatoma-derived growth factor in the developing rat heart. Dev. Dyn. 2001;222:450–458. doi: 10.1002/dvdy.1204. [DOI] [PubMed] [Google Scholar]
- 46.Everett AD, Lobe DR, Matsumura ME, Nakamura H, McNamara CA. Hepatoma-derived growth factor stimulates smooth muscle cell growth and is expressed in vascular development. J. Clin. Invest. 2000;105:567–575. doi: 10.1172/JCI7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posch M, Khoudoli GA, Swift S, King EM, DeLuca JG, Swedlow JR. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell. Biol. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y, Scott KL, Kwak SJ, Chen R, Mardon G. Sds22/PP1 links epithelial integrity and tumor suppression via regulation of myosin II and JNK signaling. Oncogene. 2011;30:3248–3260. doi: 10.1038/onc.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hutten S, Flotho A, Melchior F, Kehlenbach RH. The Nup358-RanGAP complex is required for efficient importin alpha/beta-dependent Nuclear Import. Mol. Biol. Cell. 2008;19:2300–2310. doi: 10.1091/mbc.E07-12-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerard B, Tait L, Nangia-Makker P, Shekhar M. Rad6B acts downstream of Wnt signaling to stabilize beta-catenin: Implications for a novel Wnt/beta-catenin target. J. Mol. Signal. 2011;6:6. doi: 10.1186/1750-2187-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortes R, Rosello-Lleti E, Rivera M, Martinez-Dolz L, Salvador A, Azorin I, Portoles M. Influence of heart failure on nucleocytoplasmic transport in human cardiomyocytes. Cardiovasc. Res. 2010;85:464–472. doi: 10.1093/cvr/cvp336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.