Abstract
Histone deacetylase inhibitors (HDACis) have been shown to potentiate hippocampal-dependent memory and synaptic plasticity and to ameliorate cognitive deficits and degeneration in animal models for different neuropsychiatric conditions. However, the impact of these drugs on hippocampal histone acetylation and gene expression profiles at the genomic level, and the molecular mechanisms that underlie their specificity and beneficial effects in neural tissue, remains obscure. Here, we mapped four relevant histone marks (H3K4me3, AcH3K9,14, AcH4K12 and pan-AcH2B) in hippocampal chromatin and investigated at the whole-genome level the impact of HDAC inhibition on acetylation profiles and basal and activity-driven gene expression. HDAC inhibition caused a dramatic histone hyperacetylation that was largely restricted to active loci pre-marked with H3K4me3 and AcH3K9,14. In addition, the comparison of Chromatin immunoprecipitation sequencing and gene expression profiles indicated that Trichostatin A-induced histone hyperacetylation, like histone hypoacetylation induced by histone acetyltransferase deficiency, had a modest impact on hippocampal gene expression and did not affect the transient transcriptional response to novelty exposure. However, HDAC inhibition caused the rapid induction of a homeostatic gene program related to chromatin deacetylation. These results illuminate both the relationship between hippocampal gene expression and histone acetylation and the mechanism of action of these important neuropsychiatric drugs.
INTRODUCTION
The acetylation of histone tails is an epigenetic modification of the chromatin associated with active loci and regulated by the opposing activities of lysine acetyltransferases (KATs) and histone deacetylases (HDACs) (1,2). Three independent lines of evidence support a role for the regulation of histone acetylation in neuronal plasticity and memory. First, the reduction of neuronal KAT activity has been associated with impaired intellectual abilities both in humans and mice (3–5), whereas reductions in specific HDACs have been associated with enhanced cognitive performance (6,7). Second, HDAC inhibitors (HDACis) increase histone acetylation and have been shown to potentiate memory and synaptic plasticity and to ameliorate cognitive deficits and neurodegeneration (3,4,8–13). Third, correlative evidence indicates that histone acetylation is dynamically regulated during memory formation (8,14). According to this correlative evidence, it has been hypothesized that the beneficial effects of HDACis in neurons are mediated by the facilitation of specific transcriptional responses (12,15). However, experiments in yeast and other systems have cast doubts about an active role of histone acetylation in the regulation of gene expression (16–19). HDACis represent excellent tools to manipulate the level of histone acetylation and assess its consequences in transcription.
Here, we first used chromatin immunoprecipitation coupled to deep sequencing (ChIPseq) and microarray technologies to determine genome-wide histone acetylation profiles in the adult mouse hippocampus and to define the relationship between key epigenetic marks and neuronal gene expression. We next explored the impact on gene expression and histone acetylation profiles of Trichostatin A (TSA), an inhibitor of class I and IIb HDACs that facilitates long-term potentiation and has been postulated as a memory enhancer (4,8–11), but whose precise mechanism of action and molecular targets in neurons remain largely unknown. Our experiments demonstrated that TSA-triggered dramatic changes in the genomic acetylation profiles that were largely restricted to loci marked with H3K4me3 and AcH3K9,14 in the basal state. TSA also caused the induction of a set of genes related to transcriptional repression and chromatin deacetylation but, like hemideficiency for the KAT CREB binding protein (CBP), had little impact on the induction of immediate early genes (IEGs) by activity. Overall, our experiments clarify both the relationship between hippocampal gene expression and histone acetylation and the mechanism of action of HDACi in neural tissue.
MATERIALS AND METHODS
Animals and treatments
Experiments were performed in adult (3–5 months) C57/DBA F1 hybrid females. TSA (Sigma Aldrich Química S.A.) 2,4 mg/kg was administered by intraperitoneal injection. Cbp+/− mice have been previously described (3,20). Exposure to novelty consisted of placing an individual animal in a white Plexiglas square box containing plastic tubing and small toys for 1 h. All experimental protocols were compliant with European regulations and approved by the Institutional Animal Care and Use Committee.
Gene expression and ChIP analyses
A summary of the different expression and ChIPseq experiments that are part of this study is presented in Supplementary Figure S1. Western blot, quantitative RT-PCR (RT-qPCR) and microarray analyses were performed as previously described (20,21). The lists of TSA- and novelty-regulated transcripts were retrieved using unpaired t-test and two-way analysis of variance (ANOVA). ChIP was performed as described previously (20) and ChIP-qPCR assays were performed using specific primers immediately upstream of the transcription start site (TSS). For ChIPseq, whole hippocampi from three mice were pooled in each sample to perform ChIP. Diluted chromatin was incubated overnight at 4°C in the presence of specific antibody against AcH3K9,14 (Millipore, 06-599), AcH4K12 (abcam, ab46983), AcH2B (21) or H3K4me3 (Millipore, 07-473) or rabbit-derived pre-immune serum (see Supplemental Methods and Supplementary Figure S2 for additional detail). The SICER algorithm (22) was used to extract acetylation-enriched chromatin regions (pooled input and control libraries was used as control library). See Supplementary Table S1 for information on library size, preparation and additional details. The files generated in the microarrays and ChIPseq screens are available at the Gene Expression Omnibus database with the accession number GSE44868.
See Supplemental Materials and Methods for additional details.
RESULTS
Genome-wide mapping of histone acetylation and histone H3 trimethylation in the adult mouse hippocampus
We focused on three specific aceylation marks: (i) AcH3K9,14 that is considered to be a reliable marker for active gene expression and has been associated with memory processes (8); (ii) AcH4K12 that is associated with age-related memory impairment (23); and (iii) AcH2B that is highly dependent on the KAT activity encoded by the intellectual disability gene CREBBP (3,5) and has been involved in memory consolidation (24). We also determined, for the first time in the adult hippocampus, the genomic profile of H3K4me3 that labels active promoters (25).
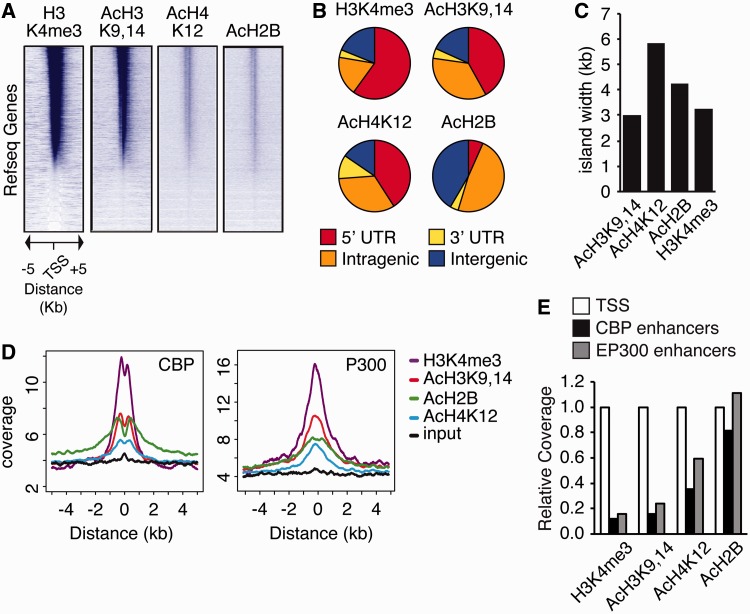
ChIPseq profiles were highly reproducible (Supplementary Figure S3) and revealed thousands of discrete genomic regions (referred to as islands) that were enriched in these epigenetic marks and that preferentially mapped in promoters and intragenic regions (Figure 1A and B and Supplementary Figure S4A, Supplementary Data Set S1). AcH3K9,14 and H3K4me3 defined sharp peaks that overlapped with the TSS, whereas AcH2B enrichment was frequently observed onto the coding sequence. AcH4K12 presented an intermediate profile with islands that were broader and blunter than those of AcH3K9,14 and that frequently included the TSS (Figure 1C). Despite the differences, the enrichment around TSS for the three acetylation marks showed a highly significant correlation (Supplementary Figure S4B), and their overlap, especially when we considered their coincidence at the level of gene loci, was high (Supplementary Figure S4C). The three acetylation marks were also enriched at putative enhancer regions (26,27) bound by the KATs p300 and CBP (Figure 1D). Interestingly and consistently with the substrate preference reported for these KATs (28), the relative enrichment of AcH2B at these regions was double than for AcH4K12 and 5–7-fold higher than for AcH3K9,14 or H3K4me3 (Figure 1E).
Figure 1.
Histone acetylation marks are enriched in gene and enhancer sequences. (A) Density heatmap showing the coverage for each epigenetic mark across 10 kb centered at the TSS of each RefSeq gene. Individual sequences were binned in 25 bp windows (400 bins per sequence), and coverage was computed and plotted as a relative color intensity scale. In all representations, RefSeq genes were ranked according to their H3K4me3 levels. (B) Pie charts present the distribution of H3K4me3, AcH3K9,14, AcH4K12 and AcH2B islands among different gene features: 5′UTR, overlapping with TSS (including islands comprising the whole gene); 3′UTR, overlapping with gene end; intragenic, inside exons or introns excluding the islands that overlapped with the 5′ or 3′-end of the gene; intergenic, upstream and downstream of genes. (C) Median width of H3K4me3, AcH3K9,14, AcH4K12 and AcH2B islands. (D) Mean enrichment profiles presenting the genomic distribution of reads for H3K4me3, AcH3K9,14, AcH4K12 and AcH2B along putative neuronal CBP enhancers (left) and brain-specific p300-bound sites (right). (E) Relative coverage of TSSs and enhancers by the different epigenetic marks.
Histone acetylation in the adult mouse hippocampus correlates with gene expression
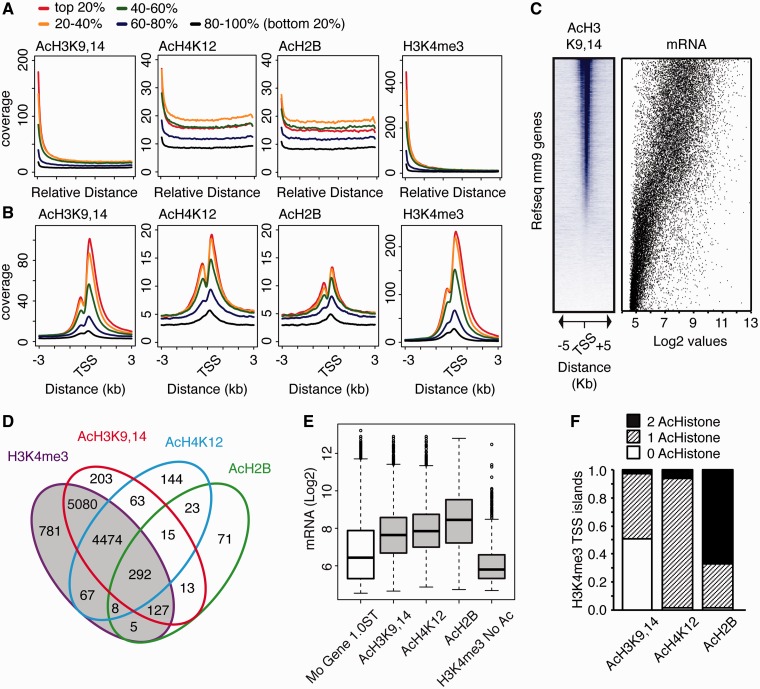
Acetylation at TSSs and mRNA expression levels showed good correlation. Highly transcribed genes according to the estimation provided by the microarray analysis exhibited strong acetylation in both intragenic sequences (Figure 2A) and around the TSS (Figure 2B), whereas silent genes were depleted of the three acetylation marks (Supplementary Figure S5). Gene ranking by acetylation level led to the same conclusion (Figure 2C). Interestingly, above a given level of expression, the correlation between expression level and enrichment was weaker.
Figure 2.
Histone acetylation is associated with active transcription. (A) Distribution of reads along gene length for the four epigenetic marks. Genes were ranked according to relative mRNA expression levels and split into five equal sets. (B) Mean enrichment profiles similar to those in A, but around all annotated TSSs. (C). AcH3K9,14 density heatmap for all RefSeqs (ranked according to their AcH3K9,14 levels around the TSSs) is presented along with relative mRNA values (scatter plot). (D) Overlap between TSSs harboring islands for H3K4me3, AcH3K9,14, AcH4K12 and AcH2B. (E) Boxplot showing the relative mRNA expression values for genes harboring an H3K4me3 island at their TSS either alone (H3K4me3 No Ac) or in combination with any of the acetylation marks (AcH3K9,14, AcH4K12 and AcH2B). Boxes correspond to the intersection between each hyperacetylation mark and H3K4me3 (gray colored area) shown in (D). The distribution of expression values for the whole array (Mo Gene 1.0ST) is also shown. (F) Ratio of TSSs with an H3K4me3 island that contain one or more acetylation marks.
The comparison of expression levels in the intersection groups for the different marks (Figure 2D) revealed a striking pattern: genes that were trimethylated at H3K4 but not acetylated at their proximal promoter (TSS) exhibited low expression levels, whereas genes in which both trimethylation and acetylation concurred at the TSSs belonged to the highly expressed group (Figure 2E). This trend was especially significant for the ∼400 genes in the overlap of H3K4me3 and AcH2B, which were on average positioned above the top quartile of genes ranked according to expression in hippocampal tissue. Almost 100% of those genes were also labeled with AcH3K9,14, and most were labeled with AcH4K12. More than 95% of the islands for AcH4K12 and AcH2B mapped to loci enriched for AcH3K9,14, whereas many genes exhibiting AcH3K9,14 did not show the other acetylation marks (Figure 2F). Two conclusions were drawn from this analysis. First, although H3K4me3 is generally associated with active transcription, this mark does not determine by itself the transcriptional activity of the locus. Second, we observed a hierarchy between the different acetylation marks, in which AcH3K9,14 seems to instruct the other two acetylation marks.
TSA-induced hyperacetylation targets active loci marked with H3K4me3 and AcH3K9,14 and associated with NF-κB binding sites
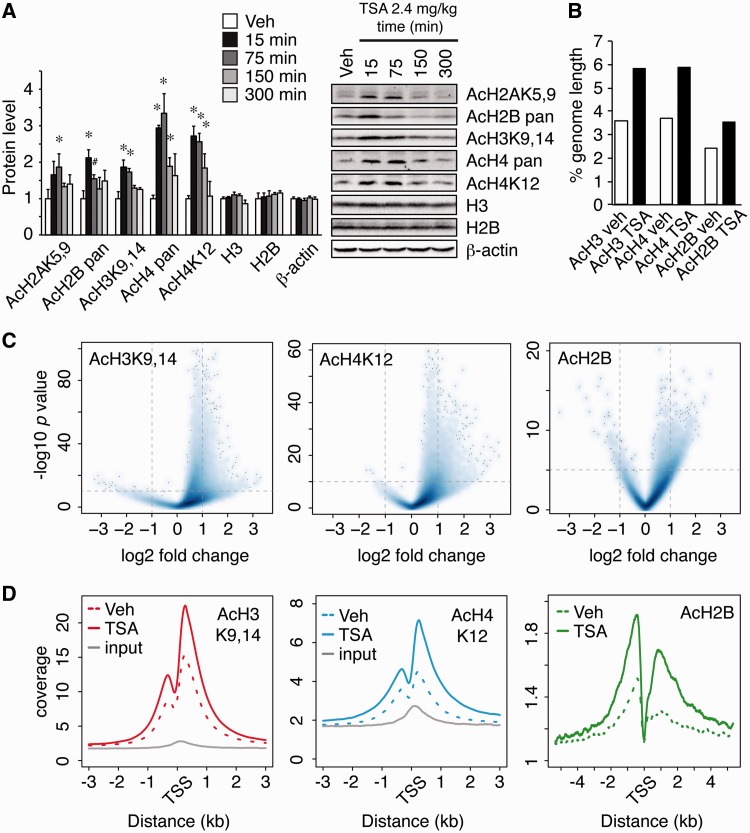
TSA causes a dramatic but transient increase in bulk chromatin acetylation in the hippocampus in vivo (Figure 3A), indicating that this compound crosses the blood–brain barrier and is then efficiently cleared by the organism. We next investigated the histone acetylation profiles 30 min after TSA injection when histone hyperacetylation was at its peak (Supplementary Figure S6). TSA increased the number of acetylation islands by 51, 128 and 18% for AcH3K9,14, AcH4K12 and AcH2B, respectively, which represents a prominent increase in the coverage of the mouse genome by these marks (Figure 3B). A differential enrichment screen identified thousands of islands significantly altered in the comparison of vehicle- and TSA-treated mice (Figure 3C and Supplementary Dataset S2). The impact of TSA was clearly observed at TSSs (Figure 3D), intragenic regions (Supplementary Figure S7A) and putative enhancers (Supplementary Figure S7B). Furthermore, our screen identified hundreds of acetylation islands that responded to the drug and were not associated with annotated genes (>10 kb from RefSeq genes) nor overlapped with CBP/p300 putative enhancers (Supplementary Figures S7C and D).
Figure 3.
TSA targets transcriptionally active genes. (A) Densitometric quantification of western blot data (left) and representative western blot images (right) of hippocampal bulk histone acetylation levels at different time points after TSA or vehicle (Veh) injection. *P < 0.05, Student’s t-test versus vehicle; n = 4–5 per group. Data are expressed as mean ± S.E.M. (B) Percentage of effective genome length showing enrichment in AcH3K9,14, AcH4K12 and AcH2B in the basal condition (Veh) and on TSA administration (TSA). (C) Volcano plots for AcH3K9,14, AcH4K12 and AcH2B showing the magnitude of differential enrichment (log2 FC) versus statistical significance (−log10 P-value) for each individual island detected in the comparison of vehicle- and TSA-treated mice. (D) Mean enrichment pattern for AcH3K9,14, AcH4K12 and AcH2B were profiled across all annotated TSSs for vehicle (dotted lines) and TSA treated mice (solid lines). Solid gray lines in AcH3K9,14 and AcH4K12 graphs show coverage in input condition.
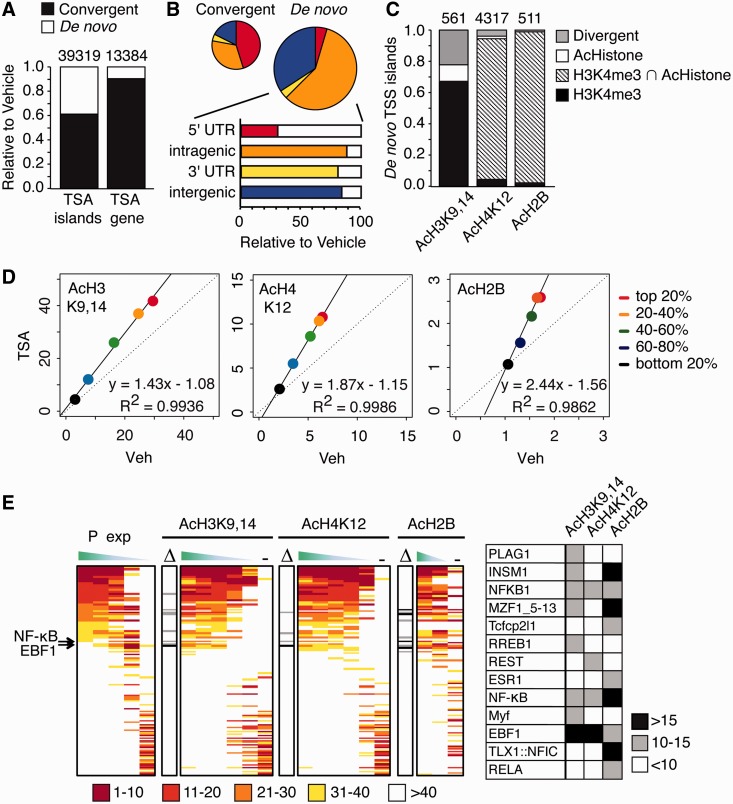
Interestingly, the mapping of de novo acetylation revealed that most of the AcH3K9,14 islands that appeared in response to TSA were located in intragenic regions of genes that already had this mark at the promoter in the basal state (Figure 4A and B). The rare de novo AcH3K9,14 islands labeling new genes mapped onto promoters pre-marked with H3K4me3 but lacking any acetylation mark (Figure 4C). In contrast, the presence of de novo AcH2B and AcH4K12 islands coincided in almost 100% of the cases with the presence of both AcH3K9,14 and H3K4me3 at the TSS (Figure 4C). In agreement with the role of AcH3K9,14 and H3K4me3 marks as anchorages for TSA-induced hyperacetylation and their association with active transcription, we found that the magnitude of hyperacetylation at the TSS correlated with the transcriptional rate of the gene. Thus, highly expressed genes underwent strong hyperacetylation, whereas low or non-expressed genes were basically not affected by HDAC inhibition (Figure 4D, slopes >1). Interestingly, AcH4K12 and AcH2B hyperacetylations were similar in the top 20% and the 20–40% group, indicating that above a given level of expression, TSA did not cause a greater increase in these marks.
Figure 4.
The presence of H3K4me3 and AcH3K9,14 drives TSA-induced chromatin hyperacetylation. (A) Bar graph shows the percentage of AcH3K9,14 islands (left bar) and annotated genes associated with those islands (right bar) in the TSA condition that are present in the vehicle condition (convergent; black) or generated de novo (de novo; white). (B) Pie charts show the annotation of AcH3K9,14 islands in TSA samples relative to their nearest gene, distinguishing between islands already present in basal condition (convergent) and those that were unique to TSA samples (de novo). The lower bar chart depicts the percentages of de novo islands according to their relative position to the nearest TSS. (C) Ratio of de novo islands for each acetylation mark that mapped to genes associated to H3K4me3 in the vehicle condition, to genes that bore both H3K4me3 and a histone acetylation mark, to genes that only had acetylation marks and those completely new (divergent). (D) Scatter plot shows mean coverage around TSSs in vehicle condition (Veh) versus TSA-treated mice (TSA). To generate the graph, sequence tags were determined at the TSS of all RefSeq (+/−1 Kb), RefSeqs were ranked according to relative mRNA expression levels and then split into five equal sets. Solid black lines in the plots denote the linear regression line. (E) Pscan software (159.149.160.51/pscan/) was used to scan for TFBSs in the promoter regions (−950/+50) of gene sets classified according to different parameters. First, genes were ranked according to their expression level in hippocampus (P exp) in five equal-size groups (from top 20% -left- to bottom 20% -right). In parallel, we also present the TFBS enrichment in the promoter regions of gene sets classified according to their acetylation changes at the TSS in response to TSA for the three acetylation marks (five equal-size groups for AcH3 and AcH4 ranked from left to right according to FDR P-values, and two equal-size groups for AcH2B ranked according to E-values; we also included an additional group (−) in each case composed with an equal-size set of randomly selected genes without any detected island for the corresponding mark). JASPAR TFBS were ranked according to Z-score values for each one of these groups. Significant enrichments (P < 0.05) were generally observed only for TFBS ranked above 40, rank values >40 were ignored. TFBS were ordered according to the top 20% expression group rank. To identify TFBS associated with TSA-dependent acetylation, we calculated the difference in rank values (Δ) between the top 20% expressed genes and the top group of differentially acetylated islands (group 1) for each histone mark. Only TFBSs with Δ > 10 in the comparison of the rank position between the top 20% group in TSA-mediated hyperacetylation for each mark and top 20% expression group are indicated.
To determine whether TSA-hyperacetylation at TSSs was associated with the presence of specific transcription factor-binding sites (TFBSs), we examined the enrichment profiles of the 130 TFBSs included in the JASPAR database. As expected, according to our acetylation/expression correlation analysis, the profiles were similar in TSA-hyperacetylated and in highly expressed genes. The deviations from this pattern indicate that TSA may preferentially target promoters with binding sites for NF-κB and the structurally related EBF1 (Figure 4E).
Uncoupling of gene transcription and histone acetylation changes after TSA treatment
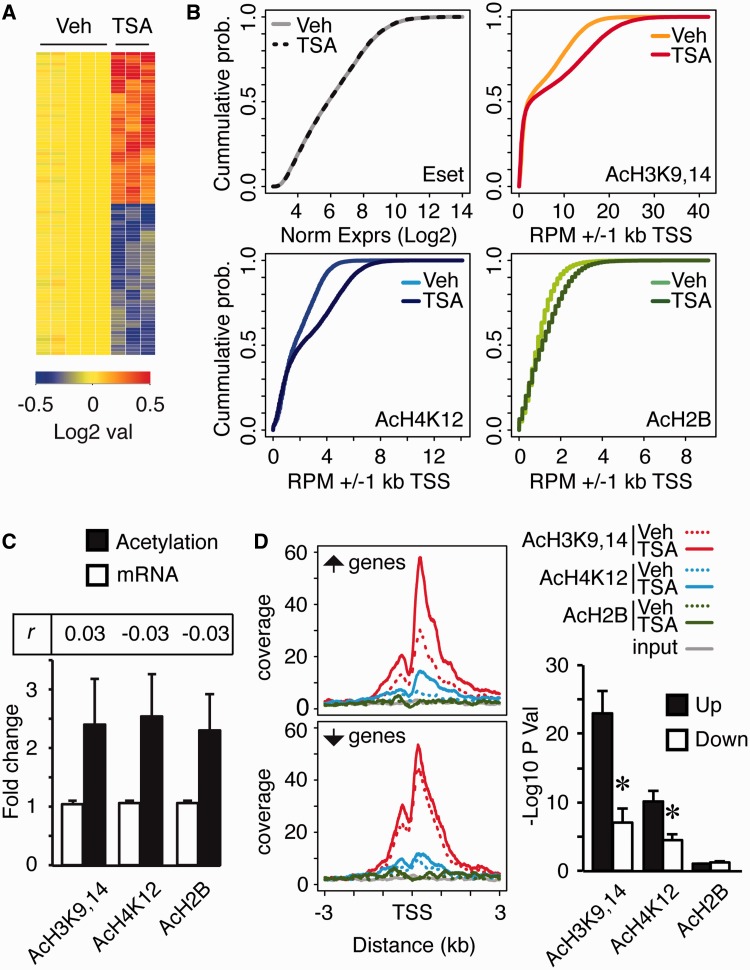
We next performed a microarray expression analysis to identify genes whose expression in the hippocampus was affected by TSA and to correlate transcriptional and histone acetylation changes. The impact of TSA in gene expression was much weaker than in acetylation profiles. TSA caused the differential expression of only 88 transcripts [adjusted P-value < 0.05; fold change (FC) > 1.2; Supplementary Table S2] including a similar number of gene upregulations and downregulations (Figure 5A).
Figure 5.
TSA causes the uncoupling between gene transcription and histone acetylation. (A) Heatmap showing the 88 Transcript Cluster IDs differentially expressed after TSA administration (adj. P < 0.05, FC > 1.2). (B) Cumulative probability distribution of expression levels (log2 values) (upper left) and normalized reads across the TSSs for AcH3K9,14 (upper right), AcH4K12 (bottom left) and AcH2B (bottom right) upon vehicle or TSA administration. The difference between vehicle and TSA treatment was statistically significant in the case of AcH3K9,14, AcH4K12 and AcH2B (P < 0.001, Kolmogorov–Smirnov test) but not for expression levels. (C) Bar chart shows the average FC of TSA-mediated histone acetylation changes (black bars) and transcript level changes (white bars) for the group of genes that exhibited a stronger hyperacetylation response to the drug (AcH3K9,14 and AcH4K12: FC > 2, FDR P < 10−10; AcH2B: FC > 2, FDR P < 10−5). Pearson correlation coefficients for each correlation are also shown (see full chart in Supplementary Figure S8B). Data are represented as mean ± SD. (D) Left: mean enrichment pattern for AcH3K9,14 (red), AcH4K12 (blue) and AcH2B (green) were profiled across upregulated and downregulated genes after TSA (solid lines) or vehicle (dotted lines) administration. Solid gray lines show coverage in the input condition. Right: bar chart depicts the P-value for histone acetylation changes occurring at upregulated or downregulated genes after TSA treatment (see also the correlation graphs in Supplementary Figure S8C). *P < 0.05 (Student’s t-test). Data are expressed as mean ± S.E.M.
Importantly, the occurrence of a large change in acetylation at the TSS did not translate into significant changes in transcript levels. Thus, the genes exhibiting the largest changes for each acetylation mark were, in average, not upregulated (Figures 5B and C and Supplementary Figure S8A and B), which uncouples the correlation between histone acetylation and gene expression levels observed at the basal state. Although we did not observe a positive correlation between the largest changes in acetylation and transcriptional alterations, the analysis of target genes identified in our microarray screen revealed a moderate correlation between changes in transcript level and changes in AcH3K9,14 at the promoter (Supplementary Figure S8C, Pearson correlation index rAcH3K9,14 = 0.53). We also observed a weak correlation with AcH4K12 changes (rAcH4K12 = 0.38), but no correlation with AcH2B changes (rAcH2B = −0.14). Overall, TSA caused the hyperacetylation of the promoters of both up- and downregulated genes, but upregulated genes showed larger and more significant increases of AcH3K9,14 and AcH4K12 than downregulated genes (Figure 5D).
TSA causes the rapid induction of a gene program related to chromatin deacetylation
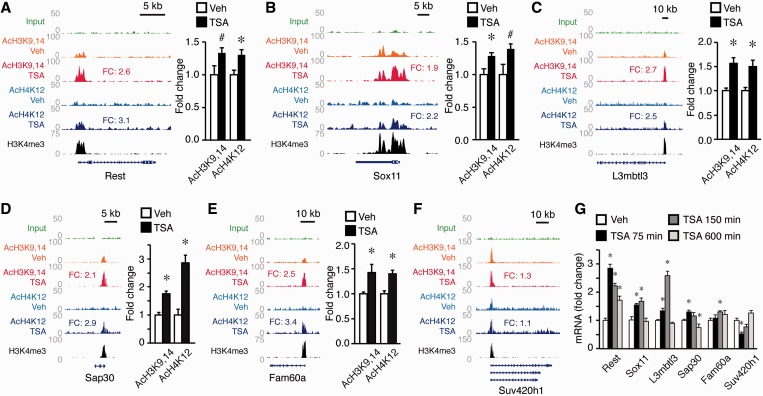
The group of genes differentially expressed shortly after TSA treatment was significantly enriched for nuclear proteins (Gene Ontology enrichment P = 0.03) and EBF1 and NF-κB-binding sites (PSCAN, P = 0.01 and 0.04, respectively), two motifs that were also associated with the genes that exhibited a greater acetylation response (Figure 4E). Among the upregulated factors, we found the polycomb group protein L3mbtl3, the transcriptional repressor Rest and the associated subunits of the Sin3-HDAC complex Sap30 and Fam60a, the regulators of transcriptional elongation Paf1 and eleven-nineteen lysine-rich leukemia and the transcription factors Sox11 and Gadd45a. Notably, some of these genes, such as Rest, L3mbtl3, Sox11 and Fam60a, belonged to the reduced group of genes that exhibited both strong acetylation at their promoter and expression upregulation upon TSA treatment.
We validated the results of our screen through ChIP and RT-qPCR assays. Genes such as Rest, L3mbtl3 or Sox11 were overexpressed in parallel with the increase of the acetylation marks (Figure 6A–E). In contrast, genes that were downregulated by TSA, such as Suv420h1, exhibited a weak response to the drug (Figure 6F). These assays also revealed the transient nature of most changes (Figure 6G).
Figure 6.
TSA induces genes involved in chromatin deacetylation. (A–E) Left: ChIPseq data at the Rest, Sox11, L3mbtl3, Sap30 and Fam60a loci. Horizontal rows display the number of normalized reads across the loci, with ‘Veh’ and ‘TSA’ denoting the treatment condition. Enrichment increases after TSA are indicated as FC. Right: Confirmation of hyperacetylation data through independent ChIP assay using oligo pairs that amplify a genomic sequence contained into the island detected by ChIPseq. Data are represented as mean ± S.E.M. *P < 0.05; #P < 0.1 (Student’s t-test), n = 4 per group. (F) ChIPseq data at Suv420h1 locus. (G) Time course and biological validation of candidate genes by RT-qPCR analysis. *P < 0.05 (Student’s t-test), n = 4–5 per group.
Histone acetylation changes do not interfere with novelty-driven gene induction
The memory-enhancing effects of HDACis might depend on the facilitation or priming of learning-induced transcription. To examine the impact of transient histone hyperacetylation in activity-driven promoters, we explored the effect of TSA on the transcriptional wave induced in the hippocampus by exposure to a novel environment (NE), which affects many genes involved in neuronal plasticity and memory consolidation (29). We performed this analysis in both wild-type (WT) animals and in mice with reduced KAT activity and histone acetylation (3,20).
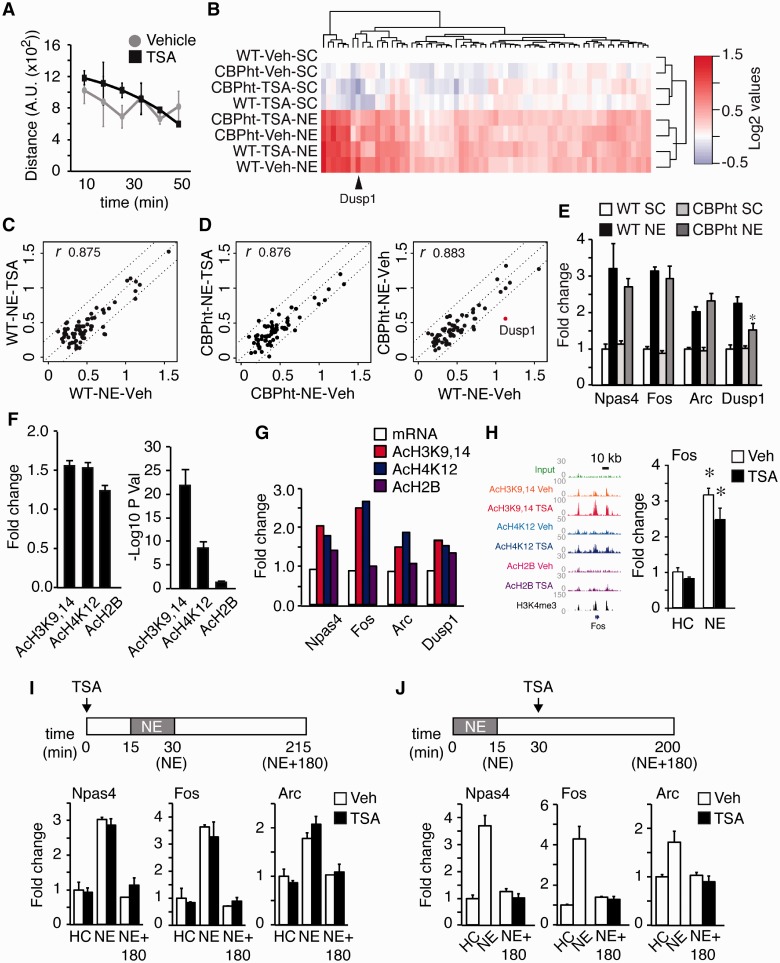
TSA did not affect the exploratory activity of the animals (Figure 7A) nor the transcriptional response to 1 h of NE (Figure 7B). The induction of IEGs, including Fos, Egr1, FosB, Npas4, Nr4a1, Arc and other genes related to memory, was remarkably similar in mice treated with vehicle or with TSA (Figure 7C and Supplementary Table S3). The induction of IEGs by novelty was also normal in mice with reduced KAT activity (Figure 7D). Three-way ANOVA of differential hippocampal gene expression in CBP-deficient mice treated with TSA or vehicle and exposed or not to novelty revealed no interaction between TSA treatment and novelty, as well as no interaction between either one of these conditions and genotype. Independent RT-qPCR assays confirmed these results (Figure 7E).
Figure 7.
TSA-induced hyperacetylation does not alter the transcriptional response to novelty. (A) TSA administration did not affect the exploration of a novel environment. (B) Heatmap showing the hierarchical clustering of transcriptional changes in response to novelty exposure of wild-type and CBP-deficient mice treated with vehicle or TSA. (C) Scatter plot showing the FC (log2) of differentially expressed transcripts on NE in WT mice treated with TSA or vehicle. Pearson correlation coefficient is shown. (D) Scatter plots showing the FC (log2) of differentially expressed transcripts on NE in CBP-deficient mice treated with TSA or vehicle (left) and comparing the induction of novelty genes in WT and Cbp+/− mice (right). The red dots represent the genes showing significant interaction between housing and genotype (non-corrected P-values). Pearson correlation coefficients are shown. (E) RT-qPCR array validation for Fos, Arc, Npas4 and Dusp1. Two-way ANOVA revealed a significant novelty effect for the four genes (P < 0.001), but only Dusp1 induction [also affected in CBP cKO mice (28)] exhibited a significant genotype effect (P = 0.04) and genotype x housing interaction (P = 0.04). n = 3 mice per group. (F) Bar chart depicts the average FC (left) and P-values (right) of histone acetylation changes occurring in NE-induced genes after TSA administration. (G) Summary of ChipSeq and array data for the IEGs Fos, Dusp1, Arc and Npas4. (H) ChIPseq data and RT-qPCR validation of gene induction at the Fos locus. * P < 0.05 (Student’s t-test), n = 3–4 per group. (I) Hippocampal mRNA expression of IEGs Npas4, Fos and Arc in mice that were injected with vehicle (Veh) or TSA (TSA) and 15 min later exposed to NE for 15 min. n = 3–4 except for control NE+180 min. (J) Hippocampal mRNA expression of IEGs Npas4, Fos and Arc in mice that were exposed to NE for 15 min and 15 min later injected vehicle (Veh) or TSA (TSA). n = 3–4. In all cases, data are expressed as mean ± S.E.M.
Of note, the promoter of most novelty-induced genes are labeled with H3K4me3 and acetylation marks at the basal state (Supplementary Figure S9A) and histone acetylation at the TSS increased in response to TSA, particularly in the case of AcH3K9,14 (Figure 7F). However, as for most genes, we did not observe a positive correlation between the acetylation increase in response to TSA and the expression change in response to this drug (Figures 7G and Supplementary Figure S9B). Particularly intriguing was the situation of Fos, which ranked among the genes most affected by TSA, but still did not show significant expression changes either at the basal state or in its induction in response to novelty (Figure 7H). Additional experiments in which TSA was administered either before (Figure 7I) or after (Figure 7J) a brief exposure to novelty demonstrated that this drug neither enhanced nor extended the induction of relevant IEGs by NE.
DISCUSSION
The appreciation of epigenetic complexity has dramatically increased for the past few years thanks to ChIPseq technology (30). Although much effort is still needed, our study represents an important initial step to describe the epigenomic landscape in the hippocampus and its regulation by neuronal activity or drugs. More than 100 posttranslational modifications of the histone tails have been reported, including >20 acetylations; we contributed here to the draft of the hippocampal epigenomic landscape with the profiles of four histone marks of particular relevance. The overlap and correlation between these epigenetic marks was high, but their profiles at the basal state, response to TSA and relationship with neuronal gene expression exhibited relevant differences. Strikingly, although H3K4me3 is generally associated with active transcription, our data indicate that loci that contain this mark but are depleted for the three acetylation marks rank among the least expressed genes in the hippocampus, which is consistent with a recent study, suggesting that H3K4me3 marks both active genes and silent or poised genes that will activate on the arrival of the right stimulus (31). AcH4K12 was the acetylation mark most affected by TSA, but AcH3K9,14 presented the best correlation with gene expression and appears to be a requirement, along with H3K4me3, for the positioning of the other two acetylation marks both in the basal state and on TSA administration. AcH2B concurred with AcH3K9,14 and to a lesser degree with AcH4K12 in highly expressed genes, which is consistent with early research on this mark (32), and exhibited a large relative enrichment in CBP- and p300-bound regions in agreement with the reported substrate preferences of these KATs (2,5,28).
Our differential acetylation profile screen led to a number of conclusions regarding the topography of TSA-induced hyperacetylation that are highly relevant for understanding the mechanisms of action and specific genomic impact of HDACis: (i) TSA treatment did not result in a global increase of histone acetylation. The changes had a precise topography and affected both specific gene sequences and putative enhancers but not intergenic regions. Moreover, TSA effects were largely restricted to loci that were enriched in AcH3K9,14 and H3K4me3 at the basal state. The concurrence of these two marks appears to be a requirement for TSA-mediated H4 and H2B hyperacetylation, which is consistent with experiments in human T-cells indicating that genes primed with H3K4me3 are more sensitive to HDACi-dependent H3K9 and H4K16 acetylation (31). (ii) TSA preferentially targeted transcriptionally active loci. AcH3K9,14 and H3K4me3 are both associated with active transcription and act as anchors for TSA action, consequently the susceptibility to TSA correlates with the transcriptional activity of the locus. This result indicates that HDACs reside with KATs on active genes or that both enzymatic activities visit frequently the same promoters to respectively add and remove acetyl groups from histones (31). This dynamic equilibrium, in turn, explains the rapid and transient effects of HDACis. (iii) TFBS analysis revealed that the promoters that contain binding sites for the transcription factors NF-κB and EBF1 are more likely to be targeted by TSA. These two transcription factors are structurally related (33) and have been recently reported to co-associate for regulating transcription (34), although it is uncertain that EBF1 expression plays a role in the adult hippocampus. NF-κB, however, has a well-known role in synaptic plasticity and learning and memory (9,35–37) and might also play a pivotal role in HDACi-dependent neuroprotection (38).
Complementary to ChIPseq, the microarray analyses revealed that, like in other cell types, TSA had a limited and cell-type-specific impact on transcription. Thus, whereas HDACis primarily affect genes related to cell cycle/apoptosis and DNA synthesis in tumor cells (39) and genes involved in differentiation and cell fate in stem cells (40,41), in the hippocampus, a tissue largely made up of non-dividing cells, the target genes are involved in transcriptional regulation and chromatin remodeling. Interestingly, among the factors upregulated by TSA, there were different components [Sap30, Rest and the recently identified Fam60a subunit (42)] of the Sin3-HDAC complex (43) that is involved in the repression of neuronal gene transcription in both non-neuronal and neuronal cells through the binding of Rest to RE1/NRSE sites (44). Experiments in cultured cortical neurons have also revealed a robust induction of Rest by TSA, suggesting that HDACs mediate its repression in neurons (45). Therefore, the upregulation of Rest and the other components of the Sin3 complex may represent the primary homeostatic response of hippocampal cells for restoring histone acetylation levels and prevent the transcriptional dysregulation that could result from prolonged chromatin hyperacetylation. The gene downregulations detected in our screen could also result from the enhanced expression of these transcriptional repressors.
Intriguingly, our experiments indicate that the dramatic change in histone acetylation profiles triggered by HDAC inhibition had a remarkably modest immediate impact on hippocampal gene expression. Consistently with this result, studies in yeast, flies and mouse fibroblasts have shown that the posttranslational modification of histones may not be as essential for gene expression as their correlative behavior with transcription initially suggested (16–18). For instance, the systematic analysis in yeast of hundreds of mutations in histone genes, including all the major acetylation sites at the histone tails, produced surprisingly moderate phenotypes and gene expression changes (19,46). In T-cells, a similar uncoupling between changes in transcription and histone acetylation caused by HDACis was interpreted considering that de novo acetylation may allow the binding of Pol II to the promoter, but this binding by itself is not sufficient to initiate transcription (31). Our ChIPseq experiments provide additional clues for understanding the modest effect of HDACis in transcription. On the one hand, TSA preferentially hyperacetylates active loci that are premarked with H3K4me3 and AcH3K9,14 at the TSS. On the other hand, the presence of these marks seems to be correlative with transcription rather than causative; they favor a transcriptionally permissive state but do not influence the transcriptional rate. As a result, the dramatic increase of histone acetylation triggered by HDACis is well tolerated by the cell and does not cause a general dysregulation of transcription.
Although further research is still needed to clarify the mechanism of action of these compounds in the nervous system, our experiments indicate that enhancing the transcription of plasticity-related genes might not be the primary mechanism. Not only TSA did not greatly affect basal gene expression but also the induction of IEGs by neuronal activity was apparently unaffected by HDAC inhibition or CBP deficiency, which is consistent with previous studies, indicating that histone acetylation levels are largely negligible for activity-dependent gene induction (10,28,47). An emerging view in the epigenetics field is that histone modifications are not involved in recruiting TFs and chromatin regulators to specific genes but may instead play a role as allosteric regulators of chromatin complexes (18), thereby having a subtle impact on transcription. According to this model, histone-dependent mechanisms would have a slow impact on gene expression that would not be detected by our microarray screen performed shortly after TSA treatment. The transient nature of both TSA-induced hyperacetylation and IEG induction by novelty would also prevent the detection of hyperacetylation-dependent changes in the expression of these genes. These results, together with our recent finding that altered neuronal histone acetylation interfered with the transcriptional neuroadaptation to environmental enrichment (20), suggest that histone acetylation changes in neurons are more relevant for the maintenance of epigenetic states than for the regulation of rapid transcriptional changes. This view also contributes to explain the ameliorative effects of HDACi in mouse models of progressive neurodegenerative disorders, such as Huntington’s and Alzheimer’s diseases, in which a chronic treatment is required (12). We cannot, however, discard that the transcriptional programs induced by more salient learning-related experiences, like training in a fear-conditioning task, could be enhanced by TSA treatment or attenuated by CBP deficiency, nor that the induction of IEGs in other brain areas were affected by these manipulations of histone acetylation levels. It is also possible that the increase of other, unexplored, acetylation marks showed a better correlation with transcriptional changes than the three marks explored here. However, recent genomic studies exploring the genomic profiles of KATs recruitment and histone acetylation have revealed a low level of specificity (25,31). In resting somatic tissues, most acetylation marks seem to co-occur in the same loci (48), and it has been suggested that the large panel of acetylation marks could redundantly contribute to maintain activation states while acetylation specificity would not be an important feature for gene regulation (49).
Interestingly, experiments in other systems indicate that HDACi actions may depend on histone-independent and even transcription-independent mechanisms (50–52). For instance, the neuroprotective effects of TSA in the nervous system are, at least, partially mediated by the acetylation of cytoskeletal proteins that restores vesicular transport and favors the release of growth factors (53). Consistently with the relevance of non-histone substrates, we observed a significant enrichment for NF-κB-binding sites in the promoter of TSA-regulated genes. Given that NF-κB is a direct target of TSA-dependent acetylation (9), its activation by acetylation can be responsible for the upregulation of Rest (54) and other rapid changes in expression detected in our array screen. Further studies should investigate the specific contribution of NF-κB acetylation to TSA-driven transcriptional changes.
To our knowledge, this is the first study that directly assesses the relationship between histone acetylation and neuronal gene expression and the mechanism of action of HDACis in the adult nervous system at the genomic level. Our results suggest alternative hypotheses for the mechanism of action of this important family of neuropsychiatric drugs and have important implications for the understanding and treatment of cognitive, psychiatric and neurodegenerative disorders related to impaired histone acetylation or in which these compounds have been proven effective.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [55–64].
ACKNOWLEDGEMENTS
The authors thank Román Olivares for excellent technical assistance in the maintenance of the mouse colony. J.L.A. holds a postdoctoral contract (JAE-doc) from the Program ‘Junta para la Ampliación de Estudios’ co-funded by the Fondo Social Europeo (FSE). S.I. holds a Juan de la Cierva contract from the Spanish Ministry of Economy and Competitiveness. L.M.V. holds a Ramón y Cajal contract from the Spanish Ministry of Economy and Competitiveness.
FUNDING
[CSD2007-00023, SAF2008-03194-E] (part of the coordinated ERA-Net NEURON project Epitherapy), [SAF2011-22855 and SAF2011-22506 to L.M.V.], Spanish Ministry of Economy and Competitiveness; [Prometeo/2012/005] Generalitat Valenciana; and the Fundació Gent per Gent. Funding for open access charge: Spanish Ministry of Economy and Competitiveness.
Conflict of interest statement. None declared.
REFERENCES
- 1.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A. Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein-Taybi syndrome and its amelioration. Neuron. 2004;42:947–959. doi: 10.1016/j.neuron.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lopez-Atalaya JP, Gervasini C, Mottadelli F, Spena S, Piccione M, Scarano G, Selicorni A, Barco A, Larizza L. Histone acetylation deficits in lymphoblastoid cell lines from patients with Rubinstein-Taybi syndrome. J. Med. Genet. 2012;49:66–74. doi: 10.1136/jmedgenet-2011-100354. [DOI] [PubMed] [Google Scholar]
- 6.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJ, Zhou Y, Wang X, Mazitschek R, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, Mullican SE, Jones S, Rusche JR, Lazar MA, et al. HDAC3 is a critical negative regulator of long-term memory formation. J Neurosci. 2011;31:764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levenson JM, O'Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 9.Yeh SH, Lin CH, Gean PW. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol. Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 10.Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, Cabrera SM, McDonough CB, Brindle PK, Abel T, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behav. Neurosci. 2007;121:1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 2010;31:605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 13.Kazantsev AG, Thompson LM. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat. Rev. Drug. Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 14.Sweatt JD. Experience-dependent epigenetic modifications in the central nervous system. Biol. Psychiatry. 2009;65:191–197. doi: 10.1016/j.biopsych.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graff J, Tsai LH. Histone acetylation: molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013;14:97–111. doi: 10.1038/nrn3427. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Bedford DC, Brindle PK. Is histone acetylation the most important physiological function for CBP and p300? Aging. 2012;4:247–255. doi: 10.18632/aging.100453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rando OJ. Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev. 2012;22:148–155. doi: 10.1016/j.gde.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. The specificity and topology of chromatin interaction pathways in yeast. Mol. Cell. 2011;42:536–549. doi: 10.1016/j.molcel.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Atalaya JP, Ciccarelli A, Viosca J, Valor LM, Jimenez-Minchan M, Canals S, Giustetto M, Barco A. CBP is required for environmental enrichment-induced neurogenesis and cognitive enhancement. EMBO J. 2011;30:4287–4298. doi: 10.1038/emboj.2011.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchis-Segura C, Lopez-Atalaya JP, Barco A. Selective boosting of transcriptional and behavioral responses to drugs of abuse by histone deacetylase inhibition. Neuropsychopharmacology. 2009;34:2642–2654. doi: 10.1038/npp.2009.125. [DOI] [PubMed] [Google Scholar]
- 22.Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peleg S, Sananbenesi F, Zovoilis A, Burkhardt S, Bahari-Javan S, Agis-Balboa RC, Cota P, Wittnam JL, Gogol-Doering A, Opitz L, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 24.Bousiges O, Vasconcelos AP, Neidl R, Cosquer B, Herbeaux K, Panteleeva I, Loeffler JP, Cassel JC, Boutillier AL. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valor LM, Pulopulos MM, Jimenez-Minchan M, Olivares R, Lutz B, Barco A. Ablation of CBP in forebrain principal neurons causes modest memory and transcriptional defects and a dramatic reduction of histone acetylation, but does not affect cell viability. J. Neurosci. 2011;31:1652–1663. doi: 10.1523/JNEUROSCI.4737-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Myers FA, Chong W, Evans DR, Thorne AW, Crane-Robinson C. Acetylation of histone H2B mirrors that of H4 and H3 at the chicken beta-globin locus but not at housekeeping genes. J. Biol. Chem. 2003;278:36315–36322. doi: 10.1074/jbc.M305822200. [DOI] [PubMed] [Google Scholar]
- 33.Treiber N, Treiber T, Zocher G, Grosschedl R. Structure of an Ebf1:DNA complex reveals unusual DNA recognition and structural homology with Rel proteins. Genes Dev. 2010;24:2270–2275. doi: 10.1101/gad.1976610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karczewski KJ, Tatonetti NP, Landt SG, Yang X, Slifer T, Altman RB, Snyder M. Cooperative transcription factor associations discovered using regulatory variation. Proc. Natl Acad. Sci. USA. 2011;108:13353–13358. doi: 10.1073/pnas.1103105108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boersma MC, Dresselhaus EC, De Biase LM, Mihalas AB, Bergles DE, Meffert MK. A requirement for nuclear factor-kappaB in developmental and plasticity-associated synaptogenesis. J. Neurosci. 2011;31:5414–5425. doi: 10.1523/JNEUROSCI.2456-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boccia M, Freudenthal R, Blake M, de la Fuente V, Acosta G, Baratti C, Romano A. Activation of hippocampal nuclear factor-kappa B by retrieval is required for memory reconsolidation. J. Neurosci. 2007;27:13436–13445. doi: 10.1523/JNEUROSCI.4430-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Federman N, de la Fuente V, Zalcman G, Corbi N, Onori A, Passananti C, Romano A. Nuclear factor kappaB-dependent histone acetylation is specifically involved in persistent forms of memory. J. Neurosci. 2013;33:7603–7614. doi: 10.1523/JNEUROSCI.4181-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang LX, Zhao Y, Cheng G, Guo TL, Chin YE, Liu PY, Zhao TC. Targeted deletion of NF-kappaB p50 diminishes the cardioprotection of histone deacetylase inhibition. Am. J. Physiol. Heart Circ. Physiol. 2010;298:H2154–H2163. doi: 10.1152/ajpheart.01015.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glaser KB, Staver MJ, Waring JF, Stender J, Ulrich RG, Davidsen SK. Gene expression profiling of multiple histone deacetylase (HDAC) inhibitors: defining a common gene set produced by HDAC inhibition in T24 and MDA carcinoma cell lines. Mol. Cancer Ther. 2003;2:151–163. [PubMed] [Google Scholar]
- 40.Karantzali E, Schulz H, Hummel O, Hubner N, Hatzopoulos A, Kretsovali A. Histone deacetylase inhibition accelerates the early events of stem cell differentiation: transcriptomic and epigenetic analysis. Genome Biol. 2008;9:R65. doi: 10.1186/gb-2008-9-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen B, Cepko CL. Requirement of histone deacetylase activity for the expression of critical photoreceptor genes. BMC Dev. Biol. 2007;7:78. doi: 10.1186/1471-213X-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith KT, Sardiu ME, Martin-Brown SA, Seidel C, Mushegian A, Egidy R, Florens L, Washburn MP, Workman JL. Functional characterization of the human FAM60A protein: a new subunit of the Sin3 deacetylase complex. Mol. Cell. Proteomics. 2012;11:1815–1828. doi: 10.1074/mcp.M112.020255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grzenda A, Lomberk G, Zhang JS, Urrutia R. Sin3: master scaffold and transcriptional corepressor. Biochim. Biophys. Acta. 2009;1789:443–450. doi: 10.1016/j.bbagrm.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roopra A, Sharling L, Wood IC, Briggs T, Bachfischer U, Paquette AJ, Buckley NJ. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 2000;20:2147–2157. doi: 10.1128/mcb.20.6.2147-2157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 46.Dai J, Hyland EM, Yuan DS, Huang H, Bader JS, Boeke JD. Probing nucleosome function: a highly versatile library of synthetic histone H3 and H4 mutants. Cell. 2008;134:1066–1078. doi: 10.1016/j.cell.2008.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kasper LH, Lerach S, Wang J, Wu S, Jeevan T, Brindle PK. CBP/p300 double null cells reveal effect of coactivator level and diversity on CREB transactivation. EMBO J. 2010;29:3660–3672. doi: 10.1038/emboj.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anamika K, Krebs AR, Thompson J, Poch O, Devys D, Tora L. Lessons from genome-wide studies: an integrated definition of the coactivator function of histone acetyl transferases. Epigenetics Chromatin. 2010;3:18. doi: 10.1186/1756-8935-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnstone RW, Licht JD. Histone deacetylase inhibitors in cancer therapy: is transcription the primary target? Cancer Cell. 2003;4:13–18. doi: 10.1016/s1535-6108(03)00165-x. [DOI] [PubMed] [Google Scholar]
- 51.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 52.Gaub P, Tedeschi A, Puttagunta R, Nguyen T, Schmandke A, Di Giovanni S. HDAC inhibition promotes neuronal outgrowth and counteracts growth cone collapse through CBP/p300 and P/CAF-dependent p53 acetylation. Cell Death Differ. 2010;17:1392–1408. doi: 10.1038/cdd.2009.216. [DOI] [PubMed] [Google Scholar]
- 53.Dompierre JP, Godin JD, Charrin BC, Cordelieres FP, King SJ, Humbert S, Saudou F. Histone deacetylase 6 inhibition compensates for the transport deficit in Huntington's disease by increasing tubulin acetylation. J. Neurosci. 2007;27:3571–3583. doi: 10.1523/JNEUROSCI.0037-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ravache M, Weber C, Merienne K, Trottier Y. Transcriptional activation of REST by Sp1 in Huntington's disease models. PLoS One. 2010;5:e14311. doi: 10.1371/journal.pone.0014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanaka Y, Naruse I, Maekawa T, Masuya H, Shiroishi T, Ishii S. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl Acad. Sci. USA. 1997;94:10215–10220. doi: 10.1073/pnas.94.19.10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lopez de Armentia M, Jancic D, Olivares R, Alarcon JM, Kandel ER, Barco A. cAMP response element-binding protein-mediated gene expression increases the intrinsic excitability of CA1 pyramidal neurons. J. Neurosci. 2007;27:13909–13918. doi: 10.1523/JNEUROSCI.3850-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Planet E, Attolini CS, Reina O, Flores O, Rossell D. htSeqTools: high-throughput sequencing quality control, processing and visualization in R. Bioinformatics. 2012;28:589–590. doi: 10.1093/bioinformatics/btr700. [DOI] [PubMed] [Google Scholar]
- 62.Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, Green MR. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 2010;11:237. doi: 10.1186/1471-2105-11-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 2011;39:e35. doi: 10.1093/nar/gkq1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.