Abstract
In eukaryotic cells, gene expression is mediated by enhancer activation of RNA polymerase at distant promoters. Recently, distinctions between enhancers and promoters have been blurred by the discovery that enhancers are associated with RNA polymerase and are sites of RNA synthesis. Here, we present an analysis of the insulin-like growth factor 2/H19 muscle enhancer. This enhancer includes a short conserved core element that is organized into chromatin typical of mammalian enhancers, binds tissue-specific transcription factors and functions on its own in vitro to activate promoter transcription. However, in a chromosomal context, this element is not sufficient to activate distant promoters. Instead, enhancer function also requires transcription in cis of a long non-coding RNA, Nctc1. Thus, the insulin-like growth factor 2/H19 enhancer is an active transcriptional complex whose own transcription is essential to its function.
INTRODUCTION
Promoters and enhancers are generally thought of as two distinct regulatory elements. Functionally, promoters have been defined as the regions where RNA transcription initiates, whereas enhancers are DNA elements that work over distance to activate transcription at promoter elements (1). Furthermore, genomic analyses have defined and distinguished promoters and enhancers by their distinctive epigenetic marks, specifically their unique patterns of histone methylation (2–4). More recently, these functional and structural distinctions between enhancers and promoters have become somewhat blurred with the identification of enhancers with promoter-like chromatin features (2,5,6) and also with the realization that enhancer regions are frequently enriched for RNA Polymerase II (RNAP) and are sites for transcription of all kinds of RNAs including bidirectional transcripts (eRNAs) and multi-exonic polyadenylated RNAs (7–13). However, the functional significance of enhancer associated RNAs remains unclear (14).
Insulin-like growth factor 2 (Igf2) and H19 are linked co-regulated genes on the distal end of mouse chromosome 7. In humans, mis-expression of these genes on chromosome 11p15.5 is associated with developmental disorders and with several types of cancer including rhabdosarcoma (15,16). Igf2 and H19 are co-ordinately regulated in that they share tissue and developmental specificities that are dependent on a series of shared tissue-specific enhancer elements.
The enhancer required for in vivo expression of Igf2 and H19 in muscle has been defined by mouse knockout studies (17). The ΔME mutation, a 20 kb deletion, centred 25 kb downstream of H19 (or 105 kb downstream of Igf2) (Figure 1A) that reduces Igf2 and H19 expression in myocytes to essentially undetectable levels (17,18). Recently, transient transfection analyses identified a 294 bp myocyte-specific core enhancer region [here called the core muscle enhancer (CME)] within the sequences defined by the ΔME deletion (19).
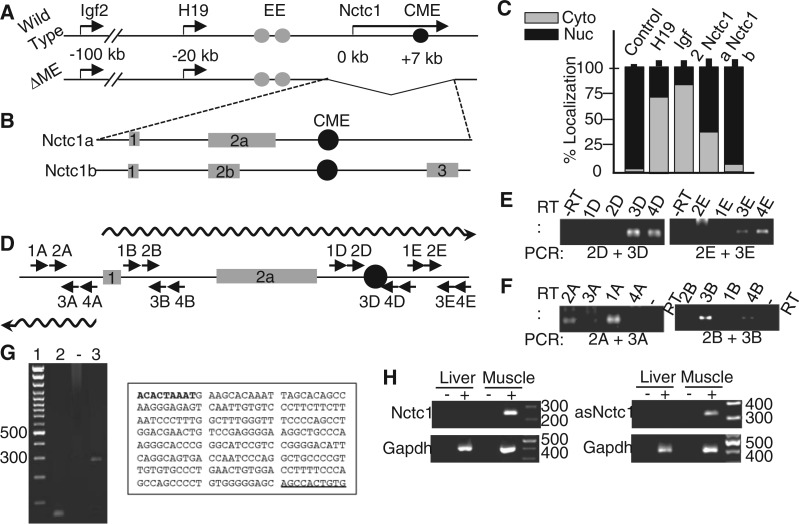
Figure 1.
Nctc1 lncRNA and the Igf2/H19 mesodermal enhancer. (A) Cartoon depiction of the Igf2/H19 locus on wild-type and ΔME chromosomes. EE, core endodermal enhancers (grey circles) (24); CME, core muscle-specific enhancer (black circle) (19) (and this study). The ΔME chromosome carries a 20 kb deletion that eliminates H19 and Igf2 expression specifically in skeletal muscle (17). (B) Genomic structures for Nctc1 isoforms 1a and b including the CME (black circle) located in intron 2. Exons are depicted as grey rectangles. (C) Cellular localization of Igf2, H19 and Nctc1 transcripts. cDNAs were generated from RNAs isolated from nuclear and cytosolic fractions of primary myoblasts and quantitated for gene expression by qRT-PCR to determine the fractional composition. As a control, we also assayed localization of unspliced Nctc1 heteronuclear RNA (hnRNA) and saw that it was 97 ± 2% nuclear. (D) Transcription at the Nctc1 locus. Nctc1a exons 1 and 2 and the CME are depicted as described above. One sense (squiggled arrow on top of the cartoon) and one major antisense transcript (squiggled arrow below the cartoon) were identified by RT-PCR using the primers depicted. (E) RT-PCR analyses to detect sense and antisense transcription across the CME. For each experiment, the gene-specific primer used for reverse transcription (RT primer) is depicted above the panel. PCR primer pairs used to detect the presence or absence of each cDNA species are depicted below the panels. Primer sequences are listed in Supplementary Table 1. (F) Sense and antisense transcription across the Nctc1 promoter were analysed as described in panel E. Results using additional primer pairs that span the locus (Supplementary Table S1) confirm the summary diagram in panel D. (G) 5′ Rapid amplification of cDNA ends identifies a single major start for the antisense Nctc1 transcription. Lane 1, 100 bp ladder; Lane 2, -RT control; Lane 3, RACE amplicon. The text box shows the conserved sequences that overlap the Nctc1 promoter. The 5′ ends of the antisense (bold) and sense (underlined) transcripts are indicated. (H) Antisense Nctc1 transcription is also muscle-specific. cDNAs were generated using random hexamer primers from RNAs isolated from neonatal liver and muscle tissue and analysed for expression of Nctc1, asNctc1 and Gapdh. ‘Plus’ and ‘minus’ indicate the inclusion or absence of reverse transcriptase enzyme in the cDNA synthesis step.
In addition to carrying the CME, the minimal enhancer region, as defined by the mouse knockout and also by transgene analyses, completely coincides with the gene, Nctc1 (Figure 1A and B). The Nctc1 promoter lies 7 kb upstream of the CME and generates a spliced long non-coding RNA (lncRNA) expressed only in myocytes (18,20). In this study, we sought to identify a role for the Nctc1 gene and/or RNA in muscle enhancer function, and therefore we performed detailed molecular and genetic analyses of the enhancer. We show that the Igf2/H19 enhancer is bipartite. Enhancer activity requires the CME element that binds transcription factors, is organized into chromatin typical of an enhancer and functions in classical in vitro reporter assays to activate promoter transcription. However, in a chromosomal context, enhancer function also requires the Nctc1 promoter and its transcription in cis. Altogether, our results demonstrate that this enhancer is an active transcriptional complex and that enhancer transcription is integral to enhancer function.
MATERIALS AND METHODS
Mice
Animal work was done according to NIH policy and approved by the Institutional Animal Care and Use Committee.
Primary myoblast culture
Primary myoblasts were isolated from neonatal pups (21) and differentiated into myotubes by growth in limiting horse serum (5%) for 24–48 h.
RNA isolation and analysis
RNAs were extracted from snap-frozen muscle tissue using TriPure Extraction Reagent (Roche) or from cultured cells by the QiaShredder column (Qiagen) and then purified with the RNeasy Micro Kit (Qiagen), including the optional treatment with DNAse I. RNA integrity and concentrations were evaluated using an Agilent 2100 Bioanalyzer, and only samples with RNA integrity numbers (RINs) greater than 9 were processed further. Complementary DNAs (cDNAs) were synthesized with Transcriptor First Strand cDNA Synthesis Kit (Roche) using random hexamer or oligo dT primers as indicated. These cDNA samples were then analysed using SYBR Green on the Roche Cycler 480II. All PCR primers are listed in Supplementary Table S1. For cellular localization analyses, RNAs were purified from nuclear and cytoplasmic fractions obtained using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific). The 5′ rapid amplification of cDNA ends (RACE) was accomplished using the GeneRacer Kit (Invitrogen) with primers indicated in Supplementary Table S1.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) analysis was performed as described (22) using antibodies specific to H3K4me1 (Abcam 8895), H3K36me3 (Abcam 9050), to H3K4me3 (Upstate 17-614) and to Ser-5(P)- RNA polymerase (Abcam 5131) or non-specific immunoglobulin G (Santa Cruz 2017). ChIP-purified DNA was quantified and normalized to input controls by quantitative reverse-transcription PCR (qRT-PCR). PCR primers are described in Supplementary Table S1.
Reporter constructs
Nctc1 sequences were cloned into the multiple cloning site of the pβgal-promoter plasmid (Clontech). For simplicity, Figure 3 presents plasmid structures and expression data for only one orientation. However, the results for the alternative orientation were essentially the same. DNA sequence endpoints for each construct are listed relative to the Nctc1 sense start site (bp 1 of exon 1). The Construct 1 insert is a 12 728 bp EcoRI-EcoRI fragment spanning from -1751 to 10980 bp. The Construct II insert is 8841 bp ScaI-ScaI-EcoRV fragment spanning from -1042 to 7799 bp. The Construct III insert is a 7715 bp ScaI-EcoRV fragment spanning from 84 to 7799 bp. The Construct IV insert is a 7540 bp ScaI-ScaI fragment spanning from -1042 to 6498 bp. The Construct V insert is a 1301 ScaI-EcoRV fragment spanning from 6498 to 7799 bp. The Construct VI insert is a 386 bp PCR amplicon spanning from 7313 to 7699 bp. The plasmids described in Figure 6 are derivatives of Construct II and were generated by insertion of transcriptional terminator fragments into the unique NsiI site at 2332 bp.
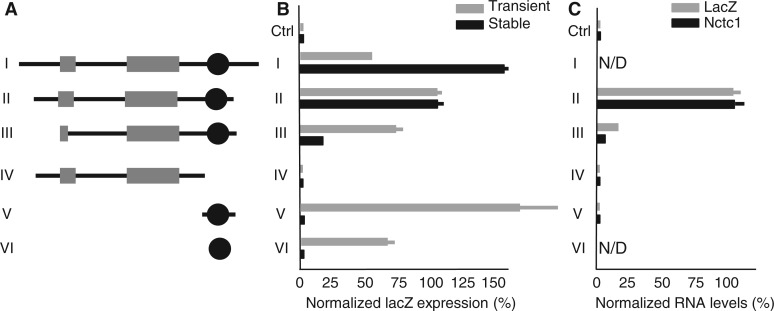
Figure 3.
In a chromatin complex, enhancer activity is dependent on both the CME and the Nctc1 Promoter. (A) Reporter constructs were generated by cloning Nctc1 sequences into the multiple cloning site of plasmid pβgal-promoter as described in ‘Materials and Methods’ section. The reporter carriers a minimal SV40 promoter fused to the lacZ reporter so that high levels of expression depend on enhancer activity from the Nctc1 insertion fragments. Depicted are key Nctc1 inserts with Nctc1 exons 1 and 2 (grey rectangles) and the CME (black circle). (B) The CME is not sufficient for enhancer function in a chromosomal context. Primary myocytes isolated from wild-type mice were transfected with the constructs depicted in panel A. Expression of lacZ in transiently (grey bars) and in stably (black bars) transfected cells is normalized as described in ‘Materials and Methods’ section and reported relative to the expression observed using Construct II. Essentially identical results were obtained using mouse C2C12 myoblasts. (C) Reporter gene activation in stably transfected cells correlates with Nctc1 promoter activity. Primary myocytes isolated from ΔME/ΔME mice were transfected with constructs depicted in (A). Expression of Nctc1 and of lacZ is normalized to that seen in cells transfected with construct II carrying the full Nctc1 gene. Nctc1 expression is quantitated using primers specific for spliced message. The ΔME deletion removes the entire Nctc1 coding region (see Figure 1A) so that no Nctc1 RNA can be generated by the endogenous locus. For panels B and C, results are reported as average values with standard deviations calculated using at least three independent samples. In panel C, N/D means not determined.
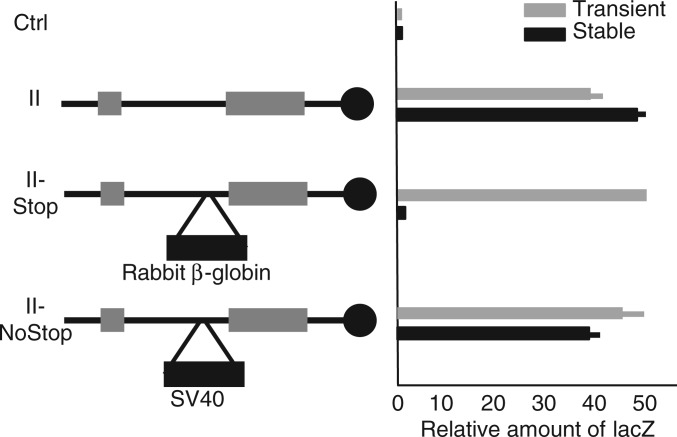
Figure 6.
Transcriptional termination in intron 1 blocks reporter gene activation specifically in stably transfected cell lines. Left panel, construct II (see Figure 3A) and derivatives carrying a transcriptional terminator (II-Stop) (28) or a similarly sized insert with no terminator activity (II-NoStop) are depicted. Right panel, lacZ expression in transiently (grey bars) and stably (black bars) transfected myocytes isolated from wild-type mice. Expression is normalized to levels from the Ctrl plasmid (the reporter plasmid with no Nctc1 insert). Results are depicted as average values with standard deviations from at least three independent transfections.
Transfection studies
DNAs were introduced into C2C12 lines by electroporation (Amaxa) or into primary mouse myoblasts by lipofection (Lipfectamine 2000 Invitrogen). Stable cell transfections included pTK-Hyg (Clontech) at 1:5 molar ratios relative to the reporter construct. Hygromycin-resistant clones were analysed as described (23).
‘Statistical significance’ was evaluated using two-tailed Student t-tests.
RESULTS
We used qRT-PCR and DNA sequencing to confirm the structures of the two Nctc1 isoforms, Nctc1a and Nctc1b, described on the UCSC Genome Browser (NBI37/mm9) (Figure 1B and Supplementary Figure S1A). Although Nctc1b represents only a minor RNA (Supplementary Figure S1B), its presence demonstrates that transcription occurs across the CME. Both Nctc1a and Nctc1b are predominantly nuclear (65 ± 9% and 92 ± 7%, respectively, n = 3) (Figure 1C).
Recent genome-wide studies indicate that many (or even most) enhancers are associated with activated RNAP enzyme and are sites of active RNA transcription (7–10). We performed extensive strand-specific RT-PCR analyses to catalogue transcription in the enhancer region. Results are summarized in Figure 1D and representative data for some of the key reactions are presented in Figure 1E and F. Consistent with the known gene structures for Nctc1, we identified sense transcription beginning at Nctc1 exon 1 and extending across the entire locus (Figure 1E and F). In contrast, antisense transcripts were highly restricted in this locus. Specifically, we could not identify any antisense transcripts in the CME region (Figure 1E). Thus, the CME is not associated with bidirectional eRNAs as is typical of many enhancers.
The one region where we did identify antisense transcription was at the 5′ end of the Nctc1 gene (see Figure 1D). RT-PCR analyses readily identified antisense transcription upstream of Nctc1 exon 1 (Figure 1F). Furthermore, Rapid Amplification of cDNA Ends (5′RACE) shows that the major start for antisense transcription is just upstream of the major sense transcription start (Figure 1G). Antisense transcription is muscle specific (Figure 1H) and is entirely dependent on the CME (Supplementary Figure S1C). Steady-state levels of sense and antisense Nctc1 RNAs are roughly comparable (Supplementary Figure S1D). In sum, the Nctc1 promoter is bidirectional, but there is no evidence for eRNA-like transcription originating from the CME region.
Consistent with our RNA analyses, published ChIP-Seq data made publically available by the ENCODE project show muscle-specific binding of RNAP across the locus but with the highest accumulations at the CME and the next highest accumulations near the Nctc1 promoter (25). Among mammalian species, the CME along with the Nctc1 promoter are the two best conserved sequences (Supplementary Figure S1A) (20). The conservation of these cis regulatory elements is in sharp contrast to the complete lack of sequence conservation of the Nctc1 RNA coding sequences.
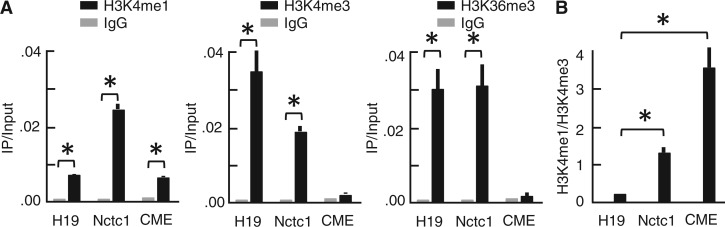
Genomic studies have distinguished promoter and enhancer elements based on their distinct chromatin structures. In particular, promoters have been associated with high levels of trimethylation at H3K4 and H3K36 and low levels of H3K4 monomethylation while the converse appears to be true for many enhancers (2–4). We used ChIP to determine structures at the Nctc1 promoter and CME (Figure 2A). We also analysed chromatin at the 5′ end of H19 exon 1 to represent the epigenetic structures of a typical promoter. The CME looks like a stereotypical enhancer: relatively high H3K4me1 and low H3K4me3 and H3K36me3. The Nctc1 promoter, however, is a curious hybrid. Like the H19 promoter, it is enriched for trimethylation of H3K4 and H3K36. However, the Nctc1 promoter also displays relatively high levels of H3K4 monomethylation so that overall, the me1/me3 ratio is like that of a classical enhancer (Figure 2B). Thus, the Nctc1 promoter appears structurally similar to the enhancer/alternative promoter hybrid element recently defined at the Nprl3 gene in the α-globin cluster (11).
Figure 2.
Epigenetic marks at the Nctc1 locus. Primary myoblasts were isolated from wild-type mice, differentiated in vitro for 24 h and analysed by ChIP. (A) H3K4me1, H3K4me3 and H3K36 at the H19 and Nctc1 promoters and at the CME Primer pairs are described in Supplementary Table S1. (B) Normalized H3K4me1/H3K4me3 ratios. N = 3, mean ± standard deviations; *P < 0.0001.
In sum, DNA conservation, chromatin structure and RNA transcription patterns all mark the Nctc1 promoter as an interesting DNA element. We next wanted to determine whether the promoter was a functionally important element. Specifically, we wanted to establish whether the Nctc1 promoter was important for the in vivo enhancer activity already mapped to the region by the analysis of the ΔME deletion.
To characterize enhancer function, we performed transfection analyses using both mouse C2C12 and primary mouse myoblast lines. Differentiation of myoblasts into myotubes is readily accomplished by transferring growing cells to medium containing reduced serum supplement and results in activation of Igf2 and H19 transcription via the shared muscle enhancer (17,18,23). To identify which DNA sequences are important for enhancer activity, we generated a series of reporter constructs by cloning DNA fragments spanning different parts of the Nctc1 locus into the multiple cloning site of plasmid, pβgal-Promoter (Figure 3A). This vector carries an SV40 promoter inserted just upstream of the lacZ gene. The promoter is only minimally productive on its own so that obtaining high levels of lacZ transcription requires insertion of an active enhancer element. Transient transfection is the method that has been most commonly used to successfully identify and characterize enhancer sequences. However, we posited that transient assays might be limited in their ability to identify all sequences critical for enhancer function in vivo because transient transfection cannot account for the role of chromatin and chromosomal confirmation in the regulation of gene expression. Therefore, we also analysed reporter expression in pools of stably transfected cell lines.
Transient transfections (Figure 3B, grey bars) confirmed the essential findings of Alzhanov et al. (19). We found a core enhancer element within intron 2 that is both necessary (Figure 3B, compare constructs II and IV) and sufficient (Figure 3B, construct VI) to drive high levels of reporter activity. The 386 bp core defined by Construct VI includes a 219 bp CpG island and a cluster of highly conserved e-boxes that bind muscle-specific transcription factors including MyoD (26) as well as myogenin, and Myf5 in vivo (MLS, data not shown). Chromosome conformation capture (3C) assays have shown that this core element physically interacts with distal H19 and Igf2 promoters (22,18) and also specifically interacts with the adjacent Nctc1 promoter (18).
Stable transfections (Figure 3B, black bars) confirm that the CME is necessary for enhancer activity (Figure 3B, compare constructs II and IV). However, in stably transfected lines, the core is no longer sufficient for enhancer function (Figure 3B, constructs V and VI). Rather, the Nctc1 promoter is also required for high levels of transcriptional enhancement (Figure 3B, compare constructs II and III). Enhancer activation of the reporter correlates well with Nctc1 transcription (Figure 3C). Thus, both conserved elements, the core enhancer and the Nctc1 promoter, are important for enhancer function in a chromosomal context.
We considered three alternative models to explain the requirement for both the Nctc1 promoter and the CME in enhancer function. Model 1 (RNA only) suggests that Nctc1 RNA is the essential product of the CME and that once made, the Nctc1 RNA acts independently of the CME to help activate target genes. This model predicts that Nctc1 provided in trans would bypass the need for the CME. Model 2 (RNA + CME) suggests that Nctc1 transcription is important, but it functions only through its interactions with the CME. In model 3 (DNA only), Nctc1 transcription and RNA synthesis are just coincidences and are not relevant to enhancer function. Instead, this model suggests the Nctc1 promoter sequences carry an independent cis-acting classical enhancer element that acts synergistically with the CME to activate target genes. We assumed that any of the three models were possible and tested them directly with genetic and molecular approaches.
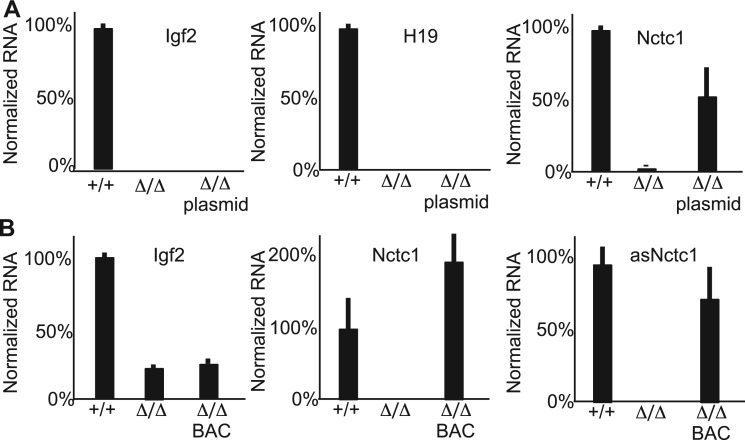
ΔME is the deletion mutation shown in Figure 1A that removes the entire Nctc1 locus. Accordingly, differentiated myotubes generated from myoblasts isolated from ΔME/ΔME mice cannot express Igf2 or H19. In Figure 4A, we show that expression of Igf2 and H19 in ΔME/ΔME primary muscle cells is not restored by the action of Construct II plasmid introduced by stable transfection, even though Construct II provides high levels of both sense (Figure 4A, panel 3) and of antisense (Supplementary Figure S1C) Nctc1. Similarly, expression of Igf2 in muscle tissue of ΔME/ΔME animals is not rescued by transgene constructs carrying the Nctc1 locus (Figure 4B). In this mouse experiment, the Nctc1 transgene is single copy bacterial artificial chromosome (BAC) that carries the entire Nctc1 locus plus 27 kb of upstream sequence and 100 kb of downstream sequence (23,27). This BAC faithfully restores expression of all Nctc1 transcripts with no effect on Igf2 RNA levels (Figure 4B). (The presence of H19 on the BAC transgene precludes its use as a marker for Nctct1 activity in this experiment.) Together, these in vitro and in vivo complementation assays indicate that Nctc1 RNA on its own does not contribute to target gene activation and cannot bypass the need for a functional CME.
Figure 4.
The Nctc1 RNA does not work in trans to drive enhancer activity. (A) Nctc1-expressing plasmids do not rescue Igf2 or H19 expression in ΔME/ΔME myotubes. RNA was isolated from wild-type cell lines (+/+), from ΔME/ΔME mutant cell lines (ΔME/ΔME) and from mutant cells stably transfected with a Construct II (see Figure 3) (Δ/Δplasmid). ΔME is a chromosomal deletion that removes the entire Nctc1 locus including the CME and the Nctc1 coding sequences (Figure 1). Expression of Nctc1, H19 and Igf2 were each normalized to Gapdh. (B) A BAC carrying the Nctc1 gene does not rescue Igf2 expression in ΔME/ΔME muscle tissue. RNA was isolated from muscle tissue dissected from wild-type neonates (+/+) and from ΔME/ΔME mutant neonates (Δ/Δ and from ΔME/ΔME littermates transgenic for a BAC carrying the Nctc1 gene plus 20 kb of upstream and 100 kb of downstream sequences (Δ/ΔBAC). The BAC transgene supplies normal levels of Nctc1 transcripts but does not rescue Igf2 expression.
Our next analyses tested the ability of Nctc1 RNAs to work in trans with the CME to establish enhancer function. Specifically, we reanalysed the stable cell transfection data from Figure 3 to compare enhancer activity of key Nctc1 reporter constructs transfected into +/+ (Figure 3B) and into ΔME/ΔME myoblasts (Figure 3C). The overall levels of expression and also patterns of expression for each plasmid in the construct series are the same in both genetic backgrounds. For example, the CME only plasmid (Construct V) is as completely defective for enhancer function in +/+ as in ΔME/ΔME myocytes. Similarly, a smaller deletion that abrogates the Nctc1 promoter (Construct III) results in the same 7-fold reduction of enhancer function in both cell types. Thus, Nctc1 RNA provided by transcription from the endogenous loci cannot rescue enhancer function of reporter constructs carrying the CME but lacking the Nctc1 promoter.
As an alternative approach to test for Nctc1 RNA function, we used siRNA to reduce Nctc1 RNA levels in +/+ myoblasts by up to 75% without decreased expression of the enhancer’s target genes, H19 and Igf2 (Figure 5). Thus, altogether, molecular and genetic studies indicate that Nctc1 RNA does not work in trans to drive CME enhancer function.
Figure 5.
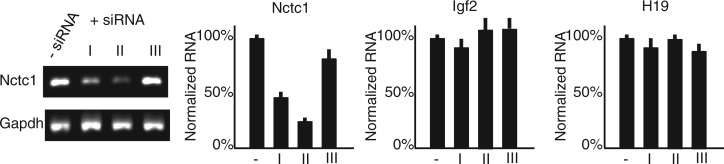
Reducing Nctc1 by siRNA does not depress enhancer activity as measured by Igf2 and H19 expression. Nctc1 siRNAs (see Supplementary Table S1 for sequences) reduce steady-state levels of Nctc1 by up to 75% with no effect on Igf2 or H19. RNA levels were analysed by qRT-PCR, normalized for Gapdh levels and then normalized to expression in the absence of siRNA. For each panel, results are depicted as average values with standard deviations from at least three independent transfections.
Finally, we tested for a role of Nctc1 transcription in cis by inserting a 2.2 kb rabbit β-globin transcriptional terminator (28) into Nctc1 intron 1 of reporter construct II (Figure 6, left panel, construct II-Stop). This insertion effectively stops Nctc1 transcription progression but not initiation (Supplementary Figure S2). To determine the effect of transcriptional termination on enhancer function, we measured activation of the lacZ reporter in transiently and in stably transfected cells (Figure 6, right panel). Enhancer activity in transient transfection is unaffected by the insertion indicating that the inserted sequences do not directly interfere with CME function. However, in stably transfected cell lines where the Nctc1 promoter region is necessary, enhancer activity is reduced >20-fold. The effect of the insertion appears dependent on its terminator activity, as a non-terminating insertion of equal size does not block enhancer activity (Figure 6, construct II-NoStop). Together, these results suggest that the promoter DNA sequence alone is not sufficient for full enhancer function, but that active transcription of the Nctc1 gene is necessary either because the process of sense transcription through the core enhancer is essential or because the Nctc1 RNA has a role in cis in activating the core enhancer.
One hypothesis is that transcription through the CME is required to establish an enhancer-like chromatin structure. However, the DNA–protein structures associated with the core enhancer were not altered when Nctc1 transcription was blocked. That is, H3K4 monomethylation and accumulation of activated RNAP at the CME were equivalent in chromatin isolated from ΔME/ΔME cells stably transfected with either wild-type construct II or with plasmids where Nctc1 transcription was blocked by promoter mutation (Supplementary Figure S3).
DISCUSSION
Igf2 and H19 are linked co-regulated genes whose RNAs are highly abundant during foetal and neonatal development. Expression of these genes is dependent on a series of downstream tissue-specific enhancers spread over a >140 kb region (17). Expression in muscle cells is particularly high and is dependent on a shared enhancer defined in vivo by a 20 kb deletion mutation (ΔME). In ΔME/ΔME muscle cells, expression of Igf2 and H19 is reduced >4000-fold to undetectable levels. Within the large region defined by the ΔME deletion, Alzanhov and colleagues had already identified a small region (CME) capable of muscle cell-specific enhancer activity as measured by reporter gene activation in transient transfection assay (19). This element binds muscle-specific transcription factors and acts as a reservoir to accumulate RNAP required for activation of the distal Igf2 and H19 promoters (18). The chromatin associated with the CME is typical of classic enhancers.
Besides carrying the CME, the DNA sequences deleted by the ΔME deletion also completely coincide with Nctc1, an lncRNA. No biochemical function for Nctc1 RNA has yet been established, and ΔME/ΔME deletion mice do not display any phenotypes that cannot be explained by the loss of H19 and Igf2 expression in mutant muscle tissue (17). Here, we tested the possibility that Nctc1 might play a role in enhancer function. Our results are clear that although the CME acts as a strong enhancer in transient assay, it is not sufficient to drive gene expression in stably transfected cells where reporter constructs have integrated into the genome and organized into chromatin. Instead, the Nctc1 promoter region is also required for strong enhancer function and enhancer activity correlates well with Nctc1 transcription.
To understand why the Nctc1 promoter is so important, we performed several genetic analyses using the ΔME mutation and a molecular knockdown of Nctc1 RNA by siRNA. These results all indicate that Nctc1 RNA itself does not play a role in trans in mediating enhancer function. Instead, transcription of Nctc1 across the region is needed in cis. Thus, a critical feature of the Igf2/H19 enhancer is that it is an active transcriptional complex. The synthesis of Nctc1 RNA is not a side effect or a by-product of enhancer activity but instead is fundamentally important to enhancer function. Nctc1 promoter sequences, like CME sequences, are well conserved among mammals, whereas Nctc1 RNA-coding sequences show essentially no conservation (20). This DNA conservation pattern, along with the lack of phenotype on siRNA knockdown, supports the idea that the act of transcription, not the Nctc1 RNA molecule, is the critical product of the Nctc1 promoter.
Although our analyses altogether clearly demonstrate a need for transcription of the sense Nctc1 RNA, they are less definitive in regard to a potential role for the antisense Nctc1 transcription. From our complementation analyses (Figure 4), we know that antisense RNA supplied in trans cannot overcome the need for the Nctc1 bidirectional promoter. However, because we were unable to find siRNAs that effectively abrogate antisense Nctc1 RNA, possibly because antisense RNA is only nuclear and transient anyway, we cannot rule out the possibility that CME function requires antisense RNA in addition to sense Nctc1 transcription in cis.
Several recent genomic analyses have established that both RNA Polymerase binding and RNA transcription (of eRNAs and also of lncRNAs) are each commonly associated with enhancers (7–12,29,30). The ncRNAs in particular are associated with cell-type-specific enhancers (12) and levels of ncRNA synthesis correlate with enhancer function as measured by the likelihood of DNA loop formation or by the levels of expression of nearby promoters (8,30,31). However, with limited exceptions (see later in the text), a functional role for these RNAs is not well established, and therefore, it has remained a reasonable hypothesis that RNA transcription is only an inconsequential side effect of the accumulation of RNA Polymerase enzyme that occurs on functioning enhancers. However, the experiments in this study provide strong genetic and molecular evidence that RNA transcription at the Igf2/H19 muscle enhancer is not a by-product of enhancer activity. Rather, lncRNA transcription is necessary for full enhancer function. Because the requirement for the Nctc1 promoter and RNA transcription was uncovered only in stable and not in transient transfection assays, we surmise that chromatin structures are implicated in Nctc1 transcription/enhancer interactions. As previously speculated, RNA Polymerase progression may be necessary to keep the core enhancer in the appropriate chromatin configuration (30,32). However, we noted here that Nctc1 transcription was not required to establish the H3K4 monomethylation nor the accumulation of Ser-5-Phophorylated RNA Polymerase that are associated with enhancer activity. These results are consistent with Hah et al. who showed recently that RNA Polymerase accumulation and enhancer-like chromatin marks already established at estrogen receptor-binding sites are not dependent on continued transcription of the enhancer-associated eRNAs (33). Thus, we favour alternative proposals that mRNA synthesis may necessarily recruit protein complexes that facilitate enhancer activity in a chromatin context (32,34–37). In this regard, Nctc1 (like many other enhancer-associated lncRNAs) is a processed RNA. Thus, for example, one result of Nctc1 transcription is a recruitment of splicing complexes to the enhancer region.
Based on their activity in transient transfection, enhancers are classically defined as orientation independent. However, in their normal chromosomal locations, enhancers are restricted in their promoter targets and typically show directional bias (31). For example, the original analysis of the ΔME mutant mice showed that the loss of enhancer phenotype was unidirectional. That is, although transcription of the upstream H19 and Igf2 genes was lost on the ΔME mutant chromosome, expression of even adjacent downstream genes, such as Mrpl23 and skeletal muscle troponin-T (Tnnt3), was unaffected (17). Certainly, several mechanisms might explain this specificity including enhancer-promoter specificity or the presence of tissue-specific boundary elements at the distal end of Nctc1. However, the requirement for the Nctc1 promoter activity in cis adds a directional aspect to the muscle enhancer that could contribute to its directionality and therefore its specificity in vivo. Future genetic studies will clarify this issue.
This study establishes that maximal enhancer performance by the CME depends on Nctc1 promoter activity. Paradoxically, Nctc1 promoter activation absolutely requires the CME (18). The interdependence of the Nctc1 promoter and the CME enhancer function is a complexity that we cannot entirely explain at this time. We note that both elements are entirely cell-type specific. Thus, it is possible that this positive feedback loop between promoter activation and full enhancer function may be a self-enforcing way to keep transcription cell-type specific and also allow Nctc1 promoter activity to function as a rheostat that regulates Igf2 and H19 levels.
Recent genome-wide studies emphasize the prevalence of tissue-specific long non-coding RNAs and have postulated their importance in regulating expression of coding RNAs. There are now well-documented cases where lncRNAs with high sequence conservation are demonstrated to interact with protein cofactors and act in trans as transcriptional co-activators. For example, in mammalian cells, steroid receptors (13), Dlx-2 (38) and heat-shock transcription factor 1 (39) all work, at least in part, through ncRNA cofactors. In Drosophila, RNA–protein complexes mediate dosage compensation by directing hyper-transcription of the single X chromosome in male cells (40). More recently, lncRNAs have been also shown to play a role in gene repression in trans. In human cells, HOTAIR RNA is expressed from the HOXC locus and acts in trans to repress expression across a 40 kb region of the HOXD cluster (41). Curiously, however, HOTAIR is not well conserved in mice, and ablation of the HoxC locus has no effect on HoxD gene expression (42).
There is also now clear experimental proof that some lncRNAs act in cis to regulate gene expression. Most prominently, in female mammals, XIST RNA is synthesized specifically from the inactive X chromosome and is required for that chromosome’s transcriptional repression (43). Long ncRNAs are also implicated in imprinting at several loci (44–47). [RNA-based mechanisms do not appear to play a role in imprinting at the Igf2/H19/Nctc1 locus (47).]
Finally, although ENCODE studies show widespread association of lncRNAs with enhancers in mammalian cells, experimental support for their function is limited so that the true importance of these RNAs remains controversial (14). However, some studies have already suggested a role for lncRNAs (or of lncRNA transcription) in gene activation in cis. Paro and colleagues used a transgene model to show that transcription through a polycomb response element prevents Polycomb group-mediated silencing (48). Most relevant to this report, Shiekhattar and colleagues focused on a subset of cell-type-specific lncRNAs and identified a several with enhancer like function (36). That is, loss of these RNAs resulted in 2-fold decreased expression of select neighbouring genes. In their Discussion, the authors speculated that many ncRNAs and their promoters would often correspond to mammalian enhancers. Similarly, based on human transcriptome analyses, Gingeras and colleagues speculated on roles for ncRNA in cell-type-specific enhancer function (12). Here, we provide strong genetic evidence that these speculations are correct. At least in this instance, the active Nctc1 promoter is a part of the Igf2/H19 muscle enhancer.
In sum, the Igf2/H19 enhancer is a transcriptional complex, and its transcriptional activity is of critical importance for its function as an enhancer. Future analyses will focus on genetic studies to understand whether there are separate roles for the Nctc1 promoter in establishing and in maintaining enhancer function and whether the promoter provides directionality and otherwise contributes to target specificity.
ACCESSION NUMBERS
ENCODE data are available through the UCSC Genome Browser (http://genome-preview.ucsc.edu/). RNAP-binding data can be found through UCSC accession numbers wgEncodeEM002117 and wgEncodeEM002118, and MyoD/myogenin binding data can be found through UCSC accession numbers wgEncodeEM00236 and wgEncodeEM002127.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD), Division of Intramural Research [1ZIAHD001804]. Funding for open access charge: Eunice Kennedy Shriver NICHD Division of Intramural Research [1ZIAHD001804].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Victoria Carter and Theresa Hernandez for animal husbandry. They thank Gerard Grosveld for guidance in establishing primary myocyte cultures.
REFERENCES
- 1.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcriptional enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Heintzman N, Hon G, Hawkins R, Kheradpour P, Stark A, Harp L, Ye Z, Lee L, Stuart R, Ching C, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman N, Stuart R, Hon G, Fu Y, Ching C, Hawkins R, Barrera L, Van Calcar S, Qu C, Ching K, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 5.Ernst J, Kheradpour P, Mikkelsen T, Shoresh N, Ward L, Espstein C, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z, Zang C, Rosenfeld J, Schones D, Barski A, Cuddapah S, Cui H, Roh T, Peng W, Zhang M, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi B, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA Pol II transcription sites overlap enhancers. PLoS Bio. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim T, Hemberg M, Gray J, Costa A, Bear D, Wu J, Harmin D, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulating enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch F, Andrau JC. Initiating RNA Polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity. Transcription. 2011;2:263–268. doi: 10.4161/trns.2.6.18747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch F, Fenouil R, Gut M, Cauchy P, Albert T, Zacarias-Cabeza J, Spicuglia S, de la Chapelle A, Heidemann M, Hintermair C, et al. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 2011;18:956–963. doi: 10.1038/nsmb.2085. [DOI] [PubMed] [Google Scholar]
- 11.Kowalczyk M, Hughes J, Garrick D, Lynch M, Sharpe J, Sloane-Stanley J, McGowan S, De Gobbi M, Hosseini M, Vernimmen D, et al. Intragenic enhancers act as alternative promoters. Mol. Cell. 2012;45:447–458. doi: 10.1016/j.molcel.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 12.Djebali S, Davis C, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanz R, McKenna N, Onate S, Albrecht U, Wong J, Tsai S, Tsai MJ, O'Malley B. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 14.Kowalczyk M, Higgs D, Gingeras T. Molecular biology: RNA discrimination. Nature. 2012;482:310–311. doi: 10.1038/482310a. [DOI] [PubMed] [Google Scholar]
- 15.Feinberg A, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 16.Weksberg R, Shen DR, Fei YL, Song QL, Squire J. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat. Genet. 1993;5:143–150. doi: 10.1038/ng1093-143. [DOI] [PubMed] [Google Scholar]
- 17.Kaffer C, Grinberg A, Pfeifer K. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 2001;21:8189–8196. doi: 10.1128/MCB.21.23.8189-8196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eun B, Sampley M, Good A, Gebert C, Pfeifer K. Promoter cross-talk via a shared enhancer explains paternally biased expression of Nctc1 at the Igf2/H19/Nctc1 imprinted locus. Nucleic Acids Res. 2013;42:817–826. doi: 10.1093/nar/gks1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alzhanov D, McInerney S, Rotwein P. Long range interactions regulate Igf2 gene transcription during skeletal muscle differenitation. J. Biol. Chem. 2010;285:38969–38977. doi: 10.1074/jbc.M110.160986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihara K, Hatano N, Furuumi H, Kato R, Iwaki T, Miura K, Jinno Y, Sasaki H. Comparative genomic sequencing identifies novel tissue-specific enhancers and sequence elements for methylation-sensitive factors implicated in in Igf2/H19 imprinting. Genome Res. 2000;10:664–671. doi: 10.1101/gr.10.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bois P, Grosveld G. FKHR (FOX01a) is required for myotube fusion of primary mouse myoblasts. EMBO J. 2003;22:1147–1157. doi: 10.1093/emboj/cdg116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoon Y, Jeong S, Rong Q, Park KY, Chung J, Pfeifer K. Analysis of the H19ICR insulator. Mol. Cell. Biol. 2007;27:3499–3510. doi: 10.1128/MCB.02170-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaffer CR, Srivastava M, Park K, Ives E, Hsieh S, Batlle J, Grinberg A, Huang SP, Pfeifer K. A transcriptional insulator at the imprinted H19/Igf2 Locus. Genes Dev. 2000;14:1908–1919. [PMC free article] [PubMed] [Google Scholar]
- 24.Yoo-Warren H, Pachnis V, Ingram RS, Tilghman SM. Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 1988;8:4707–4715. doi: 10.1128/mcb.8.11.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers R, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison R, Bernstein B, Gingeras T, Kent W, Birney E, Wold B, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao Y, Yao Z, Sarkar D, Lawrence M, Sanchez G, Parker M, MacQuarrie K, Davison J, Morgan M, Ruzzo W, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould TD, Pfeifer K. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum. Mol. Gen. 1998;7:483–487. doi: 10.1093/hmg/7.3.483. [DOI] [PubMed] [Google Scholar]
- 28.Dye M, Proudfoot N. Multiple transcript cleavage precedes polymerase release int ermination by RNA polymerase II. Cell. 2001;105:669–681. doi: 10.1016/s0092-8674(01)00372-5. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman M, Ernst J, Wilder S, Kundaje A, Harris R, Libbrecht M, Giardine B, Ellenbogen P, Bilmes J, Birney E, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2012;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang D, Garcia-Bassets I, Benner C, Wenbo L, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen M, Ohgi K, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanyal A, Lajoie B, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489:109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natoli G, Andrau JC. Noncoding transcription at enhancers; general principles and functional models. Annu. Rev. Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 33.Hah N, Murakami S, Nagari A, Danko C, Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res. 2013;23:1210–1223. doi: 10.1101/gr.152306.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai F, Orom U, Cesaroni M, Beringer M, Taatjes D, Blobel G, Shiekhattar R. Activating RNAs associate with Mediator to enhancer chromatin architecture and transcription. Nature. 2013;494:497–501. doi: 10.1038/nature11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melo C, Drost J, Wijchers P, vande Werken HD, Wit E, Oude Vrielink J, Elkan R, Melo S, Leveille N, Kalluri R, et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell. 2013;49:524–535. doi: 10.1016/j.molcel.2012.11.021. [DOI] [PubMed] [Google Scholar]
- 36.Orom U, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orom U, Shiekhattar R. Noncoding RNAs and enhancers: complications of a long-distance friendship. Trends Genet. 2011;27:433–439. doi: 10.1016/j.tig.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng J, Bi C, Clark B, Mady R, Shah P, Kohtz J. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamovsky I, Ivannikov M, Kandel E, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- 40.Gelbart M, Kuroda M. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rinn J, Kertesz M, Wang J, Squazzo S, Xu S, Brugmann S, Goodnough L, Herms J, Farnham P, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129 doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schorderet P, Duboule D. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PloS Genet. 2011;7:e1002071. doi: 10.1371/journal.pgen.1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brockdorff N. Chromosome silencing mechanisms in X-chromosome inactivation: unknown unknowns. Development. 2011;138:5057–5065. doi: 10.1242/dev.065276. [DOI] [PubMed] [Google Scholar]
- 44.Fitzpatrick G, Soloway P, Higgins M. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32:426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 45.Mancini-DiNardo D, Steele S, Levorse J, Ingram R, Tilghman S. Elongation of the Kcnq1ot transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sleutels F, Zwart R, Barlow D. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 47.Wan LB, Bartolomei M. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv. Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 48.Schmitt S, Prestel M, Paro R. Intergenic transcription through a Polycomb group response element counteracts silencing. Genes Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.