Abstract
Translational repression and deadenylation of eukaryotic mRNAs result either in the sequestration of the transcripts in a nontranslatable pool or in their degradation. Removal of the 5′ cap structure is a crucial step that commits deadenylated mRNAs to 5′-to-3′ degradation. Pat1, Edc3 and the DEAD-box protein Dhh1 are evolutionary conserved factors known to participate in both translational repression and decapping, but their interplay is currently unclear. We report the 2.8 Å resolution structure of yeast Dhh1 bound to the N-terminal domain of Pat1. The structure shows how Pat1 wraps around the C-terminal RecA domain of Dhh1, docking onto the Phe-Asp-Phe (FDF) binding site. The FDF-binding site of Dhh1 also recognizes Edc3, revealing why the binding of Pat1 and Edc3 on Dhh1 are mutually exclusive events. Using co-immunoprecipitation assays and structure-based mutants, we demonstrate that the mode of Dhh1-Pat1 recognition is conserved in humans. Pat1 and Edc3 also interfere and compete with the RNA-binding properties of Dhh1. Mapping the RNA-binding sites on Dhh1 with a crosslinking–mass spectrometry approach shows a large RNA-binding surface around the C-terminal RecA domain, including the FDF-binding pocket. The results suggest a model for how Dhh1-containing messenger ribonucleoprotein particles might be remodeled upon Pat1 and Edc3 binding.
INTRODUCTION
The fate of eukaryotic mRNAs is linked to the complement of proteins with which they associate to form messenger ribonucleoprotein particles (mRNPs) (1). The 5′ cap structure and the 3′ poly(A) tail are general hallmarks of mRNPs that are targeted, either directly or indirectly, by translation factors as well as mRNA decay factors. Translation and decay are mutually dependent and antagonistic processes. The presence of the m7G cap structure at the 5′ end, for example, is crucial for eIF4E binding and for translation initiation [reviewed in (2)]. Conversely, its removal by the decapping complex is a prerequisite for 5′–3′ degradation by the exoribonuclease Xrn1 [reviewed in (3)]. Shortening of the poly(A)-tail is also linked to the shift of mRNA from active translation to a translationally repressed state in which the transcript can either be temporarily stored or can be decapped and degraded [reviewed in (4)].
Although the exact sequence of events and interplay of the factors involved in translational repression, deadenylation and decay is currently debated, it is clear that removal of the cap structure is an irreversible step that commits the mRNA to 5′–3′ degradation [reviewed in (3)]. Studies originally in yeast have shown that decapping is catalyzed by Dcp1-Dcp2 (5,6) and is activated in vivo by a cohort of regulators, including Pat1, Edc3, Scd6, Dhh1 and the heptameric Lsm 1–7 complex (7–12). These core components of the decapping machinery are conserved from yeast to humans, suggesting the presence of common basic mechanisms (13). Additional components as well as detailed intermolecular interactions can, however, vary across species. All components of the decapping/5′–3′ decay pathway co-localize in P-bodies together with factors involved in translational repression [reviewed in (14–17)]. Two P-body components in particular, Dhh1 and Pat1 (18–24), appear to be at the intersection of translational repression and mRNA turnover [reviewed in (25,26)].
Pat1 is a conserved multidomain protein that forms a scaffold for protein–protein interactions: the N-terminal domain (NTD) binds Dhh1, the downstream region is important for P-body formation and the middle and C-terminal domains recruit a plethora of factors, including the decapping complex Dcp1–Dcp2, the Ccr4–Not complex, the Lsm1–7 complex and Xrn1 (22–24,27). The Ccr4–Not complex is a major deadenylase: it trims the mRNA 3′ end to a short oligoadenylated tail that forms the platform for the Lsm1–7 complex [reviewed in (28)]. Mechanistically, Pat1 is thus believed to link the deadenylated 3′ end with the decapping factors at the 5′ end. Indeed, Pat1 triggers deadenylation when tethered to mRNAs in human and Drosophila cells (22,24) and leads to a strong effect in decapping upon deletion in yeast (8,9). In addition to its prevalent role in mRNA degradation, in yeast Pat1 acts as a translational repressor together with Dhh1 (18).
Dhh1 (also known as RCK/p54/DDX6 in humans, Me31b in Drosophila melanogaster and CGH-1 in Caenorhabditis elegans) has been known as a decapping activator since the finding 10 years ago that its deletion in Saccharomyces cerevisiae stabilizes mRNA transcripts and inhibits decapping in vivo (10,11). However, Dhh1 does not appear to function like Pat1 and Edc3/Sdc6 by directly binding and activating the Dcp1–Dcp2 decapping complex (27,29). Evidence is instead accumulating pointing to a prevalent role of Dhh1 in translational repression in yeast (18,30) as well as in higher eukaryotes (21,31–35). Dhh1 is highly abundant in all species examined to date [yeast (36), Trypanosoma (37), Xenopus oocytes (38) and mammalian cells (39)] and is present in large excess over the expected mRNA substrates (39,40–42). Dhh1 belongs to the DEAD-box protein family of RNA-dependent ATPases, but has several unusual features. While other DEAD-box proteins bind RNA and ATP in a cooperative manner (43), Dhh1 binds RNA even in the absence of ATP (39,44). In addition, the two RecA-like domains of Dhh1 are not flexible as in most other DEAD-box proteins, but are engaged in intramolecular interactions (45). This conformational rigidity restricts the ATPase activity of Dhh1 in vitro (44,45). In vivo, the ATPase activity of yeast Dhh1 is nonetheless critical for the dissociation from P-bodies (44,46).
Dhh1 and Pat1 are emerging as crucial players in guiding the mRNPs from a translationally repressed state to a decapping state. The C-terminal RecA-like domain (RecA2) of Dhh1 is sufficient for translational repression and accumulation in P-bodies in human cells (21). Studies with the Drosophila orthologues have shown that this domain of Dhh1 binds in a mutually exclusive manner Edc3 and Scd6 (known as TraI in flies) (47), two partially redundant proteins with a similar domain organization (12). The C-terminal RecA-like domain is also required to bind Pat1 (22). In this work, we have elucidated the evolutionary conserved molecular mechanisms of the interaction between Dhh1 and Pat1, showing how Pat1 and Edc3 compete for the same surface of Dhh1 and how they impact on RNA binding. These results suggest that Dhh1 might switch protein and RNA-binding partners in the transition from translational repression to decapping.
MATERIALS AND METHODS
Protein expression and purification
S. cerevisiae Dhh130–425 and Dhh146–422 were cloned as Tobacco etc virus (TEV)-cleavable His6-GST-tag fusion proteins. They were expressed in BL21-Gold (DE3) pLysS (Stratagene) in Terrific Broth medium. Cells were resuspended in lysis buffer (20 mM Tris, pH 7.4, 200 mM NaCl) supplemented with 10 mM imidazole, DNase, lysozyme and phenylmethylsulfonyl fluoride, and lysed by sonication. Proteins (wild type and mutants) were purified using Nickel-based affinity chromatography. Point mutations were introduced by QuickChange site–directed mutagenesis according to the manufacturer’s instruction (Stratagene). The His6-GST tag was either kept or cleaved by overnight incubation with TEV. Proteins were further purified by ion exchange chromatography at pH 7.4 (Heparin, GE healthcare) followed by size-exclusion chromatography (Superdex 75, GE Healthcare).
S. cerevisiae Pat11–30, Pat11–56, Pat11–114 and Pat15–79 proteins were expressed as TEV-cleavable GST-His-tagged proteins in BL21-Gold (DE3) pLysS cells. The cells were resuspended in lysis buffer and disrupted by sonication. The proteins were purified by Nickel-based affinity chromatography and then subjected (either tagged or untagged following the addition of TEV protease) to ion exchange chromatography at pH 8.0 (MonoQ 5/50, GE healthcare) and to size exclusion chromatography (Superdex 75). For isothermal titration calorimetry (ITC), Pat1 proteins were subcloned with a His6-SUMO tag. S. cerevisiae Edc377–158 was subcloned with His6-GST tag, while Edc377–158 and Edc377–116 were subcloned as cleavable His6-SUMO proteins. All proteins were expressed and purified following similar protocols as the ones described above. For crystallization, the His6-SUMO tag was cleaved using the SUMO protease Senp2.
The complexes of yeast Dhh1 with Pat1 or Edc3 were reconstituted by incubating the individually purified proteins in a 1:1.5 molar ratio for 1 h at 4°C. Dhh1–Pat1 and Dhh1–Edc3 were further purified by size exclusion chromatography (Superdex 75) in a buffer containing 20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT.
Crystallization and structure determination
Crystallization was carried out at 18°C using the vapor diffusion method by mixing equal volumes of protein complex at 27 mg/ml and of crystallization buffer. The best diffracting crystals of Dhh146–422 K234D, V238D Pat15–79 complex were obtained in 50 mM Tris, pH 8.0, 4% MPD, 200 mM NaCl, 25% PEG 400 after 10 days. Crystals were flash-frozen in liquid nitrogen directly from the crystallization drop. Crystals of Dhh146–422 K234D, V238D Edc377–158 were obtained with 50 mM MES, pH 6.5, 5% PEG 400, 0.1 M KCl, 10 mM MgCl2. The crystals were cryoprotected by adding glycerol and diffracted to 3.5 Å resolution (data not shown). Crystals of Dhh146–422 K234D, V238D and Edc377–116 were obtained at 19 mg/ml concentration of the complex and 50 mM MES, pH 6.0, 10% MPD within 5 days. Crystals were flash-frozen by adding 25% glycerol in the crystallization buffer and diffracted to 3.25 Å resolution.
All diffraction data were collected at 100 K at the beamline PXII of the Swiss Light Source synchrotron and processed using XDS. The structures were determined by molecular replacement with the program Phaser (48) using the two RecA domains from the apo Dhh1 structure (45) as search models. The atomic model was built with the program Coot (49) and refined with PHENIX (50). The data collection and refinement statistics are summarized in Table 1.
Table 1.
Data collection and refinement statistics. The Ramachandran plot was calculated using the program Molprobity (59)
| Data set | Dhh146–422 K234D, V238D and Pat5–79 | Dhh146–422 K234D, V238D Edc377–116 |
|---|---|---|
| Space group | P4122 | P4122 |
| Cell dimensions | ||
| a, b, c (Å) | 105.6, 105.6, 122.1 | 105.8, 105.8, 124.6 |
| Data collection | ||
| Wavelength (Å) | 0.9714 | 0.9796 |
| Resolution (Å) | 48.47–2.80 (2.90–2.80) | 80.68–3.25 (3.36–3.25) |
| Rmerge (%) | 7.6 (75.8) | 9.2 (120.4) |
| I / σ(I) | 18.3 (2.8) | 23.5 (2.4) |
| Completeness (%) | 99.6 (96.17) | 99.7 (97.47) |
| Multiplicity | 6.8 | 17.3 |
| Refinement | ||
| Resolution (Å) | 53.11–2.80 | 53.70–3.25 |
| No. of unique reflections | 17 636 | 11 712 |
| Rwork / Rfree (%) | 20.42/24.71 | 20.85/24.57 |
| Average B-factors (Å2) | 48.30 | 105.60 |
| No. of atoms | ||
| Proteins | 3185 | 3115 |
| Water | 4 | |
| Stereochemistry | ||
| R.m.s.d. bond lengths (Å) | 0.003 | 0.003 |
| R.m.s.d. bond angles (°) | 0.743 | 0.790 |
| Ramachandran outliers (%) | 0.00 | 0.25 |
| Ramachandran favored (%) | 97.51 | 98.25 |
Values in parentheses are for the highest-resolution shell.
In vitro pull-down assays
Experiments were performed by mixing 3 µg of GST-tagged (bait) protein with equal molar amounts of untagged (prey) protein. Binding buffer (20 mM Hepes, pH 7.5, 250 mM NaCl, 2 mM MgAc2, 10% glycerol, 2 mM imidazole, 1 mM DTT, 0.1 % Nonidet 40) was added to a final volume of 60 µl. The reaction mixtures were incubated on ice for 1–2 h. Fifteen microliters of 50% (v/v) suspension of GSH–Sepharose beads in 200 µl of binding buffer was added to each reaction mixture and incubated at 4°C for 1 h. Beads were washed three times with 500 µl binding buffer. Bound proteins were eluted with 20 mM reduced glutathione. Samples were separated on sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie blue stain.
In vitro RNA-binding assays
The experiments were carried out essentially as previously described (52). Single-stranded 5′ biotinylated U20 RNA (Dharmacon) was mixed with 3 μg of a given protein and/or nucleotide in binding buffer (20 mM Hepes, pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 10% glycerol, 0.1% Nonidet 40) to a final volume of 60 µl and was kept at 4°C overnight. Each reaction was then supplemented with 200 µl of binding buffer and 50 µg of preblocked magnetic streptavidin beads (Dynal) for 2 h, rocking at 4°C. Beads were washed three times with 500 µl of binding buffer. Proteins were eluted from the beads with 17 µl SDS loading buffer. Eluted samples were boiled for 5 min and analyzed on SDS-PAGE.
Isothermal titration calorimetry
Dhh130–425 and His6-SUMO tagged Pat15–56, Pat15–79 or Pat15–114 proteins were dialyzed overnight in the same buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM TCEP). ITC experiments were carried out at 25°C with a VP-ITC Microcal calorimeter (Microcal, GE healthcare). The MicroCal cell was filled with Dhh130–425 at 17 μM concentration. For each titration, Pat1 was injected into the cell 45 times in 10 µl volumes per injection at the same intervals of time (5 min). The concentration of Pat1 in the syringe was ∼10 times the concentration of the protein sample in the cell. The released heat was obtained by integrating the calorimetric output curves and was corrected for the effect of dilution by carrying out a control experiment, titrating Pat1 against the buffer in the cell. The Kd values and binding ratios were calculated with the Origin (V7) software supplied with the calorimeter. We used a similar protocol to measure the Kd of Dhh130–425 and Edc377–158.
Co-immunoprecipitation assays
HEK293T cells were cultured in Dulbecco’s modified Eagle medium containing 10% fetal bovine serum (Gibco), 100 U/ml penicillin and 0.1 mg/ml streptomycin (Gibco) at 37°C/5% CO2. Plasmids were transfected with polyethyleneimine (Polysciences Inc., 1 mg/ml) for protein interaction studies. HEK293T cells were collected from confluent 6-cm dish after 24 h of transient transfection. Cells were lysed in 300 µl of lysis buffer containing 10 mM Tris, pH 7.5, 10 mM NaCl, 10 mM EDTA, 0.5% Triton X-100 and protease inhibitors (Roche). Lysate was centrifuged at 1000g for 5 min at 4°C. Thirty microliters of GFP binder protein sepharose beads were added to supernatant for 1 h at 4°C. Beads were washed five times with 1 ml of NET2 buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.05% Triton X-100) and proteins were eluted with 40 µl SDS sample buffer containing 5% β-mercaptoethanol. Eluted proteins were run on 12% polyacrylamide gels and transferred onto nitrocellulose membrane (0.45 µm pore size) (Whatman Protran BA85) for western blotting. Mouse monoclonal anti-GFP (Santa Cruz biotechnology, sc-9996) and anti-HA (Covance, MMS-101 R) antibodies and horseradish peroxidase–coupled goat anti-mouse (Millipore, AQ502A) secondary antibody were used in combination with ECL prime western blotting detection reagent (GE healthcare) for detection of HA- and GFP-tagged proteins via western blotting.
Crosslinking–mass spectrometry analysis
The protein–RNA contacts site on Dhh1 were investigated with mass spectrometry after UV-induced protein–RNA crosslinking as described in (53). The purified crosslinks were analyzed using Top10HCD method on an Orbitrap Velos instrument and the data were analyzed using OpenMS and OMSSA as a search engine (see Supplementary Methods).
RESULTS AND DISCUSSION
Biochemical and biophysical characterization of the yeast Dhh1–Pat1 interaction
S. cerevisiae Pat1 is an 88 kDa protein containing an NTD followed by a proline-rich region, a middle domain and a C-terminal domain (20,22) (Figure 1A). Dhh1 contains two RecA-like domains (RecA1 and RecA2) flanked by N-terminal and C-terminal low-complexity regions (45) (Figure 1A). The NTD of Pat1 is conserved from yeast to humans and has been shown in the case of the D. melanogaster and human proteins to mediate the interaction with the corresponding Dhh1 orthologues using co-immunoprecipitation assays (22,23). This domain is also part of a larger region found to strongly bind Dhh1 in the case of the yeast proteins (27).
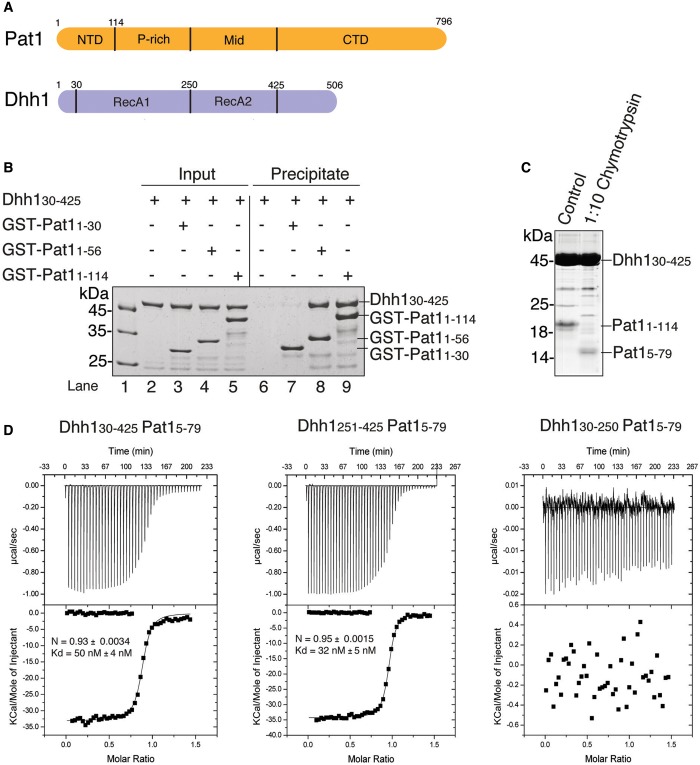
Figure 1.
Identification of the interacting regions of yeast Dhh1 and Pat1. (A) Schematic representation of the domain architecture of yeast Pat1 (in orange) and Dhh1 (in blue). The domain boundaries are derived from previous studies (45, 27, 22, 23) and from computational analysis. The NTD of Pat1 refers to the segment upstream of the proline-rich (P-rich) segment. (B) Qualitative analysis of Dhh1-Pat1 interaction by GST pull-down assays. Dhh130–425 was incubated with purified GST-tagged fragments of the Pat1 NTD (Pat11–114, Pat11–56, Pat11–30). One-sixth of the reaction mixture was used as input and the bound fractions (precipitate) were analyzed on 4–20% SDS-NuPAGE Bis-Tris gels (Invitrogen). The longest fragment of Pat1 NTD (Pat11–114) as well as Pat11–56 were precipitated Dhh130–425, while the shorter fragment Pat11–30 was unable to do so. (C) Limited proteolysis experiment. The complex of Dhh130–425 and Pat11–114 was incubated at 0.6 mg/ml with chymotrypsin (Roche) in a 1:10 (wt/wt) enzyme:protein ratio for 30 min at 4°C. The products of the proteolysis were resolved on 17% SDS-PAGE gel. (D) Quantitative analysis of Dhh1–Pat1 interaction by ITC. The MicroCal cell was filled with Dhh1 proteins at 17 μM and His6-SUMO-tagged Pat15–79 was injected at 150 μM concentration consecutively in 10 μl volumes. The three panels correspond to the ITC experiments with Dhh130–425 (RecA1-RecA2, left), with Dhh1251–425 (RecA2, central) and with Dhh130–250 (RecA1, right). Shown in the insets are the number of calculated binding sites (N), and the dissociation constant (Kd), as calculated with the program Origin.
To investigate the S. cerevisiae Dhh1–Pat1 complex, we first narrowed down the interacting domains using pull-down assays with recombinant Dhh1 and GST-tagged Pat1 proteins. The NTD of yeast Pat1 (residues 1–114) efficiently precipitated the DEAD-box core of Dhh1 (residues 30–425, including RecA1 and RecA2) (Figure 1B, lane 9). A segment of the Pat1 NTD that we engineered based on evolutionary conservation (residues 1–56) was also able to precipitate Dhh130–425 (Figure 1B, lane 8), while no binding was observed when using a fragment of Pat1 encompassing residues 1–30 (Figure 1B, lane 7). To experimentally identify a Pat1 segment suitable for structural studies, we purified the complex of Dhh130–425 and Pat11–114 and subjected it to limited proteolysis. Treatment with the protease chymotrypsin resulted in the accumulation of a fragment that we characterized using mass spectrometry analysis and N-terminal sequencing as corresponding to Pat1 residues 5–79 (Figure 1C).
Next, we assessed binding affinities quantitatively using ITC. In the presence of 150 mM salt, yeast Dhh130–425 and Pat15–79 interacted with a Kd of ∼50 nM (Figure 1D, left panel). A similar dissociation constant was obtained when using Pat15–114 or Pat15–56 (Supplementary Figure S1), indicating that most of the binding determinants reside in the evolutionary conserved segment of the Pat1 NTD. Pat15–79 and the RecA2 domain of Dhh1 interacted with a Kd of 32 nM (Figure 1D, central panel), while no binding was detected with the RecA1 domain (Figure 1D, right panel). We concluded that the Dhh1–Pat1 interaction is confined to the RecA2 domain of the DEAD-box protein.
Crystal structure determination of a yeast Dhh1–Pat1 complex
Attempts to crystallize yeast Pat15–79 or Pat15–56 with the RecA2 domain of Dhh1 failed, as the complexes appeared to be too soluble to achieve the supersaturated conditions required for nucleation. Complexes of Pat1 with Dhh130–425 yielded the same crystal form that was previously reported for apo-Dhh1 (45), and indeed contained only Dhh1. Inspection of this crystal form revealed that the RecA2 domain of a Dhh1 molecule contacts the RecA1 domain of a neighboring Dhh1 molecule in the lattice (Supplementary Figure S2A). The surface of RecA2 mediating crystal contacts is conserved and has been shown in the case of the human orthologue DDX6 to bind the Phe-Asp-Phe (FDF) motif of Edc3 (35).
We reasoned that lattice contacts might promote the dissociation of Pat1 from RecA2 during crystallization. We therefore mutated the corresponding interacting surface of the RecA1 domain (Supplementary Figure S2), which did not contribute to Pat1 binding as assessed by quantitative binding affinity measurements (Figure 1D). For crystallization purposes, we introduced the Lys234Asp, Val238Asp substitutions in Dhh146–422, a construct that includes only the ordered polypeptide region observed in the structure of apo Dhh130–425 (45). Dhh146–422 K234D, V238D and Pat15–79 crystallized as a complex and yielded crystals that diffracted to 2.8 Å resolution. The structure was determined by molecular replacement and refined to an Rfree of 24.7%, Rwork of 20.6% and good stereochemistry (Table 1). The final model consists of residues 46–421 of Dhh1 and residues 25–54 of Pat1 (hereafter referred to as Pat1N). No ordered electron density was present for residues 5–24 and 55–79 of Pat1, consistent with the results from the binding assays (Figure 1B, D and Supplementary Figure S1).
Overall structure of the yeast Dhh1–Pat1 core complex
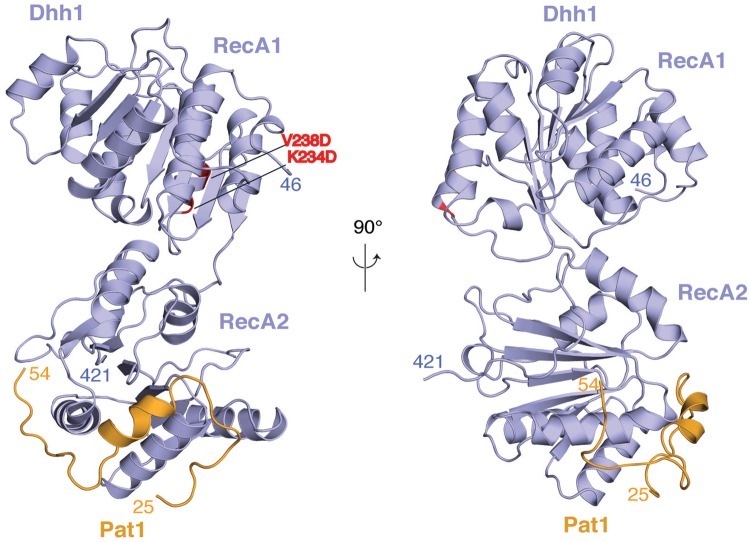
In the complex we crystallized, Dhh1 adopts an open conformation (Figure 2). The relative orientation of the two RecA domains is different from that observed in the structure of wild-type apo Dhh1 (45) (Supplementary Figure S2B). The difference probably arises from the mutations we had introduced: an intramolecular interaction between the two RecA domains observed in the structure of the wild-type protein (a salt bridge between Lys234 and Glu251) is impaired by the K234D mutation (Supplementary Figure S2B). In either conformation, the residues mutated on RecA1 are at >40 Å distance from the residues that bind Pat1N (Figure 2 and Supplementary Figure S2B). Pat1N wraps around the RecA2 domain of Dhh1, burying ∼10% (9118 Å2) of the solvent accessible surface. Pat1N folds into an α-helical segment (residues 25–42) and an extended segment (residues 43–54) (Figure 2).
Figure 2.
Structure of the yeast Dhh1–Pat1 core complex. The two RecA domains of Dhh1 are in blue. The Dhh1-binding domain of Pat1 spans residues 24–54 (in orange). The structure is shown in two views related by a 90° rotation around a vertical axis. The N- and C-terminal residues ordered in the structure are indicated. The two residues of RecA1 mutated for crystallization (K234D and V238D, shown in red) are far from the RecA2 domain, where Pat1 binds.
An extended segment of yeast Pat1 docks onto the FDF-binding site of Dhh1
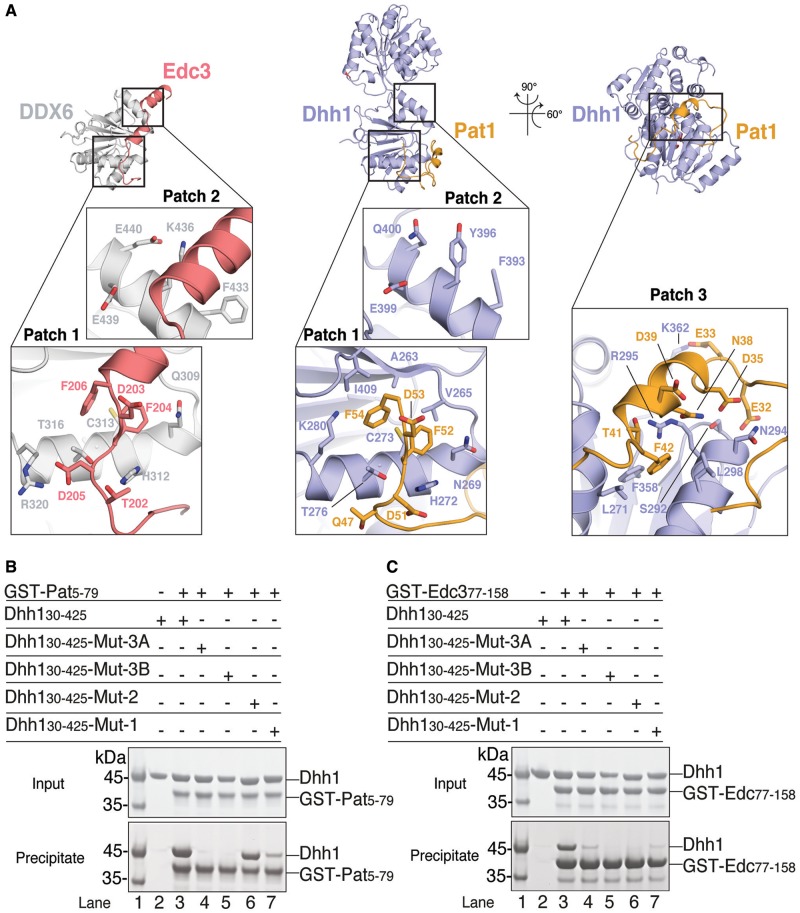
The extended segment of yeast Pat1N contains a FDF sequence motif (residues 51–53). The FDF motif binds at a shallow surface pocket on the RecA2 domain of yeast Dhh1 (that we refer to as the patch 1 surface) formed by Ala263, Val265, Cys273, Leu277 and Ile409 (Figure 3A, central panel). The motif is recognized at the same pocket and with the same conformation as the DDX6-bound FDF motif of human Edc3 (patch 1, Figure 3A, left panel) (35). Mutation of four residues surrounding the FDF-binding site in D. melanogaster Me31b has been previously shown by co-immunoprecipitation studies to abolish Edc3 binding (35). We mutated the equivalent residues at the patch 1 surface of yeast Dhh1 (Mut-1: Asn269Ala, His272Ala, Thr276Ala, Lys280Ala). In pull-down assays with purified GST-tagged Pat15–79 and Dhh146–422 proteins, the binding of Pat1 to Mut-1 was decreased as compared with wild-type Dhh1 (Figure 3B, compare lanes 3 and 7).
Figure 3.
Hotspots of interactions on Dhh1. (A) The structure of Dhh1–Pat1 (central and right panel) is shown in comparison with the previously determined structure of the RecA2 domain of human orthologue DDX6 (in gray) bound to another regulator, Edc3 (in salmon) (35) (left panel). The structures in the central and left panels are in the same orientation relative to the RecA2 domain. The orientation of the Dhh1–Pat1 structure in the right panel is related by rotations around a vertical and horizontal axis, as indicated. The close-up views show three hotspots of interactions on Dhh1 discussed in the text (patch 1, patch 2 and patch 3). Conserved residues involved in the interactions are shown in a ball-and-stick representation (at patch 1 and patch 3 for Dhh1–Pat1, and at patch 1 and patch 2 for DDX6–Edc3). Patch 1 binds FDF motifs of yeast Pat1 and human Edc3. Patch 2 binds the FDK motif of human Edc3 (35). Patch 3 binds an acidic segment of yeast Pat1. (B) Protein co-precipitations by GST pull-down assays. The experiments were performed as described in Figure 1B after incubating yeast GST-tagged Pat15–79 with Dhh130–425 wild type or mutants. Mut-1 corresponds to the N269A, H272A, T276A, K280A substitutions at Dhh1 patch1. Mut-2 corresponds to the F393A, Y396A, E399A, Q400A at patch 2. Mut-3A corresponds to S292D, N294D and Mut3-B to R295D at patch 3. The Mut-1, Mut-3A and Mut-3B mutants of yeast Dhh1 decreased the interaction with Pat1 while Mut-2 did not, consistently with the structural analysis (Figure 3A, central and right panels). (C) Similar pull-down experiments were performed with yeast GST-tagged Edc377–158 and Dhh130–425 wild type or mutants. The Mut-1, Mut-2, Mut-3A and Mut-3B mutants of yeast Dhh1 all decreased the interaction with Edc3.
In the case of human Edc3, the FDF motif is followed by two helices that contain a conserved Phe-Asp-Lys FDK motif and bind at a second surface patch (35) (patch 2, Figure 3A, left panel). In the structure of yeast Pat1N, there is no ordered extension C-terminal to the FDF motif (Figure 3A, central panel), although this part of the molecule is in principle present in the construct we crystallized. We engineered a yeast Dhh1 mutant with substitutions at patch 2 (Mut-2: Phe393Ala, Tyr396Ala, Glu399Ala, Gln400Ala) corresponding to the Me31b mutant shown to abolish Edc3 binding in the case of the D. melanogaster proteins (35). Consistently with the structure, yeast Pat15–79 was able to precipitate Mut-2 Dhh1 (Figure 3B, lane 6). As a control, we tested the Dhh1 mutants with yeast Edc377–158. This ∼80-residue long segment of yeast Edc3 includes the equivalent ∼30-residue long segment of human Edc3 that is ordered in the structure with DDX6 (35) (Figure 4A). The Mut-1 and Mut-2 Dhh1 proteins failed to interact with yeast Edc377–158 (Figure 3C, compare lane 3 with lanes 7 and 6). These results indicated that yeast Pat1 and Edc3 compete for the same FDF-binding site on Dhh1 (patch 1). Yeast Pat1N, however, lacks the additional Dhh1-binding segment at the C-terminus of the FDF motif (patch 2) that is present in human and Drosophila Edc3 (while absent in TraI) (35).
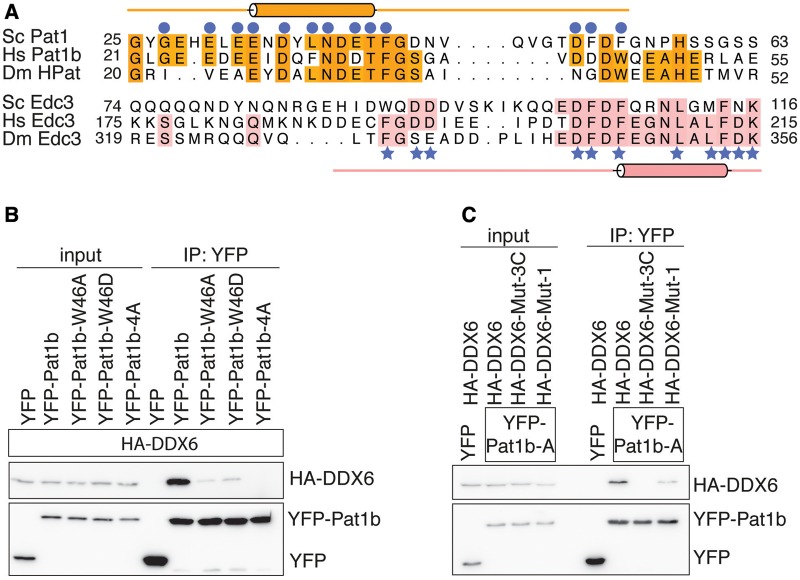
Figure 4.
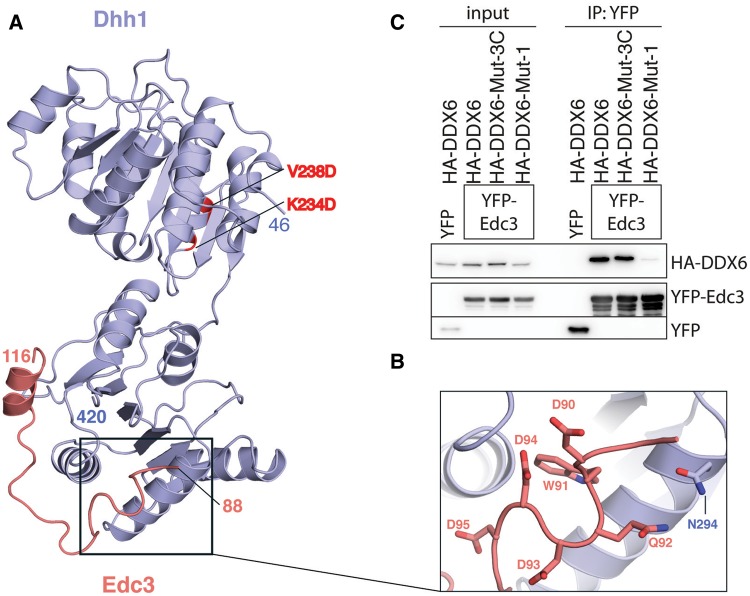
Pat1 binds Dhh1 via evolutionary conserved residues. (A) Structure-based sequence alignment of Pat1 and Edc3 orthologues from S. cerevisiae (Sc), Homo sapiens (Hs), D. melanogaster (Dm). Conserved residues are highlighted in orange for Pat1 and in salmon for Edc3. The secondary structures (helices indicated as cylinders) are shown above and below the Pat1 and Edc3 sequences and correspond to the structures of the yeast complexes reported in this manuscript. Conserved residues of yeast Pat1 and Edc3 that directly interact with Dhh1 are indicated with blue circles and stars, respectively. (B) Co-immunoprecipitation assays of H. sapiens HA-tagged DDX6 with YFP-labeled Pat1b (residues 1–84). Wild type and mutants (W46A, W46D and 4A) were transiently transfected in HEK293T cells. Trp46 of human Pat1b is predicted to reside at the corresponding FDF motif of yeast Pat1 (panel A). The human 4A mutant corresponds to the D34, D35, T36 and F37 substitutions (53). Cell lysates (input) were immunoprecipitated with GFP binder. HA-tagged and YFP-tagged proteins were detected by western blots. The Pat1b mutants (W46A, W46D and 4A) impaired the interaction with wild-type DDX6. (C) Immunoprecipitation (IP) of wild-type YFP-tagged Pat1b (residues 1–84) with HA-tagged DDX6 wild type, Mut-1 and Mut-3C. Mut-3C corresponds to the combined Mut-3A and Mut-3B substitutions (S343D, Q345D, R346D in human DDX6). The IP was done as described above. The DDX6 mutants (Mut-1 and Mut-3C) impaired the interaction with wild-type Pat1b.
The interaction at the FDF-binding site is conserved in the human Pat1–DDX6 complex
Human and Drosophila Pat1 proteins do not contain a FDF sequence motif. Structure-based sequence alignments, however, suggested the presence of an Asp-Trp (DW) motif in metazoan Pat1 proteins at the corresponding position of the yeast Pat1 FDF motif (Figure 4A). We therefore tested whether the hydrophobic residue in the DW motif of human Pat1b might dock on the FDF-binding pocket of DDX6. We transiently co-expressed full-length HA-tagged DDX6 and YFP-tagged Pat1b residues 1–84 (hereby referred to as YFP-Pat1b) in HEK293T cells and carried out co-immunoprecipitation assays with GFP-binder beads. We found that a Trp46Ala or a Trp46Asp substitution in YFP-Pat1b strongly decreased the interaction with full-length HA-tagged DDX6 (Figure 4B). The effect was similar to a Pat1-4A mutant that has been recently shown to impair Dhh1 binding (54). In this assay, mutation of the FDF-binding site in human HA-tagged DDX6 (Mut-1) significantly decreased the interaction with YFP-Pat1b (Figure 4C). We concluded that the interaction of Pat1 at the FDF-binding site of Dhh1 is evolutionary conserved in yeast and humans. Drosophila HPat is also expected to dock at the FDF-binding site of Me31b (Figure 4A), but it is possible that in this case the FDF-binding site contributes less to the overall interaction, explaining previous co-immunoprecipitation data (35).
An acidic helical segment of Pat1 docks onto a positively charged surface of Dhh1
N-terminal to the FDF motif, Pat1N features a helical segment that binds with extensive interactions at a third surface patch on Dhh1 (patch 3, Figure 3A, right panel). The helical segment of Pat1 is highly negatively charged (Figure 4A) and binds to a positively charged surface of Dhh1. Pat1 Asn34, Asn38, Thr41 and Asp45 make polar contacts with Dhh1 Ser292, Asn294 and Arg295 (Figure 3A, right panel). In addition, Pat1 Glu33 forms a salt bridge with Dhh1 Lys362 and Pat1 Phe42 makes hydrophobic contacts with Dhh1 Leu298, Phe358 and Arg295. The human Pat1b-4A mutant shown to disrupt the interaction with DDX6 (54) (Figure 4B) maps to the helical segment of Pat1 (alanine substitutions at Asp34, Asp35, Thr36, Phe37, Figure 4A). We engineered mutations in Dhh1 to disrupt patch 3. Substitutions of Ser292Asp, Asn294Asp (Mut-3A) or of Arg295Asp (Mut-3B) abolished binding to Pat15–79 in GST pull-down assays (Figure 3B, lanes 4 and 5). The interacting residues are conserved in the human orthologues. Indeed a combined mutation of the Mut-3A and Mut-3B substitutions in human DDX6 (Ser343Asp, Gln345Asp, Arg346Asp or Mut-3C) impaired the interaction with human Pat1 in co-immunoprecipitation assays (Figure 4C).
Comparison of the structure of Dhh1–Pat1 with that of the DEAD-box protein eIF4AIII in the exon junction complex (EJC) (55,56) reveals that the patch 3 surface is used in both DEAD-box proteins to recruit binding partners (Supplementary Figure S3). The EJC is an assembly formed by four proteins (eIF4AIII, Btz, Mago and Y14), RNA and ATP (57). In the EJC, eIF4AIII binds RNA with the two RecA domains in the typical closed conformation observed in all known structures of DEAD-box proteins in the active (RNA-ATP-bound) state (55,56). Btz wraps around eIF4AIII and contributes to RNA binding. The N-terminal segment of Btz contacts RNA directly (via Phe188). In addition, Btz features several positively charged residues (Arg176, Arg184 and Arg185) that form a favorable electrostatic environment for an incoming nucleic acid (Supplementary Figure S3). The helical segment of Pat1N also approaches the RNA-binding site expected for the ATP-bound conformation, but features several surface-exposed negatively charged residues (Glu28, Glu30, Glu32, Asp35, Asp39 and Glu40) that are predicted to electrostatically disfavor binding of Dhh1 to nucleic acids (Supplementary Figure S3).
Yeast Edc3 contains an acidic segment similar to Pat1
Originally as a control, we tested the Dhh1 patch 3 mutants with yeast Edc377–158 (Figure 3C). In the human DDX6-Edc3 structure (35), there is no ordered segment N-terminal to the FDF motif of Edc3 and therefore there is no binding to the corresponding patch 3 surface of DDX6. In pull-down assays, however, yeast Edc377–158 failed to interact with the Mut-3A and Mut-3B Dhh1 proteins (Figure 3C, lanes 4 and 5). As we could not rationalize these biochemical results based on the available structural information, we set out to determine the crystal structure of yeast Dhh1-Edc3. Yeast Dhh130–425 interacted with Edc377–158 with a Kd of 200 nM (Supplementary Figure S1B). The Edc377–158 construct is considerably longer than the boundaries (residues 98–127) expected from the structure of human Edc3 (35). Initial crystals of the complex of yeast Dhh146–422 K234D, V238D and Edc377–158 diffracted to 3.5 Å resolution. The electron density map was of sufficient quality to show binding of yeast Edc3 to patch 3 and to narrow down the domain boundaries (i.e. no ordered electron density beyond residue 116 of Edc3) (data not shown). We proceeded to crystallize yeast Dhh146–422 K234D, V238D and Edc377–116. The structure was determined at 3.25 Å resolution with an Rfree of 24.6% and Rfactor of 21.0%. The final model includes residues 46–420 of Dhh1 and residues 88–116 of yeast Edc3 (Figure 5A).
Figure 5.
The interaction of Dhh1 and Edc3. (A) The structure of the yeast Dhh1–Edc3 core complex is shown in a similar orientation as in Figure 3A. The two RecA domains of yeast Dhh1 are in blue and Edc3 residues 88–116 are in salmon. The two residues on RecA1 mutated for crystallization purposes are highlighted in red. (B) Close-up view of the negatively charged residues of yeast Edc3 lining the patch 3 surface of Dhh1. (C) Co-immunoprecipitation assays of human YFP-tagged Edc3 and HA-tagged DDX6 wild type, Mut-1 or Mut-3C were carried out as described in Figure 4C. With the human proteins, mutations at patch 3 (Mut-3C) do not abrogate binding.
Comparison of the yeast Dhh1-Edc3 structure with that of human DDX6-Edc3 shows a similar binding of the FDF motif at the patch 1 surface. There is no equivalent binding of a C-terminal helix to patch 2 (Figure 5A). However, this is likely an artifact of crystallization, due to a crystal contact that blocks Edc3 from accessing patch 2 of Dhh1. Indeed, binding of Dhh1 to yeast Edc377–158 was abolished by the patch 2 mutations (Figure 3C, lane 6). More importantly, yeast Edc3 features an ordered segment (residues 88–104) N-terminal to the FDF motif (Figure 5A). In this segment, Trp91 of yeast Edc3 binds in the same pocket of patch 3 as Phe42 of yeast Pat1, explaining the results of the pull-down assays with the patch 3 mutants (Figure 3C). Also similarly to yeast Pat1, this segment of Edc3 is rich in negatively charged residues (Figure 5B). While the N-terminal segment of Pat1N is evolutionary conserved, the N-terminal segment of yeast Edc3 is apparently not present in the human and Drosophila orthologues. Indeed, the Mut-3C substitutions in human DDX6 did not impair the interaction with human Edc3 in co-immunoprecipitation assays (Figure 5C). We concluded that these differences in the structures of the orthologous complexes reflect genuine differences in the interactions.
Pat1 and Edc3 disrupt RNA binding of Dhh1
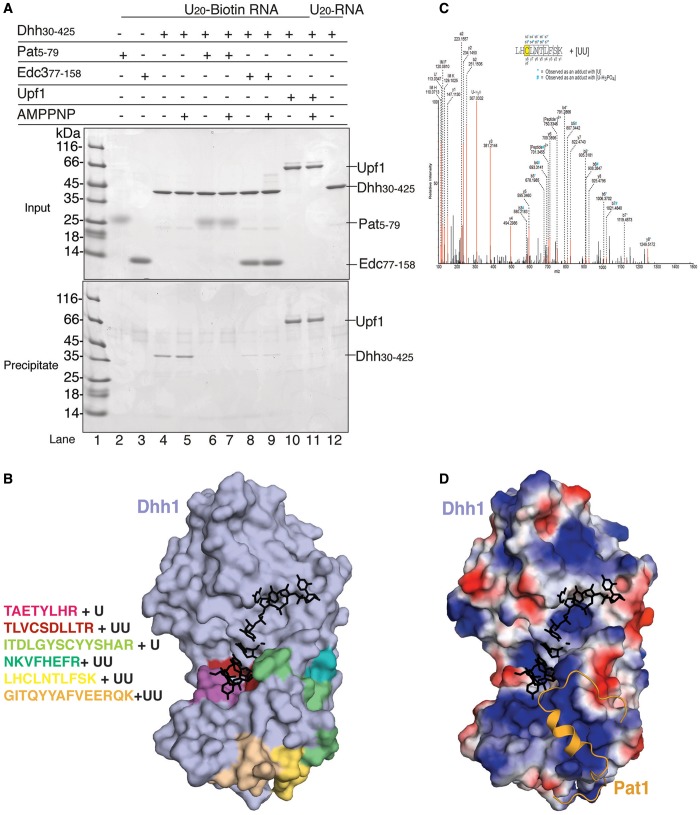
We tested the effect of yeast Pat1 and Edc3 on the RNA-binding properties of Dhh1 using a biotin-RNA pull-down assay. In agreement with previous findings, Dhh130–425 was precipitated by biotinylated RNA immobilized on streptavidin beads even in the absence of nucleotide analogs (Figure 6A, lanes 4 and 5) (44). On measuring the affinity of Dhh130–425 for a U15 RNA by fluorescence anisotropy, we found a Kd in the low micromolar range (Supplementary Figure S4A), lower than that previously reported for a full-length Dhh1 fusion protein (44), and lower than that measured for Pat1 and Edc3 using ITC (Figure 1D and Supplementary Figure S1). In the presence of either Pat15–79, or Edc377–158, Dhh130–425 failed to bind to RNA in the pull-down assay (Figure 6A, lanes 6,7 and lanes 8,9, respectively), suggesting that they interfere with RNA binding.
Figure 6.
Pat1 and Edc3 compete with RNA for binding to Dhh1. (A) Co-precipitation of 5′ biotinylated single-stranded poly-U20 RNA with yeast Dhh1 alone or in complex with Pat15–79 or Edc377–158. The pull-down experiments were performed with streptavidin beads [essentially as described in (51)] and analyzed on 4–20% NuPAGE Bis-Tris gels (Invitrogen) using 20% of the total as input and bound samples. Dhh1 bound to RNA in presence or absence of nucleotides, as previously reported (44,39). In the presence of either Pat15–79 or Edc377–158, Dhh1 binding to RNA was impaired. (B) On the left are the peptides of RecA2 identified by mass spectrometry analysis to be crosslinked to RNA (Supplementary Figure S5). On the right, surface representation of apo Dhh1 (45) (in light blue) with the RNA-crosslinked peptides shown in the same colors as on the right. Shown in black is the RNA modeled after superposition with eIF4AIII-AMPNP-RNA in EJC structure upon superposition of the RecA2 domains (54,55). Notice that, in this conformation of Dhh1, an RNA bound in the typical ATP-dependent conformation would sterically clash against the surface of Dhh1. (C) MS/MS mass spectrum of Dhh1 residues 271–280 identifies an additional mass that corresponds to two uracil nucleotides associated to a cysteine. Peptide sequence and the fragment ions are indicated on the top. The peptide fragmentation occurs with the cleavage of amide bonds resulting in b-ions and y-ions when the charge is retained by the amino-terminal and carboxy-terminal fragments, respectively. Asterisk (*) and hash (#) indicate the b-ions that were observed with a mass shift corresponding to U and U-H3PO4, respectively. IM: Immonium ions. U’: U marker ion of 113.03 Da. (D) Surface representation of apo Dhh1 (45) in the same orientation as in panel B, colored by electrostatic potential, with positively charged residues in blue and negatively charged residues in red. The binding paths of RNA (in the ATP-dependent state) and of Pat1 are shown modeled in black and in orange.
Intrigued by the observation that there was no significant difference in the RNA pull-down assays in the presence or absence of ATP, we analyzed the nucleotide-binding properties of Dhh1. First, inspection of the crystal structure of apo Dhh1 (45) indicated that its closed conformation is incompatible with ATP binding (Supplementary Figure S4B). Dhh130–425 indeed did not bind ATP in ITC experiments (Supplementary Figure S4C) and appeared to be essentially inactive in RNA-dependent ATPase assays (Supplementary Figure S4D), suggesting that Dhh1 is in an enzymatically inactive conformation.
To map how this unusual DEAD-box protein can bind RNA in the absence of ATP, we crosslinked Dhh130–425 to a U15 RNA by subjecting the complex to UV irradiation at 254 nm. We then used LC-MS/MS mass spectrometry to detect and sequence peptides conjugated to the mass of an RNA nucleotide [reviewed in (53)]. Using this approach, we identified 6 tryptic peptides mapping to the RecA2 domain of Dhh1 (Figure 6B and Supplementary Figure S5). No peptide was detected in the RecA1 domain, suggesting that this domain is not able to bind RNA when Dhh1 is in the enzymatically inactive conformation. However, the flexible N-terminal region that precedes RecA1 might contribute to RNA binding, as we detected an additional peptide encompassing residues 32–44 (Supplementary Figure S5). Within RecA2, two peptides mapped to the canonical RNA-binding surface expected for the active state of the DEAD-box protein (red and magenta patches in Figure 6B). Others mapped to the outer surface of RecA2 (green and yellow patches in Figure 6B), corresponding to the patch 1 and patch 3 sites. The mass spectrometry analysis identified a direct crosslink between a uridine nucleotide and Cys273 (Figure 6C), a residue in a central position at the FDF-binding site on patch 1 (Figure 3A, central panel). Consistently, the electrostatic properties of the RecA2 surface show patches of positive charges along the RNA-binding surface mapped by mass-spectrometry (Figure 6D). These data indicate that Dhh1 binds Pat1, Edc3 and RNA with mutually exclusive interactions at patch 1 and patch 3.
CONCLUSIONS
Dhh1 has a prominent pocket on the C-terminal RecA2 domain that recognizes short FDF linear motifs present in both yeast Pat1 and Edc3. In the case of the human orthologues, this pocket recognizes either the FDF motif of Edc3 (35) or a DW motif in Pat1b. It is possible that Dhh1 might bind other factors at this pocket, not necessarily with a FDF motif in their sequence. A hydrophobic pocket is present at the corresponding position in human eIF4AIII, where it is used to bind a Tyr-containing short linear motif of Upf3b (58). An analogous pocket is also present in yeast eIF4A and is used to bind a Trp-containing short linear motif of eIF4G (59). Other DEAD-box proteins might have evolved this structural feature to recruit short motifs present in their binding factors. While such hydrophobic pocket probably contributes binding affinity, specificity is likely provided by binding to additional surfaces. We note that binding of Pat1 or Edc3 leaves a considerable surface area of Dhh1 unoccupied and potentially available to other binding partners.
Because of their overlapping binding sites, Pat1 and Edc3 cannot bind simultaneously on Dhh1. In addition, they interfere with the RNA-binding properties of Dhh1. Mapping the RNA-binding surfaces of Dhh1 by a crosslinking–mass spectrometry approach suggests that RNA wraps around the RecA2 domain, docking also at the FDF-binding site of Dhh1 (at patch 1). In this conformation, in the absence of ATP, Dhh1 is in an enzymatically inactive state. The presence of both ATP and RNA appears to be insufficient to release the inactive conformation of Dhh1 that arises from the stabilization of an unproductive conformation of the two RecA domains by an extensive network of intramolecular interactions (44,45). Pat1 and in yeast Edc3 are expected also to approach the canonical ATP-dependent RNA-binding surface (at patch 3) with a negatively charged segment, and thus to interfere with it.
The results predict that Pat1, Edc3 and RNA bind Dhh1 as separate steps in the pathway, pointing to the presence of transitions in the architecture of an mRNP as it gets targeted to decay. A working model for the sequence and coordination of these structural rearrangements is outlined below. In the cytoplasm, Dhh1 is present in large excess and binds mRNAs in a constitutive, ATP-independent manner (39,44,45). We envision that upon triggering translational repression and deadenylation, Pat1 and Edc3 binding to Dhh1 would release it from the nonproductive ATP-independent mode of RNA binding. This displacement would allow Dhh1 to bind RNA in a productive ATP-dependent manner, triggering ATP hydrolysis and dissociation from the mRNP. This step is likely to require the binding of an activator to release Dhh1 from the enzymatically inhibited conformation. Besides the Dhh1-binding domain, Pat1 and Edc3 bridge to the Lsm complex and to the decapping complex, respectively. Discovering the precise sequel of events that lead to the swap between Pat1-containing and Edc3-containing complexes on Dhh1 as well as the identity of the Dhh1 activator are quests for future studies.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein data Bank with accession codes 4brw for the yeast Dhh1–Pat1 complex and 4bru for the yeast Dhh1–Edc3 complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online, including [60–66].
FUNDING
Max Planck Gesellschaft, the EU [ERC Advanced Investigator Grant 294371 and Marie Curie Initial Training Network RNPnet 289007]; Deutsche Forschungsgemeinschaft [SFB646, SFB1035, GRK1721, FOR1680 and CIPSM to E.C.]. Funding for open access charge: MPG internal funding.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the Max Planck Institute of Biochemistry Core Facility and Crystallization Facility; the staff members at the beamlines PXII and PXIII of the Swiss Light Source. We also thank members of our lab for useful discussions and critical reading of the manuscript. H.S. performed the biochemical, ITC and crystallography experiments; S.O. performed the coIP experiments; C.B. performed the fluorescence anisotropy experiments; K.S. and H.U. carried out the crosslinking/mass spectroscopy analysis; E.C. and H. U. supervised the project; E.C. and H.S. wrote the manuscript.
REFERENCES
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat. Rev. Mol. Cell. Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Wickens M, Goldstrohm A. Molecular biology. A place to die, a place to sleep. Science. 2003;300:753–755. doi: 10.1126/science.1084512. [DOI] [PubMed] [Google Scholar]
- 5.van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 8.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonnerot C, Boeck R, Lapeyre B. The two proteins Pat1p (Mrt1p) and Spb8p interact in vivo, are required for mRNA decay, and are functionally linked to Pab1p. Mol. Cell. Biol. 2000;20:5939–5946. doi: 10.1128/mcb.20.16.5939-5946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle J-C, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl Acad. Sci. USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decker CJ, Parker R. mRNA decay enzymes: decappers conserved between yeast and mammals. Proc. Natl Acad. Sci. USA. 2002;99:12512–12514. doi: 10.1073/pnas.212518099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell. Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 16.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol. Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 18.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:2274–2287. doi: 10.1091/mbc.E07-03-0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pilkington GR, Parker R. Pat1 contains distinct functional domains that promote P-body assembly and activation of decapping. Mol. Cell. Biol. 2008;28:1298–1312. doi: 10.1128/MCB.00936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minshall N, Kress M, Weil D, Standart N. Role of p54 RNA helicase activity and its C-terminal domain in translational repression, P-body localization and assembly. Mol. Biol. Cell. 2009;20:2464–2472. doi: 10.1091/mbc.E09-01-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas G, Braun JE, Igreja C, Tritschler F, Nishihara T, Izaurralde E. HPat provides a link between deadenylation and decapping in metazoa. J. Cell Biol. 2010;189:289–302. doi: 10.1083/jcb.200910141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozgur S, Chekulaeva M, Stoecklin G. Human Pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell. Biol. 2010;30:4308–4323. doi: 10.1128/MCB.00429-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Totaro A, Renzi F, La Fata G, Mattioli C, Raabe M, Urlaub H, Achsel T. The human Pat1b protein: a novel mRNA deadenylation factor identified by a new immunoprecipitation technique. Nucleic Acids Res. 2010;39:635–647. doi: 10.1093/nar/gkq797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weston A, Sommerville J. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 2006;34:3082–3094. doi: 10.1093/nar/gkl409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem. Soc. Trans. 2010;38:1602–1607. doi: 10.1042/BST0381602. [DOI] [PubMed] [Google Scholar]
- 27.Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping Activators in Saccharomyces cerevisiae Act by Multiple Mechanisms. Mol. Cell. 2010;39:773–783. doi: 10.1016/j.molcel.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tharun S. Lsm1-7-Pat1 complex: a link between 3′ and 5′-ends in mRNA decay? RNA Biol. 2009;6:228–232. doi: 10.4161/rna.6.3.8282. [DOI] [PubMed] [Google Scholar]
- 29.Fromm SA, Truffault V, Kamenz J, Braun JE, Hoffmann NA, Izaurralde E, Sprangers R. The structural basis of Edc3- and Scd6-mediated activation of the Dcp1:Dcp2 mRNA decapping complex. EMBO J. 2011;31:279–290. doi: 10.1038/emboj.2011.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweet T, Kovalak C, Coller J. The DEAD-box protein Dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012;10:e1001342. doi: 10.1371/journal.pbio.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamura A, Amikura R, Hanyu K, Kobayashi S. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development. 2001;128:3233–3242. doi: 10.1242/dev.128.17.3233. [DOI] [PubMed] [Google Scholar]
- 32.Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C. elegans. Development. 2001;128:3221–3232. doi: 10.1242/dev.128.17.3221. [DOI] [PubMed] [Google Scholar]
- 33.Minshall N, Standart N. The active form of Xp54 RNA helicase in translational repression is an RNA-mediated oligomer. Nucleic Acids Res. 2004;32:1325–1334. doi: 10.1093/nar/gkh303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tritschler F, Braun JE, Eulalio A, Truffault V, Izaurralde E, Weichenrieder O. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell. 2009;33:661–668. doi: 10.1016/j.molcel.2009.02.014. [DOI] [PubMed] [Google Scholar]
- 36.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 37.Kramer S, Queiroz R, Ellis L, Hoheisel JD, Clayton C, Carrington M. The RNA helicase DHH1 is central to the correct expression of many developmentally regulated mRNAs in trypanosomes. J. Cell Sci. 2010;123:699–711. doi: 10.1242/jcs.058511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Minshall N, Reiter MH, Weil D, Standart N. CPEB interacts with an ovary-specific eIF4E and 4E-T in early Xenopus oocytes. J. Biochem. 2007;282:37389–37401. doi: 10.1074/jbc.M704629200. [DOI] [PubMed] [Google Scholar]
- 39.Ernoult-Lange M, Baconnais S, Harper M, Minshall N, Souquere S, Boudier T, Bénard M, Andrey P, Pierron G, Kress M, et al. Multiple binding of repressed mRNAs by the P-body protein Rck/p54. RNA. 2012;18:1702–1715. doi: 10.1261/rna.034314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smillie DA, Sommerville J. RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci. 2002;115:395–407. doi: 10.1242/jcs.115.2.395. [DOI] [PubMed] [Google Scholar]
- 42.Haanstra JR, Stewart M, Luu VD, van Tuijl A, Westerhoff HV, Clayton C, Bakker BM. Control and regulation of gene expression: quantitative analysis of the expression of phosphoglycerate kinase in bloodstream form Trypanosoma brucei. J. Biochem. 2008;283:2495–2507. doi: 10.1074/jbc.M705782200. [DOI] [PubMed] [Google Scholar]
- 43.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Dutta A, Zheng S, Jain D, Cameron CE, Reese JC. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J. Biol. Chem. 2011;286:27454–27470. doi: 10.1074/jbc.M111.220251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carroll JS, Munchel SE, Weis K. The DExD/H box ATPase Dhh1 functions in translational repression, mRNA decay, and processing body dynamics. J. Cell Biol. 2011;194:527–537. doi: 10.1083/jcb.201007151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tritschler F, Eulalio A, Helms S, Schmidt S, Coles M, Weichenrieder O, Izaurralde E, Truffault V. A similar mode of interaction enables Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol. Cell. Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 53.Schmidt C, Kramer K, Urlaub H. Investigation of protein-RNA interactions by mass spectrometry.Techniques and applications. J. Proteomics. 2012;75:3478–3494. doi: 10.1016/j.jprot.2012.04.030. [DOI] [PubMed] [Google Scholar]
- 54.Ozgur S, Stoecklin G. Role of Rck-Pat1b binding in assembly of processing-bodies. RNA Biol. 2013;10:1–12. doi: 10.4161/rna.24086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 57.Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- 58.Buchwald G, Ebert J, Basquin C, Saulière J, Jayachandran U, Bono F, Le Hir H, Conti E. Insights into the recruitment of the NMD machinery from the crystal structure of a core EJC-UPF3b complex. Proc. Natl Acad. Sci. USA. 2010;107:10050–10055. doi: 10.1073/pnas.1000993107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl Acad. Sci. USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kramer K, Hummel P, Hsiao H, Luo X, Wahl M, Urlaub H. Mass-spectrometric analysis of proteins cross-linked to 4-thio-uracil- and 5-bromo-uracil-substituted RNA. Int. J. Mass Spectrom. 2011;304:184–194. [Google Scholar]
- 62.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sturm M, Bertsch A, Gropl C, Hidebrandt A, Hussong R, Lange E, Pfeifer N, Shulz-Trieglaff O, Zerck A, Reinert K, et al. OpenMS - an open-source software framework for mass spectrometry. BMC Bioinformatics. 2011;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bertsch A, Gröpl C, Reinert K, Kohlbacher O. OpenMS and TOPP: open source software for LC-MS data analysis. Methods Mol. Biol. 2011;696:353–367. doi: 10.1007/978-1-60761-987-1_23. [DOI] [PubMed] [Google Scholar]
- 65.Geer LY, Merkey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.