Abstract
Translation of the sigma factor RpoS is activated by DsrA, RprA and ArcA, three small non-coding sRNAs (sRNA) that expose the ribosome-binding site (RBS) by opening up an inhibitory loop. In the RpoS network, no sRNAs have been found to pair with the RBS, a most common sRNA target site in bacteria. Here, we generate Ribo-0, an artificial sRNA, which represses rpoS translation by pairing with the RBS. Ribo-0 bypasses the RNA chaperon Hfq but requires the RBS to be loosely blocked. Ribo-0 interacts with DsrA and reshapes the RpoS network. Specifically, in the intact RpoS network, DsrA activates rpoS translation by freeing up the RBS. In the modified RpoS network where Ribo-0 is introduced, the DsrA-caused RBS exposure facilitates Ribo-0 binding, thereby strengthening Ribo-0 inhibition. In other words, Ribo-0 changes DsrA from an activator to an accomplice for repressing rpoS translation. This work presents an artificial mechanism of rpoS regulation, reveals mutual effects of native and synthetic players and demonstrates genetic context-dependency of their functions.
INTRODUCTION
The alternative sigma factor RpoS is a key master regulator of stress response (1–5), governing expression of >200 genes in Escherichia coli (6,7). The rpoS gene has a long messenger RNA (mRNA) leader with complex structure carrying a cis-acting antisense element that blocks ribosome access and consequently reduces rpoS translation. In addition, the rpoS leader interacts with multiple small non-coding RNAs (sRNA) that modulate rpoS mRNA stability and translation. Thus, the rpoS mRNA provides a natural platform for characterization of cis- and trans-regulation of gene expression as well as their interplay.
rpoS translation is activated by sRNAs DsrA (8,9), RprA (10,11) and ArcZ (12), and it inhibited by OxyS (13). All the positive sRNA regulators interact with the cis-acting antisense element in the distal region of the rpoS leader, freeing up the ribosome-binding site (RBS) and derepressing rpoS translation. In contrast, there is no evidence for the direct interaction of OxyS with rpoS, and the regulatory mechanism of OxyS remains unclear (13).
Many sRNAs exert their inhibitory regulation by pairing with RBS of their target mRNAs. However, this action seems to be absent from the RpoS network or yet to be identified. Therefore, it is of interest to introduce an artificial sRNA inhibitor that directly binds to the rpoS RBS and to see whether and how the alien player interacts with the native regulators and alters the genetic network. The findings would provide new insight into how the rpoS network works and shed light on network design. Recent advance in synthetic biology makes this task possible. By combining rational design and random library screening, we have generated an artificial sRNA-designated Ribo-0 that specifically pairs with the RBS. We show that Ribo-0 significantly reduces rpoS expression and bacterial acid resistance. Interestingly, the alien sRNA alters performance of native players in the rpoS network. Specifically, Ribo-0 reverses the function of the endogenous sRNA DsrA, changing it from an activator of rpoS translation to an inhibitor that enhances the Ribo-0 repression. This is because binding of DsrA to the distal region of rpoS exposes the RBS and facilitates Ribo-0 pairing with this site. Thus, the role of DsrA is both positive and negative depending on the genetic context it resides in. These studies show how an artificial sRNA can be designed to regulate rpoS and reshape the natural genetic network by interacting with other players. Similar strategies may be extrapolated to other genetic networks or used for network design.
MATERIALS AND METHODS
Escherichia coli strains and growth conditions
The bacterial strains and plasmids used in this study are listed in Supplementary Table S1. The E. coli strain MG1655 was used for phenotypic examination throughout this study. All MG1655 mutants were grown at 37°C, with shaking at 220 rpm, in Luria-Bertani (LB) medium. The E. coli DY330 strain was used for plasmid preparation and grown at 32°C in LB medium. The antibiotics ampicillin (50 µg/ml), kanamycin (50 µg/ml) and chloramphenicol (12.5 µg/ml) were used for selection when appropriate.
DNA manipulations and sRNA library construction
Gene deletion was performed using the recombineering system (14,15). Escherichia coli MG1655 was transformed with plasmid pSim6 (a gift from Dr Donald Court) from which the expression of the λ recombination proteins is induced at 42°C. PCR fragments encompassing a loxP-cat-loxP with homology (45 nt) to the regions immediately flanking each deletion locus were transformed via electroporation into MG1655 cells harboring pSim6. After induction of λred functions, recombinants were selected for chloramphenicol resistance (encoded by the cat gene) and were further verified by colony PCR and sequencing.
For construction of an sRNA Ribo-0 library, the loxP-cat-loxP fragment was first linked to a constitutive promoter, which was used to drive sRNA expression, by overlapping PCR. The resulting PCR product was then precisely ligated to a 6-nt random sequence followed by a base pairing domain and a 6-uracil (U) string by overlapping PCR. The final PCR products with homology (45 nt) to the insertion site of a pET expression vector was transformed together with the vector into the DY330 strain after induction of λred. Recombinants were selected for chloramphenicol resistance and then collected for plasmid isolation.
rpoS-lacZ translational fusion and beta-galactosidase assays
The rpoS-lacZ translational fusion on the chromosome of E. coli MG1655 was constructed previously (16). Briefly, a lacZ-chloramphenicol resistance (cat) cassette starting with the eight codon of lacZ was linked by recombineering to the second last codon of rpoS. Cells carrying the rpoS-lacZ fusion were incubated overnight (at stationary phase) in LB medium at 37°C before quantification of the fusion expression. Expression of the rpoS-lacZ fusion was quantified using a beta-galactosidase assay as described previously (16,17). Levels of beta-galactosidase were calculated using the following formula:
Acid resistance assay
Overnight cultures were treated with acid (pH 2.0) for 2 h and then serially diluted in neutral medium. For cells deleted for rpoS, which are highly sensitive to acidic stress, they were treated with acid for 15 min. Colony forming units (CFU) were determined immediately before and after the acid treatment. Acid survival (%) was calculated with the following formula:
RNA isolation, ribonuclease protection assay and real-time PCR
Total RNAs were isolated using Trizol agent (Invitrogen) and then treated with TURBO DNaseI (Ambion, Austin, TX) to remove any DNA residuals. For determination of RNA decay, RNAs were isolated 0, 10, 20 and 30 min after rifampicin (final concentration = 500 µg/ml) was added to each culture. Ribonuclease protection assay (RPA) was performed using a RPA III kit (Ambion, Austin, TX). Specifically, biotin-labeled probes were synthesized in vitro using Biotin-14-CTP and the MAXIscript Kit from Ambion (Austin, TX), gel purified and quantified by UV spectrophotometry. Ten micrograms of total RNA and 500 pg of each biotin-labeled probe were mixed, coprecipitated and hybridized overnight at 42°C. The RNAs were then digested with RNaseA/RNase T1 mixture and run on a denaturing polyacrylamide gel (5%) in TBE buffer. The protected RNAs were transferred to a positively charged nylon membrane and visualized using the BrightStar BiotinDetect Kit (Ambion, Austin, TX).
For real-time PCR, 2 µg of total RNA was reverse transcribed in a total reaction volume of 20 µl. Each reaction was incubated at 55°C for 50 min, followed by 15 min at 70°C. The resulting reverse transcript products (complementary DNA) were then used for real-time PCR, which was carried out using an LightCycler 480II Detection System (Roche Diagnostics, Rotkreuz, Switzerland) and primers complementary to rpoS (rpoS-F, 5′-TCGCCGCCGGATGATCGAGA-3′; rpoS-R, 5′-CGCGGATCAGCCCCAGGTT-3′) and the 16 S rRNA gene (16S-F, 5′-CCCTTGAGGCGTGGCTTCC-3′; 16S-R, 5′-GCGGGCCCCCGTCAATTCAT-3′). Reactions were run in triplicate in three independent experiments. The 16S rRNA gene was used as an internal control. Expression data were normalized to that of wild-type cells carrying an empty vector pCm.
Statistical analysis
Paired t-tests were used to compare two means, and one-way ANOVA tests were used to compare multiple means obtained from beta-galactosidase, acid resistance assays and real-time PCR. P-values of <0.05 were considered statistically significant.
RESULTS
Establishment of a random library to screen for synthetic sRNAs that inhibit rpoS expression
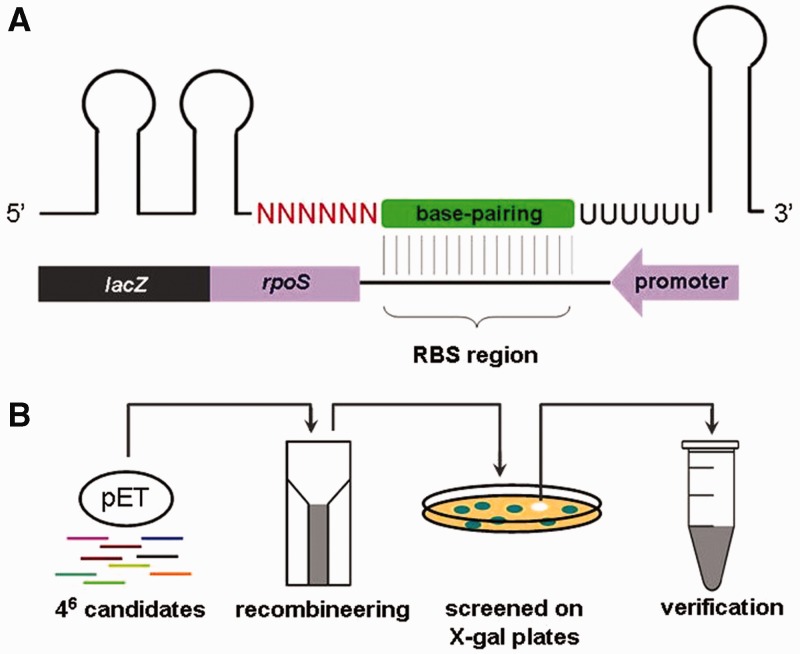
We combined rational design and random library screening to create an artificial sRNA that represses rpoS expression. The sRNA was designed to carry two stem-loop structures at the 5′ end, a 6-nt random sequence, a base-pairing domain (containing 18 nt), a 6-U stretch followed by a T7 transcriptional terminator (Figure 1A). The stem loops at the 5′ end make the sRNA highly structured and are therefore intended to enhance sRNA stability. The base-pairing domain is complementary to the RBS and intended to silence rpoS translation. The T7 terminator serves to halt sRNA transcription. The 6-U stretch has the potential to serve as an Hfq-binding site for the artificial sRNA, as the RNA chaperon Hfq binds preferentially to A/U-rich sequences (18–21). However, these modules do not guarantee generation of a functional sRNA, as regulatory mechanisms of sRNAs are not fully understood. We therefore introduced the aforementioned random sequence to establish a sRNA library, from which functionally active sRNAs could be screened for.
Figure 1.
Strategy for construction of a random sRNA library. (A) Design of a library of artificial sRNAs that repress rpoS translation by pairing with the RBS. Each putative sRNA was composed of two stem loops at the 5′ end, a 6-nt random sequence (shown in red), a base-pairing domain (shown in green), a poly-U stretch and a T7 terminator. The 6-nt random sequence allows for establishment of a library containing 46 putative sRNAs. The base-pairing domain is complementary to the RBS of the rpoS mRNA. To facilitate screening for active sRNAs, rpoS was fused in frame with lacZ. (B) Screening strategy used to search for artificial sRNAs that significantly inhibit expression of the rpoS-lacZ fusion, generating whiter colonies on LB agar plus X-gal. The chosen clones were then verified by PCR and sequencing.
We synthesized a double-stranded DNA fragment composed of all the aforementioned modules by PCR. The resulting DNA fragments containing the 6-nt random sequence were then cloned into a multi-copy vector (a pET vector deleted for the T7 promoter) under the control of a constitutive promoter of the csrB gene encoding the natural sRNA CsrB, generating a sRNA random library. The sRNA library was then introduced into a previously constructed chromosomal rpoS-lacZ translational fusion strain (the last second codon of rpoS fused with the eight codon of lacZ) (22), giving rise to a mixed population of cells harboring ∼1 × 46 putative sRNAs. Single colonies formed on X-gal agar plates after overnight culture, each overproducing a putative sRNA that has the potential to repress rpoS through the designed base-pairing domain (Figure 1B).
An artificial sRNA named Ribo-0 significantly inhibits rpoS expression
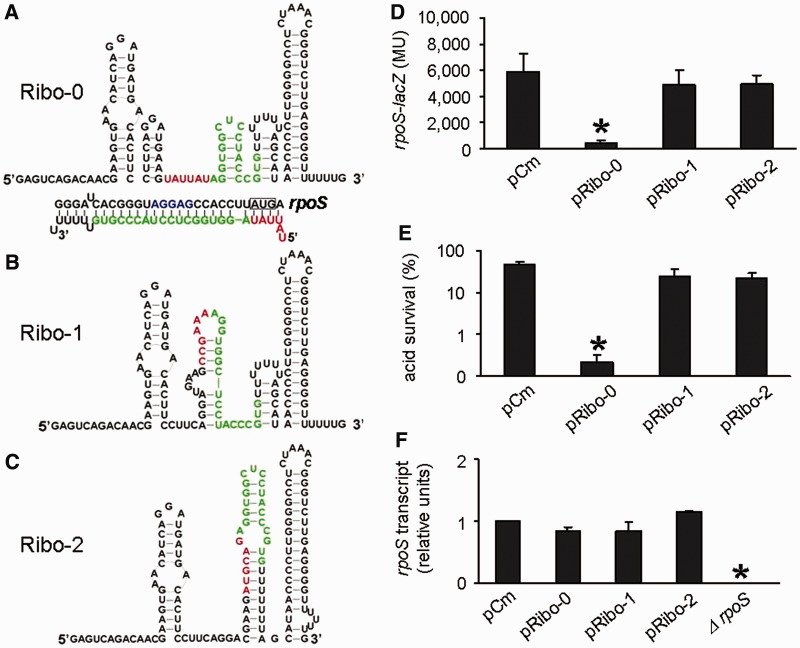
Approximately 5000 colonies were screened by color. The majority of the clones were blue, indicating that most sRNA candidates have no regulatory effects on the rpoS-lacZ fusion expression despite the rational design. This shows the necessity of introducing a random region to ensure creation of functionally active sRNAs, as we did in this study. Only two clones appeared to be whiter than others and potentially carried sRNA inhibitors of rpoS. Plasmids were isolated from the whiter clones, and sequencing analysis revealed that they carried the same sRNA construct, in which the 6-nt random sequence was UAUUAU (Figure 2A). The identified sRNA was hereafter referred to as Ribo-0. The random sequence starts with UAUU, which is complementary with the first 4 nt of the rpoS coding region. Thus, Ribo-0 is likely to interfere not only with the accessibility of the RBS but also with translational initiation. As negative controls, two blue colonies were randomly selected and their sRNAs were designated Ribo-1 and Ribo-2. Sequencing of Ribo-1 and Ribo-2 revealed that their sequence in the random region was CCGAAA (Figure 2B) and AUGCAG (Figure 2C), respectively. Secondary minimum free energy structures of Ribo-0, Ribo-1 and Ribo-2 were predicted by using the program RNAstructure5.1 (http://rna.urmc.rochester.edu/RNAstructure.html) (23). Ribo-0, Ribo-1 and Ribo-2 fold into multiple stem loops and, therefore, are all highly structured (Figure 2A–C).
Figure 2.
Repression of rpoS expression by the artificial sRNA Ribo-0. (A) Predicted secondary structures and verified sequences of Ribo-0, (B) Ribo-1 and (C) Ribo-2. The color scheme used: the 6-nt random sequence, red; the base-pairing domain, green. (D) Regulation of chromosomal rpoS-lacZ fusion by Ribo-0, Ribo-1 and Ribo-2. The rpoS fusion expression was determined using beta-galactosidase assays. (E) Regulation of acid resistance by Ribo-0, Ribo-1 and Ribo-2. Survival percentages after 2 h of acid treatment (pH 2.0) were determined by counting CFUs. (F) Effects of Ribo-0, Ribo-1 and Ribo-2 on rpoS mRNA levels. rpoS transcripts were quantified using real-time PCR. The 16S RNA was used as an internal control to ensure comparability of the samples. In experiments shown in Figures (D), (E) and (F), cells carrying pCm (an empty control) served as a negative control in addition to those overproducing Ribo-1 and Ribo-2. Error bars represent standard deviation (*P < 0.05).
To examine the putative Ribo-0 regulation of rpoS expression, we transformed the rpoS-lacZ fusion strain with the plasmid carrying Ribo-0 or the negative controls (including pRibo-1, pRibo-2 and an empty control vector pCm) and then measured beta-galactosidase activity of the resulting transformants. Compared with the negative control pCm, Ribo-0 overproduction reduced the rpoS fusion expression by 93% (Figure 2D). In contrast, neither Ribo-1 nor Ribo-2 significantly affected the rpoS fusion expression (Figure 2D). Ribo-1 and Ribo-2 may be unable to repress rpoS owing to their inactive structures or poor stability as a result of their different random sequence from that of Ribo-0. We did not dig into the underlying reason, as this is not what we set out to address. The aim of this study is to explore how artificial sRNAs like Ribo-0 reshape the RpoS regulatory network, as mentioned in the introduction section.
It is well established that RpoS is required for resistance of E. coli to extreme acidic stress (1). Consistent with this, E. coli MG1655 overproducing Ribo-0 were significantly more acid sensitive than those carrying Ribo-1, Ribo-2 or pCm (Figure 2E, note different scale for this figure), confirming the Ribo-0 repression of rpoS. Ribo-0 did not affect acid resistance in an rpoS null mutant, indicating that rpoS is the only target for Ribo-0 to regulate acid resistance (Supplementary Figure S1A). We then used quantitative real-time PCR to examine the effects of Ribo-0 on levels of rpoS transcript. Ribo-0 did not significantly reduce rpoS transcription compared with the negative controls (Figure 2F), verifying that Ribo-0 represses rpoS expression primarily at the translational level as designed.
Ribo-0 directly interacts with rpoS mRNA
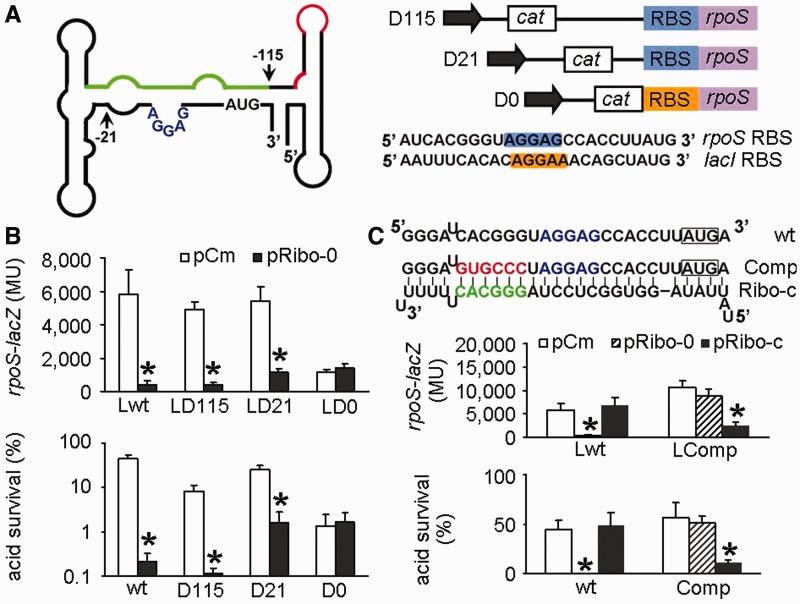
Next, we sought out to verify that Ribo-0 operates by acting on the RBS and the neighboring regions of rpoS. We inserted a cat cassette in sites with increasing distance from the translational start site. Regions upstream of the insertion sites were replaced by the cassette, resulting in 5′ leaders with different lengths (Figure 3A). The cat cassette contained a constitutive promoter so that rpoS could still be transcribed in the absence of its native promoters. The resulting mutants were named D0, D21 and D115, as 0, 21 and 115 nt of the rpoS leader upstream of the translational start site were left intact, respectively. For construction of D0, the RBS of rpoS was replaced by the RBS of the lacI gene. With D115, Ribo-0 retained the inhibitory effects on the rpoS-lacZ expression and acid resistance (Figure 3B). This was also true for D21 where the mRNA leader contained the native RBS but otherwise completely differed from the wild-type rpoS leader (Figure 3B), suggesting that the 5′ UTR regions upstream of the RBS is not essential for the Ribo-0 regulation of rpoS. In contrast, with D0 where the entire rpoS leader was moved and the rpoS RBS was replaced by the lacI RBS, Ribo-0 lost the regulation of the rpoS-lacZ expression and acid resistance, suggesting that the rpoS RBS is the target site of Ribo-0.
Figure 3.
Direct interaction between Ribo-0 and rpoS. (A) Structure of the rpoS leader and design of its truncation. The green line denotes the cis-acting antisense element that pairs with the RBS region. The red line denotes Hfq-binding sites. The arrows indicate where truncation was made in the rpoS leader. The numbers indicate the nucleotide at the 5′ end of the rpoS leader, relative to the translational start site. In mutants D115, D21 and D0, 115 nt, 21 nt and 0 nt of the rpoS leader immediately upstream of the translational start site were left intact, respectively. In D0, the rpoS RBS was replaced by the lacI RBS (shown in yellow). (B) Regulation of the rpoS-lacZ translational fusion and acid resistance by Ribo-0 in various strains. Cells carrying pCm (an empty control) served as a negative control. Error bars represent standard deviation (*P < 0.05). (C) Effects of compensatory mutations. Mutations (in red) made upstream of the RBS (in blue) in the rpoS leader (resulting in a mutant referred to as Comp or LComp in the rpoS-lacZ fusion background) and compensatory mutations (in green) made in Ribo-0 (resulting in a Ribo-0 variant referred to as Ribo-c) were highlighted in red. Error bars represent standard deviation (*P < 0.05).
To provide more evidence for the direct interaction of Ribo-0 with rpoS, we carried out compensatory mutation experiments. First, we mutated the designed binding site in the rpoS mRNA to see whether the mutations abolished the Ribo-0 repression. The RBS consensus sequence and the start codon were not mutated to maintain proper rpoS expression. Instead, 6 nt upstream of the RBS were changed from CACGGG to GUGCCC to disrupt the extensive complementarities between Ribo-0 and the rpoS mRNA. We introduced the mutations both to the rpoS-lacZ fusion and to the wild-type strain, generating mutants named LComp and Comp, respectively. The mutations impaired the regulation of the rpoS fusion expression and acid resistance by Ribo-0 (Figure 3C). We then introduced compensatory mutations to Ribo-0 so that the resulting sRNA named Ribo-c perfectly paired with the RBS and the neighboring regions of the mutated rpoS. As shown in Figure 3C, Ribo-c did not affect the unmutated rpoS but restored the regulation of the mutated rpoS. Taken together, these results clearly demonstrate that Ribo-0 directly interacts with the rpoS mRNA via the designed binding site.
Ribo-0 bypasses the requirement for Hfq
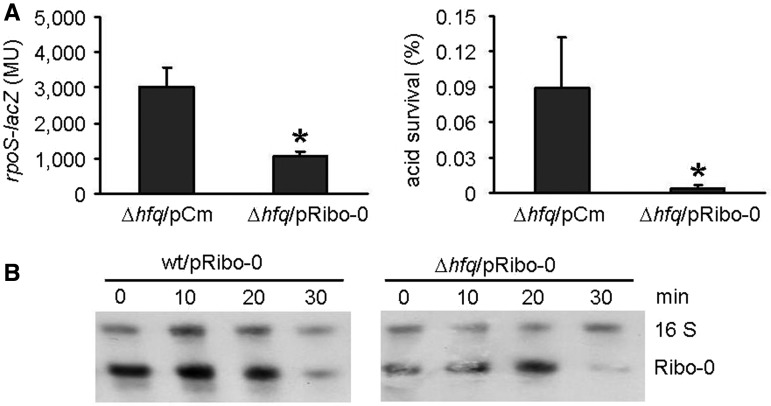
The random sequence in Ribo-0 was A/U rich (UAUUAU). It is tempting to speculate that the A/U-rich fragment may be an Hfq-binding site, as Hfq binds preferentially to single-stranded regions rich in A/U (18–21). If Hfq is required for the rpoS regulation by Ribo-0, then Ribo-0 should be unable to regulate rpoS expression in the absence of the RNA chaperon protein. However, beta-galactosidase and acid resistance assays showed that Ribo-0 retained the inhibition of rpoS expression in an hfq null mutant (Figure 4A). Many native sRNAs have longer half-lives (10–30 min) as a result of interaction with Hfq (24,25). This led us to determine whether the decay rate of Ribo-0 is affected by Hfq. No difference in the Ribo-0 stability was found between the wild-type strain and the hfq null mutant as revealed by the nuclease protection analysis of the rifampicin-treated cultures (Figure 4B). It has been previously reported that the rpoS leader contains two Hfq-binding sites [an A6 and an (AAN)4 element] (26,27). If Ribo-0 bypasses Hfq, then removing the Hfq-binding sites from the rpoS leader would not affect the repression of rpoS by Ribo-0. In accordance with this, Ribo-0 retained the rpoS regulation in D115 and D21 from which the Hfq-binding sites were absent (Figure 3B). These observations collectively suggest that Ribo-0 is functional without the need for Hfq. It has previously been demonstrated that Hfq is not essential for sRNAs when they tightly pairs with their mRNA targets (26,28). Thus, the independence of Ribo-0 on Hfq suggests that Ribo-0 and rpoS form a stable complex on their own.
Figure 4.
Independence of Ribo-0 on the Hfq protein. (A) Ribo-0 retained regulation of the rpoS-lacZ translational fusion and acid resistance in an hfq null mutant (Δ hfq). Error bars represent standard deviation (*P < 0.05). (B) Ribo-0 decay in the wild-type strain (wt) and Δ hfq. Rifampicin (final concentration: 500 µg/ml) was added to cultures in the stationary phase to stop transcription. Aliquots of cells were taken at 0, 10, 20 and 30 min after the addition of rifampicin. Total cell RNA was isolated at each time point, and RPA was performed to quantify sRNAs. The stable 16 S RNA was used as an internal control.
Ribo-0 requires a loosely blocked RBS
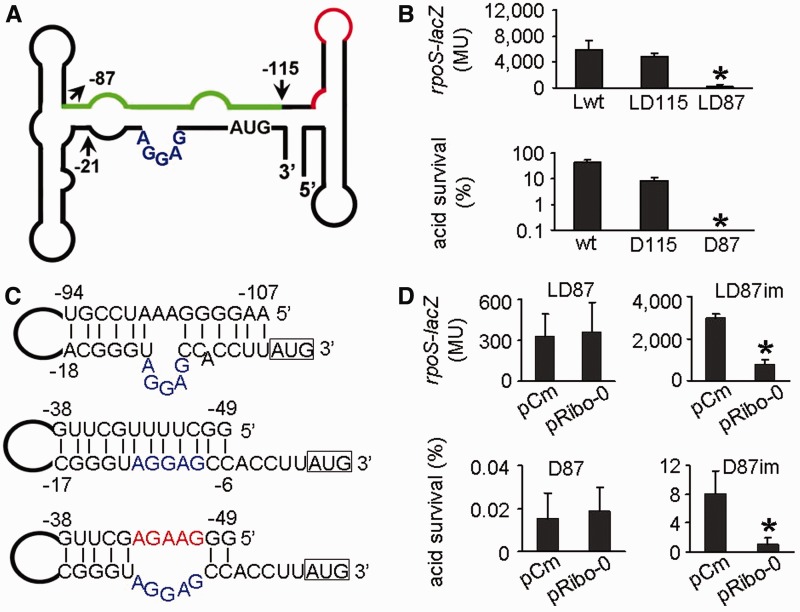
In the rpoS leader, the RBS is loosely blocked by a cis-acting antisense element. If this inhibitory element perfectly pairs with the RBS, the gene expression would be silenced, leaving no room for Ribo-0 to further repress the expression. To test this hypothesis, we sought out to construct an artificial cis-acting antisense element that binds to the RBS more tightly than the native one. We took advantage of a native element in the rpoS leader that is perfectly complementary to the RBS region. This base-pairing element is unlikely to bind to the RBS in the wild-type rpoS leader, as previous evidence did not reveal such a structure. We therefore did more serial deletions in the rpoS leader to search for a construct in which this otherwise inert element forms an inhibitory loop with the RBS region. We found that a mutant named D87, in which 87 nucleotides upstream of the translational start site was left intact (Figure 5A), showed 25% of the wild-type rpoS expression and 0.03% of the wild-type acid resistance (Figure 5B). Prediction of secondary structure of the D87 rpoS leader revealed that the 12-nt element between −49 and −38 perfectly pairs with the RBS and neighboring sequences (Figure 5C). Local free energy of this perfectly base-paired structure and its native counterpart is −14.2 kcal/mol and −10.5 kcal/mol, respectively. Thus, the 12-nt element in D87 blocks the RBS of rpoS more tightly than the antisense element in the wild-type strain. In agreement with our hypothesis, when the RBS perfectly paired with the cis-acting antisense element, Ribo-0 failed to further reduce rpoS expression and acid resistance (Figure 5D). We then introduced mutations to the rpoS fusion and the wild-type strain to disrupt the perfect base pairing. The resulting mutants were referred to as LD87im and D87im, respectively. Ribo-0 restored the repression of rpoS expression and acid resistance of the mutants (Figure 5D). Together, the RBS of rpoS has to be loosely blocked for the Ribo-0 modification of the RpoS network.
Figure 5.
Requirement of the Ribo-0 regulation of RpoS for the loosely folded inhibitory loop in the 5′ leader. (A) Structure of the rpoS leader and the location where D87 was constructed. The green line denotes the cis-acting antisense element that pairs with the RBS region. The red line denotes Hfq-binding sites. The numbers indicate the nucleotide at the 5′ end of the rpoS leader, relative to the translational start site. (B) Silenced expression of the rpoS-lacZ fusion and low levels of acid resistance in the mutant D87. (C) Predicted secondary structures of the naturally occurring inhibitory loop (upper panel) in the wild-type strain, a perfectly folded inhibitory loop in D87 (middle panel) and an artificial imperfect inhibitory loop in D87im (lower panel). Mutations made in D87im are highlighted in red and the RBS is highlighted in blue. (D) Defect of Ribo-0 in regulation of the rpoS fusion expression and acid resistance in D87, and restoration of the regulation in D87im.
Interplay of Ribo-0 and native regulation of RpoS
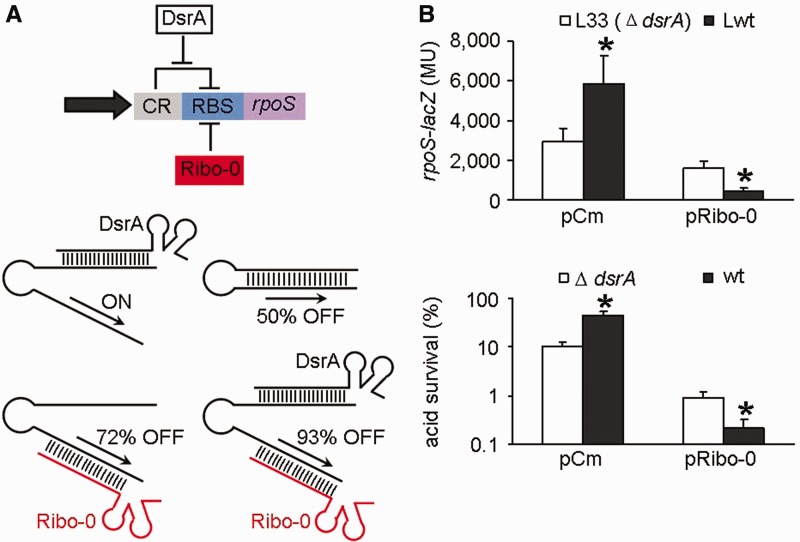
It is well established that rpoS translation is repressed by the cis-acting antisense element, which sequesters the RBS and prevents ribosome docking. As described earlier in the text, Ribo-0 represses rpoS translation by pairing with the RBS. In contrast, the native sRNA DsrA is partially complementary to the cis-repressive element, freeing up the RBS and activating rpoS translation (29,30). We speculated that the DsrA-caused exposure of the RBS would facilitate Ribo-0 binding, thereby strengthening silencing effects of Ribo-0. Conversely, Ribo-0 was expected to change DsrA from an activator to an accomplice of Ribo-0 (Figure 6A). In agreement with our hypothesis, Ribo-0 reduced the rpoS-lacZ fusion expression by 45% in a dsrA mutant background, but the percentage increased to 93% in the wild-type strain with chromosomal DsrA (Figure 6B). Consistently, Ribo-0 resulted in a 10-fold reduction in survival percentage after 2 h acid challenge in the absence of DsrA, but it led to a 200-fold reduction in acid resistance when DsrA was present (Figure 6B). The beta-galactosidase and acid resistance results demonstrate that Ribo-0 requires DsrA for its optimal silencing effects on rpoS. Conversely, Ribo-0 altered the role of DsrA in rpoS regulation. In the absence of Ribo-0, DsrA acted as an activator of rpoS expression. In the presence of Ribo-0, however, DsrA turned into reducing the rpoS-lacZ fusion expression and accordingly acid resistance (Figure 6B). Thus, Ribo-0 reverses the function of DsrA, transforming it into a repressor of rpoS. The interplay of Ribo-0 and DsrA gives rise to a new network that achieves stepwise regulation of rpoS. That is, rpoS expression is ON in the presence of chromosomal DsrA; 50% of rpoS expression is inhibited when DsrA is absent; this figure is increased to 72% when Ribo-0 is overproduced and DsrA is absent; the optimal silencing effects (93%) are achieved in the presence of DsrA when Ribo-0 is overproduced (Figure 6A).
Figure 6.
Interplay of Ribo-0 and DsrA. (A) Model for stepwise regulation of rpoS by Ribo-0 and DsrA. Abbreviations in the upper panel: CR, cis-acting repressive element in the rpoS leader; RBS, ribosome binding site. (B) Mutual effects of Ribo-0 and DsrA in their regulation of the rpoS fusion expression and acid resistance. Error bars represent standard deviation (*P < 0.05).
Chromosomal DsrA requires the Hfq chaperon to be functional (31). We therefore speculated that the interplay between Ribo-0 and DsrA would disappear in the D115 strain, as the Hfq-binding sites are absent from its rpoS leader. In accordance with this, deleting the dsrA gene from the D115 background did not affect rpoS expression or acid resistance and had no impact on the Ribo-0 regulation (Supplementary Figure S1B). It has been previously demonstrated that overexpressed sRNA OxyS competes with some endogenous sRNAs for binding to Hfq, thereby lowering their accumulation and regulatory activity (32). This led us to ask whether Ribo-0 regulates rpoS expression and interacts with DsrA by sequestering Hfq. If Ribo-0 repressed rpoS expression by competing with DsrA for Hfq, then Ribo-0 overproducers with chromosomal DsrA should produce more (or no less) RpoS than those deficient in DsrA. Our data, however, showed the opposite (Figure 6B). This together with the aforementioned finding that Ribo-0 functioned independently of Hfq suggests that the interplay between Ribo-0 and DsrA is not due to competition for Hfq.
DISCUSSION
Bacteria possess diverse means of gene regulation using RNA regulators such as cis-acting antisense mRNA leaders that sequester the RBS to prevent ribosome docking (8,33), cis-acting riboswitches that sense and respond to the availability of specific ligands, trans-acting sRNA that pair with target RNAs or bind to proteins and CRISPR RNAs that repress the uptake of foreign DNA (34). Among these, the trans-acting sRNAs are the largest set of RNA regulators, and hundreds of naturally occurring sRNAs have been identified. Most of them act through pairing with target mRNAs, modulating their translation and stability. An sRNA can target multiple mRNAs. For instance, Spot42 regulates at least 15 operons involved in uptake and catabolism of diverse carbon sources (35). An mRNA can also be regulated by multiple sRNAs. One example is rpoS that encodes the alternative sigma factor RpoS implicated in stress resistance (1–5) and virulence (36). rpoS translation is activated by DsrA (8,9), RprA (10,11) and ArcZ (12) and repressed by OxyS (13). Although the regulatory mechanisms of DsrA, RprA and ArcA have been intensively investigated, the OxyS inhibition of rpoS translation remains poorly understood. It is now known that OxyS does not repress rpoS translation by binding to the RBS region and that no sRNA has been identified to interact with the RBS region of rpoS. Here, we filled the gap by creating a trans-acting sRNA Ribo-0 that inhibits rpoS translation by pairing with the RBS region. The pre-designed action of Ribo-0 allows us to explore the implication of multiple sRNA regulators for a single target (rpoS in this context) and how they interact with each other by binding to different domains of the mRNA leader.
Combining rational design and random library screening, we have created an artificial sRNA named Ribo-0 that was designed to inhibit rpoS translation. Although all the sRNA candidates carried the base-pairing domain as well as other pre-designed modules, the majority of them had little regulatory functions. This suggests that inclusion of these regulatory modules is necessary but not sufficient for creating a functionally active sRNA owing to our incomplete understanding of their regulatory mechanisms. This also demonstrates the necessity of combining random library screening and rational design for creation of gene-specific sRNAs.
Physiological, biochemical and genetic evidence has been provided for the direct interaction between Ribo-0 and the rpoS gene. The only difference between Ribo-0 and thousands of inactive variants such as Ribo-1 and Ribo-2 is that the latter do not pair with the translational AUG start site of rpoS, suggesting that the Ribo-0 complementarity to the AUG start site is essential. Furthermore, the compensatory mutation assays revealed that complementarity to the region flanking the RBS is also indispensable. Collectively, the RBS and the neighboring regions including the AUG start site is a minimal base-pairing domain required for the Ribo-0 regulation of rpoS. Disruption of the complementarities between Ribo-0 and any elements of this base-pairing domain would impair the regulation of rpoS by Ribo-0.
The successful design and creation of the rpoS-specific sRNA indicate that artificial sRNAs with user-defined performance can be used to manipulate cell biology in a programmatic fashion. More interestingly, we show that Ribo-0 cross-talks with the rpoS regulatory network and accomplishes more complex gene regulation. With the wild-type rpoS network, a cis-acting antisense element sequesters the rpoS RBS and inhibits translation, whereas the trans-acting DsrA binds to and opens the RNA operator, relieving the repression. Now, the incorporation of Ribo-0 reshapes this network as follows. On one hand, when Ribo-0 is introduced into the network, DsrA turns from an rpoS activator into an inhibitor; on the other, DsrA is required for the optimal silencing effects of Ribo-0. When both DsrA and Ribo-0 are present, optimal repression of rpoS is achieved, forming a NAND logic gate. By this means, the rpoS gene can be activated and repressed by DsrA under different conditions (i.e. with and without Ribo-0). In a broad sense, the interplay between sRNAs demonstrated here may also exist in other genetic networks and contribute to differential gene expression in response to different extracelllular signals or growth phases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Grants Council Collaborative Research Fund [HKU1/CRF/10 to J.D.H.]; University of Hong Kong small grant funding [201007176077 to Y.J.]. Funding for open access charge: Research funds from the University of Hong Kong and from the local government.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski JL. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang FC, Libby SJ, Buchmeier NA, Loewen PC, Switala J, Harwood J, Guiney DG. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc. Natl Acad. Sci. USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stokes NR, Murray HD, Subramaniam C, Gourse RL, Louis P, Bartlett W, Miller S, Booth IR. A role for mechanosensitive channels in survival of stationary phase: regulation of channel expression by RpoS. Proc. Natl Acad. Sci. USA. 2003;100:15959–15964. doi: 10.1073/pnas.2536607100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hengge-Aronis R, Klein W, Lange R, Rimmele M, Boos W. Trehalose synthesis genes are controlled by the putative sigma factor encoded by rpoS and are involved in stationary-phase thermotolerance in Escherichia coli. J. Bacteriol. 1991;173:7918–7924. doi: 10.1128/jb.173.24.7918-7924.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sammartano LJ, Tuveson RW, Davenport R. Control of sensitivity to inactivation by H2O2 and broad-spectrum near-UV radiation by the Escherichia coli katF locus. J. Bacteriol. 1986;168:13–21. doi: 10.1128/jb.168.1.13-21.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong T, Kirchhof MG, Schellhorn HE. RpoS regulation of gene expression during exponential growth of Escherichia coli K12. Mol. Genet. Genomics. 2008;279:267–277. doi: 10.1007/s00438-007-0311-4. [DOI] [PubMed] [Google Scholar]
- 7.Schellhorn HE, Audia JP, Wei LI, Chang L. Identification of conserved, RpoS-dependent stationary-phase genes of Escherichia coli. J .Bacteriol. 1998;180:6283–6291. doi: 10.1128/jb.180.23.6283-6291.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 11.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 12.Mandin P, Gottesman S. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J. 2010;29:3094–3107. doi: 10.1038/emboj.2010.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharan SK, Thomason LC, Kuznetsov SG, Court DL. Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc. 2009;4:206–223. doi: 10.1038/nprot.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin Y, Watt RM, Danchin A, Huang JD. Small noncoding RNA GcvB is a novel regulator of acid resistance in Escherichia coli. BMC Genom. 2009;10:165. doi: 10.1186/1471-2164-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin Y, Watt RM, Danchin A, Huang JD. Use of a riboswitch-controlled conditional hypomorphic mutation to uncover a role for the essential csrA gene in bacterial autoaggregation. J. Biol. Chem. 2009;284:28738–28745. doi: 10.1074/jbc.M109.028076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Y, Huang JD. Engineering a portable riboswitch-LacP hybrid device for two-way gene regulation. Nucleic Acids Res. 2011;39:e131. doi: 10.1093/nar/gkr609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 25.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 26.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol. Microbiol. 2008;70:100–111. doi: 10.1111/j.1365-2958.2008.06392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vecerek B, Beich-Frandsen M, Resch A, Blasi U. Translational activation of rpoS mRNA by the non-coding RNA DsrA and Hfq does not require ribosome binding. Nucleic Acids Res. 2010;38:1284–1293. doi: 10.1093/nar/gkp1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon K, Gottesman S. Competition among Hfq-binding small RNAs in Escherichia coli. Mol. Microbiol. 2011;82:1545–1562. doi: 10.1111/j.1365-2958.2011.07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andre G, Even S, Putzer H, Burguiere P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beisel CL, Storz G. The base-pairing RNA spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol. Cell. 2011;41:286–297. doi: 10.1016/j.molcel.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong T, Schellhorn HE. Role of RpoS in virulence of pathogens. Infect. Immun. 2010;78:887–897. doi: 10.1128/IAI.00882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.