Abstract
We have developed a new screening methodology for identifying all genes that control the expression of a target gene through genetic or metabolic interactions. The screen combines mutant libraries with luciferase reporter constructs, whose expression can be monitored in vivo and over time in different environmental conditions. We apply the method to identify the genes that control the expression of the gene acs, encoding the acetyl coenzyme A synthetase, in Escherichia coli. We confirm most of the known genetic regulators, including CRP–cAMP, IHF and components of the phosphotransferase system. In addition, we identify new regulatory interactions, many of which involve metabolic intermediates or metabolic sensing, such as the genes pgi, pfkA, sucB and lpdA, encoding enzymes in glycolysis and the TCA cycle. Some of these novel interactions were validated by quantitative reverse transcriptase-polymerase chain reaction. More generally, we observe that a large number of mutants directly or indirectly influence acs expression, an effect confirmed for a second promoter, sdhC. The method is applicable to any promoter fused to a luminescent reporter gene in combination with a deletion mutant library.
INTRODUCTION
The adaptation of bacteria to changes in their environment is controlled on the molecular level by a large and complex regulatory network involving genes, mRNAs, proteins and metabolites (1). Understanding, and therefore eventually predicting, the dynamics of this regulatory network is a general aim of biology, and in particular of the field of systems biology. A prerequisite of this type of research is knowledge of the topology of the underlying regulatory network. An increasing number of such regulatory connections are documented in bioinformatics databases such as EcoCyc (http://EcoCyc.org), which lists > 5300 regulatory interactions for Escherichia coli (2).
Different methods are used to identify and characterize these interactions. Evidence for specific interactions can come from in vitro measurement of physical DNA–protein interactions (footprinting, gel retardation assay). Computational tools can predict interactions based on consensus sequence. Transcriptomics (DNA microarrays) is a popular approach to study transcription on a global scale (3) . Typically, this technique allows detecting the targets of a particular regulator by studying the effect of the deletion of this regulator on the expression of all genes of a genome. Chromatine Immuno-Precipitation–Chip is another global approach, identifying direct interactions of a specific transcription factor through the detection of chromosome-wide DNA binding (4). All these tools aim at identifying regulons, i.e. all the targets of a given regulator (5). However, with few exceptions (6), no global method exists to determine the regulators that control, directly or indirectly, the expression of a particular target gene. We present such a method in this article.
We use luminescent reporters to measure promoter activities in living cells (7,8). The luciferase system is particularly useful because it allows measuring gene expression overtime in a colony growing on solid medium. We have developed a technique for efficiently transforming a genome-wide library of E. coli single-gene knockout mutants (9) with the appropriate reporter plasmid. The screening strategy consists in comparing the luminescence of mutant colonies with the luminescence of a wild-type (WT) colony: a significant difference suggests that the deleted gene controls, directly or indirectly, the expression of the target gene. The results are highly reproducible. Moreover, artifacts due to metabolic influences on luciferase activity can be detected using a second reporter system, based on gfp expression (10).
We have applied this method to identify regulators that control the transcription of the acs gene, coding for the acetyl coenzyme A synthetase (11). This enzyme converts acetate to acetyl coenzyme A. We have chosen this gene for three reasons. First, the enzyme plays a key role in the ‘acetate switch’: when bacteria grow rapidly on glucose, they excrete acetate, which is subsequently used as a carbon source when glucose is exhausted (12). Acetyl coenzyme A synthetase is thus at the center of an inversion of metabolic flux from glycolysis to gluconeogenesis. Second, the accumulation of acetate in the culture medium is an important problem in industrial fermentations because this organic acid inhibits cell growth and recombinant protein production. Different approaches are currently developed to reduce acetate accumulation by modifying central metabolic pathways (13). Lastly, the acs gene was also selected because its complex regulation has been extensively described and thus allows a thorough validation of the method (14–16). Briefly, transcription of this gene is repressed by the nucleoproteins Fis and IHF and positively regulated by the cAMP receptor protein (CRP)–cAMP complex. The latter activation implies the well-studied carbon catabolite repression mechanism where the glucose concentration controls the formation of the CRP–cAMP complex by adjusting the cAMP production via the phosphotransferase system (PTS) (17). The acs gene is thus at the end of a well-known complex regulatory cascade, an ideal property for our purpose.
Our screen confirms known regulatory mechanisms of acs expression. In particular, we confirm the necessity of a functional CRP–cAMP complex for the expression of acs, as well as the modulation of acs promoter activity by IHF. The screen also highlights that glucose repression of the acs promoter cannot be completely explained by cAMP-mediated mechanisms and inducer exclusion, a phenomenon already observed for other E. coli promoters (18). A major finding is that acs transcription is significantly modified in a large number of mutants, most notably genes encoding enzymes in glycolysis and the tricarboxylic acid cycle (TCA) cycle, such as pgi, pfkA, sucB and lpdA. This tight link between metabolism and gene expression was also observed for a second target gene, sdhC. We confirmed some of these novel regulators of acs by means of independent quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) experiments.
MATERIALS AND METHODS
Strains
The screen uses the Keio collection, comprising 3985 single-gene deletions of the E. coli K-12 strain BW25113, called WT strain (9). The crp mutant in the Keio collection was shown to be incorrect (19), so we used a deletion mutant obtained from the Sauer lab.
Plasmids
The nrfA-acs intergenic region was amplified by PCR using primers 5′-TCCTCTCGAGAGGGGCTTCATCCGAAT-3′ (Xho1 site underlined) and 5′-TTTGGGATCCGCTTTTGTTCTCCTTGTAGG-3′ (BamHI site underlined) and using the BW25113 strain as a template. The PCR product, digested with Xho1 and BamHI, was inserted into a luxCDABE plasmid backbone without promoter (20) to give p-acs-lux. The plasmid has a colE1 origin of replication and a strong ribosome-binding site driving efficient expression of the reporter gene. The gltA-sdhC intergenic region was amplified by PCR from the BW25113 strain, using primers 5′-TATCCTCGAGTTAAGGTCTCCTTAGCGCCTT-3′ (Xho1 site underlined) and 5′-TTCTGAATTCGAATAACGCCCACATGCTGTTC-3′ (EcoRI underlined), and inserted into the luxCDABE plasmid backbone without promoter to give p-sdh-lux. The bla gene (conferring ampicillin resistance) and the acs promoter region of the p-acs-lux plasmid were amplified by PCR using primers 5′-TTTGGGATCCGCTTTTGTTCTCCTTGTAGG-3′ (BamHI site underlined) and 5′-TCTAAAGTGAGCTCGAGTAAACTTGGTCTGACAG-3′ (SacI site underlined). The PCR product, digested with SacI and BamHI, was inserted into pUA66 (10), to give p-acs-gfp. This plasmid has the pSC101 origin of replication and carries a gene coding for a stable and fast-folding GFP. The complete sequences of these plasmids are available on request.
High-throughput transformation and luminescence quantification
All steps were performed in 96-well plates. The entire Keio collection was transformed with the p-acs-lux plasmid, demanding 135 plates, 6 days and four persons. In addition, we transformed 517 selected mutants for a detailed kinetic analysis with the p-acs-gfp plasmid and 15% of the Keio collection, corresponding to 650 mutants on seven plates, with the p-sdhC-lux plasmid. Mutants were inoculated from frozen stock in sterile microplates containing 200 µl per well of Luria-Bertani broth (LB) with kanamycin (50 µg/ml) and placed at 37°C under shaking (200 rpm) overnight. All volumes below are indicated for one well. The next day, 4 µl of the preculture were added to new plates containing 400 µl of LB with kanamycin (50 µg/ml) and placed at 37°C under shaking for 3 h. We then transferred 50 µl of these exponentially growing cells to new plates containing 50 µl of ice-cold TSS 2× (LB containing 10% PEG, 5% DMSO and 20–50 mM Mg2+ at a final pH of 6.5) (21) and the mixture was kept on ice for 45 min before adding 4 µl of plasmid DNA (50 ng/ml). The plates were left on ice for another 10 min before applying a heat shock (90 s at 42°C) and returned on ice for an additional 10 min of incubation. We finally added 1 ml of LB containing ampicillin (100 µg/ml) and the plates were placed at 37°C under shaking overnight. The following day, we spotted the transformed mutants onto two different petri dishes containing either LB-agar-ampicillin (100 µg/ml) or minimal medium M9-agar-0.3% glucose (p/v)-ampicillin (100 µg/ml). Petri dishes were incubated at 37°C overnight and the luminescence of the colonies was measured the following days using an intensified camera (Photonic Science). The quantification of the luminescence of the colonies was performed semiautomatically by means of a custom-made Matlab® program (Supplementary Information S8). Briefly, we identified semiautomatically the location of each growing colony on the agar plate and quantified the luminescence intensity of each colony. Here we present the median luminescence intensity of each colony, given that different measures of light intensity (mean, maximum, median) gave essentially identical results.
Measuring the dynamics of gene expression
The dynamics of gene expression were measured for cells growing for 10 h in a 96-well microplate. The microplate reader (Perkin Elmer, Fusion Alpha FP-HT) allows growth with shaking and temperature control (37°C). We measured the cell density (absorbance at 600 nm) every 30 min and luminescence or fluorescence (480 nm/520 nm) every 3 min. Unless otherwise specified, the wells in the microplate contain 180 µl of minimal medium (M9) supplemented with 0.03% glucose (p/v), 0.03% sodium acetate (p/v) and ampicillin (100 µg/ml). When indicated, we added cyclic AMP (5 mM) at the beginning of the experiment. The wells were inoculated with 20 µl of stationary phase cultures (10× dilution) grown in LB-ampicillin (100 µg/ml). The fluorescence background of the bacterial cells was assessed by measuring the fluorescence of mutant bacteria transformed with a promoterless reporter plasmid. The promoter activities were computed from the fluorescence and absorbance data as previously described (20).
qRT-PCR
The strains were grown in a microplate containing M9 0.03% glucose and 0.03% acetate (preculture LB). The cells (∼108) were harvested either before glucose exhaustion (exponential growth on glucose) or after glucose exhaustion (growth on acetate). Total mRNA was protected using the RNAprotect® Bacteria Reagent (Qiagen) and then isolated using the RNeasy mini kit (Qiagen) according to the recommendations of the manufacturer. Potential trace quantities of DNA were removed using the turbo DNAse (Ambion). Samples were protected from RNase using RNaseOut ribonuclease inhibitor (Invitrogen). One microgram of RNA was reverse transcribed using SuperScript™ II Reverse Transcriptase (Invitrogen) according to the protocol of the manufacturer. Briefly, a 25 µl reaction mixture was incubated in a T3000 thermocycler (Biometra) for 10 min at 25°C, 50 min at 42°C and 15 min at 70°C. The primers (Eurogentec) used in the present study were as follows: acs: 5′-ACAGTTCTGGTGACGGTTCC-3′ and 5′-ACAGTTCTGGTGACGGTTCC-3′. We have used the rrsD gene as internal control for relative quantification: 5′-GCTACAATGGCGCATACAAA-3′ and 5′-TTCATGGAGTCGAGTTGCAG-3′. Quantitative PCR was performed in a StepOnePlus Real-Time PCR System (Applied Biosystems) using MESA Green qPCR Master Mix (Eurogentec) according to the instructions of the manufacturer. Briefly, 25 µl reactions mixtures were incubated for 10 min at 95°C and 40 PCR cycles (15 s at 95°C and 1 min at 60°C). PCRs were run in triplicate. Raw data were transformed into threshold cycle (Ct) values. The relative strength of acs expression for each mutant, compared with WT, was calculated by the comparative Ct method (ΔΔCt).
Statistical model
The measured expression levels (in units of luminescence intensity) can be described by the following linear model: yi = xi + εi, where yi represents the observed expression level for mutant i, xi its noiseless expression level and εi the measurement error, a random variable with mean 0 and variance σ2. We thus assume the error variance to be constant for all i. Moreover, the error variables are assumed to be mutually independent and independent of xi. We consider xi to be invariant across replicates, but generally xi ≠ xj for different mutants i and j. For every individual experiment, we can write: var(yi) = var(xi) + var(εi) = var(xi) + σ2. For two replicate experiments, with measurements yi and yi’, because xi = xi’, we have var(yi’ − yi) = var(εi' − εi) = var(εi') + var(εi) = 2σ2. That is, we can estimate the error variance σ2 by computing (the square of) the standard deviation of the empirical distribution of the replicate differences yi’ − yi. The total variance of the measured expression levels yi, var(yi), can equally be estimated from the data, thus providing an estimate of the relative contribution of the measurement error to the total variance, σ2/var(yi). The data set yi' − yi was checked a posteriori to verify the assumption that the error variance is constant.
RESULTS
A screening strategy for identifying the regulators of acs
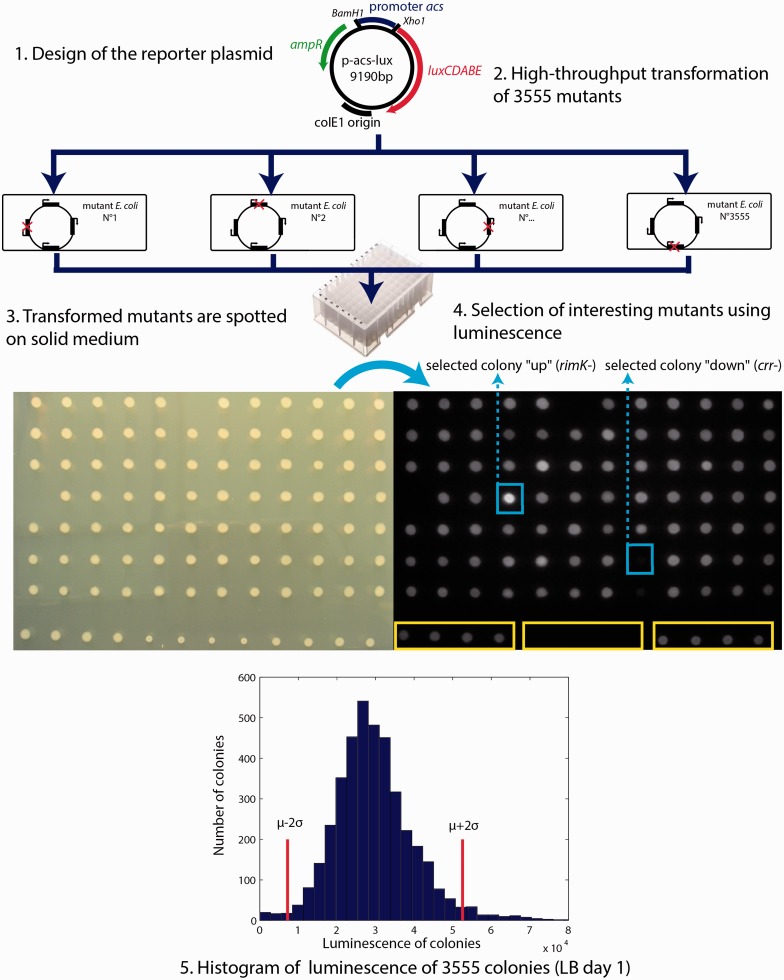
Our screening strategy is outlined in Figure 1. To measure acs promoter activity, we have constructed a reporter plasmid (p-acs-lux) containing a transcriptional fusion of the acs promoter region (394 bp) to the luxCDABE operon. This operon codes for the heterodimeric luciferase protein (luxAB) and the enzymes (luxCDE) necessary for producing the luciferase substrate, a long-chain aldehyde (22). We have chosen this reporter system because of the highly sensitive luminescence signal, the absence of background, the continuous production of bioluminescence without added substrate and the possibility to measure luminescence at the colony level. The reporter plasmid was transformed into the 3985 single-gene knockouts of E. coli (Keio collection) using our high-throughput transformation procedure (see ‘Materials and Methods’ section). Transformation efficiency was 89%: we have thus obtained 3555 transformants. We spotted the transformed bacteria on solid media and recorded the luminescence images of the colonies that appeared on the plates.
Figure 1.
A screening strategy for identifying new regulators: The acs promoter region was cloned upstream of the luxCDABE operon on a low-copy number plasmid. This plasmid is transformed into 3555 single knockout mutants of E. coli (top). The transformed bacteria were spotted onto solid LB-agar medium and the luminescence of the colonies was quantified (LB day 1) (middle). The histogram (bottom) shows the observed distribution of luminescence emitted by all colonies. The red vertical lines indicate two standard deviations above and below the mean. The names of the mutants with luciferase activity differing by more than two standard deviations from the mean are listed in Supplementary Information S4. The coefficient of variation (CV) of the observed distribution, obtained by dividing the standard deviation by the mean, equals 0.38.
The luminescence of the colonies changes as a function of culture time and growth condition. The powerful features of our screen are that we can monitor the dynamics of gene expression at the colony level and find the growth conditions that are most relevant to the gene under investigation (see Supplementary Information S1 and S2, and Supplementary Data S1). The acs promoter is activated after exhaustion of the preferred carbon source. We therefore measured the luminescence intensities of colonies after 24 and 48 h of growth on two different media: rich LB medium and minimal glucose medium. We thus obtained four complete sets of luminosity images of the plates and quantified the luminescence intensities of the 3555 colonies using a custom-made Matlab program (see ‘Materials and Methods’ section). Figure 1 shows the corresponding histogram of luminescence intensities after 24 h of growth on LB-agar plates (LB Day 1, the complete data sets are available as Supplementary Data S2 and S3). Among the 3555 transformants, 87 auxotrophic mutants did not grow on minimal medium-agar-glucose (Supplementary Information S3).
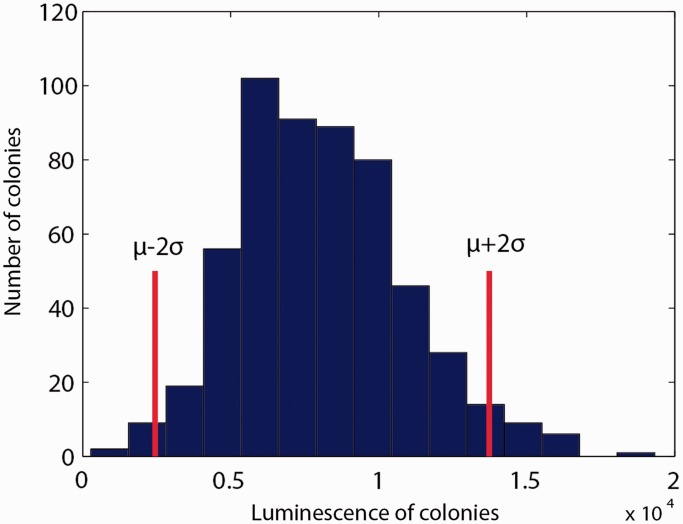
Figure 1 shows that the measured luminescence intensities are broadly distributed, with 165 mutants having an expression level that differs by more than two standard deviations from the mean, which is close to the WT expression level (Supplementary Information S4 lists these mutants). To check if this observation also holds for other promoters, we constructed a transcriptional fusion of the sdhC promoter region (708 bp) to the luxCDABE operon in the same plasmid backbone. The sdhC promoter directs the transcription of the sdhCDAB-sucABCD operon, which encodes enzymes involved in the TCA cycle and the respiratory chain (2). In particular, the sdh transcripts encode the succinate dehydrogenase complex and the suc transcripts the 2-oxoglutarate dehydrogenase and succinyl-CoA synthetase complexes. We transformed 15% of the Keio collection (552 transformants) with the p-sdhC-lux plasmid and, after spotting the strains on solid LB-agar medium, recorded the luminescence intensities observed on Day 1. Like for acs, we obtained a large variability of luminescence intensities, with similar coefficients of variation (0.38 for acs and 0.37 for sdhC, see Figure 2).
Figure 2.
Histogram with luminescence intensities obtained in partial screen for a second promoter: The sdhC promoter region was cloned into the same plasmid backbone as used for acs, giving rise to p-sdhC-lux. The luminescence emitted by colonies grown on solid LB-agar medium was quantified, as described in ‘Materials and Methods’ section. The histogram shows the results on Day 1 (LB day 1). As for acs, the activity is broadly distributed (552 mutants considered), with a coefficient of variation (CV) equal to 0.37.
Among the mutants with the lowest luciferase activity in Figure 1, we find genes involved in energy supply (atpC, atpD, atpE, atpF, atpG, sucA, sucB, sdhB, lpdA) and in catabolite repression (cyaA, crp, crr, ptsI). Among the mutants with the highest luciferase activity, our screen detects a transcription regulator (hha) whose deletion has been shown to affect catabolite repression (23) and the genes of the nuo operon, which encode the proton-translocating NADH dehydrogenase (24). Application of Fisher’s exact test shows that genes in carbon and energy metabolism (MultiFun category 1.3) are significantly overrepresented among the 155 mutants leading to a low or high activity of the acs promoter (P < 10−9). Transcription factors (MultiFun category 3.1.2) are also overrepresented among the mutants in the tails of the histogram in Figure 1 (P < 0.05).
Reproducibility of the results of the screen
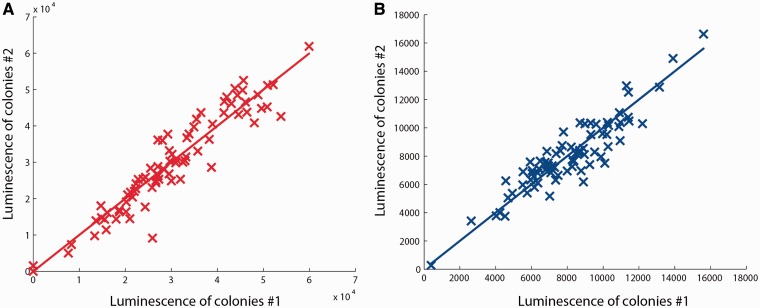
The observed broad distribution of luminescence intensities in different mutant backgrounds suggests that many genes, directly or indirectly, affect acs transcription. To verify that the broad distribution is not due to a lack of precision of the measured luciferase activity, we twice repeated the screen for 79 randomly selected mutants from the Keio collection growing on solid LB-agar medium. Figure 3A shows a scatter plot of the luminescence signals emitted by the colonies on Day 1 in the two replicate experiments. As can be seen, the reproducibility of the results is high, in the sense that the measured intensities in the two experiments are strongly correlated (correlation coefficient r = 0.94). A similar result, shown in the same plot, is obtained for the second reporter plasmid (p-sdhC-lux, r = 0.91).
Figure 3.
Reproducibility of the results of the screening strategy: (A) Scatter plot of luminescence intensities (red crosses) emitted by colonies of 79 randomly selected mutants from the Keio collection, grown on solid LB-agar medium and carrying the plasmid p-acs-lux, for two independent repetitions of the screening method (LB Day 1). (B) Idem for 77 mutants carrying p-sdhC-lux plasmid (blue crosses).
The results in Figure 3 allow us to compute an estimate of the variance of the measurement error, as explained in the ‘Materials and Methods’ section, and thus to estimate the relative contribution of the measurement error to the observed variance of the expression levels. The error variance was found to account for only 6% of the total variance of the acs expression levels observed in Figure 3A (9% for sdhC, Figure 3B). A similar observation can be made for the entire screen. The error variance contributes only 8% to the total variance of acs expression in Figure 1. We therefore conclude that the broad distribution of luminescence intensities is not due to measurement uncertainty, but mainly reflects differences in acs transcription in mutant backgrounds.
The dynamics of acs expression
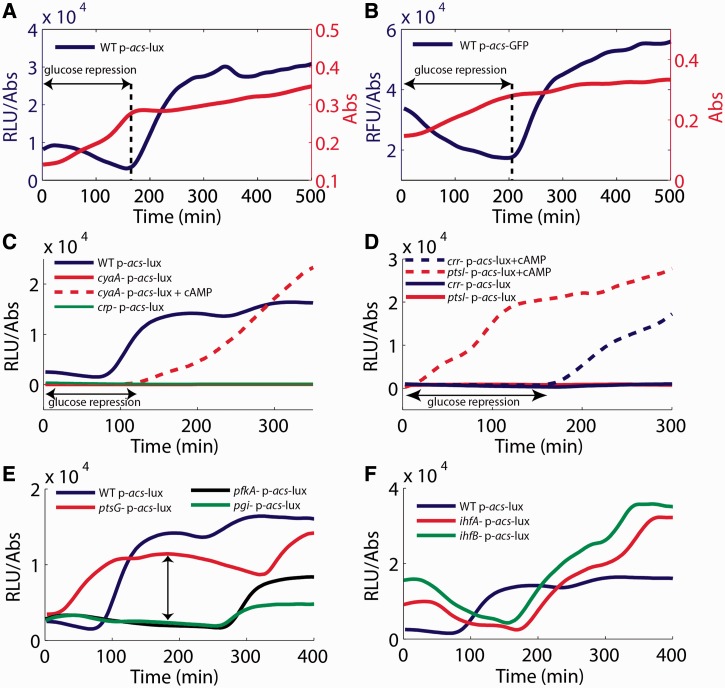
We selected 517 colonies (15% of the transformed mutant strains) with luminescence intensities significantly different from the WT level, in at least one of the four conditions, for a detailed kinetic analysis. We measured the dynamics of acs expression in these mutants in liquid cultures grown in minimal medium containing glucose and acetate, to focus on the physiologically important transition between those two carbon sources. The expression patterns were measured in an automated microplate reader and analyzed as previously described (20). Figure 4A shows the expression profile of acs in the WT strain. We observe a sharp increase of acs transcription when glucose is exhausted, in agreement with previous reports (12).
Figure 4.
Dynamics of gene expression of known mutants in liquid minimal medium containing glucose and acetate: (A) The expression profile of the acs luciferase reporter plasmid shows that acs expression is induced when glucose is exhausted. (B) The acs expression profile using GFP as a reporter is identical to the profile observed with luciferase (A). (C) acs expression is completely absent in crp and cyaA strains. cAMP added to the medium (5 mM) at the beginning of the experiment complements the cyaA strain but glucose repression is maintained. (D) Low expression of acs in the crr and ptsI strains. cAMP added to the medium at the beginning of the experiment directly complements the ptsI strain during growth on glucose, whereas the crr strain only responds once glucose is exhausted. (E) In the ptsG mutant, acs induction is delayed because glucose is consumed more slowly. Moreover, acs expression during glucose consumption is much stronger than in the WT strain. The glycolysis mutants pgi and pfkA also grow more slowly, but acs expression remains low in exponential phase. (F) The expression profiles of the ihfA and ihfB strains show stronger acs expression than the WT during growth on acetate. The highly similar expression kinetics of the two mutants (with genes located at different sites on the chromosome) shows the high internal consistency of the method.
The emission of light by luminescent reporters does not only depend on transcription from the acs promoter, but is also influenced by the metabolic state of the cell, such as concentrations of oxygen, FMNH2, ATP and NADPH. These metabolites are involved in the enzymatic reactions that lead to light emission (25). The nuo operon (13 genes) is a striking example of these metabolic effects on luciferase activity: all corresponding colonies shown are more luminescent on LB plates (Supplementary Information S5), an effect also observed for the sdhC promoter. The nuo operon codes for the NADH dehydrogenase involved in the respiratory chain. In the absence of a functional enzyme, reducing power accumulates (26), thus stimulating the luciferase reaction. To verify that the WT expression pattern is robust and independent of the reporter system, we also measured acs activity by means of a plasmid carrying a transcriptional fusion of the promoter region to a gfp reporter gene and having a different origin of replication (p-acs-gfp, see ‘Materials and Methods’ section). Figure 4B shows that an identical expression profile is obtained with this second reporter in a WT strain.
To systematically detect any artifacts due to metabolic effects affecting light production rather than acs expression, and to control for mutants affecting the plasmid copy number, we transformed all of the above-mentioned 517 mutants with the p-acs-gfp plasmid. We acquired the expression profile of acs for all mutants using the two reporter systems. Mutants for which acs activity is different in the two reporter systems likely represent artifacts of at least one of the reporter systems (Supplementary Information S6). We thus excluded 14 mutants with acs expression in a gfp reporter plasmid and no induction in a lux reporter, and 9 mutants, which for unknown reasons show no acs expression in a gfp reporter plasmid. Although fluorescence reporters are useful for detecting artifacts, note that luminescence is preferable for screening. Bacteria have a significant autofluorescence background, probably due to flavin mononucleotide or riboflavin (27), which reduces the sensitivity of the screen.
Confirmation of known regulators of acs expression
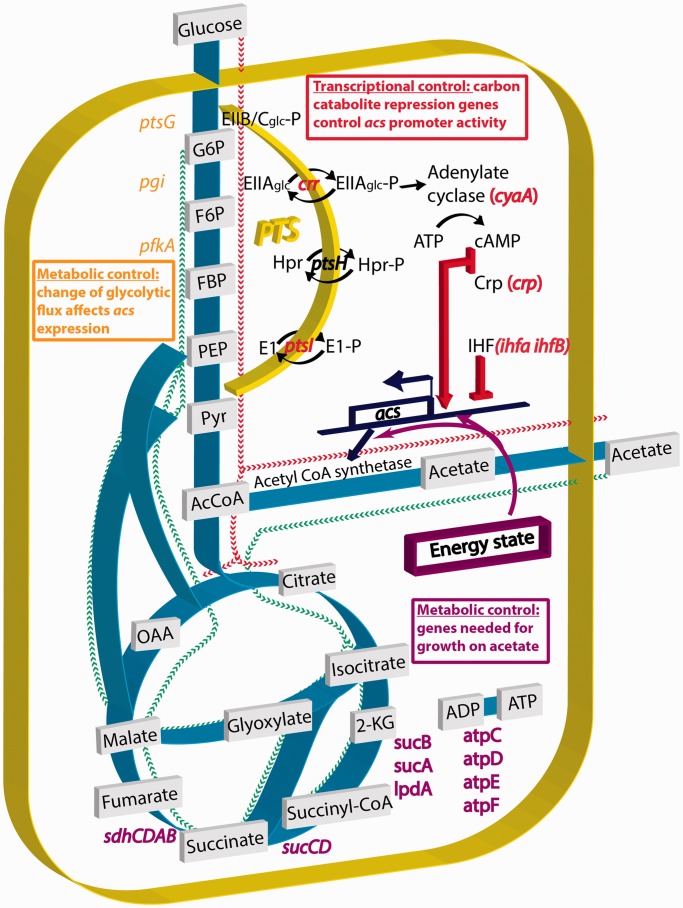
An important criterion for the validity of a screen is the confirmation of known interactions. The acs gene is positively regulated by the CRP–cAMP complex (crp gene) and thus by the well-studied carbon catabolite repression mechanism. The influx of glucose through the PTS controls adenylate cyclase (cyaA gene) activity and thus cAMP production. The PTS is composed of enzymes EI (ptsI gene), HPR (ptsH gene), EIIAglc (crr gene) and EIIB/Cglc (the glucose transporter encoded by the ptsG gene). When glucose is exhausted, the four PTS enzymes are phosphorylated and EIIAglc binds to and activates adenylate cyclase. The concentration of cAMP quickly increases and the CRP–cAMP complex activates target promoters such as the acs promoter (17) (Figure 6).
Figure 6.
The regulation of acs expression. The expression of acs is regulated by genes involved in glucose repression (cyaA, crp, crr and ptsI). A modification of the glycolytic flux has indirect consequences on acs expression (ptsG, pgi, pfkA). Genes of the TCA cycle are needed for acetate utilization and mutants in these enzymes shut down acs transcription by a yet unknown mechanism. The red and green routes are schematic representations of the flux direction when cells grow on glucose and acetate, respectively.
The analysis of the genes involved in glucose repression confirms that our screen reliably identifies dramatic, but also subtle changes in the regulation of the target gene. In the cyaA and crp strains, there is no transcription of acs at all, demonstrating the strict dependence of acs transcription on the CRP–cAMP complex. The activity of the acs promoter can be restored in the cyaA strain by adding cAMP to the medium but, interestingly, only after glucose exhaustion (Figure 4C). A similar phenomenon was observed before for other E. coli promoters (18). The crr and ptsI strains show the expected phenotype: low acs expression, although significantly higher than in the cyaA and crp strains. cAMP added to the medium clearly restores acs expression in these two strains, confirming that the mutants cannot activate adenylate cyclase. As in the cyaA strain, cAMP fails to complement the crr strain, while glucose is still present in the growth medium. In contrast, the ptsI strain is directly complemented by cAMP even in presence of glucose (Figure 4D). This difference of reactivity is not explained by the current model of catabolite repression: adding cAMP to the medium should bypass the glucose repression. Consequently, the cyaA and crr mutants should behave as the ptsI strain, with a direct activation of acs even in presence of glucose.
Expression of acs is reported to be negatively regulated by two histone-like proteins Fis and IHF (28). The Fis protein is abundant in exponential phase (29). Because acs expression is already low in this growth phase, the observed effect of the fis deletion is rather subtle: the acs expression profile is rather similar to that of the WT strain (data not shown). IHF acts mainly in stationary phase (12) and we accordingly observe a higher acs expression in late stationary phase (Figure 4F). The perfect superposition of acs expression profiles observed in the ihfA and ihfB strains (each coding for a subunit of IHF and transcribed from different chromosomal locations) confirms the high accuracy and internal consistency of the method.
As a validation of the results, we measured the transcription of several genes with an independent gene expression assay, quantitative RT-PCR. Mutants in genes coding for the PTS (ptsG, crr, ptsI), cAMP production (cyaA) and regulatory proteins (ihfB) precisely reproduce the effects observed with the reporter genes (Figure 5B).
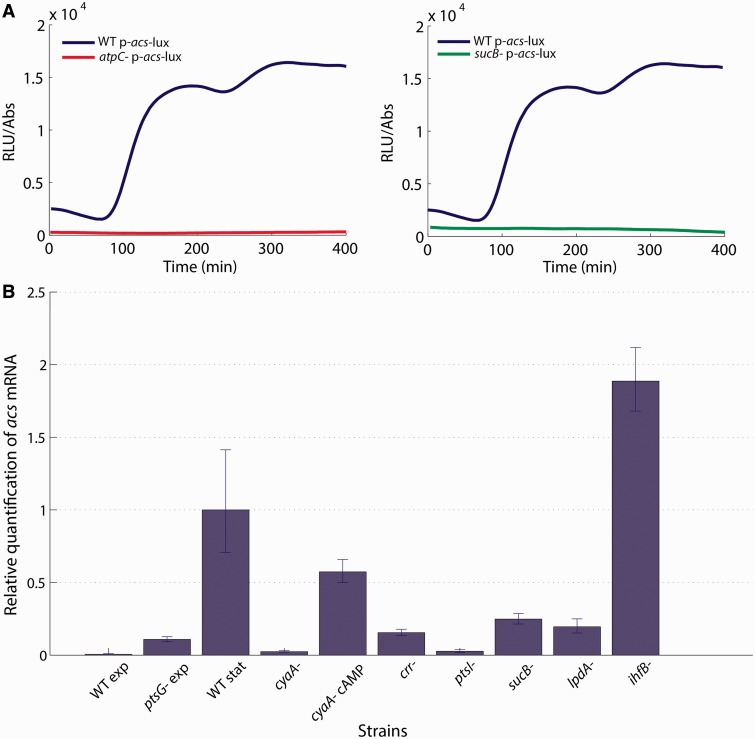
Figure 5.
Validation of existing and novel regulators of acs using qRT-PCR: (A) The atpC and sucB mutants do not induce acs (are not luminescent) when glucose is exhausted, an effect confirmed by the fluorescent reporter (Supplementary Information S6). (B) Direct measurement of relative mRNA levels of selected mutants using qRT-PCR. All mRNA levels were measured 30 min after glucose exhaustion except for the first two samples, marked WT exp and ptsG exp, which were measured during growth on glucose. All concentrations were normalized to the expression level of the induced WT strain. The acs mRNA is almost undetectable in the WT strain in exponential phase, whereas the ptsG mutant shows a clear signal. No acs expression is observed in the cyaA mutant (cyaA), but expression is restored when the medium is complemented with cAMP (cyaA cAMP). As expected, the crr and ptsI strains show weaker acs expression and the ihfB mutant stronger acs expression in comparison with the WT strain. The effect of the sucB and lpdA mutants on acs expression, revealed by the screen, are confirmed by qRT-PCR.
Metabolic regulation of acs expression
An interesting observation is that many of the regulators of acs expression returned by the screen are genes encoding enzymes in carbon metabolism. This reminds that gene expression in bacteria is tightly interlinked with the metabolic state of the cell.
A first example of this connection revealed by the screen are mutants affecting glycolytic fluxes. According to the classical model of carbon catabolite repression, a diminution of glycolytic fluxes increases the phosphorylation of the PTS leading to an increase of the concentration of cAMP and ultimately the expression of acs (30). The ptsG mutant (which lacks the glucose transporter) follows the classical model: colonies grow more slowly on glucose in the ptsG strain than in the WT strain, and the dynamical analysis shows a higher acs expression level in exponential phase (Figure 4E). Interestingly, our analysis reveals that the deletion of downstream glycolytic enzymes leads to a different and counter-intuitive expression phenotype. For example, while pgi and pfkA mutants grow more slowly on glucose, acs is not derepressed in exponential phase (Figure 4E).
As a second example, several of the mutants with the lowest colony luminescence carry deletions of genes involved in the energy supply of the cell (Figure 1), such as the genes coding for ATP synthetase (atpC, atpD, atpE, atpF) and the genes coding for enzymes of the TCA cycle: the oxoglutarate dehydrogenase complex (sucA, sucB and lpdA) and, to a lesser extent, succinyl-CoA synthetase (sucCD) and succinate dehydrogenase (sdhCDAB). In these mutants, the luminescence profile shows an almost total absence of acs expression, as illustrated for the sucB and lpdA strains in Figure 5A. While it might be argued that this is due to a failure in energy supply, the effect is reproduced with the fluorescence reporter (Supplementary Information S6), thus suggesting the existence of genuine, hitherto unknown interactions. This metabolic regulation of acs was confirmed by qRT-PCR, as we observed a significantly lower expression level of acs in the sucB and lpdA mutants (Figure 5B). Interestingly, we found that the above mutants also fail to grow on acetate (Supplementary Information S7).
DISCUSSION
A new method for identifying the regulators of a target gene
The usual way to dissect a regulatory network, closely linked to classical approaches in genetics, consists in disrupting a gene of interest and observing the effects on the organism. This approach identifies the downstream targets of the gene of interest. Here, we have developed a method to do the opposite: given a gene of interest, we identify all factors, genetic and metabolic, that affect the expression of this gene. Our method relies on high-throughput transformation of a luciferase reporter plasmid into an extensive mutant collection. In comparison with earlier work on the characterization of regulators of the activation of a protein complex, which was based on a similar principle (6), we follow expression of a target gene in vivo and dynamically overtime. Even though we have focused on transcriptional regulation, the method can certainly be extended to include translational fusions. We use the Keio mutant collection, but the scope of the screen could be extended to include, for example, mutants of non-coding RNAs and intergenic regions possibly containing unknown open reading frame (ORFs). The increasing availability of mutant collections (31–33) makes our method applicable for other organisms as well.
We have chosen luciferase as a reporter because it allows easy screening at the colony level without a significant background signal and gives highly reproducible results (Figure 3). The enzymatic reaction of luciferase depends on cellular factors such as ATP concentration or redox potential. For a given medium, deletions that seriously affect the concentration of these factors may generate false positives. We have used a second (fluorescent) reporter system to identify mutants giving rise to such artifacts (Supplementary Information S6). These mutants should be excluded in any future application of the screen.
An important novel feature of our screen is the possibility to directly measure the dynamics of gene expression during the screen or by high-throughput measurements of selected mutants in an automated plate reader. Furthermore, the original screen, as well as these additional analyses, can be carried out in many different environmental and growth conditions.
The regulation of acs transcription
The effects of some interesting mutants identified by the screen are summarized in Figure 6. We have confirmed known interactions that regulate the transcription of acs and we have discovered interesting new directions to explore. The acs promoter activity is controlled by IHF and, most importantly, the CRP–cAMP complex: transcription is completely abolished without CRP or cAMP. We also confirm the involvement of the PTS, which sets the intracellular concentration of cAMP: the crr and ptsI mutants have limited acs expression except when cAMP is added to the medium. Interestingly, even though part of the same glucose uptake machinery, we found that the ptsG mutant behaves differently from the other PTS mutants: it does not require the addition of exogenous cAMP to activate the acs expression.
We failed to induce acs expression in cyaA or WT strains by adding cAMP while glucose was still present in the growth medium. Those results could possibly be explained by another well-known mechanism of glucose repression: inducer exclusion. In this control system, the dephosphorylated form of EIIAglc, the dominant form in the presence of glucose, binds and inhibits different sugar transport systems, thus preventing the entry of the inducing sugar into the cell (34). However, inducer exclusion does not account for the glucose repression of the acs transcription in the presence of exogenous cAMP. To date, no inducer of acs expression has been described, and acetate can enter the cell by passive diffusion (35). Furthermore, the central component of inducer exclusion is EIIAglc, encoded by the crr gene. Yet, we still observe glucose repression in a crr mutant in the presence of exogenous cAMP (Figure 4D). This experiment demonstrates that neither variations of cAMP concentration nor variations of the phosphorylated state of EIIAglc are sufficient to explain the glucose repression that we observe. Consequently, while confirming the involvement of the CRP–cAMP complex and the PTS in the transcription of the acs gene, our results show that the current two main models of carbon catabolite repression are incomplete (18). Despite more than four decades of research, our understanding of the precise mechanism that underlie the glucose effect (and more generally the carbon catabolite repression) is still far from complete and remains the subject of debate (36–39).
Metabolism and gene expression
An important result of the screen for regulators of acs, which was confirmed for sdhC, is that a large number of mutants affect the expression level of the target gene. Many of these mutants are genes involved in metabolism, thus revealing novel connections between the metabolic state of the cell and gene expression. For instance, enzymes involved in growth on acetate have a genuine impact on acs expression, as verified by means of fluorescent reporters and qRT-PCR (sucB, lpdA, … ). Even though such regulatory interactions make sense from a physiological point of view, the mechanisms relating the expression of these enzymes remain to be discovered. Another example involves the glycolytic flux. Reducing glucose inflow by deleting the glucose transporter (ptsG gene) increases the basal acs expression level during growth on glucose, probably by increasing the intracellular concentration of cAMP. However, the reduction of the glycolytic flux observed during growth of the pgi and pfkA mutant strains does not lead to an enhancement of acs expression in exponential phase (Figure 4E). This result shows that neither acs promoter activity nor cAMP concentration are a simple function of the glycolytic flux, the latter being roughly the same for the ptsG, pgi and pfkA strains, but that they depend on the exact location where glycolysis is interrupted. We do not yet know the metabolic indicator nor the sensing mechanism that transmits the state of glycolysis to the activation of the acs promoter.
These different observations clearly show a strong connection between the metabolic state of the cell and acs transcription and confirm our prediction that the genetic regulatory network of E. coli is densely connected and that a majority of these connections pass through metabolism (40).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: EU FP6 grant EC-MOAN (FP6-2005-NEST-PATH-COM/043235), the ANR BioSys grant MetaGenoReg (ANR-06-BYOS-0003) and the Joseph Fourier University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Eugenio Cinquemani, Omaya Dudin, Stephan Lacour, Yves Markowicz, and Valentin Zulkower for useful discussion and comments on the manuscript. Laure Giannone helped with the analysis of the data.
REFERENCES
- 1.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 2.Keseler IM, Collado-Vides J, Santos-Zavaleta A, Peralta-Gil M, Gama-Castro S, Muñiz-Rascado L, Bonavides-Martinez C, Paley S, Krummenacker M, Altman T, et al. EcoCyc: a comprehensive database of Escherichia coli biology. Nucleic Acids Res. 2011;39:D583–D590. doi: 10.1093/nar/gkq1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown PO, Botstein D. Exploring the new world of the genome with DNA microarrays. Nat. Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 4.Grainger DC, Busby SJW. Methods for studying global patterns of DNA binding by bacterial transcription factors and RNA polymerase. Biochem. Soc. Trans. 2008;36:754–757. doi: 10.1042/BST0360754. [DOI] [PubMed] [Google Scholar]
- 5.Neidhardt FC, Savageau M. Escherichia coli and Salmonella: Cellular and Molecular Biology. Vol. 1. Washington, DC: ASM Press; 1996. pp. 1310–1324. [Google Scholar]
- 6.Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 2009;5:e1000515. doi: 10.1371/journal.pgen.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Dyk TK, Wei Y, Hanafey MK, Dolan M, Reeve MJ, Rafalski JA, Rothman-Denes LB, LaRossa RA. A genomic approach to gene fusion technology. Proc. Natl Acad. Sci. USA. 2001;98:2555–2560. doi: 10.1073/pnas.041620498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hakkila K, Maksimow M, Karp M, Virta M. Reporter genes lucFF, luxCDABE, gfp, and dsred have different characteristics in whole-cell bacterial sensors. Anal. Biochem. 2002;301:235–242. doi: 10.1006/abio.2001.5517. [DOI] [PubMed] [Google Scholar]
- 9.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 11.Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolfe AJ. The acetate switch. Microbiol. Mol. Biol. Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosset G. Improvement of Escherichia coli production strains by modification of the phosphoenolpyruvate:sugar phosphotransferase system. Microb. Cell Fact. 2005;4:14. doi: 10.1186/1475-2859-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumari S, Beatty CM, Browning DF, Busby SJ, Simel EJ, Hovel-Miner G, Wolfe AJ. Regulation of acetyl coenzyme A syntheses in Escherichia coli. J. Bacteriol. 2000;182:4173–4179. doi: 10.1128/jb.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sclavi B, Beatty CM, Thach DS, Fredericks CE, Buckle M, Wolfe AJ. The multiple roles of CRP at the complex acs promoter depend on activation region 2 and IHF. Mol. Microbiol. 2007;65:425–440. doi: 10.1111/j.1365-2958.2007.05797.x. [DOI] [PubMed] [Google Scholar]
- 16.Shin S, Chang D-E, Pan JG. Acetate consumption activity directly determines the level of acetate accumulation during Escherichia coli W3110 growth. J. Microbiol. Biotechnol. 2009;19:1127–1134. doi: 10.4014/jmb.0902.0097. [DOI] [PubMed] [Google Scholar]
- 17.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat. Rev. Microbiol. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 18.Danchin A, Ullmann A. Role of cAMP in bacteria. Adv. Cyclic Nucleotide Res. 1983;15:1–53. [Google Scholar]
- 19.Yamamoto N, Nakahigashi K, Nakamichi T, Yoshino M, Takai Y, Touda Y, Furubayashi A, Kinjyo S, Dose H, Hasegawa M, et al. Update on the Keio collection of Escherichia coli single-gene deletion mutants. Mol. Syst. Biol. 2009;5:335. doi: 10.1038/msb.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Jong H, Ranquet C, Ropers D, Pinel C, Geiselmann J. Experimental and computational validation of models of fluorescent and luminescent reporter genes in bacteria. BMC Syst. Biol. 2010;4:55. doi: 10.1186/1752-0509-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung CT, Niemela SL, Miller RH. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc. Natl Acad. Sci. USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol. Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balsalobre C, Johansson J, Uhlin BE, Juárez A, Muñoa FJ. Alterations in protein expression caused by the hha mutation in Escherichia coli: influence of growth medium osmolarity. J. Bacteriol. 1999;181:3018–3024. doi: 10.1128/jb.181.10.3018-3024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falk-Krzesinski HJ, Wolfe AJ. Genetic analysis of the nuo locus, which encodes the proton-translocating NADH dehydrogenase in Escherichia coli. J. Bacteriol. 1998;180:1174–1184. doi: 10.1128/jb.180.5.1174-1184.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koga K, Harada T, Shimizu H, Tanaka K. Bacterial luciferase activity and the intracellular redox pool in Escherichia coli. Mol. Genet. Genomics. 2005;274:180–188. doi: 10.1007/s00438-005-0008-5. [DOI] [PubMed] [Google Scholar]
- 26.Prüss BM, Nelms JM, Park C, Wolfe AJ. Mutations in NADH:ubiquinone oxidoreductase of Escherichia coli affect growth on mixed amino acids. J. Bacteriol. 1994;176:2143–2150. doi: 10.1128/jb.176.8.2143-2150.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Billinton N, Knight AW. Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal. Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 28.Browning DF, Beatty CM, Sanstad EA, Gunn KE, Busby SJW, Wolfe AJ. Modulation of CRP-dependent transcription at the Escherichia coli acsP2 promoter by nucleoprotein complexes: anti-activation by the nucleoid proteins FIS and IHF. Mol. Microbiol. 2004;51:241–254. doi: 10.1046/j.1365-2958.2003.03824.x. [DOI] [PubMed] [Google Scholar]
- 29.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogema BM, Arents JC, Bader R, Eijkemans K, Inada T, Aiba H, Postma PW. Inducer exclusion by glucose 6-phosphate in Escherichia coli. Mol. Microbiol. 1998;28:755–765. doi: 10.1046/j.1365-2958.1998.00833.x. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherens B, Goffeau A. The uses of genome-wide yeast mutant collections. Genome Biol. 2004;5:229. doi: 10.1186/gb-2004-5-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewenza S, Falsafi RK, Winsor G, Gooderham WJ, McPhee JB, Brinkman FSL, Hancock REW. Construction of a mini-Tn5-luxCDABE mutant library in Pseudomonas aeruginosa PAO1: a tool for identifying differentially regulated genes. Genome Res. 2005;15:583–589. doi: 10.1101/gr.3513905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roseman S, Meadow ND. Signal transduction by the bacterial phosphotransferase system. Diauxie and the crr gene (J. Monod revisited) J. Biol. Chem. 1990;265:2993–2996. [PubMed] [Google Scholar]
- 35.Gimenez R, Nuñez MF, Badia J, Aguilar J, Baldoma L. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 2003;185:6448–6455. doi: 10.1128/JB.185.21.6448-6455.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crasnier-Mednansky M. Is there any role for cAMP-CRP in carbon catabolite repression of the Escherichia coli lac operon? Nat. Rev. Microbiol. 2008;6:954. doi: 10.1038/nrmicro1932-c1. author reply 954. [DOI] [PubMed] [Google Scholar]
- 37.Görke B, Jörg S. Is there any role for cAMP–CRP in carbon catabolite repression of the Escherichia coli lac operon? Reply from Görke and Stülke. Nat. Rev. Microbiol. 2008;6:954. doi: 10.1038/nrmicro1932-c1. [DOI] [PubMed] [Google Scholar]
- 38.Inada T, Kimata K, Aiba H. Mechanism responsible for glucose-lactose diauxie in Escherichia coli: challenge to the cAMP model. Genes Cells. 1996;1:293–301. doi: 10.1046/j.1365-2443.1996.24025.x. [DOI] [PubMed] [Google Scholar]
- 39.Narang A. Quantitative effect and regulatory function of cyclic adenosine 5′-phosphate in Escherichia coli. J. Biosci. 2009;34:445–463. doi: 10.1007/s12038-009-0051-1. [DOI] [PubMed] [Google Scholar]
- 40.Baldazzi V, Ropers D, Markowicz Y, Kahn D, Geiselmann J, De Jong H. The carbon assimilation network in Escherichia coli is densely connected and largely sign-determined by directions of metabolic fluxes. PLoS Comput. Biol. 2010;6:e1000812. doi: 10.1371/journal.pcbi.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.