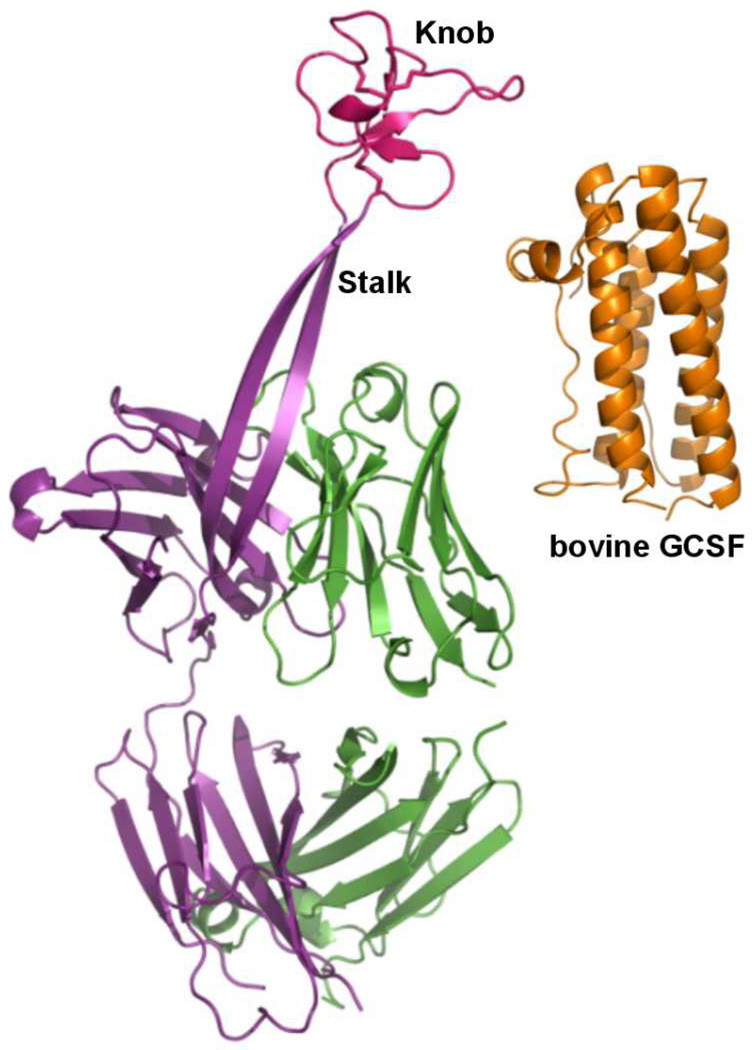
Most mammalian antibodies have complementarity determining region (CDR) loops of 8–16 residues, while some human antibodies have longer, protruding CDR loops that play a role in virus neutralization.[1] The bovine antibody repertoire features a subgroup of antibodies with exceptionally long heavy chain complementarity determining region 3 (CDR3H) loops. The lengths of CDR3H in these bovine antibodies range from 40 to 67 amino acid residues.[2] We recently solved the X-ray crystal structure of bovine antibody BLV1H12 which has a CDR3H of 61 residues (Figure 1).[3] The crystal structure revealed a novel CDR3H structure that forms a solvent exposed 2-stranded antiparallel sheet ~20 Å (7 amino acids) in length (the stalk region). This β-sheet terminates in a folded protein domain that is stabilized by three pairs of disulfide bonds (the knob region). This knob region was shown to play a critical role in recognition of a viral antigen in the case of one such bovine antibody.[3] Herein we asked whether the knob region of BLV1H12 can be substituted with other native protein and peptide ligands to create chimeric antibodies with new or enhanced properties. For example, such fusion proteins may have improved serum half-lives relative to the native polypeptide,[4] improved binding affinity or specificity due to interactions of the cognate receptor with the other bovine CDRs (or simply through bivalent binding), or improved expression and/or stability in the context of the antibody framework. To begin to explore these possibilities, we first attempted to substitute bovine granulocyte colony-stimulating factor (bGCSF) for the knob region in CDR3H of antibody BLV1H12.[5] The resulting bovine antibody-bGCSF (Ab-bGCSF) fusion protein was stably expressed in mammalian cells, exhibits potency similar to bGCSF in proliferating GCSF-dependent cells, and can significantly increase and sustain neutrophil populations for over three weeks in rodents.
Figure 1.
X-ray crystal structures of bovine antibody BLV1H12 Fab fragment (PDB ID: XXX) and bGCSF (PDB ID: 1BGC).
GCSF is a 20 kDa cytokine known to augment antimicrobial defenses by stimulating the production of white blood cells, and is used to accelerate recovery from neutropenia after chemotherapy.[6] Because GCSF has a relatively short plasma half-life (2–4 hours),[7] polyethylene glycol (PEG)- and Fc-fused GCSF variants have been synthesized with improved half-lives.[8] Similarly, we hypothesized that substitution of GCSF into CDR3H of BLV1H12 should also extend its half-life due to the large size of the chimeric protein and its binding to neonatal Fc-receptor (FcRn).[9] Fusion of bGCSF into the CDR3H may also prevent proteolysis by circulating exopeptidases. To synthesize a chimeric GCSF-BLV1H12, the BLV1H12 Fab fragment was first genetically fused with human IgG1 Fc to create a chimeric BLV1H12 full-length IgG. The gene encoding bGCSF was then inserted into the CDR3H region by fusing the N- and C-terminal of bGCSF with the ascending and descending β-strands of the stalk, respectively, to replace the knob domain (Cys108-Tyr146) (Figure S1A). To optimize folding and stability of the antibody-bGCSF fusion protein, a flexible GGGGS linker was also inserted at each end of the bGCSF fragment to increase flexibility. The resulting antibody-bGCSF fusion proteins with and without the N- and C-terminal linkers were designated as Ab-bGCSF L1 and Ab-bGCSF L0, respectively. As a control, bGCSF with a C-terminal 6×His-tag was cloned into the pFuse vector for mammalian expression and purification.
Full-length BLV1H12 IgG, the Ab-bGCSF fusion proteins and His-tagged bGCSF were transiently expressed in freestyle HEK293 cells and secreted into culture medium. Purified IgGs and His-tagged bGCSF were analyzed by SDS-PAGE gel (Figure S1B). Under non-reducing conditions, bovine BLV1H12 full-length IgG (Ab) migrates as a single band above 160 kDa due to N-glycosylation in the Fc region, and bGCSF migrates at 20 kDa. The BLV1H12-bGCSF fusion proteins with or without linker (Ab-bGCSF L1 and Ab-bGCSF L0) migrate as single bands above 190 kDa under non-reducing conditions. In the presence of 50 mM dithiothreitol (DTT), the light chains of Ab, Ab-bGCSF L0 and Ab-bGCSF L1 migrate at 23 kDa. The heavy chains of Ab and the Ab-bGCSF fusion proteins migrate at 55 kDa and 70 kDa, respectively (Figure S1B). Mass spectral analysis following treatment with peptide-N-glycosidase and DTT indicates mass increases of 15650 Da for the heavy chain of Ab-bGCSF L0 and 16280 Da for the heavy chain of Ab-bGCSF L1 relative to that of Ab (Figure S2–S5). This corresponds to two fused bGCSF proteins per IgG. The final yields of Ab, Ab-bGCSF L0, Ab-bGCSF L1 and His-tagged bGCSF are similar (~17 mg/L), and both the Ab and the Ab-bGCSF fusion proteins have solubility over 10 mg/mL in PBS (pH 7.4). These results are consistent with correct folding of the fusion proteins. In addition, the similar yields and solubilities of the Ab-bGCSF L0 and Ab-bGCSF L1 fusion proteins suggests that the GGGGS linkers do not significantly affect the expression or stability of fusion proteins.
Next, we examined the proliferative activities of the Ab-bGCSF fusion proteins using mouse NFS-60 cells which are responsive to GCSF stimulation.[10] The Ab-bGCSF fusion proteins proliferate NFS-60 cells in a dose-dependent manner (Figure 2), whereas Ab by itself has no proliferative activity, indicating that the observed activities of the Ab-bGCSF fusion proteins result from the grafted bGCSF. The potencies of the Ab-bGCSF fusion proteins (EC50: 3.1 ± 1.7 ng/mL for Ab-bGCSF L0, and 3.3 ± 1.8 ng/mL for Ab-bGCSF L1) are comparable to that of bGCSF and hGCSF (EC50: 1.6 ± 1.1 ng/mL for bGCSF, and 1.6 ± 0.2 ng/mL for hGCSF). Thus, grafting of bGCSF onto the protruding stalk of antibody BLV1H12 (Figure 1) does not appear to affect the activity of this cytokine. In addition, the Ab-bGCSF L0 and Ab-bGCSF L1 fusion proteins have nearly identical proliferative activities, again suggesting that the engineered flexible linkers are not required for folding and activity of the fused bGCSF.
Figure 2.
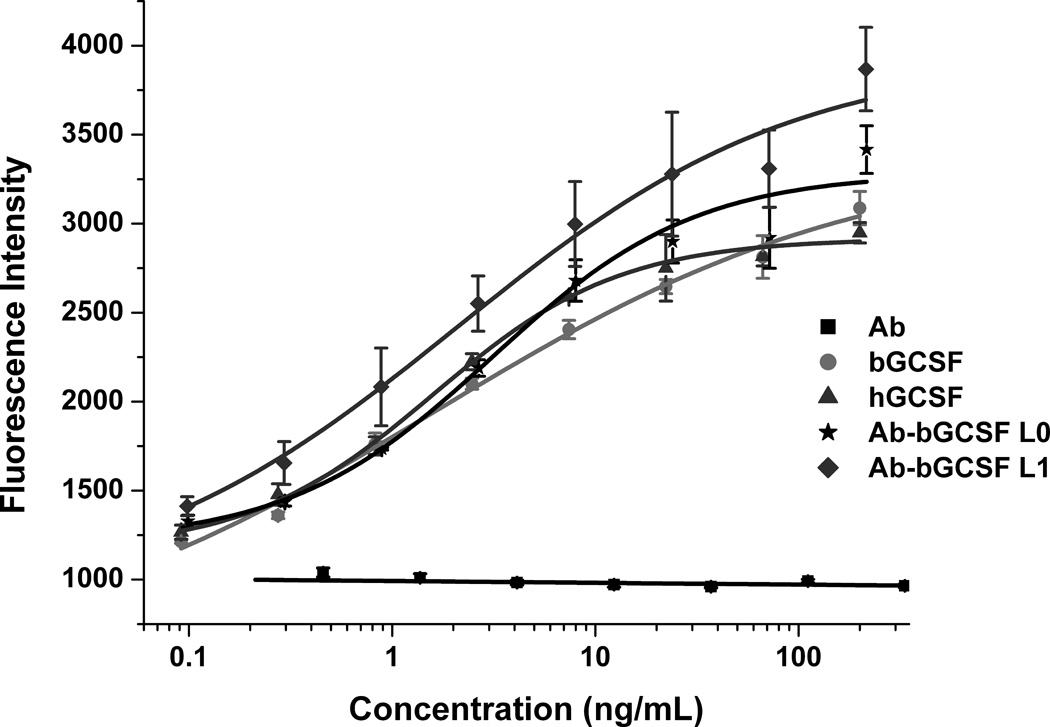
Ab-bGCSF fusion proteins stimulate proliferation of mouse NFS-60 cells in a dose-dependent manner. Cells were treated with various concentrations of bGCSF, hGCSF, BLV1H12 full-length IgG (Ab), Ab-bGCSF L0, and Ab-bGCSF L1. Cell viability was quantified using an AlamarBlue (Invitrogen) assay.
We next examined the ability of the Ab-bGCSF fusion proteins to stimulate granulocyte progenitors in cultures of human hematopoietic stem and progenitor cells.[11] Both Ab-bGCSF fusion proteins increase the number of cells in the granulocyte lineage in a dose-dependent manner, similar to bGCSF and hGCSF (Figure S6). Ab itself has no such stimulating activity on granulocyte progenitors. The EC50 values of Ab-bGCSF L0 and Ab-bGCSF L1 for human granulocyte progenitors are 11.4 ± 1.9 ng/mL and 14.6 ± 3.8 ng/mL, respectively. These activities are similar to that of hGCSF (EC50: 20.6 ± 3.1 ng/mL), but slightly lower than that of bGCSF (EC50: 1.9 ± 0.4 ng/mL), which possibly results from unfavorable interactions of the GCSF receptor with other CDR loops. The GGGGS linkers again had no significant effect on proliferative activity, consistent with the earlier results.
To determine if grafting GCSF into CDR3H of the bovine antibody increases serum half-life, we examined the pharmacokinetics (PK) of Ab, bGCSF and the Ab-bGCSF fusion proteins in mice. Intravenously injected bGCSF was quickly cleared within 24 hours, whereas Ab and the Ab-bGCSF fusion proteins remained above 20% of the maximal concentration even after 14 days (Figure 3). The estimated half-lives are 14 days for Ab, 10 days for Ab-bGCSF L0, 9 days for Ab-bGCSF L1, and 3 hours for bGCSF, by assuming a one-compartment model with first-order elimination. Thus, fusion of bGCSF into the ultralong CDR3H region significantly extends the plasma half-life of Ab-bGCSF fusion proteins.
Figure 3.
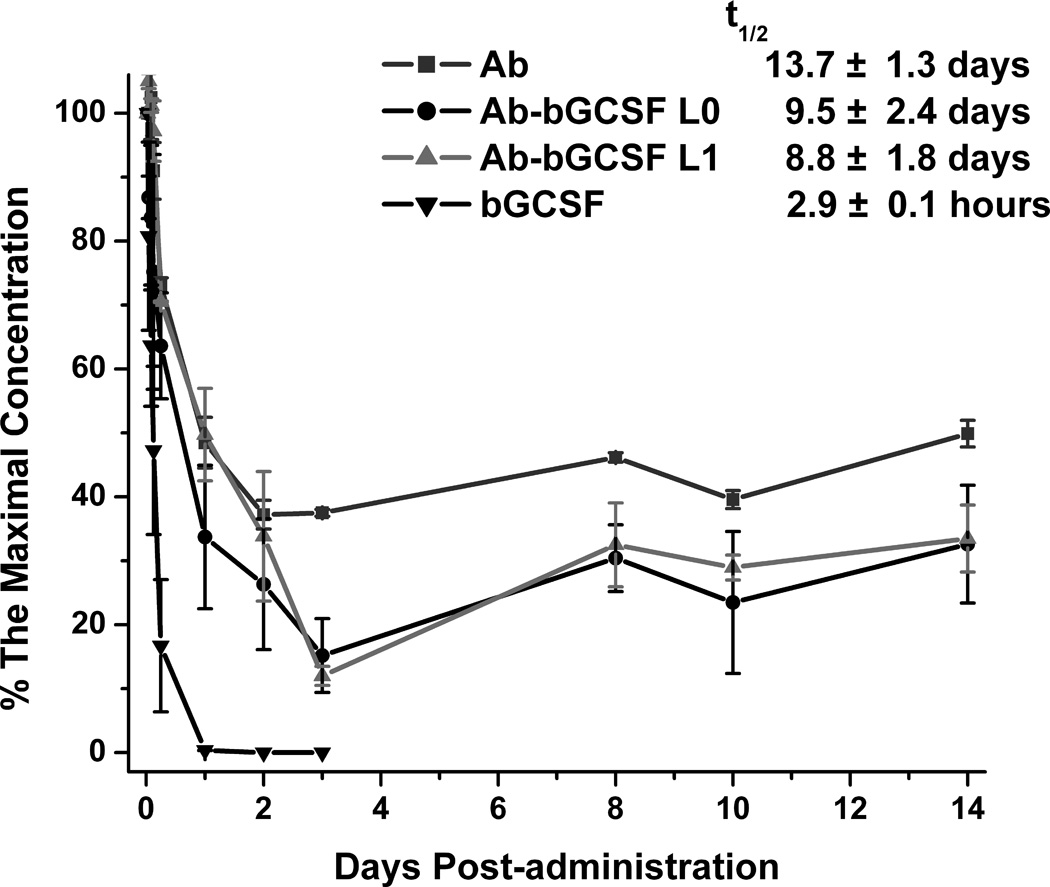
Pharmacokinetics in mice. bGCSF (8 mg/kg), BLV1H12 full-length IgG (Ab) (2.8 mg/kg), and Ab-bGCSF L0 and Ab-bGCSF L1 (2.8 mg/kg) were administrated by intravenous (i.v.) injection into BALB/c mice (three per group). Blood was collected at day 0 to day 14 and analyzed by ELISA using anti-human IgG Fc and anti-bGCSF antibodies. Data were normalized by taking maximal concentration at the first time point (30 minutes).
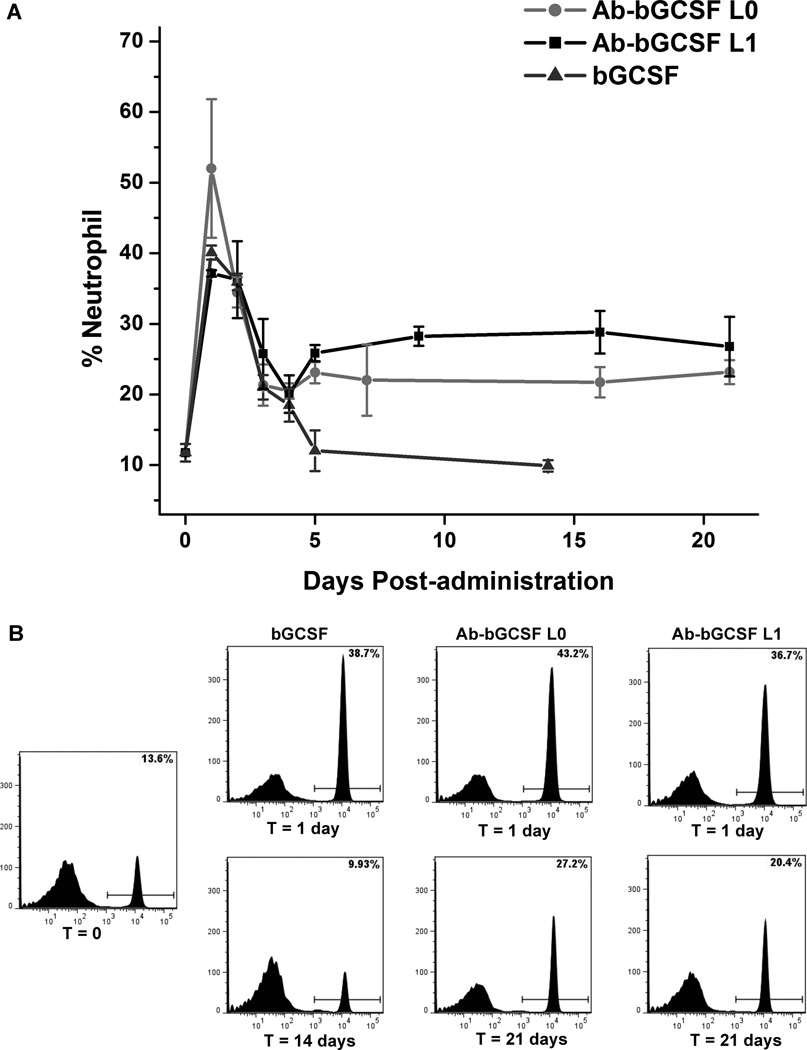
To test the activity of the Ab-bGCSF fusion proteins in vivo, white blood cells from treated mice were stained using Diff-Quick and counted under microscope to analyze neutrophil levels at day 10 after administration of Ab, bGCSF and Ab-bGCSF fusion proteins. As shown in Figure S7, neutrophil populations in white blood cells are 25% for mice treated with the Ab-bGCSF fusion proteins, which is 60% higher than that of the untreated group. For comparison, neutrophil levels in mice treated with bGCSF are 15%, which is similar to the untreated group. We next examined the time-dependent neutrophil-stimulating activity of the Ab-bGCSF fusion proteins in mice. Upon treatment with bGCSF and the Ab-bGCSF fusion proteins, flow cytometry revealed that the neutrophil populations in each group increased significantly at 24 hours (Figure 4A) — neutrophil numbers increased to 39% for bGCSF, 43% for Ab-bGCSF L0 and 37% for Ab-bGCSF L1 (Figure 4B). Thus, the Ab-bGCSF fusion proteins have similar potency to bGCSF in proliferating neutrophils in vivo, and again the GGGGS linkers do not appear to significantly affect activity. This suggests that linkers are not involved in modulating bGCSF interactions with its receptor through either a direct or indirect mechanism. Importantly, groups treated with single doses of the Ab-bGCSF fusion proteins exhibited sustained high levels of neutrophils (over 22% for L0 and 26% for L1) for more than 21 days. In comparison, the neutrophil populations in mice treated with bGCSF decreased to normal levels in 5 days, consistent with the short half-life of bGCSF. These results are consistent with the serum half-life measurements and demonstrate the ability to generate long-acting protein therapeutics by fusion of polypeptides into this novel ultralong CDR3H motif of bovine antibodies.
Figure 4.
Pharmacodynamics in mice. (A) Single doses of bGCSF (10 µg/kg) and Ab-bGCSF L0 and Ab-bGCSF L1 (50 µg/kg) were administrated by subcutaneous (s.c.) injection into BALB/c mice (3 per group). Blood was collected and analyzed for neutrophils by FACS using fluorophore-labeled anti-CD45, anti-CD11b, and anti-Ly-6G antibodies. (B) Representative flow cytometric analyses of mouse neutrophil populations after treatments with bGCSF, Ab-bGCSF L0 and Ab-bGCSF L1.
In conclusion, we have shown that the Ab-bGCSF fusion proteins, which are stably expressed in mammalian cells, significantly increase and sustain neutrophil populations for more than three weeks in rodents. Future experiments include structural characterization of the fusion proteins, fusion of other therapeutic cytokines, growth factors and peptide toxins into the ultralong CDR3H motif, humanization of the fusion proteins and mutagenesis of the other CDR loops of the fusion proteins to enhance activity and selectivity.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (NIH) Grant R01 GM062159 (to P.G.S.). This manuscript is number 23082 of The Scripps Research Institute.
Footnotes
Supporting information for this article is available on the WWW under http://www.angewandte.org
Contributor Information
Yong Zhang, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Danling Wang, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Lorenzo de Lichtervelde, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Sophie B. Sun, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA)
Vaughn V. Smider, Department of Molecular Biology, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA)
Peter G. Schultz, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
Feng Wang, Department of Chemistry, The Scripps Research Institute, 10550 N. Torrey Pines Road, La Jolla, CA 92037 (USA).
References
- 1.a) Saphire EO, Parren PW, Pantophlet R, Zwick MB, Morris GM, Rudd PM, Dwek RA, Stanfield RL, Burton DR, Wilson IA. Science. 2001;293:1155–1159. doi: 10.1126/science.1061692. [DOI] [PubMed] [Google Scholar]; b) Pejchal R, Walker LM, Stanfield RL, Phogat SK, Koff WC, Poignard P, Burton DR, Wilson IA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.a) Berens SJ, Wylie DE, Lopez OJ. Int. Immunol. 1997;9:189–199. doi: 10.1093/intimm/9.1.189. [DOI] [PubMed] [Google Scholar]; b) Lopez O, Perez C, Wylie D. Immunol. Rev. 1998;162:55–66. doi: 10.1111/j.1600-065x.1998.tb01429.x. [DOI] [PubMed] [Google Scholar]; c) Saini SS, Allore B, Jacobs RM, Kaushik A. Eur. J. Immunol. 1999;29:2420–2426. doi: 10.1002/(SICI)1521-4141(199908)29:08<2420::AID-IMMU2420>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]; d) Saini SS, Farrugia W, Ramsland PA, Kaushik AK. Int. Immunol. 2003;15:845–853. doi: 10.1093/intimm/dxg083. [DOI] [PubMed] [Google Scholar]; e) Zhao Y, Jackson SM, Aitken R. Dev. Comp. Immunol. 2006;30:175–186. doi: 10.1016/j.dci.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 3.Wang F, Ekiert DC, Ahmad I, Li W, Zhang Y, Bazirgan O, Torkamani A, Raudsepp T, Mwangi W, Criscitiello MF, Wilson IA, Schultz PG, Smider VV. Cell. 2013 doi: 10.1016/j.cell.2013.04.049. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Lobo ED, Hansen RJ, Balthasar JP. J. Pharm. Sci. 2004;93:2645–2668. doi: 10.1002/jps.20178. [DOI] [PubMed] [Google Scholar]; b) Ternant D, Paintaud G. Expert Opin. Biol. Ther. 2005;5(Suppl 1):S37–S47. doi: 10.1517/14712598.5.1.s37. [DOI] [PubMed] [Google Scholar]
- 5.Lovejoy B, Cascio D, Eisenberg D. J. Mol. Biol. 1993;234:640–653. doi: 10.1006/jmbi.1993.1617. [DOI] [PubMed] [Google Scholar]
- 6.Cooper KL, Madan J, Whyte S, Stevenson MD, Akehurst RL. BMC Cancer. 2011;11:404. doi: 10.1186/1471-2407-11-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morstyn G, Campbell L, Souza LM, Alton NK, Keech J, Green M, Sheridan W, Metcalf D, Fox R. Lancet. 1988;1:667–672. doi: 10.1016/s0140-6736(88)91475-4. [DOI] [PubMed] [Google Scholar]
- 8.a) Cox GN, Smith DJ, Carlson SJ, Bendele AM, Chlipala EA, Doherty DH. Exp. Hematol. 2004;32:441–449. doi: 10.1016/j.exphem.2004.01.012. [DOI] [PubMed] [Google Scholar]; b) de Haan G, Ausema A, Wilkens M, Molineux G, Dontje B. Br. J. Haematol. 2000;110:638–646. doi: 10.1046/j.1365-2141.2000.02252.x. [DOI] [PubMed] [Google Scholar]
- 9.Roopenian DC, Akilesh S. Nat. Rev. Immunol. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- 10.a) Bai Y, Ann DK, Shen WC. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7292–7296. doi: 10.1073/pnas.0500062102. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shirafuji N, Asano S, Matsuda S, Watari K, Takaku F, Nagata S. Exp. Hematol. 1989;17:116–119. [PubMed] [Google Scholar]
- 11.de Lichtervelde L, Antal CE, Boitano AE, Wang Y, Krastel P, Petersen F, Newton AC, Cooke MP, Schultz PG. Chem. Biol. 2012;19:994–1000. doi: 10.1016/j.chembiol.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.