Abstract
The site-specific incorporation of unnatural amino acids (UAAs) into proteins in bacteria is made possible by the evolution of aminoacyl-tRNA synthetases that selectively recognize and aminoacylate the amino acid of interest. Recently we have discovered that some of the previously evolved aaRSs display a degree of polyspecificity and are capable of recognizing multiple UAAs. Herein we report the polyspecificity of an aaRS evolved to encode a comarin containing amino acid. This polyspecificity was then exploited to introduce several UAAs into the fluorophore of GFP, altering its photophysical properties.
Keywords: Unnatural amino acids, Polyspecificity, Green Fluorescence Protein, Aminoacyl-tRNA Synthetase, Fluorescence modulation
Genetically encoded unnatural amino acids (UAAs) are useful probes of protein structure and functions, and can be used to engineer proteins with novel function.1 UAAs can be encoded in bacterial, yeast, and mammalian cells in response to a nonsense or frameshift codon by means of an orthogonal aminoacyl-tRNA synthetase (aaRS)/tRNA pair evolved to encode the desired UAA.2 Using this approach over 70 UAAs with various distinct functions have been incorporated into proteins. The evolution of an aaRS with the desired specificity involves a double sieve selection in which the UAA is first positively selected for incorporation based on chloramphenicol acyltransferase expression, followed by a negative selection based on barnase expression in the absence of the UAA to eliminate aaRSs capable of charging any of the endogenous 20 amino acids. It was recently reported, by us and others, that some aaRSs possess a degree of polyspecificity allowing them to accept multiple UAAs.3 This polyspecificity arises due to the nature of the selection strategy, as all other UAAs are absent from the selection. This allows evolved aaRSs to be cross-reactive with other UAAs, while still maintaining their orthogonality to endogenous host amino acids and enables the incorporation of multiple amino acids in response to an amber codon using only a single aaRS. Herein we describe the characterization of another polyspecific aaRS and its use to alter the fluorescent properties of GFP.
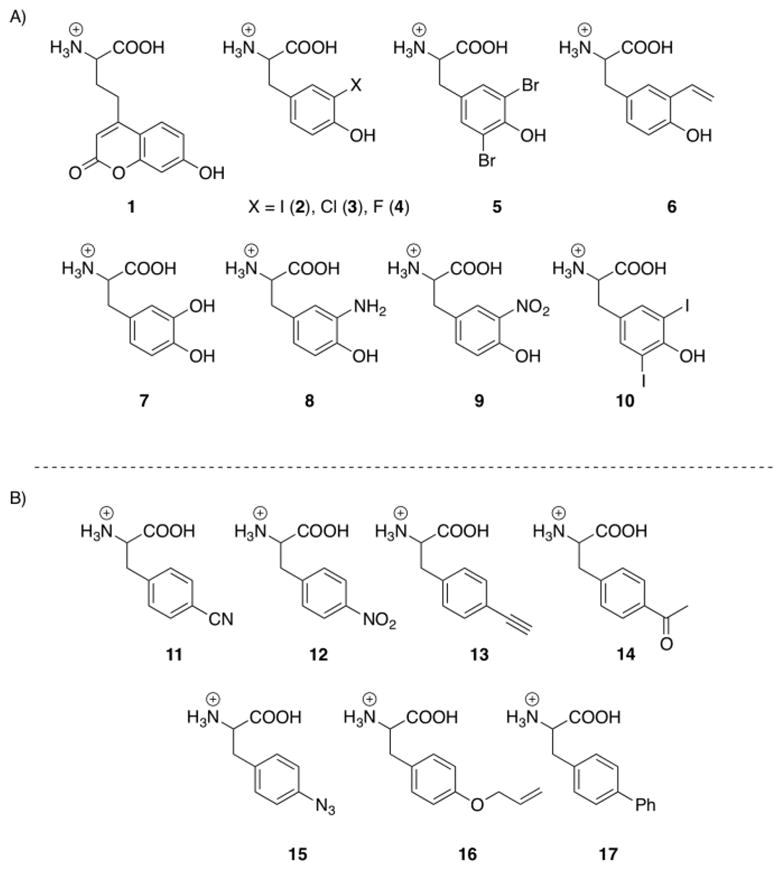
We previously employed a rapid screen to assess the polyspecificity of 15 evolved aaRSs based on GFP fluorescence.3a A GFP gene with an amber mutation at Tyr151, GFPY151X (a surface residue that can be substituted without alteration of the fluorophore) was placed under control of the T7 promoter in a pET101 vector.4 When co-transformed with a pEVOL plasmid which encodes the desired aaRS/tRNAcua pair, the amber mutation is suppressed in the presence of an unnatural amino acid.4 To determine if a particular aaRS is capable of recognizing other amino acids, E. coli harbouring the two plasmids can be cultured in a 96 well format, with each well containing a different unnatural amino acid. Using this screen the p-cyanophenylalanine aaRS (pCNFRS)5 was found to be polyspecific, incorporating over 18 UAAs.3a Additionally, the synthetase (CouARS) evolved to incorporate the coumarin (1) unnatural amino acid6 displayed a novel promiscuity for substituted tyrosyl residues (3-iodotyrosine and 3-chlorotyrosine; see Supporting Information). These results prompted us to carry out a secondary screen against structurally similar UAAs (Scheme 1a). Employing the previously described GFP assay, the CouARS was found to efficiently encode 6 unnatural amino acids including 3-iodotyrosine (2), 3-chlorotyrosine (3), 3-fluorotyrosine (4), 3,5-dibromotyrosine (5), and 3-vinyltyrosine (6) (Scheme 1a). While 2–4 have been previously encoded,7 UAAs 5 and 6 have not been incorporated in bacterial by an orthogonal aaRS/tRNA strategy. No incorporation of UAAs 7–10 was observed, potentially due to either their size (10) or the nucleophilicity/ionic character of their substituents (7–8). The polyspecificity of the CouARS may arise from the increased size of the amino acid binding pocket, as key active site residues are mutated to glycine (A67G, H70G, D158G, and L162G). Additionally, the replacement of Y32 with a glutamic acid residue facilitates a greater degree of structural flexibility while still able to engage in hydrogen bonding with the hydroxyl group of tyrosine derivatives.
Scheme 1.
Unnatural incorporation was confirmed in triplicate for GFP assays, and also by incorporation of the unnatural amino acid into a his-tagged myoglobin mutant containing an F107TAG amber mutation (MyoF107X).4 Myoglobin was purified using a Ni-NTA resin and analyzed by SDS-PAGE. The calculated mass of the purified mutant protein was confirmed by liquid chromatography/mass spectroscopy (LCMS), and in the presence of any of these amino acids (>1mM), no incorporation of a common twenty amino acid was observed within the detection limits of LCMS (Table 1). While suppression yields are somewhat low (Table 1), they are comparable to the suppression efficiencies with 1 for myoglobin expression (data not shown).
Table 1.
Unnatural amino acid incorporation by the CouARS
| Synthetase | UAA | Expecteda | Observedb | Relative % Incorporationc |
|---|---|---|---|---|
| WT | Phe | 18352 | 18353 | 100 ± 8.2 |
| CouA | 2 | 18495 | 18496 | 27 ± 2.6 |
| CouA | 3 | 18404 | 18405 | 19 ± 1.9 |
| CouA | 4 | 18387 | 18387 | 9.7 ± 0.8 |
| CouA | 5 | 18528 | 18527 | 10.1 ± 1.4 |
| CouA | 6 | 18395 | 18396 | 17.5 ± 2.1 |
LC/MS expected mass for UAA incorporated myoglobin.
Determined by ESI and deconvolution with ChemStation Software (Rev. B.03.02)
Based on GFP-Y151TAG expression, and normalized to WT GFP expression.
This polyspecificity can be employed to subtly modulate the acidity, electronic nature, and size of a single residue of a protein, by simply including the desired UAA during expression of the amber mutant of interest. This obviates the need for a separate aaRS system for each UAA, and may even preclude the necessity for aaRS selection. As a proof-of-principle experiment we selected GFP, and introduced a TAG codon at Tyr66 (pET-GFPY66X). This tyrosine residue is integral to the flurorophore, and thus alterations in its structure and electron donating capacity have previously been demonstrated to alter the fluorescence of the protein.1e, 8 Co-transformation of pEVOL-pCNF or pEVOL-CouA with pET-GFPY66X into BL21(DE3) cells, followed by expression in the presence of 1 mM UAA (induction with 1 mM IPTG, 0.02% arabinose at OD600 0.8; 16h, 30 °C) resulted in production of several previously uncharacterized GFP mutants possessing a UAA within the fluorophore. After purification using a Ni-NTA column and dialysis, the fluorescence spectra of the GFP mutants were measured (Figure 1).
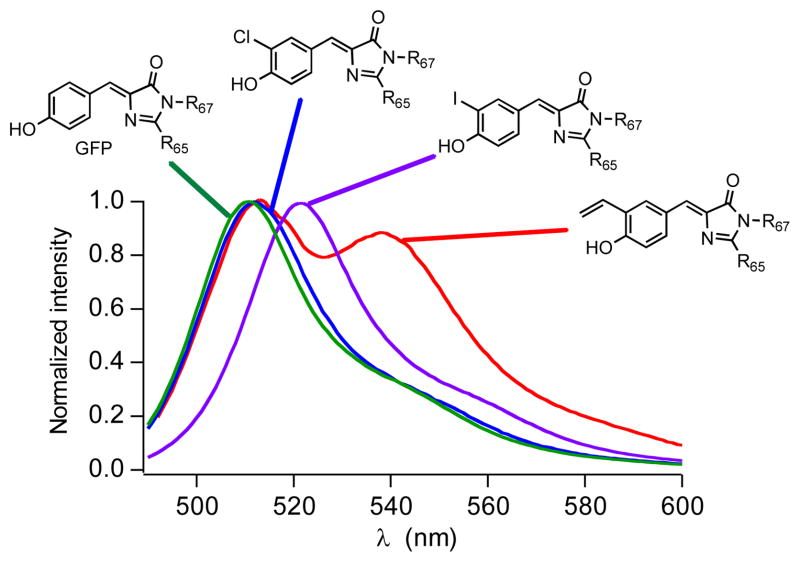
Figure 1.
Normalized fluorescence spectra of GFP and modified GFP containing unnatural amino acids 2, 3, and 6 in phosphate buffer solution solution (0.1 M NaCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) after excitation at 480 nm.
Gratifyingly, the fluorescent properties of GFP can easily be modulated by incorporating different UAAs at position 66 (a fact previously demonstrated).1e However, this can now rapidly be achieved using a single aaRS, rather than requiring expression with multiple distinct UAA/aaRS systems. Using the CouARS we were able to incorporate tyrosine derivatives 2, 3, and 6 at position 66 in GFP (Scheme 1). The fluorescence spectra of these analogs exhibited a bathochromic shift, with the most substantial shift occurring with the 3-vinyltyrosine 6 (Figure 1). The fluorescence lifetimes and quantum yields for derivatives 2, 3, and 6 are similar to GFP with 6 showing the highest quantum yield (0.53) and longest fluorescence lifetime (3.3 ns) (Table 2 and Supporting Information).
Table 2.
Fluorescent properties of GFPTAG66 Mutants
| Synthetase | UAA | λmax (nm) | φf |
|---|---|---|---|
| pCNF | 11 | 442 | 0.039 |
| pCNF | 12 | -- | ≤ 0.005 |
| pCNF | 13 | 439 | 0.038 |
| pCNF | 14 | 435 | 0.035 |
| pCNF | 15 | 501 | 0.11 |
| pCNF | 16 | 461 | 0.036 |
| pCNF | 17 | 459 | 0.066 |
| CouA | 2 | 520 | 0.43 |
| CouA | 3 | 512 | 0.45 |
| CouA | 6 | 538 | 0.53 |
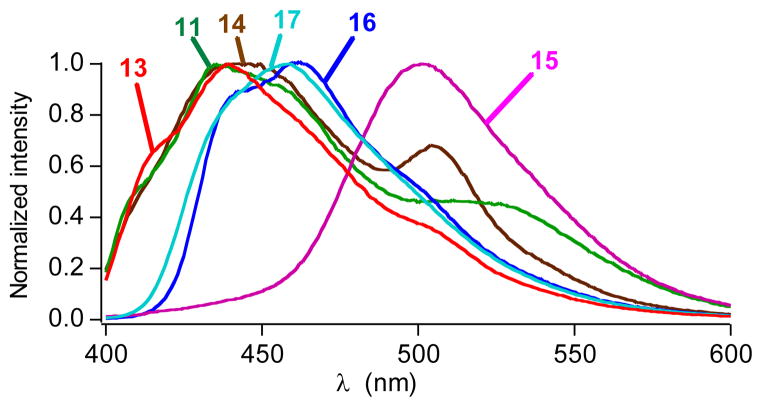
Next the pCNFRS was used to incorporate a series of p-substituted phenylalanine derivatives (11–17, Scheme 1), resulting in a hypsochromic shift of the GFP fluorescence (Figure 2). The electron donating ability of these functional groups is lower than the natural hydroxyl group of tyrosine accounting for the blue-shifted spectra. This trend is also apparent within the series of derivatives as the most strongly donating azide substituent (15) affords the least shifted emission spectrum, followed by the phenyl (16) and alloxy (17) substituents (Figure 1). In addition, the replacement of the hydroxyl group of tyrosine reduces the fluorescence quantum yields (Table 2). Interestingly, the nitro substituent (12) appears to quench fluorescence almost completely (φf ≤ 0.005).
Figure 2.
Normalized fluorescence spectra of modified GFP containing unnatural amino acids in phosphate buffer solution (0.1 M NaCl, 8 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) after excitation at 365 nm.
In summary, we have successfully demonstrated the ability to modulate protein function by the site-specific incorporation of a variety of UAAs using a single aaRS. With this technology it is feasible to both red- and blue-shift the emission spectra of GFP as a means to confer new photophysical properties. Additionally, by exploiting the polyspecificity of the coumarin aaRS, we have also been able to incorporate two previously unencoded UAAs (5 and 6). These represent potentially useful probes for the alteration of tyrosine electrochemical properties, and in the case of 6, can serve as a radical trap or metathesis precursor. Thus, aaRS polyspecificity represents a novel mechanism to rapidly encode a variety of amino acids using a singular expression system allowing for a systematic study of the effect of UAA incorporation on protein structure and function.
Supplementary Material
Acknowledgments
We would like to acknowledge Virginia Seely and Emily Remba for their assistance in manuscript preparation. This is paper # 21362 from The Scripps Research Institute. This work was funded by Grant DE-FG03-00ER46051 from the Division of Materials Sciences, Department of Energy (P.G.S.). D.D.Y. acknowledges a National Institutes of Health Ruth L. Kirchstein Postdocotoral Fellowship F32CA144213.
Footnotes
Supplementary material associated with this article can be found in the online version at doi XXXXX
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and notes
- 1.(a) Young TS, Young DD, Ahmad I, Louis JM, Benkovic SJ, Schultz PG. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1108045108. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu CC, Mack AV, Brustad EM, Mills JH, Groff D, Smider VV, Schultz PG. J Am Chem Soc. 2009;131(28):9616–7. doi: 10.1021/ja902985e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mills JH, Lee HS, Liu CC, Wang J, Schultz PG. Chembiochem. 2009;10(13):2162–4. doi: 10.1002/cbic.200900254. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Ngo JT, Champion JA, Mahdavi A, Tanrikulu IC, Beatty KE, Connor RE, Yoo TH, Dieterich DC, Schuman EM, Tirrell DA. Nat Chem Biol. 2009;5(10):715–7. doi: 10.1038/nchembio.200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wang L, Xie J, Deniz AA, Schultz PG. J Org Chem. 2003;68(1):174–6. doi: 10.1021/jo026570u. [DOI] [PubMed] [Google Scholar]; (f) Deiters A, Groff D, Ryu Y, Xie J, Schultz PG. Angew Chem Int Ed Engl. 2006;45(17):2728–31. doi: 10.1002/anie.200600264. [DOI] [PubMed] [Google Scholar]
- 2.(a) Young TS, Schultz PG. J Biol Chem. 2010;285(15):11039–44. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Liu CC, Schultz PG. Annu Rev Biochem. 2010;79:413–44. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]; (c) Liu W, Brock A, Chen S, Schultz PG. Nat Methods. 2007;4(3):239–44. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]; (d) Deiters A, Cropp TA, Mukherji M, Chin JW, Anderson JC, Schultz PG. J Am Chem Soc. 2003;125(39):11782–3. doi: 10.1021/ja0370037. [DOI] [PubMed] [Google Scholar]; (e) Santoro SW, Wang L, Herberich B, King DS, Schultz PG. Nat Biotechnol. 2002;20(10):1044–8. doi: 10.1038/nbt742. [DOI] [PubMed] [Google Scholar]
- 3.(a) Young DD, Young TS, Jahnz M, Ahmad I, Spraggon G, Schultz PG. Biochemistry. 2011;50(11):1894–900. doi: 10.1021/bi101929e. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Brustad E, Bushey ML, Brock A, Chittuluru J, Schultz PG. Bioorg Med Chem Lett. 2008;18(22):6004–6. doi: 10.1016/j.bmcl.2008.09.050. [DOI] [PubMed] [Google Scholar]; (c) Miyake–Stoner SJ, Refakis CA, Hammill JT, Lusic H, Hazen JL, Deiters A, Mehl RA. Biochemistry. 2010;49(8):1667–77. doi: 10.1021/bi901947r. [DOI] [PubMed] [Google Scholar]; (d) Stokes AL, Miyake–Stoner SJ, Peeler JC, Nguyen DP, Hammer RP, Mehl RA. Mol Biosyst. 2009;5(9):1032–1038. doi: 10.1039/b904032c. [DOI] [PubMed] [Google Scholar]; (e) Polycarpo CR, Herring S, Berube A, Wood JL, Soll D, Ambrogelly A. FEBS Lett. 2006;580(28–29):6695–700. doi: 10.1016/j.febslet.2006.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Mukai T, Kobayashi T, Hino N, Yanagisawa T, Sakamoto K, Yokoyama S. Biochem Biophys Res Commun. 2008;371(4):818–22. doi: 10.1016/j.bbrc.2008.04.164. [DOI] [PubMed] [Google Scholar]
- 4.Young TS, Ahmad I, Yin JA, Schultz PG. J Mol Biol. 2009 doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 5.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. J Am Chem Soc. 2006;128(43):13984–5. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 6.Wang J, Xie J, Schultz PG. J Am Chem Soc. 2006;128(27):8738–9. doi: 10.1021/ja062666k. [DOI] [PubMed] [Google Scholar]
- 7.Kiga D, Sakamoto K, Kodama K, Kigawa T, Matsuda T, Yabuki T, Shirouzu M, Harada Y, Nakayama H, Takio K, Hasegawa Y, Endo Y, Hirao I, Yokoyama S. Proc Natl Acad Sci U S A. 2002;99(15):9715–20. doi: 10.1073/pnas.142220099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Groff D, Wang F, Jockusch S, Turro NJ, Schultz PG. Angew Chem Int Ed Engl. 2010;49(42):7677–9. doi: 10.1002/anie.201003797. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yoo TH, Link AJ, Tirrell DA. Proc Natl Acad Sci U S A. 2007;104(35):13887–90. doi: 10.1073/pnas.0701904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.