Abstract
Background
Despite selenium's toxicity in plants at higher levels, crops supply most of the essential dietary selenium in humans. In plants, inorganic selenium can be assimilated into selenocysteine, which can replace cysteine in proteins. Selenium toxicity in plants has been attributed to the formation of non-specific selenoproteins. However, this paradigm can be challenged now that there is increasingly abundant evidence suggesting that selenium-induced oxidative stress also contributes to toxicity in plants.
Scope
This Botanical Briefing summarizes the evidence indicating that selenium toxicity in plants is attributable to both the accumulation of non-specific selenoproteins and selenium-induced oxidative stress. Evidence is also presented to substantiate the claim that inadvertent selenocysteine replacement probably impairs or misfolds proteins, which supports the malformed selenoprotein hypothesis. The possible physiological ramifications of selenoproteins and selenium-induced oxidative stress are discussed.
Conclusions
Malformed selenoproteins and oxidative stress are two distinct types of stress that drive selenium toxicity in plants and could impact cellular processes in plants that have yet to be thoroughly explored. Although challenging, deciphering whether the extent of selenium toxicity in plants is imparted by selenoproteins or oxidative stress could be helpful in the development of crops with fortified levels of selenium.
Keywords: Selenium toxicity, oxidative stress, selenoproteins, glutathione, diselenide bonds, iron–selenium cluster, electron transport, proteasome, misfolded proteins
INTRODUCTION
Selenium (Se) is a naturally occurring trace element that typically ranges from 0·01 to 2 p.p.m. in soils. Se is essential to many organisms, including some archaea, bacteria, protozoans, green algae and nearly all animals. In contrast, higher plants are not thought to have a requirement for Se, although this is still controversial (Pilon-Smits and Quinn, 2010). The chemical similarity between Se and sulphur – an essential macronutrient to all organisms – is well known. Therefore, vascular plants can still accumulate Se, which occurs when selenate is non-specifically transported into root tissue via sulphate transporters (El Kassis et al., 2007). Once inside plant cells, Se metabolites can compete at the active site of many enzymes involved in essential sulphur metabolism. Once selenate is transported into plastids, it can utilize the sulphate assimilation pathway and become reduced to organic metabolites (Pilon-Smits and Quinn, 2010). Although a concentration of Se greater than 0·1 % of dry weight in most crops is typically rare, some plants including Astragulus bisulcatus and Stanleya pinnata have evolved on Se-rich soils and can hyperaccumulate Se up to 1 % of its dry weight (Zhu et al., 2009).
Even though crops are limited in the amount of Se that can accumulate in their tissue, a plant-based diet still provides most of the essential dietary intake of Se in humans. Consequently, negligible Se accumulation in staple crops can result in low levels of dietary Se in geographically disparate areas (Zhu et al., 2009). Organisms that have a demand for Se require it for the biosynthesis of the 21st amino acid selenocysteine, which in humans is needed to make 25 specific selenoproteins. A deficiency in dietary Se decreases the abundance of selenoproteins, which can impair the immune system, thyroid processes and spermatogenesis; ultimately, a lack of Se can lead to Keshin-Beck and Keshan disease, which can be treated with Se supplementation (Papp et al., 2007). Although the daily Se requirement in adults is 55–70 µg, augmenting the requirement via Se supplementation may be beneficial. Numerous in vitro studies have noted the protective properties of Se compounds, particularly against cancer as recently reviewed (Davis, 2012). Therefore, the development of crops with fortified levels of Se is appealing, as a source of both nutrition and Se-based therapeutics. Differences in Se accumulation among crop varieties exist, such as Lactuca sativa (Ramos et al., 2010), and could lead to successful breeding programmes to boost Se levels in crops. Additionally, genetic manipulation can also increase Se levels in plants; this approach often targets the overexpression of enzymes involved in sulphur metabolism (Zhu et al., 2009).
However, similar to many essential trace elements, the difference between Se deficiency and toxicity is narrow. Selenosis (diagnosed Se toxicity) continues to remain a persistent threat to livestock in the western USA (Davis et al., 2012) where high levels of Se can naturally accumulate during the weathering of Se-rich bedrock or be influenced by anthropogenic activities (e.g. irrigation and mining). Selenosis in sheep and cattle occurs during the ingestion of Se-hyperaccumulating plants, such as Astragulus bisulcatus and Stanleya pinnata. Tragically, the most infamous case of selenosis in humans occurred in the 1960s in Hubei Province, China, where at least 100 people succumbed after digesting plants with high levels of Se (Fordyce et al., 2000). In short, a better understanding of Se toxicity is particularly salient if Se-rich nutraceutical foods are administered as therapeutics to treat or prevent disease.
With the exception of rare Se-hyperaccumulating plants, Se is toxic in plants if it accumulates to a high enough concentration (> 0·1 % plant dry weight). The physiological effects of Se stress in plants have been well documented and include stunted root growth, reduced biomass, chlorosis, reduced photosynthetic efficiency, and ultimately plant death. By contrast, the protective benefits of Se in plants have been reported and recently reviewed in both an ecological (El Mehdawi and Pilon-Smits, 2012) and a physiological context (Feng et al., 2013). The aim of this review is to discuss the two distinct types of Se stress in plants. The misincorporation of selenocysteine into non-specific selenoproteins has been recognized as a form of toxicity in plants for over 30 years. Although this paradigm is still reflected in the current literature, it is also now becoming increasingly apparent that inorganic Se is a pro-oxidant, which can cause oxidative stress in plants. Therefore, Se stress in plants is driven by two different types of Se-induced toxicities. How plants respond to these two distinct modes of Se stress will be addressed, along with the challenges of deciphering if Se stress is mainly attributable to oxidative stress or non-specific selenoproteins.
SELENIUM METABOLISM IN PLANTS
The toxicity of different Se metabolites in plants varies. Although plant Se metabolism has been thoroughly reviewed elsewhere (Terry et al., 2000; Pilon-Smits and Quinn, 2010), it is nonetheless relevant to the discussion on Se toxicity, and a schematic diagram of Se metabolism in presented in Fig. 1. Selenate is the most oxidized form of Se in well-aerated soils, and is the most bioavailable form of Se to plants. Selenate competes with sulphate at the site of sulphate transporters localized to plant roots; in Arabidopsis plants, most selenate uptake is mediated by the high-affinity sulphate transporter SULTR1;2 (El Kassis et al., 2007). The assimilation of inorganic selenate to selenocysteine occurs in plastids. The reduction of selenate to selenite is ATP-consumptive and the rate-limiting step in the assimilation of selenate to the organic metabolite, selenocysteine (Sors et al., 2005), and requires the concerted action of two enzymes. First, the enzyme ATP sulphurylase hydrolyses ATP to form adenosine phosphoselenate, or activated selenate (Dilworth and Bandurski, 1977). The reduction of adenosine phosphoselenate to selenite probably occurs via adenosine phosphosulphate reductase (APR), which is supported by the observation that knockout of the dominant APR isozyme in Arabidopsis increased the amount of selenate and decreased the concentration of selenite (Grant et al., 2011). The reduction of selenite to selenide might be catalysed by sulphite reductase, although this also has not been experimentally demonstrated. Alternatively, reduction of selenite in plants might be mediated by glutathione or glutaredoxins (Wallenberg et al., 2010), as it is in human cells, which produces the intermediate metabolite selenodiglutathione. Lastly, the enzyme cysteine synthase, which has higher affinity for selenide than for sulphide, can couple selenide with o-acetylserine to produce selenocysteine (Ng and Anderson, 1978). Selenocysteine can further be metabolized to methylselenocysteine and methylselenomethionine. Both of these methylated non-proteinaceous amino acids can ultimately be converted into volatile Se compounds (e.g. dimethyl selenide and dimethyl diselenide) that can rid the plant of toxic Se (Pilon-Smits and Quinn, 2010).
Fig. 1.
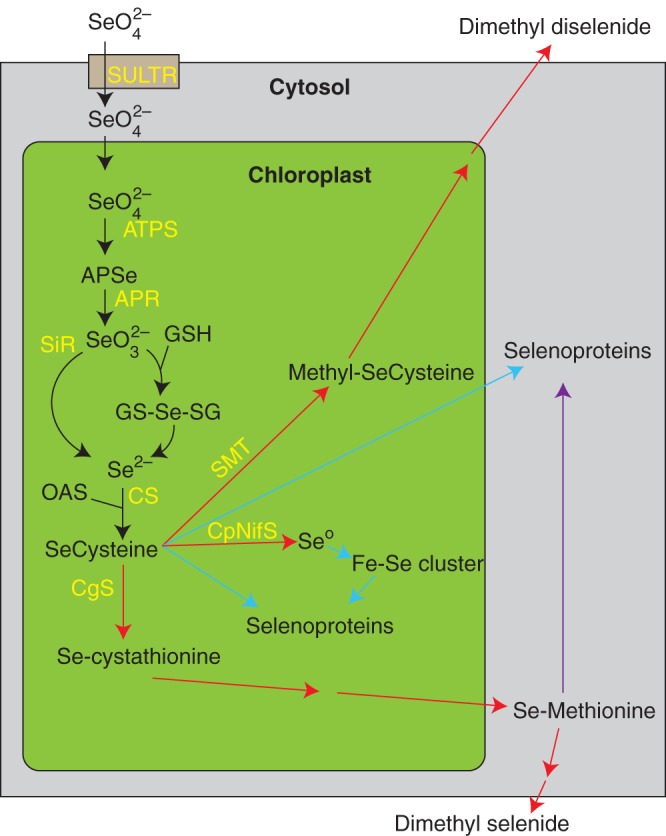
Schematic diagram of Se metabolism in plants. Selenium metabolites are depicted in black, and proteins involved in Se transport and metabolism are in yellow. Arrows in blue and red are considered, respectively, to promote and to alleviate Se toxicity associated with the assimilation of selenocysteine. The purple arrow is not considered toxic. SULTR, sulphur transporter; ATPS, ATP sulphurylase; APR, adenosine 5-phosphosulfate reductase; SiR, sulphite reductase; CS, cysteine synthase; SMT, selenocysteine methyltransferase; CgS, cysthathionine gamme synthase; APSe, adenosine phosphoselenate; GSH, glutathione; GS-Se-SG, selenodiglutathione; OAS, o-acetylserine.
THE MALFORMED SELENOPROTEIN HYPOTHESIS
Pioneering work led by Brown and Shrift (1980) led to the discovery that selenocysteine can be misincorporated into non-specific selenoproteins in Vigna radiata. After it was later revealed that Se-tolerant Astragulus species had nearly a 10-fold decrease in the concentration of Se in protein compared with non-tolerant Astragulus species (Brown and Shrift, 1981), Se toxicity in plants was explained to be driven by the formation of non-specific selenoproteins. The malformed selenoproteins hypothesis asserts that Se toxicity occurs when a tRNAcys inadvertently binds to selenocysteine instead of cysteine during translation to form non-specific and toxic selenoproteins. Astragulus bisulcatus' tolerance to Se is attributable to the presence of a chloroplastic enzyme with selenocysteine methyltransferase activity (Neuhierl and Bock, 1996), which methylates selenocysteine and prevents its random incorporation into protein. Additionally, nearly 90 % of the total Se found in the leaves of the Se-tolerant Stanleya pinnata is in the form of methylselenocysteine (Freeman et al., 2006). Notably, a selenocysteine methyltransferase is not believed to exist in most non-Se-tolerant plants.
In support of the malformed selenoprotein hypothesis, evidence in transgenic plants also suggests that diverting selenocysteine away from non-specific protein incorporation is associated with increased Se tolerance. Overexpression of an Astragulus bisulcatus selenocysteine methyltransferase in Arabidopsis and Brassica juncea increased Se tolerance, which was explained by the observed increase in methylselenocysteine and volatilization of dimethyl-diselenide (Leduc et al., 2004). Similarly, overexpression of cystathionine gamma-synthase, which converts selenocysteine into Se-cystathionine, in Brassica juncea enhanced tolerance to selenite by doubling the amount of volatilized Se (Van Huysen et al., 2003). Lastly, overexpression of an Arabidopsis chloroplastic NifS-like protein with selenocysteine lyase activity decreased the amount of non-specific selenoproteins and was associated with a two-fold increased Se tolerance (Van Hoewyk et al., 2005). These data further support the view that diverting selenocysteine away from protein misincorporation minimizes the risk of accumulating non-specific selenoproteins, and thereby increases Se tolerance in plants.
However, whether non-specific selenoproteins in plants are malformed and misfolded, as usually presumed, remains to be determined. In response to selenate treatment in Arabidopsis, 16 transcripts encoding heat shock proteins were up-regulated (Van Hoewyk et al., 2008). Plant heat shock proteins act as chaperones that assist in proper protein folding during stress (Wang et al., 2004), and perhaps their up-regulation is in response to misfolded selenoproteins. Two studies have recently implicated the proteasome's involvement in the degradation of misfolded selenoproteins in plants. In one study, a proteomic analysis revealed that Se treatment increased the abundance of proteasomal subunits in rice roots, which the authors attributed to the necessity to remove misfolded selenoproteins (Wang et al., 2012). This speculation coincided with the observation that selenate treatment in Stanleya pinnata increased protein ubiquitination (Sabbagh and Van Hoewyk, 2012). In this study, inhibition of the proteasome in Se-treated plants increased the concentration of Se in protein, which supports the view that non-specific selenoproteins in plants are misfolded and selected for proteasomal proteolysis. Despite the progress made in elucidating Se tolerance in plants since the discovery of non-specific selenoproteins in plants over 30 years ago, direct evidence that plant selenoproteins are impaired and misfolded is lacking, although this is assumed and likely to be the case. The next section draws primarily from prokaryotic and animal organisms and aims to provide evidence that the replacement of cysteine with selenocysteine is probably detrimental to protein function.
EVIDENCE THAT NON-SPECIFIC SELENOPROTEINS ARE IMPAIRED AND POSSIBLE RAMIFICATIONS IN PLANTS
Selenomethionine and selenocysteine are both selenoamino acids that can be misincorporated into proteins in plants. Methionine is not as reactive as cysteine. Therefore, the non-specific accumulation of selenomethionine in protein is not considered as deleterious as the more reactive selenocysteine, despite the possibility that selenomethione might inhibit protein synthesis (Eustice et al., 1981). Generally, however, selenomethione in proteins is not considered deleterious. In fact, in cereals such as rice, more than half of the total Se can be in the form of selenomethione (Stadlober et al., 2001). Although the abundance of cysteine residues in proteins is relatively low in all species, they have critical roles in protein structure and function, including catalysis, redox regulation, formation of disulfide bridges and metal binding sites. Despite being structurally similar, cysteine and selenocysteine have different properties. Compared with cysteine, selenocysteine is (1) larger, (2) more easily deprotonated given its lower pKa value and (3) typically more reactive due to its higher nucleophilicity (Hondal et al., 2012). Taking the physical and chemical differences between these two amino acids into account, a cysteine to selenocysteine substitution in plant protein is potentially detrimental. The direct effects of a cysteine to selenocysteine substitution in protein are briefly presented below.
Cysteines often are found in the active site of enzymes where they play a catalytic role. A cysteine to selenocysteine substitution was studied in a human and mouse methionine sulfoxide reductase, which repairs oxidized methionine residues in proteins. A selenocysteine substitution abolished activity when supplied with thioredoxin, the enzyme's natural reductant, compared with the wild-type protein containing cysteine in its active site (Kim and Gladyshev, 2005). In plants, chloroplastic methionine sulfoxide reductase has a well-known role in preventing oxidative stress, and its activity has recently been shown to positively correlate with seed longevity in Arabidopsis and Medicago sativa (Châtelain et al., 2013). In light of this evidence, the physiological ramifications of a cysteine to selenocysteine substitution in methionine sulfoxide reductase in plants could impair stress physiology and seed viability. However, not all selenocysteine substitutions are detrimental to enzyme activity. One example of a beneficial replacement of cysteine to selenocysteine comes from a glutathione-dependent peroxidase found in Citrus sinensis. The citrus' peroxidase was engineered to contain a selenocysteine in its active site, which enhanced activity four-fold when expressed in Escherichia coli (Hazebrouck et al., 2000). However, the rate of activity was still much lower than mammalian glutathione peroxidase homologues that naturally contain a selenocysteine in their active site. Yet because there are so few known selenoprotein homologues, the beneficial substitution of selenocysteine is expected to be rare in plants.
Cysteine in proteins can also function as a metal-binding residue capable of stabilizing cofactors such as iron–sulphur (Fe–S) clusters, heme groups and ions (e.g. zinc or copper). A cysteine to selenocysteine substitution at a site which coordinates metal binding would have an increased bond length due to the larger atomic radius of Se, which might alter the strength of the bond between the protein and the cofactor. Recently, the impact of a cysteine to selenocysteine substitution at a heme-binding site was studied in a bacterial cytochrome P450. In this case, a two-fold decreased monooxygenase activity of the selenocysteine variant was observed, which was associated with its redox potential being 48 mV more negative than that of the cysteine-containing control protein (Aldag et al., 2009). Therefore, metal cofactors, such as iron, complexed to selenocysteine binding sites may hinder electron transfer. If this were extended to plants, a selenocysteine at a metal binding site could alter the function of mitochondria and chloroplast, both of which contain iron-binding proteins to mediate electron transport.
The tertiary structure of many proteins is dependent upon the formation of disulfide bridges formed during the oxidation of two neighbouring cysteines. A cysteine to selenocysteine substitution in non-specific selenoproteins could create either a diselenide bridge or a mixed selenide–sulphide bridge (or selenosulfide bridge) with altered properties. For example, a diselenide bridge will be longer (about 0·2 Å) compared with a disulfide bridge. Despite the relatively subtle differences in diselenide bond length, proteins with diselenide or selenosulfide bonds are also likely to have much lower redox potentials (70–250 mV) than proteins with disulfide bridges, which can affect enzyme kinetics (Hondal et al., 2012). For example, the replacement of two cysteines with selenocysteine in thioredoxin produced a novel E. coli Se-thioredoxin with a diselenide bridge. In this case, the Se-thioredoxin could not be reduced under native conditions, which was explained by the altered redox potential of the diselenide bond (Müller et al., 1994).
Lastly, there is a chance that Fe–Se clusters can substitute for an Fe–S cluster. Fe–S clusters are prosthetic groups that become inserted into apoproteins to form functional Fe–S proteins, which are abundant in the chloroplastic and mitochondrial electron transport chain found in plants (Balk and Pilon, 2011). The sulphur that is used for the maturation of Fe–S clusters is derived exclusively from cysteine via NifS-like proteins that have cysteine desulfurase activity. It is noteworthy that the chloroplastic NifS-like protein has nearly 300-fold higher affinity for selenocysteine in vitro (Pilon-Smits et al., 2002). Thus, it is certainly plausible that NifS-like proteins can liberate elemental Se from selenocysteine, which can then be assembled into an Fe–Se cluster. In a study on spinach ferredoxin, replacement of an Fe–S cluster with an Fe–Se resulted in lower yields of reconstituted holoprotein, possibly because the Fe–Se clusters are unstable and experience difficulty inserting into an apoprotein due to its increased size (Meyer et al., 1986). In a more recent study using Klebsiella pneumoniae, the activity and electron transport of nitrogenase containing an Fe–Se cluster decreased roughly five-fold compared with the control protein with an Fe–S cluster (Hallenbeck et al., 2009). Given the role of Fe–S proteins in electron transport, it is speculated that in planta Fe–Se proteins might hinder photosynthetic or mitochondrial electron transport. However, whether photosynthetic impairment during Se stress is attributed to malformed selenoproteins or Se-induced oxidative stress remains of debate; this latter mode of Se toxicity is addressed in the next section.
INORGANIC SELENIUM INDUCES OXIDATIVE STRESS
Se compounds have pro-oxidant properties, as reviewed elsewhere (Spallholz, 1994). In plants, the toxicity of selenate and selenite has been widely reported and is dose dependent. Most plants are more sensitive to selenite than to selenate, although there are exceptions, including the green algae (Geoffroy et al., 2007). Evidence indicating that selenate and selenite induce oxidative in plants is presented below.
Glutathione is a tripeptide that is central to the cellular redox status in plants, and its importance in plant development and physiology has been extensively reviewed (Noctor et al., 2011). In Arabidopsis plants, selenate decreases levels of glutathione in both a dose- and time-dependent manner; in fact, the concentration of glutathione decreased nearly two-fold after 6 h in plants treated with 50 µm selenate (Hugouvieux et al., 2009). It was therefore not surprising when tolerance to selenate in Arabidopsis was later reported to strongly correlate with glutathione concentration (Grant et al., 2011). In this same study, manipulating glutathione content demonstrated its impact during Se stress. Plants grown on buthionine sulfoximine, an inhibitor of glutathione biosynthesis, have a two-fold increased sensitivity to selenate. In agreement with this finding, cad2-1 mutant plants with a defect in the glutathione biosynthetic pathway have a 60 % reduction in root length compared with wild-type plants grown on 20 µm selenate. Given glutathione's important roles in plant cells – including signalling and redox homeostasis – its rapid depletion would probably challenge plant growth. In fact, glutathione depletion is linked to perturbation of auxin homeostasis and decreased root growth (Koprivova et al., 2010). With this in mind, selenate-induced depletion of glutathione could explain why root growth is so severely restricted during Se treatment. The myriad roles of glutathione in plant processes seem to be ever-expanding, and its depletion during selenate stress could be a stronger determinant driving Se toxicity than previously expected.
Glutathione is a major antioxidant in plants, and its depletion during selenate stress is expected to increase the accumulation of reactive oxygen species (ROS) and thereby induce oxidative stress. Several studies have supported this view in plants. Stanleya albescens plants challenged with selenate had decreased glutathione, which resulted in increased superoxide and hydrogen peroxide accumulation compared with the more tolerant Se-hyperaccumulator Stanleya pinnata, which had elevated levels of glutathione (Freeman et al., 2010). The link between glutathione depletion and superoxide accumulation was also evident in an Arabidopsis mutant with a knockout in APR2, the dominant APR isozyme that completes sulphate reduction and probably mediates selenate reduction as well. Despite apr2-1 plants being two-fold less tolerant to selenate, they accumulated fewer selenoproteins than wild-type plants, suggesting that Se toxicity and selenoprotein accumulation can be uncoupled (Grant et al., 2011). Instead, the apr2-1 Se-sensitive phenotype was explained by elevated levels of selenate, which decreased the content of glutathione and increased the accumulation of superoxide. Se-induced superoxide, similar to other types of ROS, can damage cellular components such as membranes and proteins. For example, Triticum aestivum seedlings treated with 100 µm selenate had a two- to three-fold increase in lipid peroxidation compared with control (Łabanowska et al., 2012). Similarly, selenate treatment also compromised membrane integrity in Hordeum vulgare, which was accompanied by increased activity of ROS-scavenging enzymes, including superoxide dismutase, catalase and ascorbate peroxidase (Akbulut and Cakir, 2010). Similarly, increased activity of ROS-scavenging enzymes has also been reported in Se-treated Ulva species (Schiavon et al., 2012). Selenate treatment is also likely to oxidize proteins in plants, as observed in Stanleya pinnata plants. However, a majority of the oxidized proteins were safely removed by the proteasome, thus protecting the plants against Se toxicity (Sabbagh and Van Hoewyk, 2012).
In addition to selenate, it is now evident that selenite treatment also induces oxidative stress and the accumulation of ROS in plants. For example, in vitro experiments have confirmed that selenite reacts with glutathione to produce superoxide (Spallholz, 1994). In photosynthetic organisms, the accumulation of selenite-induced ROS can also be partially mitigated by an increase in the activity of ROS-scavenging enzymes, as reported in Coffea arabica cells (Gomes-Junior et al., 2007) and the cyanobacterium Spirula platensis (Chen et al., 2008). Coinciding with this observed selenite response, superoxide and hydrogen peroxide can accumulate in the leaves of Arabidopsis plants treated with selenite. Compared with wild-type plants, the vtc1 mutant with a defect in ascorbic acid biosynthesis accumulates more superoxide and hydrogen peroxide during selenite treatment; this suggests that the antioxidant properties of ascorbic acid help alleviate ROS accumulation in plants challenged with selenite (Tamaoki et al., 2008). ROS can act as signalling molecules, and it was proposed that selenite-induced ROS mitigate a selenite-defensive response. Additionally, the role of ROS in plant necrosis and programmed cell death is well known (De Pinto et al., 2012). In fact, two recent studies have linked selenite-induced ROS accumulation to cell death in roots. In one study, hydrogen peroxide accumulation in the primary roots of Arabidopsis was positively correlated with both the concentration of selenite and cell mortality (Lehotai et al., 2012). In another study, superoxide was also shown to accumulate in the root tips of Vicia faba (Mroczek-Zdyrska and Wójcik, 2012), and resulted in increased cell mortality and lipid peroxidation in root tips. Together, these data would suggest that Se-induced oxidative stress could be a major factor influencing Se toxicity in plants.
The reduction of selenite to selenocysteine may produce additional inorganic Se metabolites that could also induce oxidative stress in plants. The non-enzymatic reduction of selenite mediated by glutathione generates selenodiglutathione, which has been shown to be more toxic than selenite and capable of inducing mitochondrial superoxide (Wallenberg et al., 2010). Additionally, selenide has been recently shown to induce chromosome fragmentation in yeast (Peyroche et al., 2012). How selenodiglutathione and selenide directly affect Se toxicity in plants remains to be determined.
FUTURE PERSPECTIVES: UNEXPLORED TARGETS OF SELENIUM TOXICITY
As suggested above, the physiological ramifications of non-specific selenoproteins in plants could possibly hinder electron transport. In addition to mitochondrial respiration, photosynthesis is dependent upon an electron transport chain containing numerous Fe–S proteins and a few cysteine-rich proteins that are at increased risk of containing selenoproteins. Therefore, it is proposed that the accumulation of non-specific selenoproteins in the chloroplast could also hinder photosynthesis. Perhaps the protein in the chloroplastic electron transport chain that is the most likely to contain a non-specific selenoprotein is the essential and conserved protein PsaC. This small protein contains two Fe–S clusters and is cysteine-rich; in Arabidopsis, nine out of the 81 amino acids contain cysteine, eight of which coordinate Fe–S clusters. In PsaC, a cysteine to selenocysteine replacement could affect the binding of the Fe–S cluster. It is also possible that PsaC would contain an Fe–Se cluster. With this in mind, the formation of non-specific selenoproteins and Fe–Se clusters might decrease the rate of photosynthetic electron transport, as observed in selenate- treated Triticum aestivum (Łabanowska et al., 2012) and Chlamydomonas reinhardtii (Geoffroy et al., 2007). In conclusion, the abundance of Fe–S proteins coordinated by cysteine residues in the photosynthetic electron transport chain makes it susceptible to the replacement of sulphur with Se in protein.
Although the detrimental effects of Se on photosynthesis are known, the impact of Se stress on respiration is not well studied and probably deserves investigation. About 80 % of the glutathione pool is localized to the mitochondria (Zechmann et al., 2008), where it participates in the import and folding of mitochondrial proteins. Given that selenate can rapidly decrease the pool of glutathione, it is certainly likely that Se stress can quickly perturb mitochondrial processes in plants. In support of this hypothesis, human cells treated with selenite accumulate mitochondrial superoxide (Wallenberg et al., 2010); in Arabidopsis, superoxide is known to decrease the activity of aconitase (Sweetlove et al., 2002), an enzyme which participates in the tricarboxylic acid cycle. If the major culprit of Se toxicity in plants is oxidative stress induced by glutathione deficiency or aconitase inhibition, then it is expected that respiration would be adversely affected.
It is predicted that the replacement of cysteine with selenocysteine in non-specific selenoproteins would invoke the unfolded protein response, which would elicit the removal of these malformed proteins via the cytosolic proteasome. Both malformed and Se-induced oxidized proteins could form protein aggregates in the cytosol if not removed by the proteasome. If selenoproteins are unfolded, then they could also overwhelm the endoplasmic reticulum (ER) where secreted proteins are folded. In support of this view, Arabidopsis plants with a knockout of bip2 (an ER chaperone protein responsible for proper protein folding) could not survive past germination when grown on selenocysteine, perhaps because proper protein folding was compromised due to the formation of diselenide bonds in the ER (Sabbagh and Van Hoewyk, 2012). Thus, perhaps cytotoxicity of Se in plants also targets the ER. The story on protein misfolding induced by diselenide bonds is compelling but far from complete. Direct evidence pointing to an accumulation of misfolded selenoproteins in the ER could strengthen the case that non-specific selenoproteins are impaired as predicted.
Conclusions
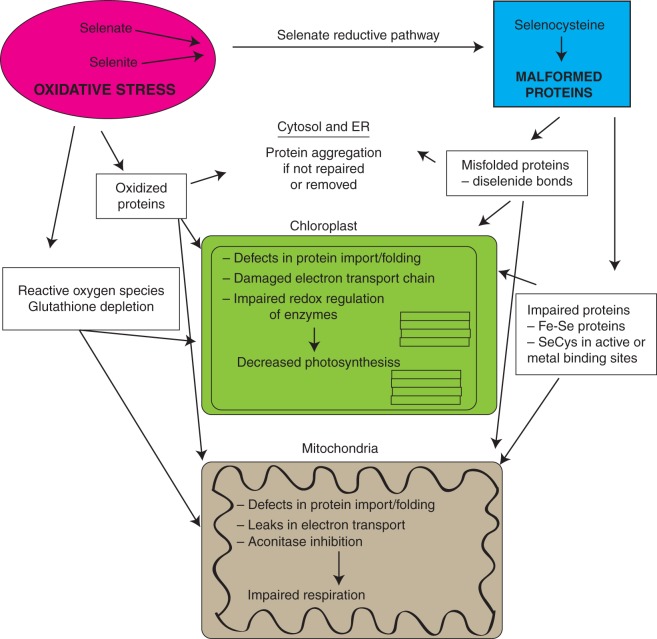
A central tenet in biology is that structure and function are inseparable, and this certainly applies to plant proteins. Subtle changes in protein folding can greatly affect protein function, and consequently the accumulation of non-specific selenoproteins is associated with Se toxicity in plants. So do malformed selenoproteins act in concert with oxidative stress to impart Se toxicity, or is one mechanism the key factor governing Se toxicity? Evidence is presented to suggest both play a role in plant Se toxicity and perhaps target the same cellular compartments in plants (Fig. 2). Therefore, challenges will probably remain in deciphering between Se toxicity induced by oxidative stress and non-specific selenoproteins. On the one hand, treating plants with selenocysteine can lead to non-specific selenoproteins, and yet free selenocysteine might be reactive and deplete NADPH (Lu and Holmgren, 2009), although it is not known if free selenocysteine induces oxidative stress in plants. Treating plants with inorganic Se also does not provide clear answers on the dual mechanisms of Se toxicity, because it can directly produce ROS such as superoxide, but it can also be assimilated into selenocysteine and readily misincorporated into non-specific selenoproteins. The accumulation of selenoproteins in mitochondria and the chloroplast could impair and cause leaks in electron transport, which also can result in the generation of superoxide (Staniek et al., 2002). Therefore, treating plants with inorganic Se could potentially lead to oxidative stress and the accumulation of non-specific selenoproteins that are malformed. As a case in point, Se-tolerant transgenic Arabidopsis plants overexpressing a Brassica oleracea methyltransferase had significantly lower amounts of hydrogen peroxide and superoxide when treated with selenite, despite accumulating the same concentration of total Se as the wild-type (Zhou et al., 2009). The increased Se tolerance in these plants was explained by the increased volatilization of dimethyl diselenide, which presumably redirected selenocysteine away from protein misincorporation; alternatively, it seems plausible that the increased Se tolerance could be explained by the significant decrease in ROS accumulation. This observation begs the question as to whether the decreased accumulation of ROS in the transgenic plants is attributable to a diminished pool of inorganic Se or a decreased accumulation of malformed selenoproteins in mitochondria or chloroplasts.
Fig. 2.
A proposed model describing how the two distinct types of Se toxicity might affect plant physiology. Inorganic Se contributes to oxidative stress, while its reduction to selenocysteine can inadvertently replace cysteine to create malformed selenoproteins. The possible targets and ramifications of Se-induced oxidative stress and non-specific selenoproteins are proposed, and illustrate the challenges of deciphering if Se stress is more attributable to just one mode of toxicity.
Some Se compounds are known to be anticarcinogenic in vitro, which could one day prompt the development of Se-based therapeutics or nutraceutical crops with elevated levels of Se. Determining the driving force of plant Se toxicity is important if augmenting Se tolerance and accumulation in crops is desirable. If it is discovered that malformed and unfolded selenoproteins exert most of Se toxicity in crops, Se stress could be alleviated by the creation of plants with a finely tuned unfolded protein response, including increased chaperones or proteasomal activity. Rather, if Se stress is mostly dictated by oxidative stress in plants, strategies to increase the ROS scavenging capacity in crops could be envisaged. It is hoped that future studies will consider both malformed selenoproteins and oxidative stress as two distinct mechanisms driving Se stress and aim to distinguish these different modes of Se toxicity in plants.
ACKNOWLEDGEMENTS
D.V.H. thanks Elizabeth Pilon-Smits and Marinus Pilon for reading a previous version of the manuscript and kindly providing helpful suggestions. Support for this research was provided by the National Science Foundation (MCB-0950648).
LITERATURE CITED
- Akbulut M, Çakır S. The effects of Se phytotoxicity on the antioxidant systems of leaf tissues in barley (Hordeum vulgare L.) seedlings. Plant Physiology and Biochemistry. 2010;48:160–166. doi: 10.1016/j.plaphy.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Aldag C, Gromov IA, García-Rubio I, et al. Probing the role of the proximal heme ligand in cytochrome P450cam by recombinant incorporation of selenocysteine. Proceedings of the National Academy of Sciences USA. 2009;106:5481–5486. doi: 10.1073/pnas.0810503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Pilon M. Ancient and essential: the assembly of iron-sulfur clusters in plants. Trends in Plant Science. 2011;16:18–26. doi: 10.1016/j.tplants.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Brown TA, Shrift A. Identification of selenocysteine in the proteins of selenate-grown Vigna radiata. Plant Physiology. 1980;66:758–761. doi: 10.1104/pp.66.4.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TA, Shrift A. Exclusion of selenium from proteins of selenium-tolerant Astragalus species. Plant Physiology. 1981;67:1051–1053. doi: 10.1104/pp.67.5.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Châtelain E, Satour P, Laugier E, et al. Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proceedings of the National Academy of Sciences USA. 2013;110:3633–3638. doi: 10.1073/pnas.1220589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TF, Zheng WJ, Wong YS, Yang F. Selenium-induced changes in activities of antioxidant enzymes and content of photosynthetic pigments in Spirulina platensis. Journal of Integrative Plant Biology. 2008;50:40–48. doi: 10.1111/j.1744-7909.2007.00600.x. [DOI] [PubMed] [Google Scholar]
- Davis CD. Selenium supplementation and cancer prevention. Current Nutrition Reports. 2012;1:16–23. [Google Scholar]
- Davis TZ, Stegelmeier BL, Panter KE, Cook D, Gardner DR, Hall JO. Toxicokinetics and pathology of plant-associated acute selenium toxicosis in steers. Journal of Veterinary Diagnostic Investigation. 2012;24:319–327. doi: 10.1177/1040638711435407. [DOI] [PubMed] [Google Scholar]
- De Pinto MC, Locato V, De Gara L. Redox regulation in plant programmed cell death. Plant, Cell and Environment. 2012;35:234–244. doi: 10.1111/j.1365-3040.2011.02387.x. [DOI] [PubMed] [Google Scholar]
- Dilworth GL, Bandurski RS. Activation of selenate by adenosine 5-triphosphate sulphurylase from Saccharomyces cerevisiae. Biochemical Journal. 1977;163:521–529. doi: 10.1042/bj1630521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kassis E, Cathala N, Rouached H, et al. Characterization of a selenate-resistant Arabidopsis mutant: root growth as a potential target for selenate toxicity. Plant Physiology. 2007;143:1231–1241. doi: 10.1104/pp.106.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mehdawi AF, Pilon-Smits EAH. Ecological aspects of plant selenium hyperaccumulation. Plant Biology. 2012;14:1–10. doi: 10.1111/j.1438-8677.2011.00535.x. [DOI] [PubMed] [Google Scholar]
- Eustice DC, Kull FJ, Shrist A. Selenium toxicity: aminoacylation and peptide bond formation with selenomethionine. Plant Physiology. 1981;67:1054–1058. doi: 10.1104/pp.67.5.1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng R, Chaoyang W, Tu S. The roles of selenium in protecting plants against abiotic stresses. Environmental and Experimental Botany. 2013;87:58–68. [Google Scholar]
- Fordyce FM, Guangdib Z, Greena K, Xinping K. Soil, grain and water chemistry in relation to human selenium-responsive diseases in Enshi District, China. Applied Geochemistry. 2000;15:117–132. [Google Scholar]
- Freeman JL, Zhang LH, Marcus MA, Fakra S, Pilon-Smits EAH. Spatial imaging, speciation and quantification of selenium in the hyperaccumulator plants Astragalus bisulcatus and Stanleya pinnata. Plant Physiology. 2006;142:124–134. doi: 10.1104/pp.106.081158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman JL, Tamaoki M, Stushnoff C, et al. Molecular mechanisms of selenium tolerance and hyperaccumulation in Stanleya pinnata. Plant Physiology. 2010;153:1630–1652. doi: 10.1104/pp.110.156570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant K, Carey NM, Mendoza M, et al. Adenosine 5-phosphosulfate reductase (APR2) mutation in Arabidopsis implicates glutathione deficiency in selenate toxicity. Biochemical Journal. 2011;438:325–335. doi: 10.1042/BJ20110025. [DOI] [PubMed] [Google Scholar]
- Geoffroy L, Gilbin R, Simon O, et al. Effect of selenate on growth and photosynthesis of Chlamydomonas reinhardtii. Aquatic Toxicology. 2007;83:149–158. doi: 10.1016/j.aquatox.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Gomes-Junior RA, Gratão PL, Gaziola SA, Mazzafera P, Lea PJ, Azevedo RA. Selenium-induced oxidative stress in coffee cell suspension cultures. Functional Plant Biology. 2007;34:449–456. doi: 10.1071/FP07010. [DOI] [PubMed] [Google Scholar]
- Hallenbeck PC, George GN, Prince RC, Thorneley RN. Characterization of a modified nitrogenase Fe protein from Klebsiella pneumoniae in which the 4Fe4S cluster has been replaced by a 4Fe4Se cluster. Journal of Biological Inorganic Chemistry. 2009;14:673–682. doi: 10.1007/s00775-009-0480-1. [DOI] [PubMed] [Google Scholar]
- Hazebrouck S, Camoin L, Faltin Z, Strosberg AD, Eshdat Y. Substituting selenocysteine for catalytic cysteine 41 enhances enzymatic activity of plant phospholipid hydroperoxide glutathione peroxidase expressed in Escherichia coli. Journal of Biological Chemistry. 2000;275:28715–28721. doi: 10.1074/jbc.M004985200. [DOI] [PubMed] [Google Scholar]
- Hondal RJ, Marino SM, Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxidants and Redox Signaling. 2012;18:1675–1689. doi: 10.1089/ars.2012.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux V, Dutilleul C, Jourdain A, Reynaud F, Lopez V, Bourguignon J. Arabidopsis putative selenium-binding protein1 expression is tightly linked to cellular sulfur demand and can reduce sensitivity to stresses requiring glutathione for tolerance. Plant Physiology. 2009;151:768–781. doi: 10.1104/pp.109.144808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Gladyshev VN. Different catalytic mechanisms in mammalian selenocysteine- and cysteine-containing methionine-R-sulfoxide reductases. PLoS Biology. 2005;3:e375. doi: 10.1371/journal.pbio.0030375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A, Mugford ST, Kopriva S. Arabidopsis root growth dependence on glutathione is linked to auxin transport. Plant Cell Reports. 2010;29:1157–1167. doi: 10.1007/s00299-010-0902-0. [DOI] [PubMed] [Google Scholar]
- Łabanowska M, Filek M, Kościelniak J, Kurdziel M, Kuliś E, Hartikainen H. The effects of short-term selenium stress on Polish and Finnish wheat seedlings—EPR, enzymatic and fluorescence studies. Journal of Plant Physiology. 2012;169:275–284. doi: 10.1016/j.jplph.2011.10.012. [DOI] [PubMed] [Google Scholar]
- LeDuc DL, Tarun AS, Montes-Bayon M, et al. Overexpression of selenocysteine methyltransferase in Arabidopsis and Indian Mustard increases selenium tolerance and accumulation. Plant Physiology. 2004;135:377–383. doi: 10.1104/pp.103.026989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehotai N, Kolbert Z, Pető A, et al. Selenite-induced hormonal and signaling mechanisms during root growth of Arabidopsis thaliana L. Journal of Experimental Botany. 2012;63:5677–5687. doi: 10.1093/jxb/ers222. [DOI] [PubMed] [Google Scholar]
- Lu J, Holmgren A. Selenoproteins. Journal of Biological Chemistry. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- Meyer J, Moulis JM, Lutz M. High-yield chemical assembly of [2Fe-2X] (X=S, Se) clusters into spinach apoferredoxin: product characterization by resonance Raman spectroscopy. Biochimica et Biophysica Acta. 1986;871:243–249. [Google Scholar]
- Mroczek-Zdyrska M, Wójcik M. The influence of selenium on root growth and oxidative stress induced by lead in Vicia faba L. minor plants. Biological Trace Element Research. 2012;147:320–328. doi: 10.1007/s12011-011-9292-6. [DOI] [PubMed] [Google Scholar]
- Müller S, Senn H, Gsell B, et al. The formation of diselenide bridges in proteins by incorporation of selenocysteine residues: biosynthesis and characterization of (Se)2-thioredoxin. Biochemie. 1994;33:3404–3412. doi: 10.1021/bi00177a034. [DOI] [PubMed] [Google Scholar]
- Ng BH, Anderson JW. Synthesis of selenocysteine by cysteine synthase from selenium accumulator and non-accumulator plants. Phytochemistry. 1978;17:2069–2074. [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, et al. Glutathione in plants: an integrated overview. Plant, Cell, and Environment. 2011;35:454–484. doi: 10.1111/j.1365-3040.2011.02400.x. [DOI] [PubMed] [Google Scholar]
- Neuhierl B, Bock A. On the mechanism of selenium tolerance in selenium-accumulating plants. Purification and characterization of a specific selenocysteine methyltransferase from cultured cells of Astragalus bisulcatus. European Journal of Biochemistry. 1996;239:235–238. doi: 10.1111/j.1432-1033.1996.0235u.x. [DOI] [PubMed] [Google Scholar]
- Papp LV, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxidant Redox Signaling. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- Peyroche G, Saveanu C, Dauplais M, Lazard M, et al. Sodium selenide toxicity is mediated by O2-dependent DNA breaks. PLoS ONE. 2012;7:e36343. doi: 10.1371/journal.pone.0036343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits EAH, Quinn CF. Selenium metabolism in plants. In: Hell R, Mendel R, editors. Cell biology of metal and nutrients. Berlin: Springer; 2010. pp. 225–241. [Google Scholar]
- Pilon-Smits EAH, Garifullina GF, Abdel-Ghany S, et al. Characterization of a NifS-like chloroplast protein from Arabidopsis. Implications for its role in sulfur and selenium metabolism. Plant Physiology. 2002;139:1309–1138. doi: 10.1104/pp.102.010280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos SJ, Rutzke MA, Haynes RJ, Faquin V, Guilherme LRG, Li L. Selenium accumulation in lettuce germplasm. Planta. 2010;233:649–660. doi: 10.1007/s00425-010-1323-6. [DOI] [PubMed] [Google Scholar]
- Sabbagh M, Van Hoewyk D. Malformed selenoproteins are removed by the ubiquitin-proteasome pathway in Stanleya pinnata. Plant Cell Physiology. 2012;53:555–564. doi: 10.1093/pcp/pcs015. [DOI] [PubMed] [Google Scholar]
- Schiavon M, Moro I, Pilon-Smit EAH, Matozzo V, Malagoli M, Vecchia FD. Accumulation of selenium in Ulva sp. and effects on morphology, ultrastructure and antioxidant enzymes and metabolites. Aquatic Toxicology. 2012;122:222–231. doi: 10.1016/j.aquatox.2012.06.014. [DOI] [PubMed] [Google Scholar]
- Sors TG, Ellis DR, Na GN, et al. Analysis of sulfur and selenium assimilation in Astragalus plants with varying capacities to accumulate selenium. Plant Journal. 2005;42:785–797. doi: 10.1111/j.1365-313X.2005.02413.x. [DOI] [PubMed] [Google Scholar]
- Spallholz JE. On the nature of selenium toxicity and carcinostatic activity. Free Radical Biology and Medicine. 1994;17:45–64. doi: 10.1016/0891-5849(94)90007-8. [DOI] [PubMed] [Google Scholar]
- Stadlober M, Sager M, Irgolic KJ. Effects of selenate supplemented fertilisation on the selenium level of cereals – identification and quantification of selenium compounds by HPLC-ICP-MS. Food Chemistry. 2001;73:357–366. [Google Scholar]
- Staniek K, Gille L, Kozlov AV, Nohl H. Mitochondrial superoxide radical formation is controlled by electron bifurcation to the high and low potential pathways. Free Radical Research. 2002;36:381–387. doi: 10.1080/10715760290021225. [DOI] [PubMed] [Google Scholar]
- Sweetlove LJ, Haezlewood JL, Herald V, et al. The impact of oxidative stress on Arabidopsis mitochondria. The Plant Journal. 2002;32:891–904. doi: 10.1046/j.1365-313x.2002.01474.x. [DOI] [PubMed] [Google Scholar]
- Terry N, Zayed A, de Souza P, Tarun A. Selenium in higher plants. Annual Review of Plant Physiology and Plant Molecular Biology. 2000;51:401–432. doi: 10.1146/annurev.arplant.51.1.401. [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Freeman JL, Pilon-Smits EAH. Cooperative ethylene and jasmonic acid signaling regulates selenate resistance in Arabidopsis. Plant Physiology. 2008;146:1219–1230. doi: 10.1104/pp.107.110742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Garifullina GF, Ackley AR, et al. Overexpression of AtCpNifS enhances selenium tolerance and accumulation in Arabidopsis. Plant Physiology. 2005;139:1518–1528. doi: 10.1104/pp.105.068684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoewyk D, Takahashi H, Inoue E, Hess A, Tamaoki M, Pilon-Smits EAH. Transcriptome analyses give insight into selenium-stress responses and selenium tolerance mechanisms in Arabidopsis. Physiologia Plantarum. 2008;132:236–253. doi: 10.1111/j.1399-3054.2007.01002.x. [DOI] [PubMed] [Google Scholar]
- Van Huysen T, Abdel-Ghany S, Hale KL, LeDuc D, Terry N, Pilon-Smits EAH. Overexpression of cystathionine-gamma-synthase in Indian mustard enhances selenium volatilization. Planta. 2003;218:71–78. doi: 10.1007/s00425-003-1070-z. [DOI] [PubMed] [Google Scholar]
- Wallenberg M, Olm E, Hebert C, Björnstedt M, Fernandes AP. Selenium compounds are substrates for glutaredoxins: a novel pathway for selenium metabolism and a potential mechanism for selenium-mediated cytotoxicity. Biochemical Journal. 2010;429:85–93. doi: 10.1042/BJ20100368. [DOI] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wang YD, Wang X, Wong YS. Proteomics analysis reveals multiple regulatory mechanisms in response to selenium in rice. Journal of Proteomics. 2012;75:1849–1866. doi: 10.1016/j.jprot.2011.12.030. [DOI] [PubMed] [Google Scholar]
- Zechmann B, Mauch F, Sticher L, Müller M. Subcellular immunocytochemical analysis detects the highest concentrations of glutathione in mitochondria and not in plastids. Journal of Experimental Botany. 2008;59:4017–4027. doi: 10.1093/jxb/ern243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Yuan Y, Yang Y, et al. Involvement of a broccoli COQ5 methyltransferase in the production of volatile selenium compounds. Plant Physiology. 2009;151:528–540. doi: 10.1104/pp.109.142521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YG, Pilon-Smits EA, Zhao FJ, Williams PN, Meharg AA. Selenium in higher plants: understanding mechanisms for biofortification and phytoremediation. Trends in Plant Science. 2009;14:436–442. doi: 10.1016/j.tplants.2009.06.006. [DOI] [PubMed] [Google Scholar]