Abstract
Background and Aims
Tribe Arabideae are the most species-rich monophyletic lineage in Brassicaceae. More than 500 species are distributed in the majority of mountain and alpine regions worldwide. This study provides the first comprehensive phylogenetic analysis for the species assemblage and tests for association of trait and characters, providing the first explanations for the enormous species radiation since the mid Miocene.
Methods
Phylogenetic analyses of DNA sequence variation of nuclear encoded loci and plastid DNA are used to unravel a reliable phylogenetic tree. Trait and ancestral area reconstructions were performed and lineage-specific diversification rates were calculated to explain various radiations in the last 15 Myr in space and time.
Key Results
A well-resolved phylogenetic tree demonstrates the paraphyly of the genus Arabis and a new systematic concept is established. Initially, multiple radiations involved a split between lowland annuals and mountain/alpine perennial sister species. Subsequently, increased speciation rates occur in the perennial lineages. The centre of origin of tribe Arabideae is most likely the Irano-Turanian region from which the various clades colonized the temperate mountain and alpine regions of the world.
Conclusions
Mid Miocene early diversification started with increased speciation rates due to the emergence of various annual lineages. Subsequent radiations were mostly driven by diversification within perennial species during the Pliocene, but increased speciation rates also occurred during that epoch. Taxonomic concepts in Arabis are still in need of a major taxonomic revision to define monophyletic groups.
Keywords: Arabis, Arabideae, Brassicaceae, centre of origin, Draba, trait evolution, diversification rates
INTRODUCTION
Rapid radiations with increased rates of diversification, due to increased speciation rates and/or decreased rates of extinction, are generally considered to be influenced by intrinsic traits (so-called key innovations) or extrinsic events (so-called key opportunities) (Hodges, 1997; Moore and Donoghue, 2009; Vamosi and Vamosi, 2011). The emergence of morphological, physiological or behavioural novelties has been linked particularly to subsequent diversification [e.g. floral nectar spurs (Hodges, 1997); flower asymmetry (Sargent, 2004; Kay and Sargent, 2009); dispersal ability and feeding generalization (Phillimore et al., 2006)]. In most cases the link between increased diversification and the emergence of a specific trait or event is based on correlations and plausible explanation chains, which is an intuitive and appealing approach (Ree, 2005; Vamosi and Vamosi, 2011), especially when the foundations are laid in a well-resolved phylogenetic framework (Drummond et al., 2012).
Brassicaceae are an angiosperm family in which morphological characters are highly homoplastic and of limited diagnostic value at the level of tribes and sometimes even at the level of genera (Koch and Al-Shehbaz, 2009; Al-Shehbaz, 2012). Consequently, there are few studies attempting to link diversification and speciation in this family with trait or character evolution. Recently, there has been progress in the establishment of a fully resolved phylogenetic tree for Brassicaceae (e.g. Koch et al., 2003, 2007; Al-Shehbaz et al., 2006; Bailey et al., 2006; Beilstein et al., 2008, 2010; German et al., 2009; Couvreur et al., 2010; Warwick et al., 2010). In its current delimitation the Brassicaceae (320 genera with 3660 species) are organized in 49 monophyletic tribes and <3 % of taxa (34 genera with 90 species) remain unassigned to one of these tribes (Al-Shehbaz, 2012) because voucher material either is not suitable for molecular analysis or is unavailable. The systematic backbone of the family is represented by three major evolutionary lineages (Beilstein et al., 2006) with tribe Aethionemeae sister to the rest of the family (Franzke et al., 2011), but many relationships between tribes and core lineages remain uncertain (Warwick et al., 2010; Al-Shehbaz, 2012). Tribe Arabideae belong to an expanded lineage II (Franzke et al., 2009) with various genera being well known, for example, as ornamental plants and often cultivated (e.g. Arabis, Aubrieta or Draba). In this expanded lineage there are another 18 tribes but relationships among these tribes remain uncertain, and only tribe Stevenieae has been shown to be sister to Arabideae (German et al., 2009).
There is increasing evidence that the early diversification events in the core family are characterized by rapid radiations (Franzke et al., 2009; Couvreur et al., 2010), and polyploid evolution involving whole genome duplications (WGDs) might have triggered the radiation of many lineages (Lysak et al., 2005, 2009; Koch, 2012; Schranz et al., 2012).
The rapid radiation of Brassicaceae involved colonization and adaptation into newly available ecological niches. This evolutionary history explains the novel morphological and ecological traits associated with shifts in mating system (Hurka et al., 2005; Ansell et al., 2008; Paetsch et al., 2010; Koelling and Mauricio, 2010), flowering time, flower symmetry and pollination biology (Busch and Zachgo, 2007; Alonso-Blanco et al., 2009), diaspore dispersal and germination (Alonso-Blanco et al., 2009; Linkies et al., 2010), and shifts between annual and perennial growth (Wang et al., 2009). However, all these studies remain speculative to linking traits and characters with radiation of the respective lineage, either because of the lack of fully resolved phylogenetic trees or due to speculative uncertainties. Moreover, these studies mainly focused on a single species or genus (for details refer to Franzke et al., 2011). At higher taxonomic levels, the first correlations between trait-evolution and molecular phylogeny were accomplished on trichome branching patterns in the Brassicaceae (Beilstein et al., 2006, 2008) and fruit characters have also been analysed in a phylogenetic perspective in tribes Isatideae (Moazzeni et al., 2007) and Brassiceae (Hall et al., 2011). However, in all of these examples the trait could not be linked to any adaptive evolutionary processes. The lack of adequate experimental and observational studies means that our knowledge of the adaptive significance of these traits is tentative at best.
Reticulate evolution and polyploidization resulting in WGDs (e.g. Lysak et al., 2005, 2009; Soltis et al., 2009; Schranz et al., 2012) are considered as major factors driving diversification. Genomes in Brassicaceae seem highly conserved in structure but not in genome size. The genomes are arranged in defined genomic blocks (Schranz et al., 2006) and polyploidization occurs frequently, increasing block number and total genome size (e.g. Jordon-Thaden and Koch, 2008), but the genomes are stabilized rapidly and are efficiently downsized in most lineages (Lysak et al., 2009). The idea that WGDs initiate speciation by reciprocal gene loss or subfunctionalization (van de Peer et al., 2009) is plausible and it has been demonstrated that WGDs could be associated with the formation of novel, evolutionary important traits (van de Peer et al., 2009; Hoffmann et al., 2012; Schranz et al., 2012 and references therein; Fawcett et al., 2013).
Tribe Arabideae is the largest tribe in Brassicaceae (Al-Shehbaz, 2012), and the clades differ widely in species number and evolutionary putatively important traits such as life-cycle history, geographical and altitudinal distribution, or percentage of polyploid species. Changes in life-cycle strategy might be a particularly important factor in diversification of Arabideae (Jordon-Thaden et al., 2010). The sister relationship between species-poor annual and species-rich perennial clades has been highlighted as an interesting pattern, as it is potentially widespread in Brassicaceae (see Karl et al., 2012) and other plant families [e.g. Ehrharta, Poaceae (Verboom et al., 2003); American Lupinus, Fabaceae (Drummond, 2008; Drummond et al., 2012); subtribe Castillejinae, Orobanchaceae (Tank and Olmstead, 2008); and tribe Delphinieae, Ranunculaceae (Jabbour and Renner, 2012)]. Systematics of Arabideae has been challenging as a result of extreme convergence in almost all conceivable morphological characters (Koch et al., 2012a); and most of these characters appear in various combinations in different tribes and evolutionary lineages, making the genus Arabis in its original and past circumscription the most critical taxon of Brassicaceae (Koch et al., 1999, 2000, 2001). The totally revised tribe, currently with 17 genera, can be arranged in several phylogenetically well-defined clades (Karl et al., 2012; Koch et al., 2012a). However, these studies were missing well-resolved relationships between clades, as sequence data [nrDNA internal transcribed spacer (ITS) and plastid DNA trnL-trnF] did not resolve the backbone structure of the phylogenetic tree and therefore greatly limited any pilot study in Brassicaceae on diversification in a large monophyletic group.
Here, we aim to present a comprehensive and reliable phylogenetic framework for the tribe, which is achieved by reconstructing a backbone phylogeny focusing on a selected set of nuclear genes. The backbone is used for a tribal phylogenetic reconstruction, including all evolutionary groups and the majority of taxa described so far. The phylogenetic hypothesis is then used to correlate evolutionary processes in space and time. In particular we will highlight the questions of how an increase in speciation rates is linked to life history, and how these rates relate to different elevations in mountainous and alpine habitats, and shifts in ploidy. We present the first complete biogeographical analysis of an entire tribe of Brassicaceae and try to provide insights about its origin, which is crucial for an understanding of the family.
MATERIAL AND METHODS
Taxon sampling and phylogenetic data set composition
To resolve the backbone structure of the phylogenetics of tribe Arabideae we generated a data set of three nuclear loci from 16 representatives of the various clades and groups in the tribe: the ITS regions 1 and 2 of nuclear encoded ribosomal RNA (hereafter ITS region), the chalcone synthase (Chs) gene consisting of two exons and one intron, and the alcohol dehydrogenase (Adh) genome consisting of seven exons and six introns. For this data set Pseudoturritis turrita from the sister tribe Stevenieae was chosen as an outgroup taxon (see, e.g., Koch et al., 2012a). Hereafter, this nuclear data set is referred to as the ‘three-marker data set’.
All further analyses were performed with the ITS data set containing a total of 312 accessions. Detailed information about the various accessions is summarized in Supplementary Data Table S1. Additionally, for the same 312 accessions a data set was generated based on the plastid trnL intron-trnL-trnF intergenic spacer region (hereafter named trnL-F). These two data sets include members of all 17 Arabideae genera and 305 different species. This makes these data sets the most comprehensive ones for the whole tribe. Itemized, the data sets comprise 71 species (74 accessions) of Arabis, 13 species (14 accessions) of Aubrieta, 197 species and accessions of Draba and 24 species from the remaining and smaller 14 Arabideae genera (Abdra, Arcyosperma, Athysanus, Baimashania, Borodiniopsis, Botschantzevia, Dendroarabis, Drabella, Pachyneurum, Parryodes, Pseudodraba, Scapiarabis, Sinoarabis and Tomostima). In addition, both data sets include three taxa from the sister tribe Stevenieae (Macropodium pterospermum, Pseudoturritis turrita, and Stevenia canescens) as outgroups (Koch et al., 2012a). The annotated alignments for the three data sets are provided in Supplementary Data Figs S1–3 (three-marker data set: S1; ITS region: S2; and trnL-F: S3). For chs and adh the alignments have been trimmed for coding regions only to perform subsequent phylogenetic analysis.
Distribution ranges, life cycles and elevations/habitats of the respective species were carefully compiled from various floras by the authors (Supplementary Data Table S2). In case of the Draba species we employed a data compilation presented earlier (Jordon-Thaden, 2010).
DNA extraction, PCR amplification and sequencing
DNA extractions were carried out according to a slightly modified protocol (Karl et al., 2012) of the CTAB method (Doyle and Doyle, 1987). PCR reactions to amplify the selected regions were performed in a final volume of 25 µL, using 10 µm of each primer, a total of 2·0 mm MgCl2 and 0·5 U of MangoTaq polymerase (Bioline, Luckenwalde, Germany). The following primers were used: ITS-18F (5′-GGAAGGAGAAGTCGTAACAAGG-3′), as modified by Mummenhoff et al. (1997); ITS-25R (5′-TCCTCCGCTTATTGATATGC-3′), designed by White et al. (1990); trnLfor (5′-CGAAATCGGTAGACGCTACG-3'), published as primer ‘c’ in Taberlet et al. (1991); trnLrev (5′-GGGGATAGAGGGACTTGAAC-3′), published as primer ‘d’ in Taberlet et al. (1991); trnL-Ffor (5′-GGTTCAAGTCCCTCTATCATCCC-3′), published as primer ‘e’ in Taberlet et al. (1991); and trnL-Frev (5′-GATTTTCAGTCCTCTGCTCTA C-3′), designed by Dobeš et al. (2004). Chalcone synthase sequences were first amplified with primers CHS-PRO1-fw (5′-CATCTGCCCGTCCATCAAACCTACC-3′) and CHS-EX2-TERM-rev (5′-TTAGAGAGGAACGCTCTGCAAGAC-3′) (Koch et al., 2000). Following our initial sequencing results, we subsequently designed a new and sequence-optimized primer pair ARA-N-CHS-1-for (5′-GGCACARAGAGCTGATGGA-3′) and ARA-N-CHS-5-rev (5′- AGAGAAGATGAGAGCRACWCG-3′), which is more specific for the orthologue copy specific to tribe Arabideae. Alcohol dehydrogenase sequences were amplified with the primer pair ADH-FOR-1 (5′-ACTACTGGTCAGATTATTCGATGC-3′) and ADH-REV-7 (5′-AAGCACCCATGGTAATGATGC-3′), as designed by Koch et al. (2000). For some accessions it was not possible to amplify the whole fragment at one stretch. In these cases the fragment was amplified in two parts and additionally the primers ADH-REV-4 (5′-CTAACCCAGTAGATAAACCACAAC-3′) and ADH-FOR-4 (5′-GTTAGTTGTGGTTTATCTACTGG-3′) (Koch et al., 2000) were utilized. The respective chs and adh fragments were purified and then cloned into the chemically competent Escherichia coli strain JM109 using the pGEM-T vector system (Promega, Madison, WI, USA). Five positively tested clones (PCR test) of each accession were sequenced using the universal T7 (5′-TAATACGACTCACTATAGGG-3′), and SP6 (5′-ATTTAGGTGACACTATAGAA-3′) primers (GATC, Konstanz, Germany; or MWG Eurofins, Ebersberg, Germany).
Further sequences for ITS and trnL-F were added from our past studies (Jordon-Thaden et al., 2010; Koch et al., 2010, 2012a; Karl et al., 2012).
Phylogenetic reconstruction of the Arabideae backbone structure
A Bayesian Markov chain Monte Carlo (MCMC) analysis (Yang and Rannala, 1997) was performed for the ‘three-marker data set’ using MrBayes v. 3.1.2 (Ronquist and Huelsenbeck, 2003). The data set was divided into three unlinked partitions (adh, chs and ITS) to apply the best fitting nucleotide substitution model to each locus, which were chosen using Modeltest v. 3.7 (Posada and Crandall, 1998). In the following analysis four simultaneous runs were performed with four chains each for 1 million generations, and in each run 1001 trees were sampled. The first 25 % of these trees were discarded as burn-in. The temperature of the heated chain was set to 0·01, as this facilitated the most efficient chain-swapping. Finally, posterior probabilities of all splits between the respective runs were compared and cumulative split frequency plots were analysed using the web-based program AWTY (Nylander et al., 2007) to confirm accomplished convergence between the individual Bayesian MCMC runs.
Constraint nodes and ITS and trnL-F phylogenetic analyses
In order to assign the well supported backbone structure of Arabideae to the single marker analyses, we applied a constrained topology to the phylogenetic analyses of the ITS data set based on the nuclear encoded three-marker set. We did not apply the constrained topology to the analyses of the trnL-F region, because a different genome is represented and because nuclear and plastidic genomes have proved to show different modes of molecular evolution in Arabideae (see Karl et al., 2012; Koch et al., 2012a).
To set up the constrained topology four nodes from the backbone of Arabideae were constrained, according to the phylogenetic reconstruction of the three-marker data set (see Fig. 1): (1) Constrained node 1 (CN1) constrains the monophyly of A. alpina and A. nordmanniana; (2) CN2 constrains CN1 and the A. auriculata clade as a monophyletic group; (3) CN3 constrains CN2, Aubrieta (including Arabis verna), Draba (and its segregates) and the A. aucheri clade as a monophyletic group; and (4) CN4 constrains CN3 and the main Arabis clade as a monophyletic group.
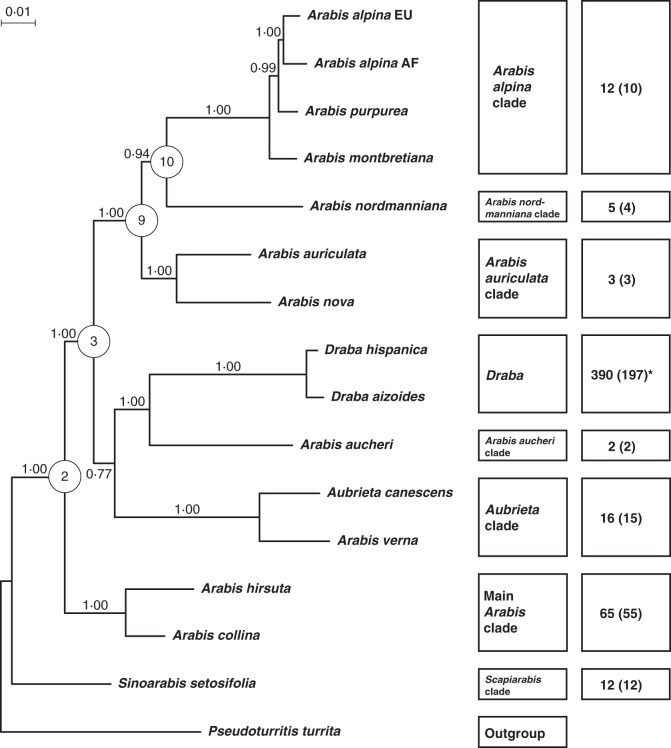
Fig. 1.
Phylogenetic tree of the Bayesian analysis based on the ‘three-marker data set’. Pseudoturritis turrita was used as the outgroup. Posterior probability values of the nodes are given along the corresponding branches. The first box on the right shows the affiliation of the individual taxa to different clades of Arabideae. The second box provides information about the clade-specific species numbers and, in parentheses, the number of sampled taxa. Four nodes (nodes 2, 3, 9 and 10), which were constrained in the subsequent analyses with the nrDNA ITS marker, are highlighted with their respective symbols of Fig. 2. *In addition, 10 (9) species of the Tomostima clade (annual segregates of Draba).
These constraints also correspond to the finding of our previous studies in the Arabideae (Karl et al., 2012; Koch et al., 2012a), making these constraints a highly reliable phylogenetic framework. However, further nodes, which connect two or more clades, remained unconstrained. We did not constrain the Aubrieta clade as sister to Draba and the A. aucheri clade because of the low posterior probability value (ppr < 0·90). Moreover, the position of the A. aucheri clade as sister of Draba also remained unconstrained because of the incongruency to the plastid DNA tree (Supplementary Data Fig. S4). This pattern is indicative of reticulate evolution and has been demonstrated to have happened in at least two clades of Arabideae (Jordon-Thaden et al., 2010; Koch et al., 2010).
Dated molecular phylogeny
Time-measured phylogenetic analyses of the ITS and trnL-F data sets were performed using the software package BEAST v. 1.6.2. (Drummond and Rambaut, 2007), which uses Bayesian MCMC to reconstruct phylogenetic trees. We performed the divergence time estimate calculations with a secondary calibration point for the tribe Arabideae introduced by Couvreur et al. (2010), which sets the age of the Arabideae between 10·0 and 23·9 Mya (mean 16·8 Mya) and fits well with our last estimates (Koch et al., 2012a). In the case of Arabideae, note that applying different divergence time estimates for the family (e.g. Beilstein et al., 2010) has only very limited effect on estimates for the Arabideae (Koch et al., 2012a). For a more detailed discussion on this topic the interested reader might refer to Franzke et al. (2011). The calculations for the ITS data set used the above-mentioned constraints, whereas the calculations for the trnL-F data set were performed without any constraints. Taxon subsets were specified for Arabideae (all taxa excluding Macropodium pterospermum, Pseudoturritis turrita and Stevenia canescens from tribe Stevenieae) and all clades of the tribe. A lognormal uncorrelated relaxed clock method was applied and a birth–death speciation model (Gernhard, 2008) was selected as tree prior. Four independent runs with 50 million generations were run for each marker on the freely available Bioportal server (www.bioportal.uio.no). In each run 10 001 generations were sampled.
The individual runs were checked in Tracer v.1.5 (Rambaut and Drummond, 2007) as having reached a stationary phase and a sufficient effective sample size (ESS > 200). All runs (log- and trees-files, respectively) were then combined with LogCombiner v.1.6.2. Finally, a single maximum clade credibility (MCC) tree with median branch lengths was produced for each marker, after burning in the first 10 000 trees using TreeAnnotator v.1.6.2.
Lineages through time plots
Lineages through time (LTT) plots were constructed with the above-mentioned single MCC trees, using the ape package (Paradis et al., 2004) of the R software environment (Ihaka and Gentleman, 1996). For each marker (ITS and trnL-F) three LTT plots were constructed. The first included the whole data set with all accessions. For the second LTT plot nearly all species of Draba and its annual segregates (Tomostima clade) were omitted, because this lineage represents >70 % of all species in tribe Arabideae and this high number might bias conclusions at the tribal level. Therefore, one single accession (D. aizoides) was included to represent the stem of Draba and the Tomostima clade. In a third LTT plot, and to differentiate between annuals and perennials, only the annual species from the total data set were considered.
Calculation of diversification rates
Clade-specific net diversification rates were estimated with BayesRate v. 1.4 (Silvestro et al., 2011). The program takes into account uncertainties of divergence times by estimating speciation and extinction rates on a distribution of trees. Furthermore, BayesRate can account for incomplete taxon sampling when computing the clade-specific diversification rates. This overcomes the problem that only half of the Draba species were included, but all other Arabideae genera were nearly complete in the LTT analyses.
Prior to the analyses, 1000 random trees were sampled from the 30 004 post burn-in trees from BEAST with the sub-sample trees feature implemented in BayesRate. For 12 specified clades and groups (see Table 2) speciation rates (S), extinction rates (E), net diversification rates (S – E) and the extinction fraction (a = E/S) were estimated under a birth–death model. Among the specified groups are two groups, which represent the annual and perennial members of Arabideae, respectively. For each marker (ITS and trnL-F) 100 million MCMC iterations (1000 runs with 100 000 iterations each) were run with a burn-in of 10 %. Results were checked and visualized in Tracer v.1·5.
Table 2.
Net diversification rates (speciation rate S – extinction rate E) and the extinction fraction (a = E/S) for 12 clades and groups of Arabideae calculated with BayesRate v. 1·4
| ITS |
trnL-F |
|||
|---|---|---|---|---|
| Net diversification rates | Extinction fraction (%) | Net diversification rates | Extinction fraction (%) | |
| Perennial clades/groups | ||||
| Core Draba | 0·402/0·894 (0·873)/1·402 | 17·8/46·0/71·8 | 0·304/0·641 (0·630)/0·998 | 15·0/43·0/69·6 |
| Main Arabis clade | 0·136/0·495 (0·469)/0·895 | 20·5/54·1/86·2 | 0·120/0·386 (0·365)/0·701 | 17·7/50·8/83·4 |
| Aubrieta clade | (0·072/0·977 (0·855)/2·149)* | 18·7/55·2/91·9 | 0·024/0·379 (0·333)/0·833 | 22·2/59·0/94·2 |
| Scapiarabis clade | 0·011/0·207 (0·182)/0·457 | 16·1/52·6/90·3 | 0·004/0·116 (0·099)/0·269 | 27·3/64·2/97·1 |
| Perennials of the A. alpina clade | (0·023/0·928 (0·758)/2·274)* | 23·3/60·8/96·0 | 0·010/0·334 (0·279)/0·795 | 20·8/57·2/94·4 |
| A. nord-manniana clade | (0·007/1·056 (0·802)/2·833)* | 27·2/64·7/97·5 | (0·006/0·892 (0·623)/2·601)* | 26·6/64·0/97·4 |
| Annual clades | ||||
| Tomostima clade | 0·009/0·218 (0·190)/0·483 | 20·5/57·2/93·6 | 0·010/0·210 (0·179)/0·481 | 18·6/55·9/92·2 |
| A. auriculata clade | 0·000/0·198 (0·128)/0·616 | 27·5/64·4/97·9 | 0·000/0·062 (0·042)/0·184 | 29·4/66·1/98·6 |
| A. aucheri clade | (0·000/0·475 (0·214)/1·881)* | 26·4/63·0/97·6 | (0·000/0·657 (0·269)/2·615)* | 25·5/62·9/96·8 |
| Arabideae | 0·177/0·381 (0·373)/0·591 | 56·1/74·0/89·3 | 0·196/0·393 (0·378)/0·603 | 32·3/56·8/79·5 |
| Annuals | 0·039/0·153 (0·147)/0·274 | 18·3/53·0/86·6 | 0·043/0·143 (0·137)/0·252 | 12·8/45·8/79·5 |
| perennials | 0·209/0·443 (0·434)/0·690 | 50·6/70·6/88·2 | 0·203/0·419 (0·411)/0·643 | 29·8/55·3/79·1 |
Mean values are indicated in bold, median values in parentheses; the lower and upper bounds of the 95 % highest posterior density (HPD) interval are given before and after these values, respectively.
* Large 95 % HPD intervals make these estimates less reliable.
Ancestral area reconstructions (AARs)
Distinct geographical regions were carefully specified and thereby roughly orientated on terrestrial ecozones and their actual outline and boundaries followed largely the major habitat types (MHTs)/ecoregions of the ‘WWF ecoregions 200’ map (Olson et al., 2001). However, the maximum number of discrete geographical units was restricted to 15 by the limitation of the RASP software (Yu et al., 2011), and in some cases adjustments had to be made to accommodate the actual distribution ranges of species and their allocation to the individual geographical regions. Finally, 14 geographical regions were defined (see Fig. 2 and Supplementary Data Table S3): North and Central America (A), South America (B), Iberian Peninsula & North Western Africa (C), Central Mediterranean & Southern Balkan (D), Europe (E), Western and Central Anatolia & Levantine coast (F), Caucasus, Eastern Anatolia & Iranian mountain ranges (G), high mountains of the Arabian peninsula and Eastern Africa (H), Central Asian mountain ranges (I), Eastern Himalaya and Tibet-Chinese mountains (J), Eastern Asia (K) and Siberia & Russian Far East (L). A more detailed characterization of these regions is given in Supplementary Data Table S3.
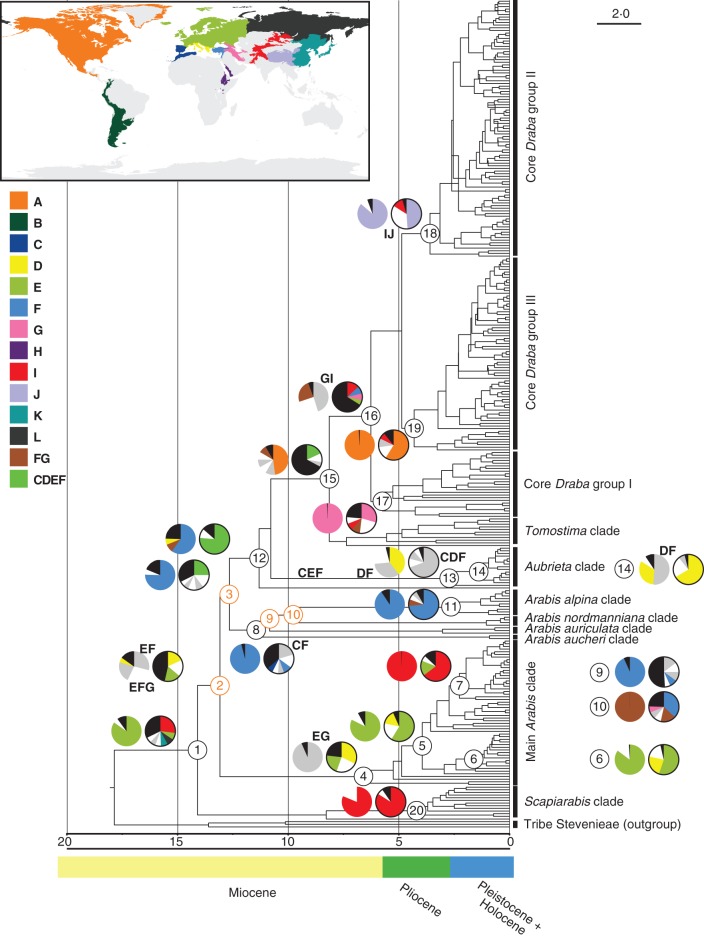
Fig. 2.
Maximum clade creditability (MCC) tree from the divergence time estimation in BEAST based on ITS sequence information containing 309 taxa of tribe Arabideae. The respective time scale is given below the tree. Vertical bars on the right show the affiliation to different clades of Arabideae. Unassigned taxa are not covered by these bars. The four constrained nodes (nodes 3, 8, 9 and 10, see Fig. 1) are indicated in orange font. For 20 nodes (nodes 1–20) the results of the ancestral area reconstructions [S-DIVA (left) and BBM (right, black-rimmed)] are displayed as pie charts, representing the possible ancestral ranges and their probabilities. Colour codes of specified geographical regions are: A: North & Central America, B: South America, C: Iberian peninsula & NW Africa, D: Central Mediterranean & Southern Balkans, E: Europe, F: Western and Central Anatolia & Eastern Mediterranean, G: Caucasus, Eastern Anatolia & Iranian mountain ranges, H: high mountains of the Arabian peninsula and Eastern Africa, I: Central Asian mountain ranges, J: Eastern Himalaya and Tibet-Chinese mountains, K: Eastern Asia, L: Siberia & Russian Far East. For the two most frequent multi-region areas (FG and CDEF) additional colour codes were specified. Black slices combine all regions with a probability <5 %. All other slices, which do not refer to one of the above mentioned areas, are alternately indicated in white or light grey. Their respective distribution area is given in some cases, when the area is the most probable area for a node or has a probability of 25 % or more.
Reconstructions of ancestral areas, i.e. the distribution area of the ancestor of a monophyletic group (Bremer, 1992), were performed using the program RASP 2.0b, which has both the Statistical DIVA (S-DIVA, Yu et al., 2010) and a Bayesian binary MCMC (BBM) approach implemented. For both approaches 10 000 trees were randomly sampled from the 30 004 post burn-in tree topologies of the BEAST analyses and frequencies of ancestral ranges were calculated and averaged over the sampled trees. The possible ancestral ranges are shown for every node present in the respective MCC trees. In the S-DIVA analysis the maximum number of regions per node was set to 3. This resulted in 300 possible area states, of which 49 area states were included; the remaining 251 area states, which contained non-adjacent regions, were excluded from the S-DIVA analyses.
In the BBM analyses four chains were run for 30 million generations. Every 30 000 generations ancestral area states were sampled. The maximum number of regions was set to 5 without any exclusion of area states, allowing more widespread and non-adjacent area states. The distribution of the virtual outgroup was set to ‘outgroup’. Stevenia canescens and Macropodium pterospermum represent their respective genera in respect of distribution and therefore are stated with a distribution range which exceeds the distribution range of the single species. Additionally, both analyses were performed for a dataset where Arabis caerulea was pruned, to test for the impact of its unresolved position at the base of the Arabideae phylogenetic tree.
Ancestral character state reconstructions
Ancestral character state analyses were run for life cycle strategy (annual versus perennial) and elevation of habitat, using the BBM approach of the program RASP 2.0b. Settings were made analogous to those of the AAR, but the maximum number of states was set to 2.
For the reconstructions of life-cycle strategy annual species were labelled with ‘A’, and perennial ones with ‘B’. Species described as either annuals or perennials were labelled with ‘AB’, indicating both life cycle strategies were possible. Some species, which are described to have an annual life cycle, can grow as short-lived perennials under favourable conditions (Arabis borealis, A. planisiliqua subsp. nemorensis, Draba albertina, D. bifurcata, D. brachystylis, D. corrugata, D. mogollonica and D. stenoloba). Nevertheless, they are stated as annuals. Stevenia canescens represents the genus Stevenia, in which both life cycles are present, and it is therefore labelled with the equivocal character ‘AB’.
For the reconstructions of habitat elevations taxa were labelled as lowland species (A) if they occur generally in lowland habitats and/or ascend to low montane elevations at most. Accordingly, taxa which occur generally in high montane/alpine habitats were labelled as alpine species (B). Occasional occurrences in higher or lower elevations were not taken into account. In contrast, taxa which generally occur both in lowland and in montane/alpine habitats were labelled with the equivocal character state ‘AB’ (e.g. arctic-alpine Arabis alpina, Draba aurea or D. fladnizensis). Stevenia canescens is labelled with the combined character ‘AB’ as it represents the whole genus, which includes several lowland species and one alpine species in Tibet. A comprehensive list presented as Supplementary Data Table S4 registers the applied distribution ranges and character states for life cycle and habitat.
RESULTS
Dataset information and phylogenetic trees
All new DNA sequences have been submitted to NCBI and accession codes are provided with Supplementary Data Table S1. The alignment of the ‘three-marker data set’ consisted of 2880 characters, of which 2224 were constant, 245 were autapomorphic and 411 were potentially parsimony-informative (Supplementary Data Fig. S1). As most appropriate evolutionary models, the TrN + I + Γ model was applied to the adh partition, the TrNef + I + Γ model to the chs partition and the GTR + I + Γ model to the ITS partition.
The resulting tree is presented in Fig. 1, and had a harmonic mean for the ln likelihood of –10 482·59 and an average standard deviation of split frequencies of 0·005. Contrary to previous phylogenetic studies in tribe Arabideae, the tree and especially its backbone is fully resolved and all nodes are strongly supported by posterior probability values (ppr > 0·9). The Arabis alpina clade (A. montbretiana, A. purpurea and two accessions of A. alpina) and A. nordmanniana are sistered by the A. auriculata clade (A. auriculata and A. nova). Draba and A. aucheri are sister to the Aubrieta clade (including Arabis verna). However, this sister group relationship has a lower support from posterior probability values (ppr = 0·77). The ‘three-marker data set’ also revealed the position of the main Arabis clade (A. collina and A. hirsuta) as sister to both groups mentioned above (A. alpina/A. nordmanniana/A. auriculata clades and Draba/A. aucheri/Aubrieta clades), and the early branching position of Sinoarabis setosifolia in Arabideae was confirmed.
The ITS alignment consisted of 656 characters, of which 318 were constant, 99 were autapomorphic and 239 were potentially parsimony-informative (Supplementary Data Fig. S2). As most appropriate evolutionary model, the GTR + I + Γ model was applied to the analyses.
The resulting tree is presented in Fig. 2 and its general outline and the respective positions of the clades are similar to the ‘three-marker tree’, mostly due to the four constrained backbone nodes. The sole exception is the position of the A. aucheri clade, which is sister to the A. alpina/A. nordmanniana/A. auriculata clades instead of being sister to Draba. Whereas the ‘three-marker tree’ is informative in resolving the Arabideae backbone structure, the ITS phylogenetic tree, based on a comprehensive data set, is highly indicative for the composition and internal structure of the individual clades. Nevertheless, support for all above-mentioned clades remained maximal in the comprehensive phylogenetic trees (Supplementary Data Figs S4 and S5). The results are congruent with previous studies in Arabideae (Draba – Jordon-Thaden et al., 2010; main Arabis – Koch et al., 2010; A. alpina clade – Karl et al., 2012; Scapiarabis clade – Koch et al., 2012a) and could affiliate numerous taxa to their respective clades. Moreover, the presented phylogenetic tree could verify several patterns which had been previously suggested: in particular, the inclusion of the monotypic genera Dendroarabis and Pachyneurum in the main Arabis clade (as mentioned in German et al., 2009) and the monophyly of Aubrieta as sister to Arabis verna (as indicated in Karl et al., 2012). Furthermore, it could be shown that Draba and its annual segregates (i.e. Abdra, Athysanus and Tomostima) form two, distinct and well-separated clades in the tribe (see Jordon-Thaden et al., 2010). Posterior probability (ppr) values for the individual nodes are given with Supplementary Data Fig. S5.
The combined trnL-F alignment (Supplementary Data Fig. S3) originally consisted of 1234 characters, but positions 1–5, 533–581 and 1209–1234 were excluded from the analyses due to the number of missing data. Furthermore, positions 219–407, a large indel in the trnL intron, which is only present in the two outgroup taxa Macropodium and Stevenia, were also excluded from the analyses. The final trnL-F alignment consisted of 965 characters, of which 638 were constant, 113 were autapomorphic and 214 were potentially parsimony-informative. As the most appropriate evolutionary model the TVM + I + Γ model was applied to the analysis.
The phylogenetic backbone of Arabideae remains unresolved in the trnL-F tree (presented in Supplementary Data Fig. S4), as most of the nodes only have negligible support from posterior probability values (ppr < 0·75). There are some further incongruencies between the plastid DNA (trnL-F) tree and the nuclear ‘three-marker tree’, mainly the structure of the Scapiarabis clade (see Koch et al., 2012a) and the position of the A. aucheri clade. However, both the sister-group relationship of the A. nordmanniana clade to the A. alpina clade and the sister-group relationship between Aubrieta and Draba are also displayed in the trnL-F tree.
Divergence time estimates and LTT plots
The evolution of the Arabideae tribe is placed in the Neocene, with its most probable origin in the mid Miocene (∼15 Mya). Divergence time estimates of the individual clades indicate that they originated in the late stages of the Miocene (6–8 Mya – Draba, main Arabis clade) or later in the Pliocene or Pleistocene. Hence, diversification in the different Arabideae clades took place in the last 5 Myr. The results of the divergence time calculations are listed in Table 1 and the phylogenetic tree is presented in Fig. 2. Mean values and confidence intervals for all major clades are congruent with previous studies of the tribe (Koch et al., 2010, 2012a; Karl et al., 2012).
Table 1.
Parameters and results of the divergence time estimate calculations using secondary calibration of node age of tribe Arabideae (Couvreur et al., 2010)
| Based on nrDNA ITS | Based on cpDNA trnL-F | |
|---|---|---|
| Runs/generations | Four independent runs with 50 million generations each | Four independent runs with 50 million generations each |
| log likelihood | –8605·87/–8568·87/–8533·48 | –6753·57/–6724·46/–6671·78 |
| tmrca of the tribe Arabideae (Ma) | 10·0/14·1/21·5 | 10·0/14·1/21·5 |
| tmrca of constrained node CN1 (Ma) | 5·00/9·80/16·2 | – |
| tmrca of constrained node CN2 (Ma) | 5·92/10·9/17·6 | – |
| tmrca of constrained node CN3 (Ma) | 8·14/12·7/19·8 | – |
| tmrca of constrained node CN4 (Ma) | 8·65/13·1/20·5 | – |
| tmrca of Draba and the Tomostima clade (Ma) | 4·73/8·14/12·9 (exclusive Drabella muralis) | 8·45/13·6/21·4 |
| tmrca of the core Draba (w/o Draba hederifolia and Draba verna) (Ma) | 3·48/6·26/10·0 | 4·51/7·79/12·4 (furthermore exclusive Draba crypthanta) |
| tmrca of Aubrieta (including Arabis verna) (Ma) | 0·91/2·74/5·14 | 1·87/5·19/9·49 |
| tmrca of the Arabis alpina clade (including its annual sister species) (Ma) | 0·96/2·65/4·90 | 1·29/3·27/5·85 |
| tmrca of the Arabis nordmanniana clade (Ma) | 0·18/1·05/2·24 | 0·13/1·43/3·34 |
| tmrca of the Arabis auriculata clade (Ma) | 0·78/3·02/5·87 | 4·00/10·2/17·5 |
| tmrca of the Arabis aucheri clade (Ma) | 0·077/0·90/2·16 | 0·007/0·83/2·26 |
| tmrca of the main Arabis clade (Ma) | 3·27/6·58/10·9 | 3·50/7·09/11·8 |
| tmrca of the Scapiarabis clade (Ma) | 1·94/4·17/7·16 | 4·26/10·6/18·6 |
| ucld.mean | 2·32 × 10−9/4·48 × 10–9/6·59 × 10−9 | 0·99 × 10−9/1·94 × 10–9/2·84 × 10−9 |
| meanRate | 2·58 × 10−9/4·77 × 10–9/6·90 × 10−9 | 1·00 × 10−9/1·92 × 10–9/2·78 × 10−9 |
Mean values are indicated in bold; the lower and upper bounds of the 95 % highest posterior density (HPD) interval are given before and after that value, respectively. Enforced bounds are indicated in italics.
Divergence times of most clades (e.g. main Arabis, Aubrieta, Arabis alpina or Draba) tend to be somewhat earlier in the analysis running the plastid trnL-F marker, but only two clades (Arabis auriculata clade, Scapiarabis clade) show considerable higher ages analysing trnL-F, which corresponds with their different position in the trnL-F tree compared with the ITS tree.
The LTT plots (Fig. 3) for the ITS marker indicate two periods of increased lineage accumulation during the evolution of Arabideae. Numbers of new lineages increased rapidly in the early stages of the evolution of Arabideae (Mid Miocene, 15–10 Mya) and more explicitly starting from the Early Pliocene (approx. 5 Mya). Between both periods the increase of new lineages is delayed. The dashed line indicates the number of lineages without the Draba radiation (Fig. 3). It is apparent that the second increase of lineage accumulation starts earlier (approx. 7–8 Mya), if Draba and its annual segregates are fully included. The red line indicates the lineage accumulation of the annual species exclusively. The increase of new annual lineages is relatively high in the Mid Miocene, but lineage accumulation declines after that period even during the Pliocene and Pleistocene.
Fig. 3.
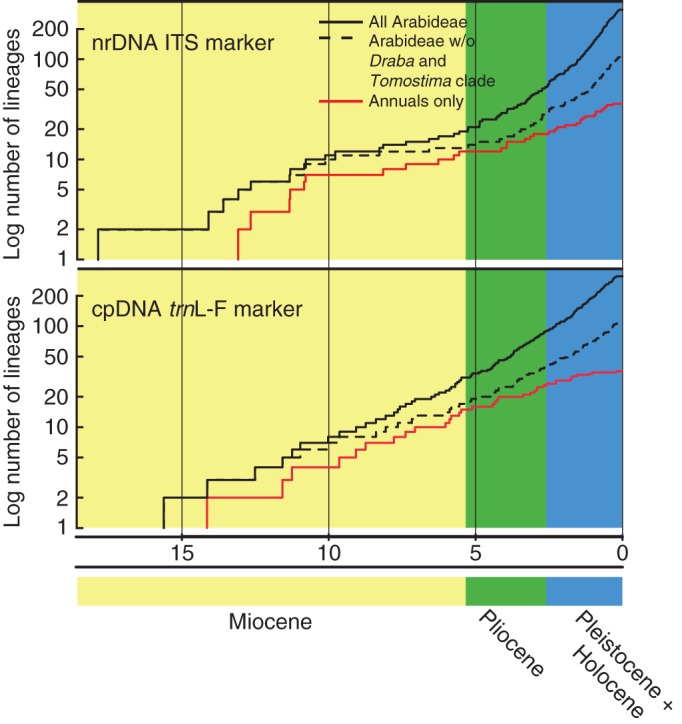
Lineages through time (LTT) plots of the respective ITS (top) and trnL-F (bottom) MCC trees. The solid line represents the increase of lineages over time for all taxa of the data set, the dashed line the increase without the Draba and Tomostima clade, and the red line the increase of the annual species of Arabideae.
For the trnL-F marker the sharp switches from steep to plain increase and vice versa in the Late Miocene and Early Pliocene are less prominent. Similarly, the earlier onset of the second increase of species diversification within Draba and the relatively steep increase of annual lineages in the Mid Miocene are less obvious than with the ITS data.
As most of the missing species belong to the core Draba clade (approx. 190 species) and the number of other missing species (15–20) is negligible, it can be concluded that inclusion of these species would result only in a even more pronounced increase in new lineages in the Pliocene and Pleistocene.
Net diversification rates
In general the calculated net diversification rates for perennial clades are higher than those for annual clades (two or more times higher). Moreover, the values for all perennials pooled in one group are approximately three times higher than for the respective annual group. Net diversification rates of the whole tribe Arabideae [0·381 net speciation events per million years (ITS) and 0·393 net speciation events per million years (trnL-F)] are intermediate between both pools of life-cycle strategy, but relatively close to those of the perennial group, considering the larger portion of perennials in the tribe. Net diversification rates and the fraction of extinction for 12 clades and groups are given in Table 2. However, for small clades with relative young crown ages (A. aucheri clade, A. nordmanniana clade, perennial members of the A. alpina clade or the Aubrieta clade) net diversification rates have extremely wide confidence intervals. Their mean values might therefore have been over-estimated, but median values are still only considerably lower in case of the A. aucheri clade.
Among the perennial clades Draba [0·894 (ITS) and 0·641 (trnL-F)] and three smaller clades (A. nordmanniana clade, perennial members of the A. alpina clade and the Aubrieta clade) have net diversification rates that are on average twice as high as the pooled perennial rates [0·443 (ITS) and 0·419 (trnL-F)]. As mentioned above the higher rates for the latter three clades might be over-estimated. Whereas Draba shows genuinely accelerated net diversification rates, the rates for the Scapiarabis clade [0·207 (ITS), 0·116 (trnL-F)] are low and comparable to those of the annual clades. Extinction fractions are generally lower in the perennial clades, but the values have large confidence intervals and are more or less similar across all clades/groups.
Ancestral area reconstruction
The ancestral area reconstructions of the S-DIVA and BBM analysis showed in principle similar results. However, in the BBM analysis the most probable area states often have lower probabilities (<50 %), and several areas with probabilities between 10 and 30 % are equally likely to circumscribe the distribution range of the most recent ancestor of a node. Moreover, the BBM analysis tends to produce larger ancestral areas by the inclusion of several adjacent regions. This effect also results in ancestral areas that seem to be shifted westwards, compared with the respective ancestral areas of the S-DIVA analysis. Results of the ancestral area reconstructions analyses for 20 nodes of particular interest are incorporated in Fig. 2. The original result files from the ancestral area reconstruction analyses are given with Supplementary Data Tables S5a and b.
Incongruencies between both analyses are present for node 1 (origin of Arabideae) and node 15 (Draba and its annual segregates). For all other nodes the results of both analyses are either congruent (e.g. nodes 5, 11, 18, 19 or 20) or inclusive (e.g. nodes 3, 12 or 16).
The S-DIVA analysis suggests that Arabideae (node 1) originated in Europe, whereas the BBM analysis favours a Central Asian origin (region I). Besides, further Asian distributions (regions K, L and IL) and a European distribution (region E) are mentioned with probabilities >5 %. Ancestral area states for node 1 are shifted towards Europe in the BBM analysis, when a different setting for the virtual outgroup (null or wide) is chosen. However, the effect of different virtual outgroup settings is negligible for all other nodes. Moreover, the vague origin of the tribe might be affected by the early branching position of the European A. caerulea, as its exclusion from the analyses increases the probabilities of an Asian origin [regions I (49·88 %), IK (13·45 %) and IL (13·27 %)] in the BBM analysis. For the S-DIVA analysis an ancestral area in the eastern Irano-Turanian region is then indicated [regions GI (51·35 %), FGI (20·67 %) and EGI (19·82 %)]. Generally, the exclusion of A. caerulea does not affect the remaining nodes, but in the S-DIVA analysis the most probable ancestral areas for some early diverging nodes (nodes 2, 3 and 12) are shifted slightly towards the Eastern Mediterranean (regions D or DF, respectively). However, these differences do not change the common picture of early distribution of Arabideae.
All nodes that diverged during the Mid- and Late Miocene (nodes 2, 3, 8, 9, 10 and 12) have their most likely ancestral areas is Anatolia and/or the (Eastern) Mediterranean. In some cases the distribution range is spreading into the adjacent Iranian/Caucasus region (nodes 2, 10 and 12) or northwards into the European region (node 2). In the BBM analysis for several nodes (nodes 3, 8, 9 and 12) the Western Mediterranean (region C) is included into the most likely ancestral areas, a region which is not mentioned for these nodes in the S-DIVA analysis.
For node 15 (Draba and its annual segregates) the S-DIVA analysis indicates a North American origin (region A, 48 %), but four areas with a distribution from the eastern Mediterranean to Iran have a combined probability of approx. 30 %. The BBM analysis suggests an ancestral area around the Mediterranean (region CDEF, CDF or CEF) for this node. For core Draba (node 16, excluding D. verna and D. hederifolia) both analyses give similar results, indicating an ancestral distribution in the Irano-Turanian floristic region (regions GI, FGI, FG or I). However, in the BBM analysis the most likely ancestral area has a probability of only 13 %. Ancestral areas for the three different groups in core Draba (see Jordon-Thaden et al., 2010) are relatively clear and seem unaffected by the reticulate evolution of several species in the genus (Jordon-Thaden et al., 2010). For core Draba group I (node 17), region G (Caucasus/Iran) is indicated as the most likely ancestral distribution in both analyses. The BBM suggests that this distribution might have been extended eastwards into Central Asia (region I). As most probable ancestral area, North America (region A) is shown for core Draba group II (node 19) and eastern Asia (regions J or IJ) for core Draba group III (node 18).
The S-DIVA analysis indicates a broad Euro-Iranian distribution (region EG) as ancestral area for the main Arabis clade (node 4), but the BBM analysis specifies it more to south-eastern Europe (regions D, DE or E). Node 6 (Arabis hirsuta agg.) has its most probable distribution in Europe (region E), whereas for node 7 (North American and Asian Arabis species) a Central Asian distribution is suggested (region I) in both analyses. Analogous to Draba, reconstruction of ancestral areas of these two major groups in the clade was not affected by species with a hybrid history between these groups (see Koch et al., 2010). In summary, the S-DIVA analysis suggests 338 dispersal, 107 vicariance and 19 extinction events, whereas the BBM analysis suggests a slightly higher number of dispersal events (368 dispersal, 69 vicariance and six extinction events).
Reconstruction of changes in life cycle and habitat preferences
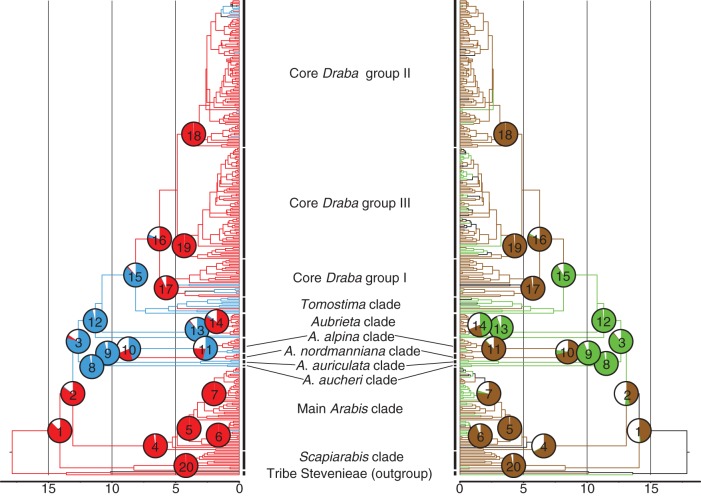
Perennialism is indicated as the ancestral life-cycle strategy for Arabideae (Fig. 4, node 1). The results of the ancestral life-cycle reconstructions are graphically presented for 20 nodes of particular interest in the left part of Fig. 4. In addition, the probabilities of the respective nodes are presented in Supplementary Data Table S6. This strategy of perennialism is maintained for node 2, but in all following early-diverging nodes (nodes 3, 8, 9, 10, 12 and 15) annualism is indicated as the most probable ancestral life-cycle strategy, whereas the probability for node 10 (includes the perennial A. nordmanniana clade and the A. alpina clade) is <50 %. However, the probabilities for the remaining five nodes are high, varying between 79·8 % (node 3) and 97·0 % (node 8). With the exception of nodes 11 (A. alpina clade) and 13 (annual A. verna and Aubrieta), all major nodes that emerged in the Late Miocene or later have high probabilities for perennialism as the ancestral life-cycle strategy. Most prominently, these are the main Arabis clade (nodes 4–7) and core Draba (nodes 16–19). Therefore, the annual life cycle of few species in these clades (e.g. A. planisiliqua subsp. nemorensis, D. brachystylis or D. nuda) can be considered as a derived character.
Fig. 4.
Graphical presentation of the results of the ancestral state reconstructions concerning life cycle (left part) and habitat elevation (right part). The results are superimposed on the BEAST MCC trees of the ITS analysis. Probability values are presented as pie charts for 20 main nodes (nodes 1–20). Uncoloured pies refer to the equivocal character states (AB), whereas single character states (A or B) are indicated in their respective colours (annual life cycle = blue, perennial life cycle = red; lowland habitats = green, montane/alpine habitats = brown). Branches are indicated in the above-mentioned colours, if the subsequent node had a probability of >50 % for the respective character state. Analogous nodes are indicated in black, if none of the single character states had a probability >50 %.
The results of the ancestral reconstructions for habitat elevation are graphically presented for 20 nodes of particular interest in the right part of Fig. 4. In addition, the probabilities of the respective nodes are shown in Supplementary Data Table S6. Ancestral species of Arabideae (nodes 1 and 2) occurred either in high montane/alpine elevations or were distributed in both lowland and alpine habitats, as the character states B and AB are indicated with similar high probabilities. Comparable to the life-cycle strategy reconstructions, for nearly all following early-diverging nodes (nodes 3, 8, 9, 12 and 15) lowlands are indicated as most probable ancestral habitats. With the exception of nodes 13 and 14 (Aubrieta clade), for all other nodes montane/alpine elevations are indicated as ancestral habitats. Although there is quite a number of species in the main Arabis clade and Draba, which occur in lowland or slightly higher elevations, this also applies for these two clades (nodes 4–7 and 16–19).
DISCUSSION
Introductionary comments on the phylogeny and systematics of the Arabideae
The presented study includes 309 taxa of the Arabideae and presents the most comprehensive phylogenetic analysis of the tribe so far. The included taxa make up >60 % of the currently recognized species of Arabideae. Most of the excluded taxa belong to the species-rich genus Draba (about 390 species in total; Al-Shehbaz, 2012), which leaves the remaining 16 genera of Arabideae with much higher taxon coverage (80–100 %). The excluded Draba species are predominantly perennials occurring in montane to subnival elevations in North and South America, Central Asia, Europe or the Caucasus, and there is no further indication that they should fall outside their generic boundaries.
The previously proposed (e.g. Koch et al., 2012a) existence of eight evolutionary separate clades (Draba and its annual segregates, Aubrieta clade, Arabis alpina clade, Arabis nordmanniana clade, Arabis auriculata clade, Arabis aucheri clade, main Arabis clade and Scapiarabis clade) in Arabideae is significantly confirmed herein. However, to accommodate the most recent taxonomic adjustments (Al-Shehbaz, 2012) Draba and its annual segregates (the Tomostima clade) are regarded as distinct clades. This is concordant with our results and previous studies on Draba (Jordon-Thaden et al., 2010). There are only two species of Arabideae which remain unaffiliated to one of the clades (Arabis caerulea and Drabella muralis) and we have no indication from morphology and/or molecular data that any of the missing taxa should form another relevant clade in the tribe. Thus, this study provides a robust phylogenetic framework of Arabideae, applicable to both detailed taxonomic work and subsequent evolutionary and comparative studies.
The aim to restore monophyly for all genera has been achieved for 16 out of the 17 genera with the latest taxonomic adjustments from 2012 (Al-Shehbaz, 2012; Koch et al., 2012a). The sole exception is the genus Arabis, the paraphyletic circumscription of which is demonstrated. The majority of Arabis species are grouped into the ‘main Arabis’ clade (55 species analysed, 65 species to be expected to belong to this group altogether). However, another 20 species (among them Arabis alpina, the type species of the genus) are grouped into four other clades, which are closely related to other genera such as Aubrieta and Draba.
As a consequence, a monophyletic circumscription of Arabis could be retained only when the species in the same clade as the generic type, A. alpina, are included in a newly circumscribed Arabis. However, this would leave the species of the main Arabis clade (with well-known species such as A. blepharophylla, A. hirsuta and A. procurrens) in need of a new generic designation. Nonetheless, A. verna and A. caerulea are in need of new taxonomic placements with the first species best integrated into Aubrieta. Finally, the monotypic genera Dendroarabis and Pachyneurum have been circumscribed and maintained as distinct genera with distinguishing morphological characters (German and Al-Shehbaz, 2008; German et al., 2012), although they cluster clearly in the main Arabis clade and their taxonomic inclusion into this group is worthy of consideration.
Biogeography and origin of the Arabideae clades
Reconstructions of past geographical patterns revealed that all early diversification events in Arabideae during the Mid and Late Miocene took place in the (western) Irano-Turanian floristic region and/or the (eastern) Mediterranean. Region F (Anatolia and the Levantine coast), in particular, is included as the most likely ancestral area for almost all nodes, whereas the Western Mediterranean, the Caucasus and the Iranian mountains are indicated as potential centres of origin with lower frequencies (Figs 2 and 5). These early diversification events gave rise to all of the presently existing clades and their respective stems. These results are congruent with past hypotheses placing not only the origin of Arabideae into the Irano-Turanian region (Hedge, 1976), but also the origin of the entire family. Species diversity in Brassicaceae is not distributed equally (Koch and Kiefer, 2006): the most important diversification centres are found in the Irano-Turanian region (approx. 150 genera and approx. 900 species with 530 endemics) and the Mediterranean region (ca. 113 genera and approx. 630 species with 290 endemics). Adjacent regions in North America (ca. 99 genera and 778 species with 600 endemics) and in the Saharo-Sindian region (ca. 65 genera and 180 species with 62 endemics) show a significant reduction in species diversity (Hedge, 1976; Al-Shehbaz, 1984; Appel and Al-Shehbaz, 2003). This reduction of species diversity is continued in southern regions (South America with 40 genera and 340 species; Southern Africa with 15 genera and at least 100 species; Australia and New Zealand with 19 genera and 114 species) (Allan, 1961; Marais, 1970; Hewson, 1982; Al-Shehbaz, 1984; Appel and Al-Shehbaz, 2003). This overall distribution pattern might also provide some evidence for the origin of the family in the Irano-Turanian region and follows the basic ideas of Hedge (1976) who claimed that the origin of Brassicaceae occurred in a region encompassing the Mediterranean to the Irano-Turanian territory. In summary, tribe Arabideae could well serve as a representative monophyletic group mirroring crucifer diversification and evolution.
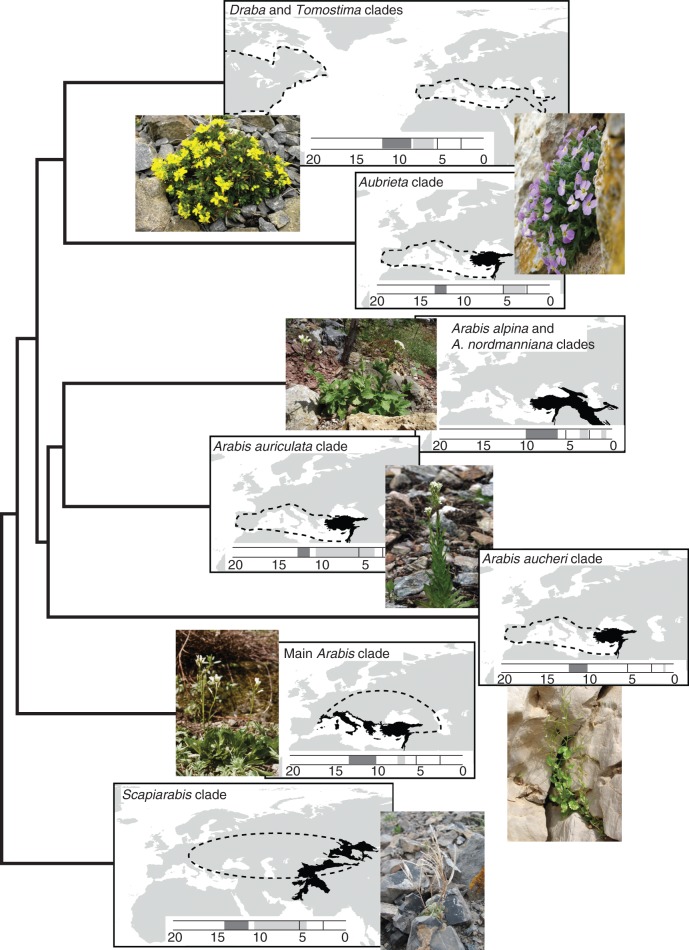
Fig. 5.
Graphical illustration of the putative areas and times of origin for the nine clades of Arabideae, as summarized in Table 3. The respective maps are connected by a sketched phylogram, based on the outline of the ITS-based phylogenetic tree of the entire tribe (Fig. 2). Their position in the horizontal direction indicates the crown age of the respective clades. Times of origin are also stated in the timeline at the bottom of each map: dark grey for the stem group and light grey for the crown group of the respective clade. The maps include areas of origin for the stem group [solid black for the most probable region(s), dashed outlines for the less probable regions].
The geomorphological outline of Eurasia during the late Miocene is comparable to today, as major barriers such as the Turgay strait or a continuous Tethys ocean no longer existed (Tiffney and Manchester, 2001; Meulenkamp and Sissingh, 2003). Furthermore, the alpide orogeny was already in progress and the resulting mountain ranges formed a continuous belt from the Balkans to Central Asia (Steininger and Rögl, 1984; Meulenkamp and Sissingh, 2003), which was only temporarily disconnected by narrow gateways (Jolivet et al., 2006) or shallow basins during phases of marine transgressions (Piller et al., 2007). Elevations of the Qinghai–Tibetian Plateau (QTP), the Himalayas (Molnar, 2005; Wang et al., 2008) and the central Alps (Campani et al., 2012) were high enough to act as effective geographical barriers for non-alpine species (Koch et al., 2012a). However, for other alpide mountain chains, such as the Tian-Shan, the Taurus and East Anatolian Plateau, the major phases of uplift took place during the the Late Miocene/Early Pliocene or even later (Güldali, 1979; Dewey et al., 1986; Bullen et al., 2003).
Consequently, the region ranging from the Mediterranean to Central Asia might have constituted a relatively open corridor during the early phases of the evolution of Arabideae, which was accessible also for non-alpine species. Furthermore, it has served as a geographical cradle for the early evolution of Arabideae and the clades within the tribe, which have all evolved in this region or more confined parts of it. In the case of Draba (node 16) the suggested origin of the genus in region G (Iran/Caucasus) is congruent with its previously proposed origin in the ‘Turkey–Caucasus–Iran region’ based on trnL-F haplotype statistics (Jordon-Thaden, 2010). Similarly, for the Arabis alpina clade (node 11) the suggested origin in region F (Western and Central Anatolia & Levantine coast) is consistent with previous studies, which determined the origin of Arabis alpina in Central Anatolia based on the distribution of the ancestral trnL-F haplotypes (Koch et al., 2006; Ansell et al., 2011; Karl et al., 2012).
Major migratory events into regions outside the ancestral distribution range of Arabideae did not start before the Pliocene. However, such migratory events occurred in two clades only (Draba, main Arabis clade), if we exclude the recent expansion of Arabis alpina with its migration to Eastern Africa approximately 0·5 Mya (Koch et al., 2006) and its postglacial extension into arctic regions (Koch et al., 2006; Ehrich et al., 2007). Draba, especially core groups II and III, has colonized vast areas in Asia, the Americas and Arctic regions. Jordon-Thaden (2010) proposed migration routes from its Turkey–Caucasus–Iran origin to the Mediterranean (group I) and Central Asia (ancestor of groups II and III). North and eventually South America were then colonized from Asia via Beringia (group III) (Koch and Al-Shehbaz, 2002). However, several more recent migratory events within Draba contributed to increased hybridization and the reticulate ties between species from different core groups (Koch and Al-Shehbaz, 2002).
Early range expansions of the main Arabis clade involved migration to Central Europe, whereas the subsequent colonization of East Asia and North America did not occur before the onset of Pleistocene cycles of glaciation and deglaciation (Koch et al., 2010). Furthermore, species of the main Arabis clade did not colonize high mountain areas and alpine habitats in the American Cordillera or the Himalayas as many Draba species did.
Driving factors of evolutionary diversification
As outlined above, distribution and colonization patterns indicate tremendous past success of various clades in Arabideae. Consequently, the question arises if there are traits and characters obviously correlated with colonization and diversification.
It is intriguing that, when superimposed on the phylogenetic trees, the patterns of life cycle and habitat elevation are almost congruent, especially for all nodes that diverged during the early phases in the evolution of Arabideae in the Mid Miocene (Fig. 4). This close correlation between life-cycle strategy and preferred habitat elevation expands the previously raised idea of the pattern of sister relationships between species-poor, annual clades and a larger groups of predominantly perennial species. In addition, the perennial species are prevalent in high montane to alpine elevations, whereas the annual species mainly occur in lowland habitats.
In Arabideae this pattern occurred at least four times: (1) the relationship between Draba (approx. 390 species, perennial dominated) and its annual segregates (Tomostima clade) with about ten species (split 8–11 Mya in the Mediterranean or the Irano-Turanien region); (2) the annual (three species) and perennial (about seven species) members of the Arabis alpina group (split 2·7–3·3 Mya in Anatolia; see Karl et al., 2012); (3) the annual species A. verna as sister to the perennial genus Aubrieta with approx. 15 species (split 2·7–5·2 Mya in the Eastern Mediterranean); and (4) the annual A. auriculata and A. aucheri clades (together five species) as sister groups to the A. nordmanniana and A. alpina clades (together approx. 15 species), which are both dominated by perennials (split 10–13 Mya in Anatolia or the Mediterranean).
Despite the generally species-richer perennial clades, the first, but more moderate phase of increase of lineage accumulation in Arabideae in the Mid Miocene was caused mostly by the formation of annual, lowland lineages. In contrast, the second, more pronounced phase with increase of new lineages, which started in the Pliocene, was almost exclusively caused by the formation of numerous perennial, montane/alpine lineages. During the first phase in the Mid Miocene climatic conditions in the Mediterranean region and neighbouring regions in the East changed from subtropical to more temperate temperature regimes (Flower and Kennett, 1994; Böhme, 2003). Moreover, East Antarctic ice sheet growth and polar cooling also had large effects on global carbon cycling and on the terrestrial biosphere, including aridification of mid-latitude continental regions (Flower and Kennett, 1994; van Dam, 2006). In concordance with life-cycle theory (Cole, 1954) annuals generally seem better adapted to arid conditions with erratic water supply (Schaffer and Gadgil, 1975) in the lowlands. However, these annual lineages remained relatively species-poor compared with the greater diversification in the perennial lineages. The perennials colonized montane and alpine habitats, which in general provide moderate and cooler climatic conditions and a more stable water supply from rainfall and condensation due to orographic effects (Roe, 2005) and are therefore considered better suited for the needs of perennials (Cole, 1954; Schaffer and Gadgil, 1975; Evans et al., 2009). Moreover, montane habitats generally provide a greater abiotic diversity, due to topographic complexity and climatic and ecological gradients (Brown, 2001; Badgley, 2010). As a result, the expansion into such habitats has provided multiple opportunities for diversification, due to the creation of a variety of new ecological niches and the existence of effective barriers to gene flow (Rahbek and Graves, 2001; Hughes and Eastwood, 2006).
In this respect, the switch between life cycle (annual to perennial or vice versa) must be considered as an important evolutionary key innovation. Key innovations are seen as adaptations that enabled the respective taxa to overcome ecological or migratory range restriction (Simpson, 1953). The change in life-cycle strategy made it possible for the perennial lineages to colonize higher elevations, and to diversify in these new habitats. The close correlation between both features has also been pointed out for other plant taxa, for example for American Lupinus species, where the switch from an annual to perennial life cycle led to a range expansion into montane/alpine habitats (Drummond et al., 2012), or the genus Anthemis, where the shift from wetter mountain areas to drier coastal plains was accompanied also by a switch from a perennial to annual life cycle (Lo Presti and Oberprieler, 2009).
Usually it is predicted that a key innovation has subsequently caused increased net diversification after the overcoming of previous restrictions (Simpson, 1953; Maynard Smith and Szathmary, 1995; see also Hunter, 1998). These key innovations could, for example, affect mechanisms linked to speciation, e.g. reproductive isolation (Kay et al., 2006) or ecological opportunity (Yoder et al., 2010). Several studies have linked this emerging ecological opportunity to subsequent macroevolutionary diversification and adaptive radiation (e.g. Losos, 2009; Marazzi and Sanderson, 2010; see also Fawcett et al., 2013). Likewise, in the case of American Lupinus the colonization of alpine habitats was followed by increased diversification rates (Drummond et al., 2012), highlighting the shift in life-cycle strategy as an adaptive key innovation. Similar results were also obtained for the perennials in Draba (Jordon-Thaden and Koch, 2008).
By contrast, the actual impact of a single trait is often hard to detect (Kay et al., 2006), as these traits have underlying correlations with other traits (Vamosi and Vamosi, 2011). Consequently, several studies have indicated that the emergence of a novel adaptive trait (even if it is considered as a key innovation) is not sufficient to directly spur diversification (Levinton, 1988; Davies et al., 2004). Although focusing on major genome duplication events (e.g. as a result of hybridization, reticulation and polyploidization) rather than key innovations, Santini et al. (2009) and Schranz et al. (2012) showed the possibility that a great proportion of diversification is not directly connected to genome duplication and its newly established adaptive key traits. Instead extrinsic events such as episodes of biogeographical or climatic change, formation of new habitats, or biotic interactions have contributed to subsequent diversifications in these groups.
Apart from the emergence of key innovations, dispersal into new environments can also provide ecological opportunity (Yoder et al., 2010), making dispersal events and the extent of geographical distribution ranges important factors in respect of diversification. Arabideae represents an excellent case in this regard. The tribe contains several well-resolved sister-group relationships of annual/lowland and perennial/alpine clades and these clades differ remarkably in species number (approx. 10 to 390 species). Species numbers are especially divergent in the perennial/alpine clades, which should all hold potential for subsequent diversification, if we consider the switch to a perennial life cycle and the ability to colonize montane habitats as key factors for diversification success. Within Arabideae all perennial/alpine clades originated in the Irano-Turanian floristic region or the adjacent parts of the Mediterranean. However, the clades which stayed mostly in their original distribution areas (Arabis alpina clade – Eastern Mediterranean, Arabis nordmanniana clade – Caucasus, Aubrieta – Eastern Mediterranean, Scapiarabis clade – Central Asian and Chinese mountain ranges) remained species-poor, whereas the species-richer clades (Draba, main Arabis clade) have undergone multiple migratory events (Jordon-Thaden, 2010; Koch et al., 2010). Analogous, the WGD radiation lag-time hypothesis of Schranz et al. (2012) involves one or more dispersal events following the evolution of key novel traits, which first instigates the diversification of the respective group, whereas its sister group remains relatively species-poor in the original distribution area. This pattern indicates the importance of dispersal and colonization success (i.e. geographical range expansions) as a vital factor to maintain high rates of diversification (see Fritz et al., 2011). As an example, for the Scapiarabis clade, which has a comparable age to Draba and the main Arabis clade, but explicitly lowered net diversification rates, it has been discussed that its inability to colonize the high alpine zones (>4000 m a.s.l.) blocked effective dispersal across the mountain chains of the QTP. Migratory routes were then possible only along mountain chains and finally restricted their habitats to a ring around the QTP between the xeric lowlands and the high alpine elevations (Koch et al., 2012a). The causal chain with perennialism as as first key innovation followed by altitudinal range expansion and secondary diversification after geographical range expansion is suggested by the annual Tomostima clade, which has colonized vast lowland areas in America (outside the geographical cradle of Arabideae), but remained relatively species-poor.
As a result it can be concluded that in most cases increased diversification cannot be attributed to a single event, e.g. the emergence of a novel adaptive trait. Instead such a key innovation must encounter the adequate environmental context, so that the respective taxonomic group will diversify (see De Queiroz, 2002). In this respect it has been demonstrated that it might be necessary to invade new habitats to play out fully the diversification potential of the acquired key innovation (see also von Hagen and Kadereit, 2003; Drummond et al., 2012).
The highly fragmented habitats of the American Cordillera and the Himalayas, which are home to numerous endemic Draba species, might therefore have contributed to vast diversification in this genus. The diversification in Draba was highly affected by hybridization, polyploidization and alternative reproductive modes (Jordon-Thaden and Koch, 2008), but the genus might have also experienced hybridization between evolutionary lineages from different continents in its early evolutionary history (Koch and Al-Shehbaz, 2002). Hybridization and polyploidization are among the most important factors in speciation (Soltis et al., 2009; Soltis and Soltis, 2009), and in particular for Draba high levels of polyploidy (>4×) are prevalent, especially for arctic or alpine species (Jordon-Thaden and Koch, 2008). Note here that arctic species of Arabideae belong almost exclusively to Draba, and here the percentage of perennial polyploids is highest (Jordon-Thaden and Koch, 2008; Jordon-Thaden et al., 2009). Among the few exceptions is Arabis alpina, which has a wide distribution including arctic regions and is a diploid perennial (Karl et al., 2012). Generally, polyploidy is considered beneficial to maintain taxa in extreme habitats, such as unstable environments (Fawcett et al., 2009), or alpine altitudes and arctic latitudes (Brochmann et al., 2004). In comparison, the main Arabis clade has a considerable number of tetraploids, but higher polyploids are the rare exception (octoploid A. eschscholtziana). In all other clades of Arabideae (independent of being annual or perennial dominated) diploid species prevail with some rare tetraploids (e.g. Arabis parvula or Aubrieta scardica). Similarly, hybridization events in the main Arabis clade are set in more recent time frames (see Koch et al., 2010). For the other clades of Arabideae there are no signs that hybridization had played a significant role in promoting species diversity. Moreover, asexual modes of reproduction such as apomixis are another way to impede admixture of derived taxa with their recent ancestors. Within Arabideae apomixis is only known from Draba; members of the other clades are either facultative selfers or outcrossers. In summary, hybridization and polyploidization have played an important role in the evolutionary history and colonization of arctic–alpine habitats in one particular genus, namely Draba, which is the most species-rich clade. For all other clades this is less obvious, and additional data might help to find further correlations.
CONCLUDING REMARKS
Utilizing a well-resolved and comprehensive phylogenetic tree for Arabideae and analyses performed subsequently, we could (1) corroborate a more clear-cut phenomenon for Brassicaceae in the relationship between annual- and perennial-dominated clades (Karl et al., 2012), and (2) reconstruct a biogeographical and evolutionary scenario for Arabideae, which is summarized diagrammatically in Fig. 5.
Early diversification in the tribe was mostly due to the emergence of new annual lineages, which were prevalent in lowland habitats of the Mediterranean and the Irano-Turanian region. The Irano-Turanian region has been suggested as the cradle of Brassicaceae (Hedge, 1976; Franzke et al., 2011) and this study has revealed further evidence to support the evolutionary importance of this region. However, annual lineages remained relatively species-poor during the following epochs and only the switch towards a perennial life-cycle strategy and the ability to colonize high montane/alpine habitats increased diversification in these respective lineages. Table 3 summarizes the nine clades of Arabideae with their presence/absence of features, which are considered as important factors of diversification. In particular, the switch to a perennial life-cycle strategy must be considered a key innovation, as it is the prerequisite for the altitudinal shift and subsequent diversification success. This type of key innovation has been documented for various angiosperm groups (e.g. Apiaceae, Sun et al., 2004; Orobanchaceae, Tank and Olmstead, 2008; Pontederiaceae, Barret and Graham, 1997). Therefore, this transition could be considered an important evolutionary phenomenon more widely than in Brassicaceae alone, and there is a widely held opinion among plant evolutionary biologists that annuals are derived from perennial ancestors and that the shift between these two strategies is unidirectional (e.g. Stebbins, 1957).
Table 3.
Summary of Arabideae and allocation of evolutionary important features
| Core Draba clade | Main Arabis clade | Aubrieta clade | Scapiarabis clade | A. alpina clade | Tomostima clade | A. nordmanniana clade | A. auriculata clade | A. aucheri clade | |
|---|---|---|---|---|---|---|---|---|---|
| No. of species | ∼390 | ∼70 | ∼15 | 12 | ∼10 | ∼8 | 5 | 3 | 2 |
| Perennial life cycle | X | X | X | X | X | – | X | – | – |
| Ancestors present in high montane/alpine environments | X | X | X | X | X | – | X | – | – |
| Major geographical range expansion (i.e. migration from centre of origin) | X | X | – | – | (X) | – | – | – | – |
| Hybridization | X | X | – | – | – | – | – | – | – |
| High polyploidization (>4×) | X | – | – | – | – | – | – | – | – |
| Asexual reproductive modes observed | X | – | – | – | – | – | – | – | – |
| Putative geographical origin (crown group) | (Eastern) Ir.–Tur. region | Europe–Caucasus region, E Med., or (SE) Europe | CE Med. (ranging to Anatolia) | C Asia | Anatolia | North America | Caucasus | broader Med., incl. Western Ir.–Tur. region | Anatolia and E Med. or W Med. |
| Putative geographical origin (stem group) | North America or Med. to W Ir.–Tur. region | E Med to Ir.–Tur. region, CE Med, or (SE) Europe | Med. to Western Ir.–Tur. region | C Asia (or Eurasia) | Ir.–Tur. region | North America or Med. to W Ir.–Tur. region | Ir.–Tur. region | Anatolia, E Med. (to W Med.) | Anatolia, E Med. (to W Med.) |
| Putative time of origin (crown group) (Mya) | 6·3–7·8 | 6·6–7·1 | 2·7–5·2 | 4·2–10·6 | 2·7–3·3 | ∼6–8 | 1·1–1·4 | 3·0–10·2 | 0·8–0·9 |
| Putative time of origin (stem group) (Mya) | ∼8–11·5 | ∼10–13 | ∼11–12·5 | ∼11·5–14 | ∼6–10 | ∼8–11·5 | ∼6–10 | ∼11–12 | ∼10–12 |
If the feature occurs regularly in a clade it is marked with ‘x’, whereas if it occurs less frequently it is marked with ‘(x)’. If the feature does not occur or only rarely occurs in a clade it is marked with ‘–’. Sources for the collected information are the data compilations of the authors (see Supplementary Data Table S2) and Jordon-Thaden (2010), as well as the knowledge and resource database for Brassicaceae BrassiBase (Koch et al., 2012b). Information about putative areas and periods of origin for the clades is derived from Fig. 2, Table 1 and Supplementary Data Fig. S4.
However, it was shown that this intrinsic factor did not directly spur the considerably accelerated diversification during the Pliocene and Pleistocene, which was mainly achieved by the emergence of new perennial lineages. Instead, the ability of lineages to colonize new environments, occupy opening niches and adapt to new conditions during the constant change of climatic conditions is emphasized to have driven most of this ‘secondary’ and more recent diversification. In this respect range expansion via migration but also hybridization and polyploidization are again indicated as major factors for plant diversification.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank the gardener team of the Botanical Garden Heidelberg and in particular Torsten Jakob for continuous support with plant cultivation. Thanks go also to Susanne Ball for her substantial help in the laboratory and handling all the material and germplasm collections and to Peter Sack, who organized and helped with the voucher exchanges. We appreciate also a critical discussion and many comments on the final draft version provided by Graham Muir and various re-determinations done by Dmitry German. We are very grateful to Ihsan Al-Shehbaz, who continuously provided comments for many of the various clades and groups of taxa dealt with. We also thank the curators of the herbaria of the Arnold Arboretum of Harvard University (A), the Botanical Garden and Botanical Museum Berlin (B), the Natural History Museum London (BM), University of Copenhagen (C), Agriculture Canada, Ottawa (DAO), Royal Botanical Garden Edinburgh (E), Gray Herbarium of Harvard University (GH), Botanical Garden and Herbarium Heidelberg (HEID), University of California, Berkeley (JEPS & UC), Royal Botanical Gardens Kew (K), Botanical Museum Lund (LD), V. L. Komarov Botanical Institute, St Petersburg (LE), Botanische Staatssammlung Munich (M), Real Jardin Botanico Madrid (MA), the Missouri Botanical Garden (MO), Botanical Museum Oslo (O), University Osnabrück (OSBU), Uppsala University (UPS), Smithsonian Institution, Washington, DC (US), University of Utah, Salt Lake City (UT), Natural History Museum Vienna (W), University of Natural Resources and Life Sciences, Vienna (WHB), and the Herbarium of the University of Washington, Seattle (WTU), for providing voucher material. Moreover, plant or seed material was received from various fellow scientists: Dmitry German (COS, University Heidelberg), Christiane Kiefer (MPIPZ Cologne), Birol Mutlu (Inönü University Malatya) and Ji-Pei Yue (Kunming Institute of Botany). This work was supported by DFG research grants (KO2302 11/1, 12/1 and 13/1) to M.A.K.
LITERATURE CITED
- Allan HH. In: Flora of New Zealand. Owen RE, editor. 1: Cruciferae. Wellington, NZ: Government Printer; 1961. pp. 174–189. [Google Scholar]
- Alonso-Blanco C, Aarts MGM, Bentsink L, et al. What has natural variation taught us about plant development, physiology, and adaptation? Plant Cell. 2009;21:1877–1896. doi: 10.1105/tpc.109.068114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shehbaz IA. The tribes of Cruciferae (Brassicaceae) in the southeastern United States. Journal of the Arnold Arboretum. 1984;65:343–373. [Google Scholar]
- Al-Shehbaz IA. A generic and tribal synopsis of the Brassicaceae (Cruciferae) Taxon. 2012;61:931–954. [Google Scholar]
- Al-Shehbaz IA, Beilstein MA, Kellogg EA. Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Systematics and Evolution. 2006;259:89–120. [Google Scholar]
- Ansell SW, Grundmann M, Russell SJ, Schneider H, Vogel JC. Genetic discontinuity, breeding-system change and population history of Arabis alpina in the Italian Peninsula and adjacent Alps. Molecular Ecology. 2008;17:2245–2257. doi: 10.1111/j.1365-294X.2008.03739.x. [DOI] [PubMed] [Google Scholar]
- Ansell SW, Stenoien HK, Grundmann M, et al. The importance of Anatolian mountains as the cradle of global diversity in Arabis alpina, a key arctic–alpine species. Annals of Botany. 2011;108:241–252. doi: 10.1093/aob/mcr134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel O, Al-Shehbaz IA. Cruciferae. In: Kubitzki K, editor. Families and genera of vascular plants. Berlin: Springer; 2003. pp. 75–174. [Google Scholar]
- Badgley C. Tectonics, topography, and mammalian diversity. Ecography. 2010;33:220–231. [Google Scholar]
- Bailey CD, Koch MA, Mayer M, et al. A global nrDNA ITS phylogeny of the Brassicaceae. Molecular Biology and Evolution. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- Barret SCH, Graham SW. Adaptive radiation in the aquatic plant family Pontederiaceae: insights from phylogenetic analysis. In: Givnish TJ, Systsma KJ, editors. Molecular evolution and adaptive radiation. Cambrigde: Cambridge University Press; 1997. pp. 225–258. [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Kellogg EA. Brassicaceae phylogeny and trichome evolution. American Journal of Botany. 2006;93:607–619. doi: 10.3732/ajb.93.4.607. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Al-Shehbaz IA, Mathews S, Kellogg EA. Brassicaceae phylogeny inferred from phytochrome A and ndhF sequence data: tribes and trichomes revisited. American Journal of Botany. 2008;95:1307–1327. doi: 10.3732/ajb.0800065. [DOI] [PubMed] [Google Scholar]
- Beilstein MA, Nagalinguma NS, Clementsa MD, Manchesterb SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proceedings of the National Academy of Sciences USA. 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhme M. The Miocene Climatic Optimum: evidence from ectothermic vertebrates of Central Europe. Palaeogeography, Palaeoclimatology, Palaeoecology. 2003;195:389–401. [Google Scholar]
- Bremer K. Ancestral areas: a cladistic reinterpretation of the centre of origin concept. Systematic Biology. 1992;41:436–445. [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, et al. Polyploidy in arctic plants. Journal of the Linnean Society. 2004;82:521–536. [Google Scholar]
- Brown JH. Mammals on mountainsides: elevational patterns of diversity. Global Ecology and Biogeography. 2001;10:101–109. [Google Scholar]
- Bullen ME, Burbank DW, Garver JI. Building the Northern TienShan: integrated thermal, structural, and topographic constraints. Journal of Geology. 2003;111:149–165. [Google Scholar]
- Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proceedings of the National Academy of Sciences USA. 2007;104:16714–16719. doi: 10.1073/pnas.0705338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campani M, Mulch A, Kempf O, Schlunegger F, Mancktelow N. Miocene paleotopography of the Central Alps. Earth and Planetary Science Letters. 2012;337:174–185. [Google Scholar]
- Cole LC. The population consequences of life-history phenomena. Quarterly Review of Biology. 1954;29:103–137. doi: 10.1086/400074. [DOI] [PubMed] [Google Scholar]
- Couvreur TLP, Franzke A, Al-Shehbaz IA, Bakker FT, Koch MA, Mummenhoff K. Molecular phylogenetics, temporal diversification and principles of evolution in the mustard family (Brassicaceae) Molecular Biology and Evolution. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Barraclough TG, Chase MW, Soltis PS, Soltis DE, Savolainen V. Darwin's abominable mystery: insights from a supertree of the angiosperms. Proceedings of the National Academy of Sciences USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Queiroz A. Contingent predictability in evolution: key traits and diversification. Systematic Biology. 2002;51:917–929. [PubMed] [Google Scholar]
- Dewey JF, Hempton MR, Kidd WFS, Saroglu F, Sengör AMC. Shortening of continental lithosphere: the tectonics of eastern Anatolia, a young collision zone. Geological Society, London, Special Publications. 1986;19:3–36. [Google Scholar]
- Dobeš C, Mitchell-Olds T, Koch M. Extensive chloroplast haplotype variation indicates Pleistocene hybridization and radiation of North American Arabis drummondii, A.×divaricarpa, and A. holboellii (Brassicaceae) Molecular Ecology. 2004;13:349–370. doi: 10.1046/j.1365-294x.2003.02064.x. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small amounts of fresh leaf tissue. Phytochemical Bulletin, Botanical Society of America. 1987;19:11–15. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:pe214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond CS. Diversification of Lupinus (Leguminosae) in the western NewWorld: derived evolution of perennial life history and colonization of montane habitats. Molecular Phylogenetics and Evolution. 2008;48:408–421. doi: 10.1016/j.ympev.2008.03.009. [DOI] [PubMed] [Google Scholar]
- Drummond CS, Eastwood RJ, Miotto STS, Hughes CE. Multiple continental radiations and correlates of diversification in Lupinus (Leguminosae): testing for key innovation with incomplete taxon sampling. Systematic Biology. 2012;61:443–460. doi: 10.1093/sysbio/syr126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrich D, Gaudeul M, Assefa A, et al. Genetic consequences of Pleistocene range shifts: contrast between the Arctic, the Alps and the East African mountains. Molecular Ecology. 2007;16:2542–2559. doi: 10.1111/j.1365-294X.2007.03299.x. [DOI] [PubMed] [Google Scholar]
- Evans MEK, Smith SA, Flynn RS, Donoghue MJ. Climate, niche evolution, and diversification of the “bird-cage” evening primroses (Oenothera, sections Anogra and Kleinia) American Naturalist. 2009;173:225–240. doi: 10.1086/595757. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, van de Peer Y. Plants with double genomes might have had a better chance to survive the Cretaceous–Tertiary extinction event. Proceedings of the National Academy of Sciences USA. 2009;106:5737–5742. doi: 10.1073/pnas.0900906106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, van de Peer Y, Maere S. Significance and biological consequences of polyploidization in land plant evolution In. In: Greilhuber J, Doležel J, Wendel JF, editors. Plant genome diversity volume 2: physical structure, behaviour and evolution of plant genomes. Vienna: Springer; 2013. pp. 277–293. [Google Scholar]
- Flower BP, Kennett JP. The middle Miocene climatic transition: East Antarctic ice sheet development, deep ocean circulation and global carbon cycling. Palaeogeography, Palaeoclimatology, Palaeoecology. 1994;108:537–555. [Google Scholar]
- Franzke A, German D, Al-Shehbaz IA, Mummenhoff K. Arabidopsis family ties: molecular phylogeny and age estimates in the Brassicaceae. Taxon. 2009;58:425–437. [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K. Cabbage family affairs: the evolutionary history of Brassicaceae. Trends in Plant Science. 2011;16:108–116. doi: 10.1016/j.tplants.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Fritz SA, Jønsson KA, Fjeldså J, Rahbek C. Diversification and biogeographic patterns of four island radiations of passerine birds. Evolution. 2011;66:179–190. doi: 10.1111/j.1558-5646.2011.01430.x. [DOI] [PubMed] [Google Scholar]
- German DA, Al-Shehbaz IA. Dendroarabis, a new Asian genus of Brassicaceae. Harvard Papers in Botany. 2008;13:289–291. [Google Scholar]
- German DA, Friesen N, Neuffer B, Al-Shehbaz IA, Hurka H. Contribution to ITS phylogeny of the Brassicaceae, with special reference to some Asian taxa. Plant Systematics and Evolution. 2009;283:33–36. [Google Scholar]
- German DA, Chen WL, Smirnov SV, et al. Plant genera and species new for China recently found in North West Xinjiang. Nordic Journal of Botany. 2012;30:61–69. [Google Scholar]
- Gernhard T. New analytic results for speciation times in neutral models. Bulletin of Mathematical Biology. 2008;70:1082–1097. doi: 10.1007/s11538-007-9291-0. [DOI] [PubMed] [Google Scholar]
- Güldali N. Geomorphologie der Türkei. Beihefte Tübinger Atlas des vorderen Orients. 1979;A 4:1–226. [Google Scholar]
- Hall JC, Tisdale T, Donohue K, Wheeler A, Al-Yahya M, Kramer EM. Convergent evolution of a complex fruit structure in the tribe Brassiceae (Brassicaceae) American Journal of Botany. 2011;98:1989–2003. doi: 10.3732/ajb.1100203. [DOI] [PubMed] [Google Scholar]
- Hedge IC. A systematic and geographical survey of the Old World Cruciferae. In: Vaughan JG, Macleod AJ, Jones BMG, editors. The biology and chemistry of the Cruciferae. London: Academic Press; 1976. pp. 1–45. [Google Scholar]
- Hewson HJ. Brassicaceae. In: Briggs BG, Barlow BA, Eichler H, et al., editors. Flora Australia. Vol. 8. Canberra: Australian Government Publishing Service; 1982. pp. 231–357. [Google Scholar]
- Hodges SA. Rapid radiation due to a key innovation in columbines (Ranunculaceae: Aquilegia) In: Givnish T, Systma K, editors. Molecular evolution and adaptive radiation. Cambridge: Cambridge University Press; 1997. pp. 391–405. [Google Scholar]
- Hoffmann FG, Opazo JC, Storz JF. Whole-genome duplications spurred the functional diversification of the globin gene superfamily in vertebrates. Molecular Biology and Evolution. 2012;29:303–312. doi: 10.1093/molbev/msr207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes C, Eastwood R. Island radiation on a continental scale: exceptional rates of plant diversification after uplift of the Andes. Proceedings of the National Academy of Sciences USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. Key innovations and the ecology of macroevolution. Tree. 1998;13:31–36. doi: 10.1016/s0169-5347(97)01273-1. [DOI] [PubMed] [Google Scholar]
- Hurka H, Paetsch M, Bleeker W, Neuffer B. Evolution within the Brassicaceae. Nova Acta Leopoldina. 2005;92:113–127. [Google Scholar]
- Ihaka R, Gentleman R. R: a language for data analysis and graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- Jabbour F, Renner SS. A phylogeny of Delphinieae (Ranunculaceae) shows that Aconitum is nested within Delphinium and that Late Miocene transitions to long life cycles in the Himalayas and southwest China coincide with bursts in diversification. Molecular Phylogenetics and Evolution. 2012;62:928–942. doi: 10.1016/j.ympev.2011.12.005. [DOI] [PubMed] [Google Scholar]
- Jolivet L, Augier R, Robin C, Suc JP, Rouchy JM. Lithospheric-scale geodynamic context of the Messinian Salinity Crisis. Sedimentary Geology. 2006;188:9–33. [Google Scholar]
- Jordon-Thaden I. Germany: PhD thesis, Ruperto-Carola University of Heidelberg; 2010. Species and genetic diversity of Draba: phylogeny and phylogeography. [Google Scholar]
- Jordon-Thaden I, Koch MA. Diversity patterns in the genus Draba: a first global perspective. Plant Ecology and Diversity. 2008;1:255–263. [Google Scholar]
- Jordon-Thaden I, Hase I, Al-Shehbaz IA, Koch MA. Molecular phylogeny and systematics of the genus Draba s.l. (Brassicaceae) and identification of its closest related genera. Molecular Phylogenetics and Evolution. 2010;55:524–540. doi: 10.1016/j.ympev.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Karl R, Kiefer C, Ansell S, Koch MA. Systematics and evolution of arctic-alpine Arabis alpina L. (Brassicaceae) and its closest relatives in the eastern Mediterranean. American Journal of Botany. 2012;99:778–794. doi: 10.3732/ajb.1100447. [DOI] [PubMed] [Google Scholar]
- Kay KM, Sargent RD. The role of animal pollination in plant speciation: integrating ecology, geography and genetics. Annual Review of Ecology, Evolution and Systematics. 2009;40:637–656. [Google Scholar]
- Kay K, Voelckel C, Yan J, Hufford K, Kaska D, Hodges SA. Floral characters and species diversification. In: Harder L, Barrett S, editors. Ecology and evolution of flowers. Oxford: Oxford University Press; 2006. pp. 311–325. [Google Scholar]
- Koch MA. Mid-Miocene divergence of Ionopsidium and Cochlearia and its impact on the systematics and biogeography of the tribe Cochlearieae (Brassicaceae) Taxon. 2012;61:76–92. [Google Scholar]
- Koch M, Al-Shehbaz IA. Molecular data indicate complex intra- and intercontinental differentiation of American Draba (Brassicaceae) Annals of the Missouri Botanical Garden. 2002;89:88–109. [Google Scholar]
- Koch MA, Al-Shehbaz IA. Molecuar systematics and evolution of “wild” crucifers (Brassicaceae or Cruciferae) In: Gupta S, editor. Biology and breeding of crucifers. London: Taylor and Francis Group. 2009. pp. 1–19. [Google Scholar]
- Koch M, Kiefer C. Molecules and migration: biogeographical studies in cruciferous plants. Plant Systematics and Evolution. 2006;259:121–142. [Google Scholar]
- Koch M, Bishop J, Mitchell-Olds T. Molecular systematics and evolution of Arabidopsis and Arabis. Plant Biology. 1999;1:529–537. [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis and related genera. Molecular Biology and Evolution. 2000;17:1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248. [DOI] [PubMed] [Google Scholar]
- Koch M, Haubold B, Mitchell-Olds T. Molecular systematics of the Cruciferae: evidence from coding plastome matK and nuclear CHS sequences. American Journal of Botany. 2001;88:534–544. [PubMed] [Google Scholar]
- Koch M, Mummenhoff K, Al-Shehbaz IA. Molecular systematics, evolution, and population biology in the mustard family (Brassicaceae): a review of a decade of studies. Annals of the Missouri Botanical Garden. 2003;90:151–171. [Google Scholar]
- Koch M, Kiefer C, Vogel J, Ehrich D, Brochmann C, Mummenhoff K. Three times out of Asia Minor – the phylogeography of Arabis alpina L. (Brassicaceae) Molecular Ecology. 2006;15:825–839. doi: 10.1111/j.1365-294X.2005.02848.x. [DOI] [PubMed] [Google Scholar]
- Koch MA, Dobeš C, Kiefer C, Schmickl R, Klimeš L, Lysak MA. SuperNetwork identifies multiple events of plastid trnF (GAA) pseudogene evolution in the Brassicaceae. Molecular Biology and Evolution. 2007;24:63–73. doi: 10.1093/molbev/msl130. [DOI] [PubMed] [Google Scholar]
- Koch MA, Karl R, Kiefer C, Al-Shehbaz IA. Colonizing the American continent: systematics of the genus Arabis in North America (Brassicaceae) American Journal of Botany. 2010;97:1040–1057. doi: 10.3732/ajb.0900366. [DOI] [PubMed] [Google Scholar]
- Koch MA, Karl R, German DA, Al-Shehbaz IA. Systematics, taxonomy and biogeography of three new Asian genera from the Brassicaceae, tribe Arabideae: an ancient distribution circle around the Asian high mountains. Taxon. 2012a;61:955–969. [Google Scholar]
- Koch MA, Kiefer M, German D, et al. BrassiBase: tools and biological resources to study characters and traits in the Brassicaceae – version 1·1. Taxon. 2012b;61:1001–1009. [Google Scholar]
- Koelling VA, Mauricio R. Genetic factors associated with mating system cause a partial reproductive barrier between two parapatric species of Leavenworthia (Brassicaceae) American Journal of Botany. 2010;97:412–422. doi: 10.3732/ajb.0900184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinton JS. Genetics, paleontology, and macroevolution. New York: Cambridge University Press; 1988. [Google Scholar]
- Linkies A, Graeber K, Knight C, Leubner-Metzger G. The evolution of seeds. New Phytologist. 2010;186:817–831. doi: 10.1111/j.1469-8137.2010.03249.x. [DOI] [PubMed] [Google Scholar]
- Lo Presti RM, Oberprieler C. Evolutionary history, biogeography and eco-climatological differentiation of the genus Anthemis L. (Compositae, Anthemideae) in the circum-Mediterranean area. Journal of Biogeography. 2009;36:1313–1332. [Google Scholar]
- Losos JB. Lizards in an evolutionary tree: ecology and adaptive radiation of anoles. Berkeley: University of California Press; 2009. [Google Scholar]
- Lysak MA, Koch MA, Pecinka A, Schubert I. Chromosome triplication found across the tribe Brassiceae. Genome Research. 2005;15:516–525. doi: 10.1101/gr.3531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Koch MA, Beaulieu JM, Meister A, Leitch IJ. The dynamic ups and downs of genome size evolution in Brassicaceae. Molecular Biology and Evolution. 2009;26:85–98. doi: 10.1093/molbev/msn223. [DOI] [PubMed] [Google Scholar]
- Marais W. Cruciferae. In: Codd LE, Winter BD, Killick DJB, Rycroft HB, editors. Flora of southern Africa. Vol. 13. Pretoria: Government Printer; 1970. pp. 1–118. [Google Scholar]
- Marazzi B, Sanderson MJ. Large-scale patterns of diversification in the widespread legume genus Senna and the evolutionary role of extrafloral nectaries. Evolution. 2010;64:3570–3592. doi: 10.1111/j.1558-5646.2010.01086.x. [DOI] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmáry E. The major transitions in evolution. Oxford: Oxford University Press; 1995. [Google Scholar]
- Meulenkamp JE, Sissingh W. Tertiary palaeogeography and tectonostratigraphic evolution of the northern and southern peri-Tethys platforms and the intermediate domains of the African-Eurasian convergent plate boundary zone. Palaeogeography. Palaeoclimatology, Palaeoecology. 2003;196:209–228. [Google Scholar]
- Moazzeni H, Zarre S, Al-Shehbaz IA, Mummenhoff K. Seed-coat microsculpturing and its systematic application in Isatis (Brassicaceae) and allied genera in Iran. Flora. 2007;202:447–454. [Google Scholar]
- Molnar P. Mio-Pliocene growth of the Tibetan Plateau and evolution of East Asian climate. Palaeontologia Electronica. 2005;8:1–23. http://palaeo-electronica.org/2005_1/molnar2/molnar2.pdf . [Google Scholar]
- Moore BR, Donoghue MJ. A Bayesian approach for evaluating the impact of historical events on rates of diversification. Proceedings of the National Academy of Sciences USA. 2009;106:4307–4312. doi: 10.1073/pnas.0807230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mummenhoff K, Franzke A, Koch M. Molecular data reveal convergence in fruit characters, traditionally used in the classification of Thlaspi s.l. (Brassicaceae) – evidence from ITS-DNA sequences. Botanical Journal of the Linnean Society. 1997;125:183–199. [Google Scholar]
- Nylander JAA, Wilgenbusch JC, Warren DL, Swofford DL. AWTY (Are we there yet?): a system for graphical exploration of MCMC convergence in Bayesian phylogenetics. Bioinformatics. 2007;24:581–583. doi: 10.1093/bioinformatics/btm388. [DOI] [PubMed] [Google Scholar]
- Olson DM, Dinerstein E, Wikramanayake ED, et al. Terrestrial ecoregions of the world: a new map of life on earth. BioScience. 2001;51:933–938. [Google Scholar]
- Paetsch M, Mayland-Quellhorst S, Hurka H, Neuffer B. Evolution of the mating system in the genus Capsella (Brassicaceae) In: Glaubrecht M, editor. Evolution in action – adaptive radiations and the origins of biodiversity. Berlin: Springer; 2010. pp. 77–100. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Phillimore AB, Freckleton RP, Orme CDL, Owens IPF. Ecology predicts large-scale patterns of phylogenetic diversification in birds. American Naturalist. 2006;168:220–229. doi: 10.1086/505763. [DOI] [PubMed] [Google Scholar]
- Piller WE, Harzhauser M, Mandic O. Miocene central Paratethys stratigraphy – current status and future directions. Stratigraphy. 2007;4:151–168. [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rahbek C, Graves GR. Multiscale assessment of patterns of avian species richness. Proceedings of the National Academy of Sciences USA. 2001;98:4534–4539. doi: 10.1073/pnas.071034898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer v. 1.5. 2007 Available from http://beast.bio.ed.ac.uk/Tracer . [Google Scholar]
- Ree RH. Detecting the historical signature of key innovations using stochastic models of character evolution and cladogenesis. Evolution. 2005;59:257–265. [PubMed] [Google Scholar]
- Roe GH. Orographic precipitation. Annual Review of Earth Planetary Sciences. 2005;33:645–671. [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Santini F, Harmon LJ, Carnevale G, Alfaro ME. Did genome duplication drive the origin of teleosts? A comparative study of diversification in ray-finned fishes. BMC Evolutionary Biology. 2009;9:e194. doi: 10.1186/1471-2148-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent RD. Floral symmetry affects speciation rates in angiosperms. Proceedings of the Royal Society B. 2004;271:603–608. doi: 10.1098/rspb.2003.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffer WM, Gadgil M. Selection for optimal life histories in plants. In: Cody M, Diamond J, editors. The ecology and evolution of communities. Cambridge, MA: Belknap Press; 1975. pp. 142–157. [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends in Plant Science. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP. Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Current Opinion in Plant Biology. 2012;15:147–153. doi: 10.1016/j.pbi.2012.03.011. [DOI] [PubMed] [Google Scholar]
- Silvestro D, Schnitzler J, Zizka G. A Bayesian framework to estimate diversification rates and their variation through time and space. BMC Evolutionary Biology. 2011;11:e311. doi: 10.1186/1471-2148-11-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG. The major features of evolution. New York: Columbia University Press; 1953. [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversification. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annual Review of Plant Biology. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Self fertilization and population variability in the higher plants. American Naturalist. 1957;91:337–354. [Google Scholar]
- Steininger FF, Rögl F. Paleogeography and palinspastic reconstruction of the Neogene of the Mediterranean and Paratethys. In: Dixon JE, Robertson AHF, editors. The geological evolution of the eastern Mediterranean. Vol. 17. London: Geological Society; 1984. pp. 659–668. [Google Scholar]
- Sun FJ, Downie SR, Hartman R. An ITS-based phylogenetic analysis of the perennial, endemic Apiaceae subfamily Apiodeae of western North America. Systematic Botany. 2004;29:419–431. [Google Scholar]
- Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Molecular Biology. 1991;17:1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tank DC, Olmstead RG. From annuals to perennials: phylogeny of subtribe Castillejinae (Orobanchaceae) American Journal of Botany. 2008;95:608–625. doi: 10.3732/ajb.2007346. [DOI] [PubMed] [Google Scholar]
- Tiffney BH, Manchester SR. The use of geological and paleontological evidence in evaluating plant phylogeographic hypotheses in the Northern Hemisphere Tertiary. International Journal of Plant Sciences. 2001;162:S3–S17. [Google Scholar]
- Vamosi JC, Vamosi SM. Factors influencing diversification in angiosperms: at the crossroads of intrinsic and extrinsic traits. American Journal of Botany. 2011;98:460–471. doi: 10.3732/ajb.1000311. [DOI] [PubMed] [Google Scholar]
- van Dam JA. Geographic and temporal patterns in the late Neogene (12–3 MA) aridification of Europe: the use of small mammals as paleoprecipitation proxies. Palaeogeography, Palaeoclimatology, Palaeoecology. 2006;238:190–218. [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nature Reviews Genetics. 2009;10:725–732. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- Verboom GA, Linder HP, Stock WD, Baum D. Phylogenetics of the grass genus Ehrharta: evidence for radiation in the summer-arid zone of the South African cape. Evolution. 2003;57:1008–1021. doi: 10.1111/j.0014-3820.2003.tb00312.x. [DOI] [PubMed] [Google Scholar]
- von Hagen KB, Kadereit JW. The diversification of Halenia (Gentianaceae): ecological opportunity versus key innovation. Evolution. 2003;57:2507–2518. [PubMed] [Google Scholar]
- Wang CS, Zhao XX, Liu ZF, et al. Constraints on the early uplift history of the Tibetan Plateau. Proceedings of the National Academy of Sciences USA. 2008;105:4987–4992. doi: 10.1073/pnas.0703595105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Farrona S, Vincent C, et al. PEP1 regulates perennial flowering in Arabis alpina. Nature. 2009;459:423–427. doi: 10.1038/nature07988. [DOI] [PubMed] [Google Scholar]
- Warwick SI, Mummenhoff K, Sauder CA, Koch MA, Al-Shehbaz IA. Closing the gaps: phylogenetic relationships in the Brassicaceae based on DNA sequence data of nuclear ribosomal ITS region. Plant Systematics and Evolution. 2010;285:209–232. [Google Scholar]
- White TJ, Burns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, CA: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Yang Z, Rannala B. Bayesian phylogenetic inference using DNA sequences: a Markov chain Monte Carlo method. Molecular Biology and Evolution. 1997;14:717–724. doi: 10.1093/oxfordjournals.molbev.a025811. [DOI] [PubMed] [Google Scholar]
- Yoder JB, Clancey E, Des Roches S, et al. Ecological opportunity and the origin of adaptive radiations. Journal of Evolutionary Biology. 2010;23:1581–1596. doi: 10.1111/j.1420-9101.2010.02029.x. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He XJ. S-DIVA (statistical dispersal-vicariance analysis): a tool for inferring biogeographic histories. Molecular Phylogenetics and Evolution. 2010;56:848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- Yu Y, Harris AJ, He XJ. RASP (Reconstruct Ancestral State in Phylogenies) 2·0 beta. 2011 doi: 10.1016/j.ympev.2015.03.008. Available at http://mnh.scu.edu.cn/soft/blog/RASP . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.