Abstract
Background and Aims
The genome size of an organism is determined by its capacity to tolerate genome expansion, given the species' life strategy and the limits of a particular environment, and the ability for retrotransposon suppression and/or removal. In some giant-genomed bulb geophytes, this tolerance is explained by their ability to pre-divide cells in the dormant stages or by the selective advantage of larger cells in the rapid growth of their fleshy body. In this study, a test shows that the tendency for genome size expansion is a more universal feature of geophytes, and is a subject in need of more general consideration.
Methods
Differences in monoploid genome sizes were compared using standardized phylogenetically independent contrasts in 47 sister pairs of geophytic and non-geophytic taxa sampled across all the angiosperms. The genome sizes of 96 species were adopted from the literature and 53 species were newly measured using flow cytometry with propidium iodide staining.
Key Results
The geophytes showed increased genome sizes compared with their non-geophytic relatives, regardless of the storage organ type and regardless of whether or not vernal geophytes, polyploids or annuals were included in the analyses.
Conclusions
The universal tendency of geophytes to possess a higher genome size suggests the presence of a universal mechanism allowing for genome expansion. It is assumed that this is primarily due to the nutrient and energetic independence of geophytes perhaps allowing continuous synthesis of DNA, which is known to proceed in the extreme cases of vernal geophytes even in dormant stages. This independence may also be assumed as a reason for allowing large genomes in some parasitic plants, as well as the nutrient limitation of small genomes of carnivorous plants.
Keywords: Genome size evolution, Cx-value, life form, spring geophytes, ephemeroids, storage organ, energy reserves, flow cytometry
INTRODUCTION
Genome size varies considerably among angiosperms (Bennett and Leitch, 2012). Within closely related taxa, a few multiple fold differences in genome size are frequently due to polyploidy (but see also Piegu et al., 2006), while over longer evolutionary time scales the >2000-fold difference in genome size across angiosperms (Greilhuber et al., 2006) is mostly accounted for by the proliferation and removal of repetitive DNA, namely of retrotransposons (Bennetzen et al., 2005). While polyploidy is rather an incidental event in plant evolution (Soltis et al., 2009; Fawcett et al., 2013), the tendency of retrotransposons to amplify seems to be a continuous and ubiquitous molecular force driving genome size increase until it became selectively disadvantageous for an organism (Petrov, 2001; Bennetzen et al., 2005; Kejnovský et al., 2013). The genome size of an organism is thus determined (1) by the capacity to tolerate genome expansion given by a species' life strategy and the limits of a particular environment (Grime and Mowforth, 1982; Leitch and Bennett, 2007; Greilhuber and Leitch, 2013) and (2) by the ability for retrotransposon suppression and/or removal (Petrov, 2001).
There are three recognized major effects of genome size increase which perhaps mostly determine its selective advantage or disadvantage: (1) prolonged duration of DNA replication and cell cycle lengths (Bennett, 1971, 1987; Francis et al., 2008); (2) increase in cell size and change of cellular surface to volume ratio (Cavalier-Smith, 2005; Gregory, 2005); and (3) increase of energetic demand needed for building extra DNA and larger cells (Leitch and Bennett, 2004; Cavalier-Smith, 2005). The tolerance of these effects thus strongly depends on the life strategy of a specific species and a particular environment, or vice versa the particular genome size may limit a species to adopt specific life strategies or colonize some environments (Bennett, 1987). For example, genome size is usually reduced in annual species (therophytes; Leitch and Bennett, 2007) to ensure short cell cycles and enable annuals to complete their life cycle before the end of the growing season. Actually, an ephemeral or annual lifestyle seems impossible with a genome size over approx. 7 Gbp or approx. 20 Gbp, respectively (Bennett, 1987). Larger genome sizes and polyploidy seem also to be prevented in woody species (phanerophytes and chamaephytes; Bennett and Leitch, 2012) in which this does not result from any temporal limitation but rather from structural and physiological constraints on the size of wood cells, ensuring proper mechanical properties of woody tissue, or the need for smaller and denser stomata ensuring enough stomatal conductance necessary for transport of water and nutrients through long xylem pathways (Stebbins, 1938; Beaulieu et al., 2008). The larger genome sizes are thus regularly found only in perennial herbs (hemicryptophytes and geophytes) which are neither temporally limited like annuals nor perhaps so strongly limited with structural constraints on the cell size as is expected to be the case of supporting woody tissue in trees and shrubs.
Within (perennial) angiosperms, the absolutely largest genomes (up to 298 Gbp) are almost always found in geophytic plants which are phylogenetically clustered, especially in several large-genomed families of monocot orders Liliales and Asparagales (Bennett and Leitch, 2012; Veselý et al., 2012). Giant genomes in groups of vernal geophytes (ephemeroids) are assumed to be produced by the tolerance of prolonged cell division which continues underground in the bulbs during the ‘dormant stage’ or even by the selective advantage of the larger cells in the rapid growth of their fleshy body formed by pumping water into the pre-divided cells during the favourable wet spring period (Grime and Mowforth, 1982; Grime, 1983). The upper limit for genome expansion in giant genome geophytes is thus thought to be determined by the size and regulability of stomata (directly related to carbon dioxide intake and transpiration) restricting their body development to cooler and stably wet spring periods (Veselý et al., 2012). Nevertheless, recent surveys also show relatively high genome sizes in other groups of geophytic plants (Baranyi and Greilhuber, 1999; Veselý et al., 2012), suggesting the potential existence of a more universal mechanism allowing for genome expansion in geophytes. Among others, this might be due to sufficient nutrient reserves needed for investment in extra DNA synthesis which are stored in the sub-terranean storage organs and available irrespective of the availability and variation of external nutrient resources. If this is true, a geophytic life strategy should regularly result in an increase in genome size in geophytic lineages compared with their sister non-geophytic lineages; this trend should appear independently of ephemeroid or any particular geophytic strategy in all geophytic lineages. While the genome size data on geophytic plants are representative for the large geophytic clades (Liliaceae, Melanthiaceae, Asparagaceae and Amaryllidaceae), they are still frequently lacking for phylogenetically isolated lineages or species needed for verifying the universality of the above-mentioned trends. The data on genome size are also largely absent for the sister non-geophytic groups of geophytic lineages, critically needed for testing the effect of geophytism on genome size in the evolutionary context.
Here we extracted data on genome sizes of geophytes from the existing literature and selectively measured the genome size in their non-geophytic relatives. The differences in monoploid genome sizes were then compared for 47 pairs of geophytic species and their close (sister) non-geophytic relatives with standardized phylogenetically independent contrasts (Felsenstein, 1985).
MATERIALS AND METHODS
Closely related pairs of geophytic and non-geophytic taxa (mostly species) were searched for across all angiosperms. Specifically we searched for the most recent sister contrasts according to the series of published phylogenies (Table 2). In five selected genera where phylogeny is not exactly known, we assumed polytomous relationships of species.
Table 2.
Comparison of geophytes and sister non-geophytes
| Node number | Geophyte | Non-geophyte | Distance | Cx (G) | Cx (N) | Relative increase | Therophyte contrast | Higher N ploidy contrast | Ephemeroid geophyte | Storage organ | Phylogeny |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Aristolochia rotunda, A. fimbriata | Aristolochia clematitis | 1 | 366 | 379 | –0.03 | No | No | No | Tuber | |
| 2 | Ranunculus bulbosus | Ranunculus polyanthemos, R. sardous | 1 | 5117 | 4522 | 0.13 | Yes | No | No | Tuber | Hörandl et al. (2005) |
| 3 | Anemone narcissiflora | Hepatica nobilis | 2 | 8776 | 15 174 | –0.21 | No | No | No | Rhizome | Ehrendorfer and Samuel (2001) |
| 4 | Anemone apennina, A. blanda, A. coronaria, A. hortensis, A. nemorosa, A. ranunculoides | Anemone sylvestris, A. virginiana | 1 | 11 186 | 8314 | 0.35 | No | No | Yes | Rhizome | Ehrendorfer and Samuel (2001) |
| 5 | Isopyrum thalictroides | Thalictrum simplex galioides | 2 | 427 | 542 | –0.11 | No | Yes | Yes | Rhizome | |
| 6 | Epimedium alpinum, Jeffersonia diphylla, J. dubia, Podophyllum emodi | Berberis vulgaris, Nandina domestica | 4 | 5364 | 1746 | 0.52 | No | No | Yes | Rhizome | Kim et al. (2004) |
| 7 | Sanguinaria canadensis | Chelidonium majus, Dicranostigma franchetianum | 4 | 971 | 883 | 0.02 | No | No | Yes | Rhizome | Gleissberg and Kadereit (1999) |
| 8 | Corydalis cava, C. intermedia, C. solida, C. pumila | Corydalis lutea | 1 | 704 | 295 | 1.39 | No | Yes | Yes | Tuber | |
| 9 | Parthenocissus himalayensis | Parthenocissus tricuspidata | 1 | 795 | 514 | 0.55 | No | No | No | Tuber | Nie et al. (2010) |
| 10 | Oxalis acetosella, O. corymbosa, O. pes-caprae, O. spiralis, O. vulcanicola, O. linearis, O. megalorrhiza | Oxalis corniculata repens, O. dillenii | 2 | 1643 | 282 | 2.41 | Yes | Yes | No | Bulb | Oberlander (2009) |
| 11 | Passiflora quadrangularis | Passiflora edulis | 1 | 2139 | 1557 | 0.37 | No | No | No | Root | Muschner et al. (2003) |
| 12 | Mercurialis perennis | Mercurialis annua, M. huetii | 1 | 787 | 669 | 0.18 | Yes | No | No | Rhizome | Krähenbühl et al. (2002) |
| 13 | Euphorbia cf apios | Euphorbia helioscopia | 1 | 717 | 438 | 0.64 | Yes | No | No | Root | Frajman and Schönswetter (2011) |
| 14 | Lathyrus tuberosus | Lathyrus heterophyllus, L. latifolius, L. annuus | 1 | 6029 | 7265 | –0.17 | Yes | No | No | Tuber | Kenicer et al. (2005) |
| 15 | Lathyrus laxiflorus | Lathyrus aphaca | 1 | 8210 | 4519 | 0.82 | Yes | No | No | Root | Kenicer et al. (2005) |
| 16 | Bryonia alba, B. dioica, B. verrucosa, Ecballium elaterium | Echinocystis lobata, Luffa cylindrica | 4 | 1883 | 767 | 0.36 | Yes | No | No | Tuber | Kocyan et al. (2007); Voltz and Renner (2008) |
| 17 | Begonia grandis, B. dregei, B. socotrana | Begonia luxurians | 1 | 584 | 313 | 0.86 | No | No | No | Tuber | Forrest et al. (2005) |
| 18 | Geranium tuberosum | Geranium columbinum, G. sanguineum | 1 | 606 | 817 | –0.26 | Yes | Yes | No | Tuber | |
| 19 | Cardamine bulbifera | Cardamine impatiens | 1 | 307 | 178 | 0.72 | Yes | No | Yes | Rhizome | Carlsen et al. (2009) |
| 20 | Tropaeolum tuberosum | Tropaeolum majus | 1 | 1239 | 1154 | 0.07 | No | Yes | No | Tuber | Andersson and Andersson (2000) |
| 21 | Impatiens omeiensis | Impatiens parviflora, I. glandulifera | 1 | 1063 | 933 | 0.14 | Yes | No | No | Tuber | Janssens et al. (2006) |
| 22 | Symphytum tuberosum | Symphytum officinale | 1 | 671 | 635 | 0.06 | No | No | No | Tuber | |
| 23 | Vinca herbacea | Vinca minor | 1 | 789 | 670 | 0.18 | No | No | No | Root | |
| 24 | Asclepias syriaca | Vincetoxicum hirundinaria | 2 | 411 | 323 | 0.14 | No | No | No | Rhizome | Sennblad and Bremer (2002) |
| 25 | Phlomis tuberosa | Phlomis russeliana | 1 | 1982 | 2311 | –0.14 | No | Yes | No | Rhizome | |
| 26 | Stachys affinis | Stachys sylvatica | 1 | 1398 | 1136 | 0.23 | No | No | No | Tuber | Bendiksby et al. (2011) |
| 27 | Scrophularia nodosa | Scrophularia umbrosa neesii, S. vernalis | 1 | 676 | 470 | 0.44 | Yes | Yes | No | Rhizome | |
| 28 | Datura inoxia | Datura stramonium, D. quercifolia | 1 | 2221 | 1795 | 0.24 | Yes | No | No | Root | |
| 29 | Solanum tuberosum, S. pinnatisectum | Solanum lycopersicum, S. etuberosum | 1 | 773 | 803 | –0.04 | Yes | No | No | Tuber | Szinay et al. (2012) |
| 30 | Ipomoea batatas, I. trifida, I. tiliacea | Ipomoea quamoclit | 1 | 799 | 619 | 0.29 | Yes | No | No | Tuber | Huang et al. (2002) |
| 31 | Adoxa moschatellina | Sambucus nigra, S. racemosa, S. ebulus | 2 | 20 106 | 10 898 | 0.42 | No | No | Yes | Rhizome | Moore and Donoghue (2009) |
| 32 | Valeriana tuberosa | Valeriana officinalis, V. dioica | 1 | 1616 | 1408 | 0.15 | No | No | No | Tuber | |
| 33 | Hacquetia epipactis | Sanicula europaea | 1 | 1346 | 1054 | 0.28 | No | No | Yes | Rhizome | |
| 34 | Doronicum hungaricum | Doronicum austriacum | 1 | 3762 | 3540 | 0.06 | No | No | No | Tuber | |
| 35 | Bellis sylvestris | Bellis perennis, B. annua | 1 | 998 | 1476 | –0.32 | Yes | No | No | Rhizome | |
| 36 | Helianthus tuberosus | Helianthus annuus, H. petiolaris, H. debilis, H. niveus | 1 | 3339 | 3250 | 0.03 | Yes | No | No | Tuber | Timme et al. (2007) |
| 37 | Dahlia pinnata | Bidens frondosa, B. radiata | 2 | 1056 | 918 | 0.08 | Yes | No | No | Root | |
| 38 | Cosmos atrosanguineus | Cosmos bipinnatus, C. sulphureus | 1 | 3588 | 2225 | 0.61 | Yes | No | No | Root | |
| 39 | Scorzonera mollis | Scorzonera austriaca | 1 | 2405 | 4979 | –0.52 | No | No | No | Root | |
| 40 | Arisaema flavum | Pistia stratiotes | 2 | 2480 | 284 | 3.87 | No | No | No | Tuber | Tam et al. (2004) |
| 41 | Homalomena rubescens | Philodendron erubescens, P. squamiferum, P. pinnatifidum | 2 | 8937 | 3621 | 0.73 | No | No | No | Tuber | Tam et al. (2004); Gauthier et al. (2008) |
| 42 | Triglochin bulbosa | Triglochin maritima | 1 | 863 | 590 | 0.46 | No | No | No | Bulb | |
| 43 | Narthecium ossifragum | Narthecium reverchonii | 1 | 404 | 358 | 0.13 | No | No | No | Rhizome | |
| 44 | Ruscus aculeatus, R. hypoglossum, R. hypophyllum | Semele androgyna | 2 | 8744 | 5921 | 0.24 | No | No | No | Rhizome | Kim et al. (2010) |
| 45 | Asphodelus microcarpus, A. albus | Asphodelus fistulosus | 1 | 3411 | 2452 | 0.39 | No | No | No | Root | Lifante (1996) |
| 46 | Arrhenatherum palaestinum | Arrhenatherum elatius | 1 | 4301 | 3474 | 0.24 | No | Yes | No | Bulb | |
| 47 | Hordeum bulbosum | Hordeum vulgare | 1 | 3808 | 4890 | –0.22 | Yes | No | No | Bulb | Blattner (2004) |
The table shows Cx-values, relative increase of genome size in geophytes, contrast with therophytes, contrast with non-geophytes, with a higher ploidy level than geophytes, presence of ephemeroid phenology in geophytes, type of storage organ in the geophyte (tuber, bulb, thick rhizome or tuberous root), and reference for the phylogeny used.
Genome sizes for 50 selected species were extracted from the Plant C-value database (Bennett and Leitch, 2012) and our previous data for 46 species (Veselý et al., 2012) were added. When more data were available for a species, the most recent and those produced by flow cytometry were preferred (Supplementary Data, Table S1). In other cases, all available data for a species were averaged. In addition to the literature data, genome size was also measured in 53 selected species to increase the number of existing geophyte/non-geophyte contrasts (Supplementary Data, Table S1).
Genome sizes of selected species were estimated by flow cytometry (ML, Partec GmbH) using a two-step procedure with propidium iodide (Otto, 1990; Doležel et al., 2007). Detailed sample preparation and dye concentrations follow Šmarda et al. (2008). As the internal standard for calculation of genome size, we used the fully sequenced cultivar of rice (Oryza sativa ‘Nipponbare’, 2C = 777·64 Mbp; International Rice Genome Sequencing Project, 2005) and a series of conventional primary internal plant standards (Doležel and Greilhuber, 2010) whose genome sizes were derived from the genome size estimate of rice (cf. Veselý et al., 2012). Our genome size estimates are thus usually slightly lower than the data reported in the Plant C-value database, mostly based on the measurements with standards whose genome size is derived from the overestimated value for genome size for human, 2C = 7 pg (Doležel and Greilhuber, 2010). Our standardization procedure provides an estimate of the human genome size (6·19 pg or 6 055 Mbp) very close to the human genome size predicted from the complete genome sequencing projects (6·29 pg or 6 153 Mbp; International Human Genome Sequencing Consortium, 2004), and we therefore hope that our genome size estimates are thus closer to biological reality. Because of the occasional peak overlaps between the sample and the primary standards, a series of secondary internal standards was also established from the already measured species (Table 1). Details on the sample and secondary standard measurements are given in Supplementary Data, Table S1.
Table 1.
Standards used for flow cytometry measurements
| Standard | Genome size 2C (Mbp) |
|---|---|
| Oryza sativa ‘Nipponbare’ | 777·64 |
| Carex acutiformis | 799·93 |
| Ipomoea quamoclit | 1238·30 |
| Solanum lycopersicum ‘Stupické polní rané’ | 1696·81 |
| Bellis perennis | 3089·89 |
| Epipremnum aureum | 7815·39 |
| Pisum sativum ‘Ctirad’ | 7841·27 |
| Ruscus aculeatus | 20 137·45 |
| Vicia faba ‘Inovec’ | 23 272·88 |
| Leucojum aestivum | 61 563·46 |
| Haemanthus albiflos | 65 112·69 |
When known with certainty, the names of cultivars are given.
The differences in genome size (tolerance to genome expansion) between geophytic and non-geophytic species or clades were compared with standardized phylogeny-independent contrasts (Felsenstein, 1985), comparing only contrasts between sister groups. In total, our search and measurements produced 47 sister contrasts (Table 2). If the clade was represented by three or more species, their mean genome size was calculated using the standard node-based method as in the standard analysis of phylogeny-independent contrasts (Felsenstein, 1985; Webb et al., 2011) considering published molecular phylogenies (Table 2). To avoid genome size increases originating due to polyploidy (i.e. not to resolve the question of whether polyploids arise more frequently in geophytic or non-geophytic lineages; this testing would require a different data sampling design), we only compared the differences in monoploid genome sizes (Cx-values; Greilhuber et al., 2005). Ploidy levels of species for calculation of Cx-values were based on comparison of absolute 2C DNA contents of species and consensus of chromosome numbers in the IPCN database (Goldblatt and Johnson, 1979–onwards) and Fedorov (1969). When variable ploidy levels were reported for a species, we preferred that from the region of the sample's origin (e.g. Kubát et al., 2002) and that with no logical conflict with already published genome sizes in the Plant C-value database (Bennett and Leitch, 2012).
The absolute size of any contrasts in genome size between two taxa increases naturally (1) with the divergence time from the common ancestor, under the assumption of a Brownian motion model of genome size evolution (followed here; Felsenstein, 1985; Garland et al., 1992) and (2) with the genome size of the common ancestor (Oliver et al., 2007). To remove these two effects and to make contrasts fully comparable, we calculated contrasts in a relative fashion and applied a divergence time-dependent standardization. The relative increase (r) of genome sizes in geophytes (G) compared with the sister non-geophytes (N) was calculated using the formula r = (G – N)/N. Because of the absence of actual divergence times of compared pairs, we adopt an alternative standardization approach by dividing contrasts by arbitrarily selected values aimed to reflect phylogenetic relatedness (distance) of compared groups. Contrasts were divided: by 1 when comparing sister or very closely related species from the same genus; by 2 when comparing closely related genera; and by 4 when comparing distantly related genera of the same family. The difference of contrasts from zero was tested by one-sample Wilcoxon signed-rank test.
To ensure that the relationship is a universal feature of geophytism (nutrient/energy storage) and that it is not only caused by the inclusion of geophytes with a ephemeroid strategy, where genome size increase is a priori expected (Grime and Mowforth, 1982; Greilhuber, 1995; Hodgson et al., 2010; Veselý et al., 2012), the analyses were also repeated omitting all eight contrasts with ephemeroid geophytes. Geophytes differ in the type of storage organs (Dafni et al., 1981) and the respective effectiveness of nutrient and energy accumulation, which might play a role in determining tolerance of genome expansion. Therefore, geophytes were also classified according to the type of storage organs (tuber, bulb, thick rhizome or tuberous root; Table 2), and the possible differences in the tendency to genome expansion among these four categories were tested by Kruskal–Wallis analysis of variance (ANOVA). Life form was also noted in all non-geophytic plants (Supplementary Data, Table S1). The identifications of life forms, ephemeroid strategy and types of storage organs were based on our personal experience with the plants measured and based on their visual inspection.
In several cases, geophytes were compared with therophytes or diploid geophytes with polyploid non-geophytes. These contrasts might provide false positives in the analyses (i.e. the larger genome size observed in a geophyte would not be due to its genome expansion but due to genome downsizing in the sister non-geophyte) because in both therophytes and polyploids a species' tendency to genome downsizing is known (see the Introduction; Bennett, 1987; Leitch and Bennett, 2004). For this reason, testing the difference of contrasts from zero was also repeated by removing either therophyte–geophyte (19 cases) or diploid geophyte–polyploid non-geophyte contrasts (eight cases) or by removing both (24 contrasts). The effect of genome downsizing may also be present in polyploid geophytes. In this case, however, polyploid genome downsizing cannot produce any false positives and may only decrease the resulting statistical significance of the expected trend of genome size increase in geophytes. Therefore, these cases were not corrected in the analyses.
Because a difference may potentially exist between the data from our laboratory and that reported in the Plant C-value database (given by a different standardization, see above), we carried out all statistical analyses either with original estimates or with the data adjusted for better comparability. This adjustment was performed with the data from the Plant C-value database by multiplying them by 6·19/7, i.e. by the expected difference ratio based on the human genome size accepted in our laboratory (6·19 pg) and by many authors included in the Plant C-value database (7 pg). This adjustment resulted in slightly better statistical significance of the results. Otherwise, however, the results were principally the same and therefore only results with original, unadjusted values are reported further for simplicity.
RESULTS
Our search and flow cytometry measurements of 53 species provided 47 sister contrast comparisons (149 species are included in total). In most cases, geophytism was associated with the increase in monoploid genome size (Table 2) and this trend was statistically significant when calculating either with the whole data set (Fig. 1; n = 47, z = 4·127, P < 0·001) or with the data for non-vernal geophytes only (n = 39, z = 3·419, P < 0·001). The tendency for a genome size increase also did not differ among geophytes with different storage organs [Kruskal–Wallis test, H (3, n = 47): H = 1·655, P = 0·647]. Omission of the geophyte vs. therophyte and the diploid geophyte vs. polyploid non-geophyte comparisons from the analyses produced significant results similar to the analyses with the complete data set (n = 28, z = 3·461, P < 0·001 by omitting contrasts with therophytes; n = 39, z = 3·963, P < 0·001 for omitting contrasts with polyploid non-geophytes; and n = 23, z = 3·315, P < 0·001 by omitting both). This indicates no or a negligible effect of false positives due to genome downsizing in therophytes or polyploids on the reported results.
Fig. 1.
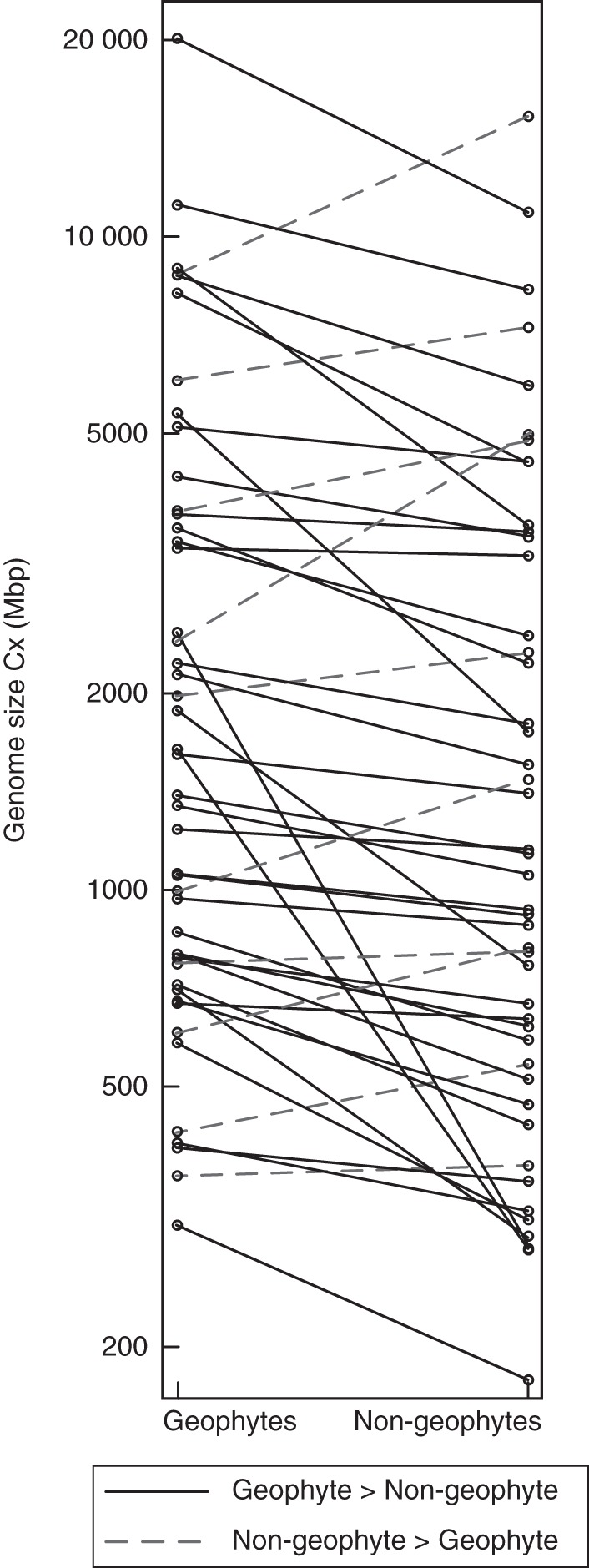
Comparison of genome sizes of geophytes and their sister non-geophytes. Cases of a larger genome in geophytes are given as solid lines; whereas cases of a larger genome in non-geophytes are given as dashed lines.
DISCUSSION
Due to the strong phylogenetic clustering and the lack of sister non-geophytes, the analysed data include only a few species from large-genomed families in the orders Liliales and Asparagales (only two families – Asparagaceae and Xanthorrhoeaceae – are represented here) in which geophytism is synapomorphous and which include the majority of geophytic species richness of angiosperms. Even though the effect of these previously studied large-genomed families (Grime and Mowforth, 1982; Zonneveld, 2010) is considerably suppressed in the present phylogenetically corrected analysis, geophytes still have larger genomes and show higher tolerance to genome expansion than their non-geophytic relatives. The universality of the observed trend is also supported by the fact that significant results are obtained regardless of whether the data are included on vernal geophytes, in which genome size increase has already been reported multiple times previously (Grime and Mowforth, 1982; Greilhuber, 1995; Hodgson et al., 2010; Veselý et al. 2012), or not. At the same time, however, this indicates that mechanisms of genome expansion tolerance known in vernal geophytes (the advantage of larger cells and their water pumping or the ability of cell to pre-divide) are either more widespread in geophytes than already expected or the increase in genome size in geophytes is primarily allowed by other favourable traits.
Looking for an alternative explanation of the increased ability of geophytes for genome expansion, energetic reserves may be of particular interest. The storage of energetic and nutrition reserves in storage organs is the key feature of geophytic plants (Dafni et al., 1981; Ruiters and McKenzie, 1994) which makes geophytes relatively independent of the availability of energetic and nutrition sources in the environment. This may be favourable in the case of increased nutrition and energetic demand needed for synthesis of extra DNA, hypothetically allowing geophytes to carry out continuous DNA synthesis irrespective of the variation of nutrients and other resources in the external environment. Energetic and nutrition independence seems to be the essential condition allowing the pre-division of the cell during the dormant period in the underground bulbs of some large-genomed geophytes. Although the ability to carry out cell pre-division is frequently assumed to be the major reason for the tolerance of the genome size increase in some large-genomed bulbous plants (Grime, 1983; Greilhuber, 1995), this would certainly not be possible without a high level of energetic and nutrient independence. Hence the explanation of genome expansion by the ability of a cell to pre-divide in some bulb geophytes (Grime and Mowforth, 1982; Greilhuber, 1995) is not in conflict with our nutrition independence hypothesis as it may be seen only as a special case in which geophytes can profit from their common energetic independence.
Support for the importance of energetic and nutrition independence in determining the tolerance of plants to genome expansion may also be found in other plants relatively independent of the fluctuation of external energetic and nutrition resources. Most noticeably, this is the case of parasitic plants which gain the majority of their nutrients and energy from their hosts (Hibberd and Jeschke, 2001). Like geophytes, indeed, the genome sizes of parasitic plants are frequently apparently higher than in their non-parasitic relatives. For example, parasitic Cuscuta has a 6-fold larger genome than the rest of Convolvulaceae, and parasitic Krameria has a >7-fold larger genome than the sister family Zygophyllaceae (Bennett and Leitch, 2012; P. Šmarda et al., unpubl. data). Large genome sizes are further observed in parasitic species of Santalales (Leitch et al., 2005), including Viscum album (2C = 201·2 Gbp) belonging to six species with the highest genome sizes known to date (Bennett and Leitch, 2012; Pellicer et al., 2010; Zonneveld, 2010). On the other hand, very small genome sizes are typical of some carnivorous plants (Greilhuber et al., 2006; A. Veleba et al., Masaryk University, Czech Republic, pers. comm.), frequently growing in extremely nutrient-poor environments and depending to a great extent on the incidental supply of nutrients and energy from the captured prey (Ellison, 2006; Karagatzides et al., 2009; Adamec, 2011).
The above evidence clearly suggests that continuous energetic and nutrition supply or independence (as typical of geophytes and parasitic plants) may be one of the important determinants of species tolerance to genome expansion. Therefore, further research seems desirable to verify this hypothesis in other groups of plants with lower genome sizes in which the trend may be masked by other selective forces or in direct experiments searching for the effect of nutrient limitation or availability on the selection of plants with small or large genome sizes. The first comparisons of genome sizes in local plant communities in field nutrition experiments indicate indeed that nutrient availability may play a role in the selection of plants with different genome size (Šmarda et al., 2013). However, further experiments are clearly needed to verify the universality and the importance of nutrient supply or independence in determining tolerance of particular organisms to genome expansion and evolution of plant genome size diversity.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We would like to thank the Botanical Garden of the Faculty of Science, Masaryk University, Brno and the Prague Botanic Garden, Prague, for providing plant material. This study was supported by the Czech Science Foundation (project nos P505/11/0881, P506/10/1363, P506/13/29362S and 526/09/H025).
LITERATURE CITED
- Adamec L. Ecophysiological look at plant carnivory: why are plants carnivorous? In. In: Seckbach J, Dubinsky Z, editors. All flesh is grass. Cellular origin, life in extreme habitats and astrobiology. Berlin: Springer-Verlag; 2011. pp. 455–489. [Google Scholar]
- Andersson L, Andersson S. A molecular phylogeny of Tropaeolaceae and its systematic implications. Taxon. 2000;49:721–736. [Google Scholar]
- Baranyi M, Greilhuber J. Genome size in Allium: in quest of reproducible data. Annals of Botany. 1999;83:687–695. [Google Scholar]
- Beaulieu JM, Leitch IJ, Patel S, Pendharkar A, Knight CA. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytologist. 2008;179:975–986. doi: 10.1111/j.1469-8137.2008.02528.x. [DOI] [PubMed] [Google Scholar]
- Bendiksby M, Thorbek L, Scheen AC, Lindqvist C, Ryding O. An updated phylogeny and classification of Lamiaceae subfamily Lamioideae. Taxon. 2011;60:471–484. [Google Scholar]
- Bennett MD. The duration of meiosis. Proceedings of the Royal Society B: Biological Sciences. 1971;178:259–275. [Google Scholar]
- Bennett MD. Variation in genomic form in plants and its ecological implications. New Phytologist. 1987;106(Suppl.):177–200. [Google Scholar]
- Bennett MD, Leitch IJ. Plant DNA C-values Database (release 6·0, December 2012) 2012 http://data.kew.org/cvalues/ [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Annals of Botany. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR. Phylogenetic analysis of Hordeum (Poaceae) as inferred by nuclear rDNA ITS sequences. Molecular Phylogenetics and Evolution. 2004;33:289–299. doi: 10.1016/j.ympev.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Carlsen T, Bleeker W, Hurka H, Elven R, Hochmann C. Biogeography and phylogeny of Cardamine (Brassicaceae) Annals of the Missouri Botanical Garden. 2009;96:215–236. [Google Scholar]
- Cavalier-Smith T. Economy, speed and size matter: evolutionary forces driving nuclear genome miniaturization and expansion. Annals of Botany. 2005;95:147–175. doi: 10.1093/aob/mci010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafni A, Cohen D, Noy-Meir I. Life-cycle variation in geophytes. Annals of the Missouri Botanical Garden. 1981;68:652–660. [Google Scholar]
- Doležel J, Greilhuber J. Nuclear genome size: are we getting closer? Cytometry. 2010;77A:635–642. doi: 10.1002/cyto.a.20915. [DOI] [PubMed] [Google Scholar]
- Doležel J, Greilhuber J, Suda J. Estimation of nuclear DNA content in plants using flow cytometry. Nature Protocols. 2007;2:2233–2244. doi: 10.1038/nprot.2007.310. [DOI] [PubMed] [Google Scholar]
- Ehrendorfer F, Samuel R. Contributions to a molecular phylogeny and systematics of Anemone and related genera (Ranunculaceae–Anemoninae) Acta Phytotaxonomica Sinica. 2001;39:293–307. [Google Scholar]
- Ellison AM. Nutrient limitation and stoichiometry of carnivorous plants. Plant Biology. 2006;8:740–747. doi: 10.1055/s-2006-923956. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Van de Peer Y, Maere S. Significance and biological consequences of polyploidization in land plant evolution. In: Leitch IJ, Greilhuber J, Doležel J, Wendel J, editors. Plant genome diversity volume 2: physical structure, behaviour and evolution of plant genomes. Wien: Springer-Verlag; 2013. pp. 277–293. [Google Scholar]
- Fedorov AA. Chromosome numbers of flowering plants. Leningrad: Nauka; 1969. [Google Scholar]
- Felsenstein J. Phylogenies and the comparative method. American Naturalist. 1985;125:1–15. doi: 10.1086/703055. [DOI] [PubMed] [Google Scholar]
- Forrest LL, Hughes M, Hollingsworth PM. A phylogeny of Begonia using nuclear ribosomal sequence data and morphological characters. Systematic Botany. 2005;30:671–682. [Google Scholar]
- Frajman B, Schönswetter P. Giants and dwarfs: molecular phylogenies reveal multiple origins of annual spurges within Euphorbia subg. Esula. Molecular Phylogenetics and Evolution. 2011;61:413–424. doi: 10.1016/j.ympev.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Francis D, Davies MS, Barlow PW. A strong nucleotypic effect on the cell cycle regardless of ploidy level. Annals of Botany. 2008;101:747–757. doi: 10.1093/aob/mcn038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Harvey PH, Ives AR. Procedures for the analysis of comparative data using phylogenetically independent contrasts. Systematic Biology. 1992;41:18–32. [Google Scholar]
- Gauthier MPL, Barabe D, Bruneau A. Molecular phylogeny of the genus Philodendron (Araceae): delimitation and infrageneric classification. Botanical Journal of the Linnean Society. 2008;156:13–27. [Google Scholar]
- Gleissberg S, Kadereit JW. Evolution of leaf morphogenesis: evidence from developmental and phylogenetic data in Papaveraceae. International Journal of Plant Sciences. 1999;160:787–794. [Google Scholar]
- Goldblatt P, Johnson DE, editors. Index to plant chromosome numbers (September 2012) 1979–onwards. Missouri Botanical Garden, St Louis. http://www.tropicos.org/Project/IPCN . [Google Scholar]
- Gregory TR. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Annals of Botany. 2005;95:133–146. doi: 10.1093/aob/mci009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J. Chromosomes of the monocotyledons (general aspects) In: Rudall PJ, Cribb PJ, Cutler DF, Humphries CJ, editors. Monocotyledons: systematics and evolution. Kew: Royal Botanic Garden; 1995. pp. 379–414. [Google Scholar]
- Greilhuber J, Leitch IJ. Genome size and the phenotype. In: Leitch IJ, Greilhuber J, Doležel J, Wendel J, editors. Plant genome diversity volume 2: physical structure, behaviour and evolution of plant genomes. Wien: Springer-Verlag; 2013. pp. 323–344. [Google Scholar]
- Greilhuber J, Doležel J, Lysák M, Bennett MD. The origin, evolution and proposed stabilization of the terms ‘genome size’ and ‘C-value’ to describe nuclear DNA contents. Annals of Botany. 2005;95:255–260. doi: 10.1093/aob/mci019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greilhuber J, Borsch T, Miller K, Worberg A, Porembski S, Barthlott W. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biology. 2006;8:770–777. doi: 10.1055/s-2006-924101. [DOI] [PubMed] [Google Scholar]
- Grime JP. Prediction of weed and crop response to climate based upon measurements of nuclear DNA content. Aspects of Applied Biology. 1983;4:87–98. [Google Scholar]
- Grime JP, Mowforth MA. Variation in genome size – an ecological interpretation. Nature. 1982;299:151–153. [Google Scholar]
- Hibberd JM, Jeschke WD. Solute flux into parasitic plants. Journal of Experimental Botany. 2001;52:2043–2049. doi: 10.1093/jexbot/52.363.2043. [DOI] [PubMed] [Google Scholar]
- Hodgson JG, Sharafi M, Jalili A, et al. Stomatal vs. genome size in angiosperms: the somatic tail wagging the genomic dog? Annals of Botany. 2010;105:573–584. doi: 10.1093/aob/mcq011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörandl E, Paun A, Johansson JT, et al. Phylogenetic relationships and evolutionary traits in Ranunculus s.l. (Ranunculaceae) inferred from ITS sequence analysis. Molecular Phylogenetics and Evolution. 2005;36:305–327. doi: 10.1016/j.ympev.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Huang J, Corke H, Sun M. Highly polymorphic AFLP markers as a complementary tool to ITS sequences in assessing genetic diversity and phylogenetic relationships of sweetpotato (Ipomoea batatas (L.) Lam.) and its wild relatives. Genetic Resources and Crop Evolution. 2002;49:541–550. [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Janssens S, Geuten K, Juan YM, Song Y, Küpfer P, Smets E. Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) using chloroplast atpB-rbcL spacer sequences. Systematic Botany. 2006;31:171–180. [Google Scholar]
- Karagatzides JD, Butler JL, Ellison AM. The pitcher plant Sarracenia purpurea can directly acquire organic nitrogen and short-circuit the inorganic nitrogen cycle. PLoS One. 2009;4:1–9. doi: 10.1371/journal.pone.0006164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kejnovský E, Hawkins JS, Feschotte C. Plant transposable elements: biology and evolution. In: Wendel J, Greilhuber J, Doležel J, Leitch IJ, editors. Plant genome diversity volume 1: plant genomes, their residents, and their evolutionary dynamics. Wien: Springer-Verlag; 2012. pp. 17–34. [Google Scholar]
- Kenicer GJ, Kajita T, Pennington RT, Murata J. Systematics and biogeography of Lathyrus (Leguminosae) based on internal transcribed spacer and cpDNA sequence data. American Journal of Botany. 2005;92:1199–1209. doi: 10.3732/ajb.92.7.1199. [DOI] [PubMed] [Google Scholar]
- Kim JD, Kim SH, Kim CH, Jansen RK. Phylogeny of Berberidaceae based on sequences of the chloroplast gene ndhF. Biochemical Systematics and Ecology. 2004;32:291–301. [Google Scholar]
- Kim JH, Kim DK, Forest F, Fay MF, Chase MW. Molecular phylogenetics of Ruscaceae sensu lato and related families (Asparagales) based on plastid and nuclear DNA sequences. Annals of Botany. 2010;106:775–790. doi: 10.1093/aob/mcq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocyan A, Zhang LB, Schaefer H, Renner SS. A multi-locus chloroplast phylogeny for the Cucurbitaceae and its implications for character evolution and classification. Molecular Phylogenetics and Evolution. 2007;44:553–577. doi: 10.1016/j.ympev.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Krähenbühl M, Juan YM, Küpfer P. Chromosome and breeding system evolution of the genus Mercurialis (Euphorbiaceae): implications of ITS molecular phylogeny. Plant Systematics and Evolution. 2002;234:155–169. [Google Scholar]
- Kubát K, Hrouda L, Chrtek J Jr, Kaplan Z, Kirschner J, Štěpánek J, editors. Key to the Flora of the Czech Republic. Praha: Academia; 2002. [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biological Journal of the Linnean Society. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Bennett MD. Genome size and its uses: the impact of flow cytometry. In: Doležel J, Greilhuber J, Suda J, editors. Flow cytometry with plant cells. Weinheim: Wiley-VCH Verlag GmBH & KGaA; 2007. pp. 153–176. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifante ZD. Pollen morphology of Asphodelus L. (Asphodelaceae): taxonomic and phylogenetic inferences at the infrageneric level. Grana. 1996;35:24–32. [Google Scholar]
- Moore BR, Donoghue MJ. A Bayesian approach for evaluating the impact of historical events on rates of diversification. Proceedings of the National Academy of Sciences, USA. 2009;106:4307–4312. doi: 10.1073/pnas.0807230106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschner VC, Lorenz AP, Cervi AC, et al. A first molecular phylogenetic analysis of Passiflora (Passifloraceae) American Journal of Botany. 2003;90:1229–1238. doi: 10.3732/ajb.90.8.1229. [DOI] [PubMed] [Google Scholar]
- Nie ZL, Sun H, Chen ZD, Meng Y, Manchester SR, Wen J. Molecular phylogeny and biogeographic diversification of Parthenocissus (Vitaceae) disjunct between Asia and North America. American Journal of Botany. 2010;97:1342–1353. doi: 10.3732/ajb.1000085. [DOI] [PubMed] [Google Scholar]
- Oberlander KC. Molecular systematic study of southern African Oxalis (Oxalidaceae). Stellenbosch University: South Africa; 2009. PhD thesis. [Google Scholar]
- Oliver MJ, Petrov D, Ackerly D, Falkowski P, Schofield OM. The mode and tempo of genome size evolution in eukaryotes. Genome Research. 2007;17:594–601. doi: 10.1101/gr.6096207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F. DAPI staining of fixed cells for high resolution flow cytometry of nuclear DNA. Methods in Cell Biology. 1990;33:105–11. doi: 10.1016/s0091-679x(08)60516-6. [DOI] [PubMed] [Google Scholar]
- Pellicer J, Fay MF, Leitch IJ. The largest eukaryotic genome of them all? Botanical Journal of the Linnean Society. 2010;164:10–15. [Google Scholar]
- Petrov AD. Evolution of genome size: new approaches to an old problem. Trends in Genetics. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- Piegu B, Guyot R, Picault N, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Research. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiters C, McKenzie B. Seasonal allocation and efficiency patterns of biomass and resources in the perennial geophyte Sparaxis grandiflora subspecies fimbriata (Iridaceae) in lowland coastal fynbos, South Africa. Annals of Botany. 1994;74:633–646. [Google Scholar]
- Sennblad B, Bremer B. Classification of Apocynaceae s.l. according to a new approach combining Linnaean and phylogenetic taxonomy. Systematic Biology. 2002;51:389–409. doi: 10.1080/10635150290069869. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, et al. Polyploidy and angiosperm diversifications. American Journal of Botany. 2009;96:336–348. doi: 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Cytological characteristics associated with the different growth habits in the Dicotyledons. American Journal of Botany. 1938;25:180–198. [Google Scholar]
- Szinay D, Wijnker E, van den Berg R, Visser RGF, de Jong H, Bai Y. Chromosome evolution in Solanum traced by cross-species BAC-FISH. New Phytologist. 2012;195:688–698. doi: 10.1111/j.1469-8137.2012.04195.x. [DOI] [PubMed] [Google Scholar]
- Šmarda P, Bureš P, Horová L, Foggi B, Rossi G. Genome size and GC content evolution of Festuca: ancestral expansion and subsequent reduction. Annals of Botany. 2008;101:421–433. doi: 10.1093/aob/mcm307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šmarda P, Hejcman M, Březinová A, et al. Effect of phosphorus availability on the selection of species with different ploidy levels and genome sizes in a long-term grassland fertilisation experiment. New Phytologist. 2013 doi: 10.1111/nph.12399. in press. doi: 10.1111/nph.12399. [DOI] [PubMed] [Google Scholar]
- Tam SM, Boyce PC, Upson TM, et al. Intergeneric and infrafamilial phylogeny of subfamily Monsteroideae (Araceae) revealed by chloroplast trnL-F sequences. American Journal of Botany. 2004;91:490–498. doi: 10.3732/ajb.91.3.490. [DOI] [PubMed] [Google Scholar]
- Timme RE, Simpson BB, Linder CR. High-resolution phylogeny for Helianthus (Asteraceae) using the 18S–26S ribosomal DNA external transcribed spacer. American Journal of Botany. 2007;94:1837–1852. doi: 10.3732/ajb.94.11.1837. [DOI] [PubMed] [Google Scholar]
- Veselý P, Bureš P, Šmarda P, Pavlíček T. Genome size and DNA base composition of geophytes: the mirror of phenology and ecology? Annals of Botany. 2012;109:65–75. doi: 10.1093/aob/mcr267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltz SM, Renner SS. Hybridization, polyploidy, and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae) American Journal of Botany. 2008;95:1297–1306. doi: 10.3732/ajb.0800187. [DOI] [PubMed] [Google Scholar]
- Webb C, Ackerly D, Kembel S. Phylocom: software for the analysis of phylogenetic community structure and character evolution. 2011 doi: 10.1093/bioinformatics/btn358. http://phylodiversity.net/phylocom/ [DOI] [PubMed] [Google Scholar]
- Zonneveld BJM. New record holders for maximum genome size in Eudicots and Monocots. Journal of Botany. 2010;2010:527357. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


